Abstract
The limited sensitivity of circulating tumor cell (CTC) detection in pancreatic adenocarcinoma (PDAC) stems from their extremely low concentration in the whole circulating blood, necessitating enhanced detection methodologies. This study sought to amplify assay-sensitivity by employing diagnostic leukapheresis (DLA) to screen large blood volumes. Sixty patients were subjected to DLA, with a median processed blood volume of ~ 2.8 L and approximately 5% of the resulting DLA-product analyzed using CellSearch (CS). Notably, DLA significantly increased CS-CTC detection to 44% in M0-patients and 74% in M1-patients, yielding a 60-fold increase in CS-CTC enumeration. DLA also provided sufficient CS-CTCs for genomic profiling, thereby delivering additional genomic information compared to tissue biopsy samples. DLA CS-CTCs exhibited a pronounced negative prognostic impact on overall survival (OS), evidenced by a reduction in OS from 28.6 to 8.5 months (univariate: p = 0.002; multivariable: p = 0.043). Additionally, a marked enhancement in sensitivity was achieved (by around 3-4-times) compared to peripheral blood (PB) samples, with positive predictive values for OS being preserved at around 90%. Prognostic relevance of CS-CTCs in PDAC was further validated in PB-samples from 228 PDAC patients, consolidating the established association between CTC-presence and reduced OS (8.5 vs. 19.0 months, p < 0.001). In conclusion, DLA-derived CS-CTCs may serve as a viable tool for identifying high-risk PDAC-patients and aiding the optimization of multimodal treatment strategies. Moreover, DLA enables comprehensive diagnostic profiling by providing ample CTC material, reinforcing its utility as a reliable liquid-biopsy approach. This high-volume liquid-biopsy strategy presents a potential pathway for enhancing clinical management in this malignancy.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12943-023-01880-1.
Keywords: Pancreatic ductal adenocarcinoma, PDAC, Circulating tumor cells, CTCs, High-blood volume analysis, Liquid biopsy, Diagnostic leukapheresis, DLA, Single cell genomics, CTC profiling
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is characterized by a high mortality-to-incidence ratio and is projected to become the second leading cause of cancer-related death in Western countries by 2030 [1]. Only 15–20% of newly diagnosed patients have localized disease and are eligible for curative surgical treatment [2]. Despite surgery with curative intent, patients can only expect a 5-year overall survival rate of around 20–25% due to occurrence of metastatic disease, the most common kind of recurrence [3–5]. Therefore, biomarkers are urgently needed to identify patients with clinically occult systemic PDAC at the timepoint of surgery in order to select high-risk patients for more intense regimens or to avoid major surgery in patients unlikely to benefit.
In this context, circulating tumor cells (CTCs) are promising biomarkers for PDAC, as they can represent active systemic cancer [6]. The current gold standard for CTC detection is the FDA cleared CellSearch (CS) system, which defines CTCs as EpCAM+/CK+/CD45−/DAPI + cells (CS-CTC). While CS-CTC detection is high in advanced prostate or breast cancer, it is surprisingly low in PDAC, with approximately 10% in M0 patients and 40% in M1 patients [7, 8]. Compared to prostate and breast cancer, only few studies with relatively small cohorts have been reported on CS in PDAC. The data suggested that CS-CTCs may identify operable PDAC patients at high risk for poor survival [7, 8]. However, the low detection frequencies clearly indicate that a significant number of high-risk patients escape detection, limiting the utility for patient selection or CTC-based diagnostic profiling. To address these issues, we employed an ultra-sensitive, high-blood-volume approach to enhance CS-CTC detection via leukapheresis.
Our objective was to demonstrate the feasibility of identifying high-risk patients in early PDAC stages and to enable the harvest of a sufficient quantity of CTCs for liquid biopsy-based profiling, thereby complementing tissue biopsies in cases of inoperable disease. According to our hypothesis, the low CTC detection is mainly due to an insufficient sample volume relative to the total body volume. Statistical models accounting for the Poisson distribution of rare CTCs calculated that the analysis of 1–5 L of blood should enable the detection of 10–50 CTCs in virtually every metastatic cancer patient [9, 10]. As a first step to address this challenge, we introduced diagnostic leukapheresis (DLA) [11], a shortened leukapheresis protocol to harvest a white blood cell (WBC) fraction (i.e. MNCs) based on their specific density from more than two liters of blood by continuous centrifugation. Since density profiles of epithelial cancer cells overlap with the targeted MNCs [12], CTCs become co-collected along the process. Previous studies have demonstrated the feasibility of using DLA for CTC enrichment in patients with advanced breast, prostate, and lung cancers [13–16]. Therefore, we sought to investigate whether DLA could improve the sensitivity of CS-CTC detection and consequently enhance identification of high-risk patients PDAC.
Results and discussion
DLA significantly improves detection of CS-CTCs in PDAC
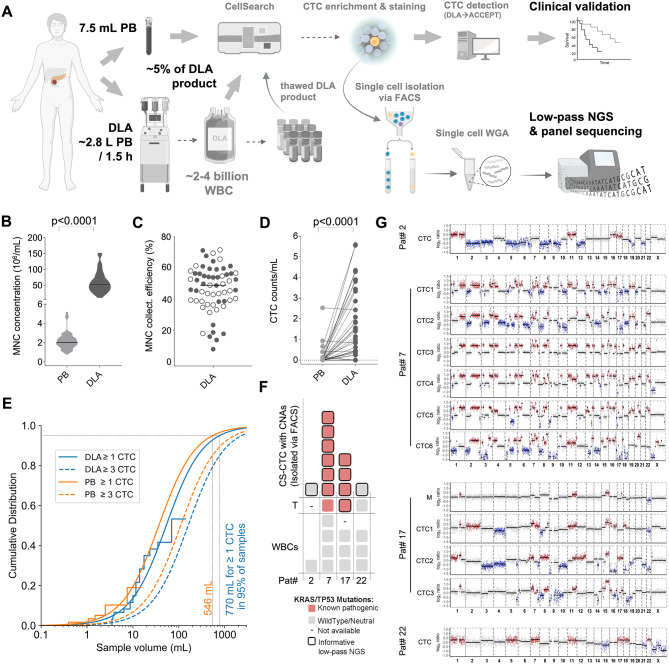
Please refer to Additional File 1, Supplementary Methods for the materials and methods of this study. To explore the effects of DLA for PDAC CTCs, we followed the workflow illustrated in Fig. 1A. DLA with sufficient MNC collection efficacy (median: 49%; range: 8–71%; Fig. 1B-C) for this purpose was performed in 60 PDAC patients (DU-DLA-cohort) without technical issues or notable adverse effects. Although lower efficacies also produced CS-CTC positive samples, DLAs with a collection efficacy equal to or above the median resulted in a higher CS-CTC-detection frequency (11/30 vs. 21/30; Fisher’s exact test: p = 0.02, Fig. 1C). From the processed median blood volume of ~ 2.8 L (2,795mL, range: 769-5,765mL) we generated DLA products with a median volume of 45mL (range: 10-100mL). The capacity of the CS-assay required limiting the sample input to around 200*106 WBC [11], which represented approximately 5% of a DLA product. The remaining material was cryopreserved as aliquots. The calculated CS-input corresponded to a median blood volume of ~ 70mL (70.6mL, range: 20.50mL-181.03mL) (Additional File 1, Supplementary Fig. 1). Applying this process, we observed a significantly increased detection frequency of 53% (32/60) in DLA compared to 19% in matched PB samples (11/58; Fisher’s Exact Test, p < 0.001). With 1.05 (±1.51) CTC/mL the mean CS-CTC concentration was 10-times higher in DLA compared to 0.10 (±0.37) CTC/mL in PB (Fig. 1D). To estimate the overall CTC-load in DLA products, we extrapolated the detected CS-CTCs to the entire DLA product, revealing a calculated median of 71.5 CTCs (range: 0-218) per DLA product and a 60-fold (mean) increase in CTC numbers compared to PB. This enrichment suggests that PDAC-CTCs fall into the density window targeted by our DLA settings. Next, we aimed to determine the DLA-product volume, required to analyze for detecting at least one CS-CTC in every PDAC patient. Fitting our PB and DLA data to a distribution function described previously for PB [9], we ascertained that the distribution function also holds for the larger volumes of the DLA product. Furthermore, we extrapolate that analyzing a DLA product with a corresponding median blood volume of 1000mL is expected to yield 1 CTC in 96% of samples, 3 CTC in 88% of samples and 10 CTC in 67% of samples (Fig. 1E). This volume is equivalent to approximately 76% of the DLA product (34mL). The 30% discrepancy between the volumes derived from PB samples (7.5mL) and DLA samples (~ 70mL) indicates the CTC loss incurred in the DLA sample preparation process as compared to PB. However, it is important to stress that forthcoming technological advancements [10, 17], proficient in managing the designated DLA volumes, should feasibly enable processing of the required DLA-product volume in an imminent time frame.
Fig. 1.
Study overview and DLA findings: A Experimental setup (created in part with BioRender.com); B MNCs per mL in peripheral blood samples (D-PB, n = 60) versus matched DLA samples (D-DLA, n = 60); C Collection efficacy achieved for the targeted MNC population (n = 60). The filled circles indicate CTC-positivity (≥ 1 CTC detected); D CTC-concentration per mL in PB samples (light grey circles) and matched DLA samples (dark grey circles); E. Extrapolation of log-logistic fit curves for peripheral blood samples (PB) and DLA samples (DLA). Stair plots show the empirical CDF and the continuous line the fit. Extrapolation indicates probability (Cumulative Distribution) for a certain analyzed sample volume to have at least 1 CTC or 3 CTCs in every sample; F Genomic analysis of individual CS-CTCs isolated from DLA products with panel sequencing of KRAS and TP53 genes (T: tumor tissue); G Plots of copy number profiles along the 22 autosomes expressed as log2 ratios. Significant copy number gains and losses are highlighted in red and blue, respectively (M: FFPE material from metastatic tissue)
Uncovering the genomic alterations of PDAC CTCs through DLA-enabled profiling
Owing to their rarity in PB, genomic data on CS-CTCs in PDAC have remained unavailable until now. To demonstrate the feasibility of employing DLA-enriched CS-CTCs as a surrogate for molecular profiling and to validate their malignant character, we re-analyzed cryopreserved material from CS-CTC-positive DLA samples. Notably, the quantity of CS-CTCs in the frozen DLA samples was not significantly deviant from those freshly and prospectively analyzed (Additional File 1, Supplementary Fig. 2). These results affirm that processing the entire DLA product could further elevate the sensitivity of our approach, as was suggested by our extrapolation. After isolation, we conducted genomic profiling of 11 single CS-CTCs and matched tumor tissue of four patients (#2, #7, #17, #22), including one patient (#22) in whom relevant copy number alteration (CNAs) were not detected in the tumor tissue (Fig. 1F-G; Additional File 1, Supplementary Table 4). In two patients (#2 and #22), KRAS and TP53 mutations were not detected in either the CTCs harboring CNAs or the matched tumor tissue. Patient #7 exhibited consistent detection of a G12V mutation in both CTCs and tumor tissue that displayed an additional G12D mutation (Fig. 1F-G; Additional File 1, Supplementary Table 4). In patient #17, CTCs showed a G12V mutation while a G12R mutation was observed in the matched tumor tissue (Additional File 1, Supplementary Table 4). As a control, we analyzed 26 matched and non-matched white blood cells isolated from CS cartridges of thawed PDAC DLA samples and did not detect any pathogenic mutations (Additional File 1, Supplementary Table 4). The different hot-spot mutations between tumor tissue and CTCs prompted us to perform subsequent short tandem-repeat analysis (STR), which authenticated the sample identity (data not shown). Collectively, this exemplary genomic CTC-profiling not only confirmed the malignant nature of DLA-enriched CS-CTCs but also demonstrated the feasibility of performing comprehensive genomic analysis on CS-CTCs from DLA-products, providing additional information compared to PDAC tissue. Considering the recognized intra-patient molecular heterogeneity, employing their versatile analysis could offer substantial advantages over classical tissue biopsies in metastatic PDAC. This is particularly pertinent in the context of emerging molecular therapies, which will necessitate diagnostic molecular tumor profiling [2, 18, 19].
DLA enriched CS-CTCs are linked to higher tumor stage and poor survival
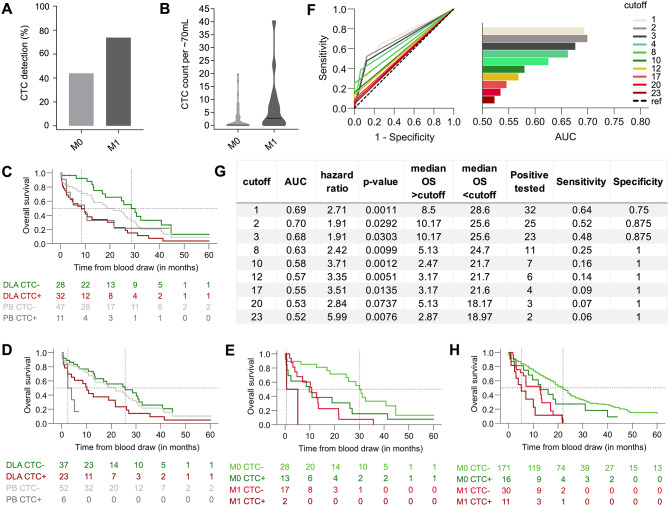
To validate the clinical relevance of CS-CTCs enriched by DLA, we examined their association with clinical parameters. Given the individual variations in assay input (Additional File 1, Supplementary Fig. 1), CTC counts were normalized to the median analyzed blood-equivalent of ~ 70mL, with resulting decimal numbers rounded to the nearest integer (range: 0–40 CTCs). Although CTC count normalization was crucial for accurate concentration and survival analyses, notable differences in detection frequencies between M0 and M1 patients were evident without this adjustment. For DLA samples, detection rates were 18/41 for M0 patients and 14/19 for M1 patients (Fisher’s exact test: p = 0.0507, see Fig. 2A). Meanwhile, for PB samples, the detection rate was 4/41 for M0 patients and 7/17 for M1 patients (Fisher’s exact test: p = 0.0099). Notably, only DLA samples demonstrated a significantly higher CS-CTC concentration in M1 patients compared to M0 patients (median: 3.0 vs. 0.0, Mann-Whitney U test: p = 0.024, Fig. 2B). Subsequently, we tested the prognostic value of DLA-enriched CS-CTCs and adopted a cut-off of ≥ 1CTC per ~ 70mL, aligning with thresholds used in existing PDAC studies involving CS [7, 8]. Additionally, to determine a more robust threshold for DLA, we incorporated ≥ 3CTC/~70mL, considering a ~ 5% probability of detecting 0 CTC with an average detection frequency/threshold of 3 CTCs per sample versus a ~ 36% probability at a mean detection frequency/threshold of 1 CTC per sample, due to the Poisson distribution. Strikingly, DLA-derived CS-CTC revealed prognostic significance at both thresholds, with one CTC per sample reducing median overall survival (OS) to 8.5 months, compared to 28.6 months without CS-CTC (log-rank test: p = 0.002; Fig. 2C), and ≥ 3CTCs reducing OS to 10.2 months vs. 25.6 months below this threshold (log-rank test: p = 0.033; Fig. 2D). Upon multivariable analysis, the adverse effect of the threshold of ≥ 1CTC retained its significance, underscoring prognostic relevance. The prognostic effects of DLA CS-CTC outperformed those observed for CS-CTC in matched PB samples (Fig. 2C and D, Additional File 1, Supplementary Table 5), especially concerning sensitivity and accuracy in identifying patients at high risk for short OS (Additional File 1, Supplementary Table 8). The superior prognostic impact of DLA CS-CTCs was also evident in M0 patients and those treated with curative intent (Additional File 1, Supplementary Fig. 3, and Supplementary Tables 6–8). Notably, a relatively high OS-rate was apparent for M0 patients without DLA CS-CTC (e.g., at 20 months), while DLA CS-CTC-positive M0 patients exhibited similarly poor OS compared with CS-CTC-negative M1 patients (Fig. 2E). As anticipated, CS-CTC-positive M1-patients displayed the worst prognosis. Interestingly, an analysis for CTC-cutoff values, which considered both hazard ratios and AUC values collectively (Fig. 2F and G), confirmed our tested cutoffs as optimal. Expectedly, elevating CTC-cutoff values enhanced specificity while diminishing sensitivity. Further validation of the prognostic value of CS-CTCs in PDAC was sought through analyzing PB samples in an independent, large cohort (HE: n = 170) and in combination with the DU-PB-cohort (n = 228; Fig. 2H, Additional File 1, Supplementary Table 9), affirming the significant and independent prognostic impacts of CS-CTCs across various subgroups (Additional File 1, Supplementary Fig. 5 and Supplementary Tables 9–11), albeit at low detection rates. Intriguingly, even with the low detection frequency, the stage-dependent effects of CS-CTCs were congruent with the observations made in DLA CS-CTCs (Fig. 2E and H). Taken together, the prognostic data suggest that CS-CTCs could serve as a useful biomarker to select M0-patients for intensified multimodal treatments and to guide cancer-directed surgery in the challenging situation of oligometastatic PDAC [20]. For CTC + M1-patients, tumor resection may be contraindicated, whereas for M1-patients negative for DLA-CS-CTCs resection may be beneficial if technically feasible. Nonetheless, this premise requires further validation through prospective clinical studies.
Fig. 2.
Association of PDAC CS-CTC with clinical parameters. A Frequency of CTC-positivity (≥ 1 CTC detected) in DLA products according to the metastatic stage; B Normalized CS-CTC-concentration in DLA samples according to the metastatic stage; C-D OS according to status of DLA CS-CTC (n = 60, CTC/~70mL) in green/red compared to PB CS-CTC (n = 58, CTC/7.5mL blood) in grey. C Cutoff for CTC positivity: ≥1 CTC; D Cutoff for CTC positivity: ≥3 CTC; E OS according to the CTC-status separated by metastatic stage in DLA products (optimal cutoffs: M0 = 3CTC/~70mL blood; M1 = 23CTC/~70mL blood, see Additional File 1, Supplementary Fig. 4); F ROC curves and corresponding AUC values for the top 10 DLA CS-CTC cutoffs normalized to ~ 70mL of blood. These cutoffs were selected based on their combined highest hazard ratio and AUC value in the DLA cohort of 60 PDAC patients (ref: reference); G Parameters for all significant (p < 0.05) DLA CS-CTCs cutoff values calculated in the cohort of 60 PDAC patients (Cutoff: CTCs/~70mL); H OS according to CS-CTC status (≥ 1CTC/7.5mL blood) in PB pooled HE/DU-PB cohorts separated by metastatic stage (M0 vs. M1)
Conclusion
Our findings emphasize the clinical significance of CS-CTCs in PDAC, highlighting the necessity of high-volume blood analysis for enhanced detection. This approach complements and potentially surpasses other circulating biomarkers in PDAC, such as ctDNA. Even with the limitation of our study restricting analysis to 5% of DLA products, our CS-CTC detection align with ctDNA detection rates even in advanced PDAC [21, 22]. This can be attributed to minimal cfDNA shedding in PDAC and the challenges associated with obtaining tissue in non-operable cases for tumor-informed approaches. While ctDNA analysis is immensely valuable, it predominantly leans towards quantification in PDAC due to these limitations. Conversely, DLA CTCs open avenues for versatile qualitative analyses, addressing clinically relevant diagnostic tests with the potential even enabling functional CTC studies [23]. Emerging technical solutions for analyzing entire DLA products will further amplify sensitivity [10, 17]. Leukapheresis, despite its complexity compared to conventional blood draws, is clinically routine and, as exploited in our study, provides a practical method to obtain high-quality CTC-material. Implementing this ultra-sensitive approach in newly diagnosed PDAC patients could enable CTC-based therapeutic decision-making, facilitating nuanced staging and risk stratification for personalized surgical treatment. Our findings pave the way for subsequent prospective clinical trials, exploring the DLA approach either independently or synergistically with other circulating biomarkers to refine liquid biopsies for PDAC patients.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Material 1. Description of data: This file contains Supplementary Methods, Supplementary Tables 1-11 and Supplementary Figures 1-5 as supporting information and data for the main manuscript.
Acknowledgements
We thank all the patients who have volunteered and donated their biomaterials for the study. We thank Maria Wecker and Swetlana Seidschner for excellent technical assistance.
Abbreviations
- AUC
area under the curve
- CNAs
copy number alterations
- CDF
cumulative distribution function
- CS
CellSearch
- CTCs
circulating tumor cells
- CS-CTCs
CS-detected EpCAM+/CK+/CD45−/DAPI + CTCs
- DU
Düsseldorf
- DLA
diagnostic leukapheresis
- EpCAM
epithelial cell adhesion molecule
- HE
Heidelberg
- HR
hazard ratio
- KRAS
Kirsten rat sarcoma viral oncogene homolog
- log-rank test
non-parametric statistical test for survival data
- M0
absence of distant metastasis
- M1
presence of distant metastasis
- MNC
Mononuclear cell
- OS
overall survival
- PB
peripheral blood
- PDAC
pancreatic ductal adenocarcinoma
- ROC
receiver operating characteristic
- SNVs
single nucleotide variants
- TP53
tumor protein p53
Author contributions
N.H.S., J.C.F., S.T. and W.T.K. designed the DLA study of D-Cohort. N.N.R., T.H. and J.W. designed the PB study of the HD-cohort; S.T., W.T.K., G.F., J.C.F., J.M.R., N.N.R., T.H., U.B. and J.W. diagnosed patients, evaluated patients, and provided samples; J.C.F. and J.M.R. were responsible for DLA and provided samples; E.B., G.V.D., R.G., C.D., D.N., H.N., U.B., N.N.R. and N.H.S. performed and analyzed CS experiments; R.G., L.H., W.G. and I.E. contributed to PDAC tissue analysis and genomic studies; R.G., R.N., A.F., D.N., H.N. performed and contributed to isolation of CTCs and their genomic single cell analysis; M.S. performed and interpreted bioinformatic analysis of NGS-data; S.C., R.G., E.B., U.B., F.A.W.C., N.N.R performed statistical analyses; PB performed STR-analyses; N.H.S., G.F., R.G., R.N., S.C., J.C.F., S.T., W.T.K., N.N.R., T.H., J.W., E.B., U.B., E.B., G.V.D., C.D., D.N., H.N., T.F., L.H. F.A.W.C., W.G., I.E. and P.B. interpreted data and contributed to figures and tables; N.H.S. and N.N.R. supervised the study and wrote the manuscript with input from all authors. The authors read and approved the final manuscript.
Funding
This research was funded by the Brigitte und Dr. Konstanze Wegener Foundation (Project #01 to N.H.S. and W.T.K.), in part by the German Cancer Foundation (Förderschwerpunkt-programm der Deutschen Krebshilfe ‘Translationale Onkologie’; Grant 70112504 to N.H.S., H.N., T.F.), in part by the German Research Foundation DFG (GZ FL865/2 − 1 to G.F.), and in part by the Innovative Medicines Initiative Joint Undertaking (IMI JU) in conjunction with CANCER-ID [Grant Agreement #115749 to N.H.S.].
Data Availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
The studies were independently approved by the Ethics Committees of the Medical Faculty of the Heinrich-Heine University Duesseldorf and the Ruprecht-Karls-University Heidelberg, respectively. All patients included into the analysis provided written informed consent.
Consent for publication
Not applicable.
Competing interests
None
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Nikolas H. Stoecklein, Georg Fluegen, Rosa Guglielmi, Rui P.L. Neves, Johannes C. Fischer, Ulrich Bork and Nuh N. Rahbari contributed equally
Contributor Information
Nikolas H. Stoecklein, Email: Nikolas.Stoecklein@med.uni-duesseldorf.de
Nuh N. Rahbari, Email: nuh.rahbari@uniklinik-ulm.de
References
- 1.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74:2913–21. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- 2.O’Kane GM, Ladak F, Gallinger S. Advances in the management of pancreatic ductal adenocarcinoma. CMAJ. 2021;193:E844–51. doi: 10.1503/cmaj.201450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Groot VP, Rezaee N, Wu W, Cameron JL, Fishman EK, Hruban RH, Weiss MJ, Zheng L, Wolfgang CL, He J. Patterns, timing, and predictors of recurrence following pancreatectomy for pancreatic ductal adenocarcinoma. Ann Surg. 2018;267:936–45. doi: 10.1097/SLA.0000000000002234. [DOI] [PubMed] [Google Scholar]
- 4.Hugenschmidt H, Labori KJ, Borgen E, Brunborg C, Schirmer CB, Seeberg LT, Naume B, Wiedswang G. Preoperative CTC-Detection by CellSearch((R)) is Associated with early distant Metastasis and impaired survival in Resected Pancreatic Cancer. Cancers (Basel) 2021, 13. [DOI] [PMC free article] [PubMed]
- 5.Jones RP, Psarelli EE, Jackson R, Ghaneh P, Halloran CM, Palmer DH, Campbell F, Valle JW, Faluyi O, O’Reilly DA, et al. Patterns of recurrence after resection of pancreatic ductal adenocarcinoma: a secondary analysis of the ESPAC-4 randomized adjuvant chemotherapy trial. JAMA Surg. 2019;154:1038–48. doi: 10.1001/jamasurg.2019.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alix-Panabieres C, Pantel K. Liquid Biopsy: from Discovery to Clinical Application. Cancer Discov. 2021;11:858–73. doi: 10.1158/2159-8290.CD-20-1311. [DOI] [PubMed] [Google Scholar]
- 7.Hugenschmidt H, Labori KJ, Brunborg C, Verbeke CS, Seeberg LT, Schirmer CB, Renolen A, Borgen EF, Naume B, Wiedswang G. Circulating Tumor cells are an Independent predictor of shorter survival in patients undergoing resection for pancreatic and Periampullary Adenocarcinoma. Ann Surg. 2020;271:549–58. doi: 10.1097/SLA.0000000000003035. [DOI] [PubMed] [Google Scholar]
- 8.Riethdorf S, O’Flaherty L, Hille C, Pantel K. Clinical applications of the CellSearch platform in cancer patients. Adv Drug Deliv Rev. 2018;125:102–21. doi: 10.1016/j.addr.2018.01.011. [DOI] [PubMed] [Google Scholar]
- 9.Coumans FA, Ligthart ST, Uhr JW, Terstappen LW. Challenges in the enumeration and phenotyping of CTC. Clin Cancer Res. 2012;18:5711–8. doi: 10.1158/1078-0432.CCR-12-1585. [DOI] [PubMed] [Google Scholar]
- 10.Mishra A, Dubash TD, Edd JF, Jewett MK, Garre SG, Karabacak NM, Rabe DC, Mutlu BR, Walsh JR, Kapur R, et al. Ultrahigh-throughput magnetic sorting of large blood volumes for epitope-agnostic isolation of circulating Tumor cells. Proc Natl Acad Sci U S A. 2020;117:16839–47. doi: 10.1073/pnas.2006388117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fischer JC, Niederacher D, Topp SA, Honisch E, Schumacher S, Schmitz N, Zacarias Fohrding L, Vay C, Hoffmann I, Kasprowicz NS, et al. Diagnostic leukapheresis enables reliable detection of circulating Tumor cells of nonmetastatic cancer patients. Proc Natl Acad Sci U S A. 2013;110:16580–5. doi: 10.1073/pnas.1313594110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stoecklein NH, Fischer JC, Niederacher D, Terstappen LW. Challenges for CTC-based liquid biopsies: low CTC frequency and diagnostic leukapheresis as a potential solution. Expert Rev Mol Diagn. 2016;16:147–64. doi: 10.1586/14737159.2016.1123095. [DOI] [PubMed] [Google Scholar]
- 13.Fehm TN, Meier-Stiegen F, Driemel C, Jager B, Reinhardt F, Naskou J, Franken A, Neubauer H, Neves RPL, van Dalum G, et al. Diagnostic leukapheresis for CTC analysis in Breast cancer patients: CTC frequency, clinical experiences and recommendations for standardized reporting. Cytometry A. 2018;93:1213–9. doi: 10.1002/cyto.a.23669. [DOI] [PubMed] [Google Scholar]
- 14.Mout L, van Dessel LF, Kraan J, de Jong AC, Neves RPL, Erkens-Schulze S, Beaufort CM, Sieuwerts AM, van Riet J, Woo TLC, et al. Generating human Prostate cancer organoids from leukapheresis enriched circulating tumour cells. Eur J Cancer. 2021;150:179–89. doi: 10.1016/j.ejca.2021.03.023. [DOI] [PubMed] [Google Scholar]
- 15.Tamminga M, Andree KC, Hiltermann TJN, Jayat M, Schuuring E, van den Bos H, Spierings DCJ, Lansdorp PM, Timens W, Terstappen L, Groen HJM. Detection of circulating Tumor cells in the diagnostic leukapheresis product of Non-small-cell Lung Cancer patients comparing CellSearch((R)) and ISET. Cancers (Basel) 2020, 12. [DOI] [PMC free article] [PubMed]
- 16.Lambros MB, Seed G, Sumanasuriya S, Gil V, Crespo M, Fontes M, Chandler R, Mehra N, Fowler G, Ebbs B, et al. Single-cell analyses of Prostate Cancer liquid biopsies acquired by Apheresis. Clin Cancer Res. 2018;24:5635–44. doi: 10.1158/1078-0432.CCR-18-0862. [DOI] [PubMed] [Google Scholar]
- 17.Stevens M, Mentink A, Nanou A, Coumans FAW, Isebia KT, Kraan J, Hamberg P, Martens JWM, Terstappen L. Improved enrichment of circulating Tumor cells from diagnostic leukapheresis product. Cytometry A 2023. [DOI] [PubMed]
- 18.Ansari D, Tingstedt B, Andersson B, Holmquist F, Sturesson C, Williamsson C, Sasor A, Borg D, Bauden M, Andersson R. Pancreatic cancer: yesterday, today and tomorrow. Future Oncol. 2016;12:1929–46. doi: 10.2217/fon-2016-0010. [DOI] [PubMed] [Google Scholar]
- 19.Park W, Chawla A, O’Reilly EM. Pancreatic Cancer: a review. JAMA. 2021;326:851–62. doi: 10.1001/jama.2021.13027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pausch TM, Liu X, Cui J, Wei J, Miao Y, Heger U, Probst P, Heap S, Hackert T. Survival benefit of resection Surgery for pancreatic ductal adenocarcinoma with liver metastases: a propensity score-matched SEER database analysis. Cancers (Basel) 2021, 14. [DOI] [PMC free article] [PubMed]
- 21.Guven DC, Sahin TK, Yildirim HC, Aktepe OH, Dizdar O, Yalcin S. A systematic review and meta-analysis of the association between circulating Tumor DNA (ctDNA) and prognosis in Pancreatic cancer. Crit Rev Oncol Hematol. 2021;168:103528. doi: 10.1016/j.critrevonc.2021.103528. [DOI] [PubMed] [Google Scholar]
- 22.Lapin M, Edland KH, Tjensvoll K, Oltedal S, Austdal M, Garresori H, Rozenholc Y, Gilje B, Nordgard O. Comprehensive ctDNA measurements improve prediction of clinical outcomes and enable dynamic Tracking of Disease Progression in Advanced Pancreatic Cancer. Clin Cancer Res. 2023;29:1267–78. doi: 10.1158/1078-0432.CCR-22-3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Faugeroux V, Pailler E, Oulhen M, Deas O, Brulle-Soumare L, Hervieu C, Marty V, Alexandrova K, Andree KC, Stoecklein NH, et al. Genetic characterization of a unique neuroendocrine transdifferentiation prostate circulating Tumor cell-derived eXplant model. Nat Commun. 2020;11:1884. doi: 10.1038/s41467-020-15426-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material 1. Description of data: This file contains Supplementary Methods, Supplementary Tables 1-11 and Supplementary Figures 1-5 as supporting information and data for the main manuscript.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.