Abstract
Pseudomonas species are plant, animal, and human pathogens; exhibit plant pathogen-suppressing properties useful in biological control; or express metabolic versatilities valued in biotechnology and bioremediation. Specific detection of Pseudomonas species in the environment may help us gain a more complete understanding of the ecological significance of these microorganisms. The objective of this study was to develop a PCR protocol for selective detection of Pseudomonas (sensu stricto) in environmental samples. Extensive database searches identified a highly selective PCR primer pair for amplification of Pseudomonas 16S rRNA genes. A protocol that included PCR amplification and restriction analysis, a general cloning and sequencing strategy, and phylogenetic analyses was developed. The PCR protocol was validated by testing 50 target and 14 nontarget pure cultures, which confirmed the selectivity to 100%. Further validation used amplification of target sequences from purified bulk soil DNA followed by cloning of PCR products. Restriction analysis with HaeIII revealed eight different fragmentation patterns among 36 clones. Sequencing and phylogenetic analysis of 8 representative clones indicated that 91.7% of the products were derived from target organisms of the PCR protocol. Three patterns, representing only 8.3% of the 36 clones, were derived from non-Pseudomonas or chimeric PCR artifacts. Three patterns, representing 61.1% of the clones, clustered with sequences of confirmed Pseudomonas species, whereas two patterns, representing 30.6% of the clones, formed a novel phylogenetic cluster closely associated with Pseudomonas species. The results indicated that the Pseudomonas-selective PCR primers were highly specific and may represent a powerful tool for Pseudomonas population structure analyses and taxonomic confirmations.
The genus Pseudomonas includes species with functions of ecological, economic, and health-related importance. Some species are pathogenic for plants (8, 42), while others are opportunistic pathogens of animals or humans (10, 33, 45). Some species exhibit plant growth-promoting and pathogen-suppressing functions and may be exploited for use in biological control (18, 31). A prominent property of some species or strains is their metabolic versatility, making them attractive candidates for use in bioremediation (35, 39). Many studies have described the potential of Pseudomonas species to degrade a variety of compounds (12, 14, 15, 17, 41). A specific, culturing-independent detection protocol for Pseudomonas would represent a valuable tool in ecological and diagnostic studies of this genus. Recent advances in molecular ecological techniques and taxonomy open ways to design highly specific PCR protocols, especially for detection of 16S (small-subunit [SSU]) rRNA genes (4, 11, 29, 49). Genus-specific 16S rRNA gene PCR primer design depends on both a well-defined molecular taxonomy and a representative collection of target sequences. Each of these issues has been extensively addressed for the genus Pseudomonas in the last few years (27, 28, 35).
The genus Pseudomonas, which was described by Migula in 1894 as a genus of gram-negative, rod-shaped microorganisms (32), has been subject to repeated taxonomic revisions (35). DNA-RNA hybridization techniques have revealed five RNA classes among these pseudomonads (24, 32, 36). This extensive degree of heterogeneity among the old genus of Pseudomonas is reflected by the presence of distantly related species which have since been placed in existing or newly defined genera. Former Pseudomonas species of RNA groups II to IV have been reclassified to genera including Brevundimonas, Sphingomonas, Burkholderia, Ralstonia, Aminobacter, Comamonas, Acidovorax, Hydrogenophaga, Telluria, and Stenotrophomonas (16, 19, 35). These analyses and taxonomic rearrangements identified the RNA group I species, including the type species P. aeruginosa and other species such as P. fluorescens, P. putida, and P. syringae, as members of a phylogenetically homogeneous group referred to as Pseudomonas (sensu stricto) (28, 34). This classification is in agreement with phylogenetic information obtained from 16S rRNA sequence data as presented in the Ribosomal Database Project (RDP [27]). Molecular taxonomy based on 16S rRNA sequences places the genus Pseudomonas, which has been referred to as the type I or fluorescent group of pseudomonads, in the group called Pseudomonas and relatives, along with the subgroups of Acinetobacter and Teredinibacter. Directly adjacent phylogenetic branches are formed by the Oceanospirillum group and the Colwellia assemblage. This molecular definition of the genus Pseudomonas may be used to develop a 16S rRNA-based detection approach specific for this genus.
The multitude of important characteristics of Pseudomonas species has inspired numerous taxonomic studies, which have led to the development of a better understanding of this genus. The approaches used include selective culturing (38), PCR amplification of rRNA genes (21, 45) in conjunction with restriction fragment length polymorphism (RFLP) analysis (5, 25) or DNA sequencing (28), DNA and RNA hybridization (3, 6, 36, 40), protein (7, 46) and fatty acid (42, 47) analyses, carbon source utilization properties (13, 17, 22), and specific antibodies (23). These techniques have been applied to characterize Pseudomonas species of both defined cultures and uncharacterized isolates obtained from a variety of sources. However, these approaches have not offered a rapid approach to specifically monitor and characterize population structures of the genus Pseudomonas (sensu stricto) in the environment.
The objective of this study was to develop a 16S rRNA gene-based PCR protocol for the selective detection of Pseudomonas (sensu stricto) in environmental samples. Sequence information of 16S rRNA sequences available from RDP and GenBank was compiled and searched for Pseudomonas-selective PCR (Ps-PC) primer sequences. Theoretical evaluation of a highly selective PCR primer pair is presented here, together with experimental validation performed on pure cultures and bulk DNA extracted from an agricultural soil sample.
MATERIALS AND METHODS
Bacterial cultures and soil DNA.
Experimental specificity testing of PCR primers was performed with pure bacterial cultures obtained from the American Type Culture Collection (ATCC; Rockville, Md.), culture DNA samples (kindly provided by B. Bochner, Biolog Inc., Hayward, Calif.), cultures obtained from the Environmental Protection Agency (EPA; Corvallis, Oreg.) culture collection, and environmental isolates from the Corvallis, Oreg., area. Information on the cultures used in this study is summarized in Table 1. Selected environmental isolates from a variety of studies were identified with the Biolog microbial identification system according to the manufacturer’s recommendations.
TABLE 1.
Primer mismatch testing in RDP with the CHECK_PROBE algorithm
| Sequence source and taxonomic descriptiona | No. of primer/target mismatchesb
|
RDP ribotype descriptionc | |
|---|---|---|---|
| Ps-for | Ps-rev | ||
| Beta (purple) bacteria | |||
| Iodobacter fluviatile ATCC 33051 | 3 | 0 | Neisseria group |
| Burkholderia solanacearum ATCC 11696 | 1 | 0 | Burkholderia subgroup I |
| Burkholderia pickettii ATCC 27512 | 1 | 0 | Burkholderia subgroup I |
| Spirillum voluntans ATCC 19554 | 0 | 3 | Spirillum voluntans group |
| Azoarcus sp. strain S5b2 | 1 | 0 | Rhodocyclus group |
| Azoarcus indigens VB32 | 1 | 0 | Rhodocyclus group |
| Gamma (purple) bacteria | |||
| Ectothiorhodospira halochloris ATCC 35916 | >3 | 0 | Ectothiorhodospira group |
| Coxiella burnetii Q177 | 1 | 0 | Legionella group |
| Methylococcus capsulatus bath | 2 | 0 | Methylomonas group |
| Oceanospirillum minutulum ATCC 19193 | 0 | 3 | Halomonas group |
| Moraxella osloensis | 0 | NS | Pseudomonas and relatives group |
| Moraxella osloensis SCB111 | 1 | 0 | Pseudomonas and relatives group |
| Pseudomonas aeruginosa ATCC 10145 | 0 | 2 | Pseudomonas and relatives groupd |
| Pseudomonas aeruginosa ATCC 25330 | 1 | 0 | Pseudomonas and relatives groupd |
| Pseudomonas aeruginosa NF13 | 0 | NS | Pseudomonas and relatives groupd |
| Flavobacterium lutescens ATCC 27951 | 0 | 0 | Pseudomonas and relatives groupd |
| Pseudomonas mendocina ATCC 25411 | 0 | 0 | Pseudomonas and relatives groupd |
| Pseudomonas flavescens B62 NCPPC 3063 | 0 | 0 | Pseudomonas and relatives groupd |
| Pseudomonas putida PaW1 isolate mt-2 | 0 | 0 | Pseudomonas and relatives groupd |
| Pseudomonas syringae A501 | 0 | 0 | Pseudomonas and relatives groupd |
| Pseudomonas fluorescens ATCC 13525 | 0 | 0 | Pseudomonas and relatives groupd |
| Azospirillum sp. strain DSM 1727 | 0 | 0 | Pseudomonas and relatives groupd |
| Pseudomonas syringae 31R-1 | 0 | 0 | Pseudomonas and relatives groupd |
| Pseudomonas syringae env. FIE8 | NS | 0 | Pseudomonas and relatives groupd |
| Pseudomonas fluorescens MS 1650 | 0 | 0 | Pseudomonas and relatives groupd |
| Shewanella putrefaciens ATCC 8071 | 3 | 0 | Alteromonas group |
| Delta (purple) Bacteria | |||
| Bdellovibrio bacteriovorus ATCC 43826 | 0 | 2 | Bdellovibrio bacteriovorus group |
| Desulfobacter sp. strain DSM 2057 | 0 | >3 | Desulfobacter subgroup |
| Nannocystis exedens ATCC 25963 | 0 | >3 | Myxobacteria subgroup |
| Epsilon (purple) bacteria | |||
| Bacteroides ureolyticus ATCC 33387 | 0 | >3 | Campylobacter and relatives group |
| Bacteroides gracilis ATCC 33236 | 0 | 2 | Campylobacter and relatives group |
| Campylobacter (13 species and 31 sequences) | 0 | >3 | Campylobacter and relatives group |
| Fuscobacteria and relatives | |||
| Fusobacterium periodonticum ATCC 33693 | >3 | 0 | Fuscobacteria and relatives group |
| Gram-positive bacteria | |||
| Haloanaerobium salsugo VS-752 | 0 | >3 | Anaerobic halophiles subgroup |
| Haloanaerobium praevalens ATCC 33744 | 0 | 3 | Anaerobic halophiles subgroup |
| Clostridium bifermentans NCIMB 10716 | >3 | 0 | Clostridium lituseburense group |
| Clostridium ghonii ATCC 25757 | >3 | 0 | Clostridium lituseburense group |
| Listeria monocytogenes ATCC 35152 | >3 | 0 | Listeria-Brochothrix group |
CHECK_PROBE searches of the RDP prokaryotic SSU database. Sequences perfectly matching with at least one primer were extracted and are listed in phylogenetic order.
The CHECK_PROBE searches allowed as many as three mismatches. The numbers of mismatches observed for the Ps-for and Ps-rev primer targets are listed for each sequence. NS, no primer target site sequence was available; >3, primer target sites exceeding the search cutoff of three mismatches.
Ribotype classifications of the listed sequences as defined by RDP.
The 13 sequences of the Pseudomonas subgroup in RDP.
Bulk DNA extracted from an agricultural field soil, purified (37, 50), and spectrophotometrically (Spectrometer Lambda 2S; Perkin-Elmer, Norwalk, Conn.) quantified as 32.9 to 34.0 μg/g (dry weight equivalent) was kindly provided by L. A. Porteous (U.S. EPA). The field soil (Hyslop Agricultural Field Laboratory, Oregon State University, Corvallis) was classified as Woodburn silt loam and was composed of 72.2% silt, 21.5% clay, 6.3% sand, and 6.3% organic matter.
Primer design.
RDP release 5 (May 1995) was used with a total of 2,849 prokaryotic entries, 13 of which were classified as members of the genus Pseudomonas (27). Specific alignments of 16S rRNA sequences were retrieved by the SUB_ALIGNMENT routine from the RDP (27), and potential PCR primer target regions were manually determined. Their specificity was theoretically evaluated by the CHECK_PROBE routine of the RDP by using the total prokaryotic SSU rRNA sequence database as a target. Primer sequences were redesigned and tested in the same way until optimal specificity was achieved. Novel 16S rRNA gene sequences were retrieved from GenBank with similarity searches with BLAST (2) and were analyzed for matches with the designed PCR primers.
PCR amplification, DNA analysis, and cloning.
The following Ps-PCR primer set was developed: forward primer Ps-for (20-mer [5′-GGTCTGAGAGGATGATCAGT-3′]) and reverse primer Ps-rev (18-mer [5′-TTAGCTCCACCTCGCGGC-3′]). According to the convention of the Oligonucleotide Probe Database (OPD [1]), the names of these primers were S-G-Psmn-0289-a-S-20 and S-G-Psmn-1258-a-A-18, respectively. The universal SSU primer set (11) included forward primer uni-for (17-mer [5′-TGCCAGCAGCCGCGGTA-3′]) and reverse primer uni-rev (18-mer [5′-GACGGGCGGTGTGTACAA-3′]). PCR cocktails for 100-μl reaction mixtures contained 1× reaction buffer (Boehringer Mannheim, Indianapolis, Ind.), 200 nM each deoxynucleoside (Boehringer Mannheim), 5 mg of bovine serum albumin (Sigma Co., St. Louis, Mo.) per ml, 200 nM each oligonucleotide primer (Center for Gene Research and Biotechnology, Oregon State University), and sample DNA in a volume of 95 μl. Approximately 105 cells or a corresponding amount of DNA from pure cultures or 0.5 μg of purified soil DNA was used as a template. PCR amplification was performed with a PTC-100 Thermal Cycler (M. J. Research, Inc., Watertown, Mass.) with a hot bonnet and was run with block temperature control. After initial denaturation for 5 min at 95°C, samples were maintained at 80°C to allow for hot start conditions and addition of 5 μl of enzyme solution containing 1 U of Taq DNA polymerase (Boehringer Mannheim) in 1× reaction buffer. PCR was performed with 30 or 40 temperature cycles under the standard high-stringency conditions listed in Table 2. The use of other annealing temperatures is indicated in the text. A 10-min final extension at 72°C was performed at the end of the cycling steps, and then samples were maintained at 4°C.
TABLE 2.
PCR temperature cycling conditions on an MJR PTC-100 operated with block control
| Method | Sample type | Temp (°C)a under the following conditions and incubation timesb:
|
|||||
|---|---|---|---|---|---|---|---|
| Denaturation
|
Annealing
|
Extension
|
|||||
| 11 s | 15 s | 8 s | 1 min | 10 s | 1 min | ||
| Ps-PCRc | Cultures | 94 | 92 | 66 | 68 | 74 | 72 |
| Ps-PCRc | Soil DNA | 94 | 92 | 63 | 65 | 74 | 72 |
| uni-PCRd | Cultures | 94 | 92 | 60 | 62 | 74 | 72 |
Target block temperature for which the temperature cycler was programmed.
Time period for which the block of the temperature cycler was maintained at the given target temperature.
PCR performed with primers Ps-for and Ps-rev.
PCR performed with the universal SSU primers uni-for and uni-rev.
Five microliters of the PCR products was analyzed on 1% agarose gels (Gibco/BRL, Life Technologies Inc., Gaithersburg, Md.), and 5 μl was reserved for cloning. The remaining 90 μl of PCR products was mixed with 90 μl of precipitation solution (20% polyethylene glycol 8000 and 2.5 M NaCl [Sigma Co.]), and the mixture was incubated for 20 min at 37°C and microcentrifuged at a high speed for 15 min. DNA pellets were washed with 70% ethanol, air dried, and resuspended in 40 μl of 1× restriction enzyme buffer M with 2 U of restriction enzyme HaeIII (Boehringer Mannheim). After overnight digestion at 37°C, restriction fragments were separated by gel electrophoresis with either 2% agarose (Gibco/BRL, Life Technologies, Inc.) or 4% MetaPhor (FMC BioProducts, Rockland, Maine). Molecular weight markers were either phage λ DNA digested with HindIII (Gibco/BRL, Life Technologies, Inc.) or phage ΦX174 DNA digested with HaeIII (Boehringer Mannheim).
PCR products were cloned with the TA cloning kit (Invitrogen Co., San Diego, Calif.) according to the manufacturer’s recommendation. For screening, small amounts of white colonies were added to a Ps-PCR with 30 cycles as described above. PCR product sizes and HaeIII RFLP patterns were analyzed as described above.
The HaeIII RFLP patterns of specific Pseudomonas PCR products were named by using the following system. Capital letters were used for patterns obtained from pure cultures or sequences retrieved from databases. Previously described RFLP patterns observed among cloned soil Ps-PCR products were named by the corresponding lowercase letters. Novel patterns observed among PCR product clones were labelled by lowercase letters of the end of the alphabet (w, x, y, and z).
DNA sequencing and sequence analyses.
Plasmid DNA was prepared with Qiagen 100 plasmid midiprep columns according to the recommendations of the manufacturer (Qiagen, Inc., Chatsworth, Calif.). DNA sequencing was performed at the Center for Gene Research and Biotechnology, Oregon State University. For sequence determination of the cloned Ps-PCR products, a generally applicable sequencing strategy was developed. The sequences for entire cloned PCR products, approximately 990 bp in length, were determined by using four consecutive sequencing reactions on either DNA strand. In addition to the vector-encoded T7 and SP6 sequencing primer sites, forward primers 240-for (5′-TGCCAGCAGCCGCGGTA-3′), 520-for (5′-GATACCCTGGTAGTCCACG-3′), and 800-for (5′-GTGCTGCATGGCTGTCGTC-3′) and reverse primers 280-rev (5′-GCTTTACGCCCAGTAATTC-3′), 520-rev (5′-CGTGGACTACCAGGGTATC-3′), and 800-rev (5′-GACGACAGCCATGCAGCAC-3′) were designed and used for sequencing. The names of these primers indicate the approximate locations and orientations within the Ps-PCR products. Sequence datum-derived HaeIII RFLP fragmentation patterns were determined with MacDNASIS Pro version 3.2 software (Hitachi Software Engineering America, Ltd., San Bruno, Calif.). Similarity searches of the GenBank database were performed with BLAST (2), and those of the RDP database were performed with SIMILARITY_RANK and CHIMERA_CHECK (27). For phylogenetic analyses, novel sequences were manually aligned to the alignments retrieved from RDP by using the multiple alignment routine of MacDNASIS Pro version 3.2. Average linkage cluster analysis of aligned sequences for construction of phylogenetic trees was performed with Treecon version 1.15 (48). Pairwise distance values were determined by the two-parameter model (20), taking into account the fraction of positions differing by transition or transversion. Clustering was determined by unweighted pair group with mathematical averages (UPGMA) analysis of pairwise genetic distance values. Unrooted phylogenetic trees were deduced from 100 bootstrap samplings (9). Maximum parsimony and maximum likelihood analyses were performed by phylogenetic analysis using parsimony (PAUP) version 4.0.0d57 (43) on a Sparc 10 UNIX workstation. For heuristic-parsimony and maximum-likelihood runs, the maximum numbers of retained trees (Maxtrees parameters) were set to 2,000 and 100, respectively. All characters were treated as unordered, and an equal substitution rate was assumed. For each analysis, the strict consensus tree was determined, with the Pseudomonas-specific clone 6 (Ps-clone 6), defined as the outgroup sequence. The resulting trees were manually compared to the UPGMA tree determined with Treecon version 1.15 as described above.
Nucleotide sequence accession numbers.
Nucleotide sequences have been deposited in the GenBank database under accession nos. AF006502 for Ps-clone 1, AF006503 for Ps-clone 3, AF006504 for Ps-clone 15, AF006505 for Ps-clone 4, AF006506 for Ps-clone 14, and AF006507 for Ps-clone 6.
RESULTS
Design and theoretical evaluation of Ps-PCR primers.
The target regions for PCR primers Ps-for and Ps-rev were identified at locations 289 to 308 and 1258 to 1275, respectively, of the P. aeruginosa ATCC 10145 (type species and type strain) rRNA gene, which are homologous to locations 292 to 311 and 1263 to 1280 in the Escherichia coli 16S rRNA gene, respectively. Theoretical hybridization targets for each primer were determined by extensively searching the 2,849 prokaryotic SSU rRNA gene sequences in RDP (Table 1). Ps-for (the forward primer) displayed a perfect match with 11 of 12 available Pseudomonas sequences and 12 of the 2,836 non-Pseudomonas sequences. Ps-rev (the reverse primer) perfectly matched 11 of 12 Pseudomonas sequences and 44 non-Pseudomonas sequences. For the non-Pseudomonas sequence matches, these values represented 0.4 and 1.5%, respectively, of the entire RDP database. In combination, the two primers exhibited a high degree of selectivity for the RDP Pseudomonas subgroup. The only exceptions were two P. aeruginosa sequences (cultures ATCC 10145 and ATCC 25330), each showing mismatches in a different primer (Table 1). The same analysis revealed that all available non-Pseudomonas sequences in the database displayed mismatches for at least one primer, confirming the Pseudomonas selectivity of these primer regions and supporting the feasibility of a 16S rRNA-based Ps-PCR protocol. Amplification may also occur with some non-Pseudomonas sequences of the beta and gamma proteobacterial subdivisions, which display only one mismatch in one of the primers (Table 1) or possibly with sequences not documented in RDP. For further testing of the Ps-PCR primers, 51 recently published Pseudomonas 16S rRNA sequences encoding the whole PCR target sequence were identified in the GenBank database, retrieved, and tested for mismatches (for information on these sequences and references, see Fig. 4). Five of these sequences, i.e., 3 of 17 P. stutzeri (ATCC 11607, LS 401, and SP 1402) sequences, the P. tolaasii (ATCC 33618) sequence, and 1 of 2 Chryseomonas luteola (IAM 13000) sequences, were found to display single mismatches in one primer region. In summary, of 64 tested Pseudomonas 16S rRNA sequences retrieved from the RDP and GenBank databases, only 8 (12.5%) displayed mismatches with the primers, whereas none of 2,836 tested non-Pseudomonas sequences displayed a perfect match for both primers.
FIG. 4.
Phylogenetic analyses of the 6 confirmed Ps-clone sequences (boldface letters) and 70 control sequences from RDP (underlined) and GenBank. The unrooted phylogenetic tree is the result of UPGMA cluster analysis with 100 bootstrap samplings. Bootstrap values larger than 50 are shown. The arrow marked Ps & Rel indicates the branch closely corresponding to the group Pseudomonas and relatives in RDP. The arrow marked Ps (RDP) indicates the cluster containing the Pseudomonas subgroup from RDP. The subbranches labelled I, II, and III are characterized by the specific occurrence of marker species such as P. putida in I, P. aeruginosa in II, and P. fluorescens and P. syringae in III. The open arrow indicates the designed target area of the Ps-PCR protocol. For reference, the GenBank accession or RDP number (∗) is listed. †, sequences within the Ps-PCR target area which displayed mismatches with one of the primers. To demonstrate the diagnostic value of HaeIII RFLP patterns for a majority of Pseudomonas identifications, the HaeIII restriction maps of the Ps-PCR target areas are shown.
Experimental testing of the Ps-PCR protocol on pure cultures.
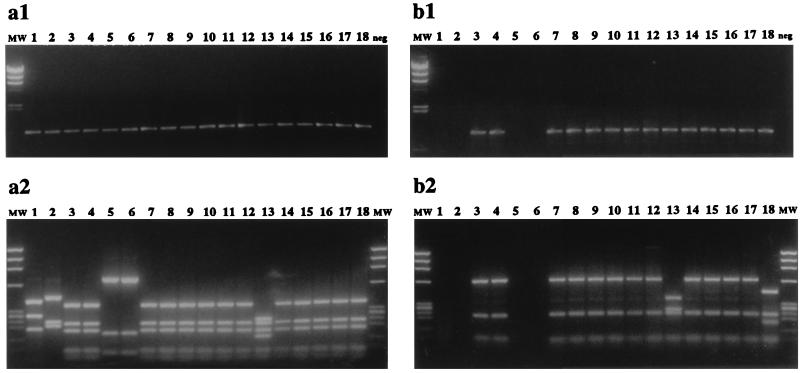
The specificities of the Ps-PCR primers on 64 pure cultures were experimentally tested (Table 3), with PCR performed at annealing temperatures of 62, 65, 68, or 72°C. The cultures analyzed included 18 Pseudomonas and 11 non-Pseudomonas cultures obtained from culture collections and 32 Pseudomonas and 3 non-Pseudomonas soil isolates identified with the Biolog system. The analyses revealed that all 50 Pseudomonas (sensu stricto) cultures tested positive by the Ps-PCR protocol, while the 14 non-Pseudomonas cultures tested negative. All cultures tested positive by the universal-PCR protocol for SSU rRNA genes (Table 3). The 16S rDNA sequence of Burkholderia pickettii (ATCC 27511) displayed 1 mismatch with the forward Ps-PCR primer (Table 1) and was the only non-Pseudomonas sequence tested that required the high annealing temperature of 68°C to prevent its amplification. All Pseudomonas cultures tested positive at this high annealing temperature, including P. aeruginosa (ATCC 10145), the published sequence of which displayed two mismatches with the reverse primer (Table 1). No amplification was observed when annealing was performed at 72°C (data not shown). Figure 1 shows the 16S rRNA gene PCR analyses of 18 pure cultures with the universal SSU primer set (Fig. 1a) or the Ps-PCR primer set (Fig. 1b). PCR products (Fig. 1a1 and 1b1) and corresponding HaeIII RFLPs (Fig. 1a2 and 1b2) are shown. The three RFLP patterns obtained from Ps-PCR products were pattern A (Fig. 1b2, lanes 3, 4, 7 to 12, and 14 to 17), pattern B (Fig. 1b2, lane 13), and pattern C (Fig. 1b2, lane 18). These RFLP patterns represented all of the patterns observed for the pure cultures tested in this study. The HaeIII RFLP patterns shown for the universal SSU primer set (Fig. 1a2) represented 5 of 13 described in this study (Table 3) and were labelled “Psm”.A (lane 1), BurII.A (lane 2), Psm.A (lanes 3, 4, 7 to 12, and 14 to 18), “Psm”.B (lanes 5 and 6), and Psm.B (lane 13). The quotation marks in Psm indicate that these patterns did not represent Pseudomonas (sensu stricto) cultures. In summary, the Ps-PCR protocol was suitable for specific amplification of 16S rRNA Pseudomonas genes, and HaeIII RFLP analysis of Ps-PCR products experimentally confirmed the specificity of the Ps-PCR method for target organisms at 100%.
TABLE 3.
Results of experimental specificity testing of pure cultures with the Ps-PCR primers
| Culture descriptiona |
HaeIII RFLP pattern
|
|
|---|---|---|
| Ps-PCRb | Universal PCRc | |
| Culture collections | ||
| P. putida type B ATCC 17527* | A | Psm.A |
| P. putida ATCC 11172 | A | Psm.A |
| P. aeruginosa ATCC 10145 | A | Psm.A |
| P. syringae pv. syringae Biolog 7350* | A | Psm.A |
| P. syringae pv. syringae EPA 22A-93 | A | Psm.A |
| P. syringae pv. tomato EPA PT-23 | A | Psm.A |
| P. syringae pv. phaseolica EPA JA-019 | A | Psm.A |
| P. cichorii ATCC 10857* | A | Psm.A |
| P. taetrolens Biolog 8570* | A | Psm.A |
| P. synxantha ATCC 9890* | A | Psm.A |
| P. fragi ATCC 4973* | A | Psm.A |
| P. resinovorans ATCC 14235* | A | Psm.A |
| P. mendocina ATCC 25411* | A | Psm.A |
| P. agarici ATCC 25941* | B | Psm.B |
| P. fulva ATCC 31418* | A | Psm.A |
| P. stutzeri ATCC 17588* | A | Psm.A |
| P. alcaligenes B ATCC 14909* | A | Psm.A |
| P. aureofaciens EPA 604 | A | Psm.A |
| Burkholderia pickettii ATCC 27511 | − | BuhI.A |
| Burkholderia andropogonis ATCC 23061* | − | BuhII.A |
| Burkholderia cepacia ATCC 25416 | − | BuhII.A |
| Comamonas testosteroni ATCC 11996 | − | Com.A |
| Acidovorax facilis ATCC 11228 | − | Acv.A |
| Hydrogenophaga flava ATCC 33667 | − | Hyp.A |
| Brevundimonas vesicularis ATCC 11426 | − | Brm.A |
| Stenotrophomonas maltophila ATCC 13637 | − | Stm.A |
| P. floridana ATCC 25616* | − | “Psm”.A |
| Pseudomonas species-like group II Biolog 1574* | − | “Psm”.A |
| P. boreopolis ATCC 3242* | − | “Psm”.B |
| Biolog-identified environmental isolates | ||
| P. putida A1 isolates E8P3D2, E12P3C1, F2P4B3, and 77ys | A | Psm.A |
| P. putida B1 isolates F2C1B1, BMD B2, and BMD B7 | A | Psm.A |
| P. fluorescens type A isolate E2P5B1 | A | Psm.A |
| P. fluorescens type B isolates 75csr, 137ymr, 182ysr, and 228csr | A | Psm.A |
| P. fluorescens type C isolate F4C3D3 | A | Psm.A |
| P. fluorescens type G isolates E2P5A3, F2C1B3, 27cm, 129,ymr, 7A, and 2A | A | Psm.A |
| P. marginalis isolates A5P4C3 and 18B | C | Psm.A |
| P. corrugata isolates E8C1D1 and 104ysr | A | Psm.A |
| P. viridilivida isolates E2P4B3 and E2P4A1 | A | Psm.A |
| P. aurantiaca isolates RES A5 and FB49H | A | Psm.A |
| P. viridiflava isolate RES A7 | A | Psm.A |
| P. fragi isolate 7C | A | Psm.A |
| P. fuscovaginae isolate 7B | A | Psm.A |
| Pseudomonas sp. isolate RES A4 | A | Psm.A |
| Pseudomonas sp. isolate BMD B36 | A | Psm.A |
| Acinetobacter sp. isolate 78 cs | − | Acb.A |
| Acinetobacter calcoacitices isolate FA18C | − | Acb.A |
| Alcaligenes xylosoxidans isolate FA30G | − | Alc.A |
Fifty Pseudomonas and 14 non-Pseudomonas cultures used to test the specificity of the Ps-PCR protocol. ∗, culture DNA sample kindly provided by Biolog Inc.
The HaeIII RFLP patterns of Ps-PCR products. The three detected patterns (as shown in Fig. 1b2) were labelled A, B, and C. −, cultures that did not yield an amplification product by the Ps-PCR protocol with 30 cycles at an annealing temperature of 68°C.
The HaeIII RFLP patterns of PCR products obtained with the universal SSU rRNA gene primer set. Patterns (as shown in Fig. 1a2) were labelled with a genus abbreviation followed by a capital letter. Psm, Pseudomonas; BuhI, Burkholderia type I; BuhII, Burkholderia type II; Com, Comamonas; Hyp, Hydrogenophaga; Brm, Brevundimonas; Stm, Stenotrophomonas; Acb, Acinetobacter; Alc, Alcaligenes; quotation marks, non-Pseudomonas (sensu stricto) cultures that are still named Pseudomonas.
FIG. 1.
PCR amplification and HaeIII RFLP patterns of pure cultures. PCR amplification (30 cycles) of 16S DNA coding for rRNA of pure cultures was performed either with a universal SSU primer set (a) or with the Ps-PCR primer set (b). (a1 and b1) Amplification products from cultures (lanes 1 to 18) and a negative control (neg) on 1% agarose gels. The molecular weight marker (MW) was λ HindIII. (a2 and b2) HaeIII RFLP patterns on 2% agarose gels. The molecular weight marker (MW) was ΦX174 HaeIII. The 18 cultures were “Pseudomonas” boreopolis (ATCC 3242) (lane 1), Burkholderia andropogonis (ATCC 23061) (lane 2), P. cichorii (ATCC 10857) (lane 3), P. syringae pv. syringae (Biolog 7350) (lane 4), “P.” floridana (ATCC 25616) (lane 5), “Pseudomonas” sp.-like group II (Biolog 1574) (lane 6), P. taetrolens (Biolog 8570) (lane 7), P. putida biotype B (ATCC 17527) (lane 8), P. synxantha (ATCC 9890) (lane 9), P. fragi (ATCC 4973) (lane 10), P. recinovorans (ATCC 14235) (lane 11), P. mendocina (ATCC 25411) (lane 12), P. agarici (ATCC 25941) (lane 13), P. fulva (ATCC 31418) (lane 14), P. stutzeri (ATCC 17588) (lane 15), P. alcaligenes (ATCC 14909) (lane 16), P. fluorescens biotype G (isolate 27cm) (lane 17), and P. marginalis (isolate A5P4C3) (lane 18). The Pseudomonas genus names that are in quotation marks are not Pseudomonas (sensu stricto) cultures.)
Ps-PCR analysis of total DNA from an agricultural soil sample.
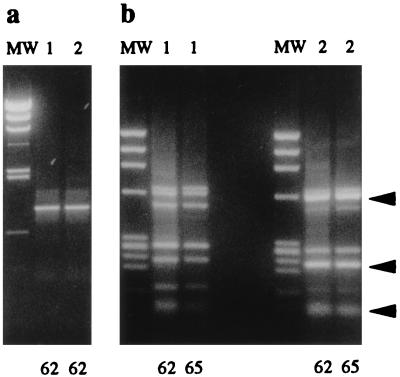
To further evaluate the specificity of the Ps-PCR protocol, purified DNA extracts from an agricultural soil sample were used as a complex source for 16S rRNA gene amplification targets of culturable and nonculturable microorganisms. Two soil DNA preparations extracted from 0.5 g of subsamples of a 100-g soil sample were used for amplification, with annealing temperatures of 62, 65, and 68°C. Amplification products were obtained only for the 62 and 65°C annealing temperatures (Fig. 2a). HaeIII RFLP analysis of the Ps-PCR amplification products revealed small variations in relative band intensities between subsamples. These variations were reproducible and stable at the two annealing temperatures (Fig. 2b). The patterns were composed of seven prominent bands, suggesting a relatively low complexity of the main amplification target in this soil. Some of the bands migrated at molecular weights identical to those for the RFLP pattern A observed for the majority of pure Pseudomonas cultures (arrows in Fig. 2b; Fig. 1b2, lanes 3, 4, 7 to 12, and 14 to 17), suggesting that Pseudomonas 16S rRNA sequences represented a highly enriched component in the amplification product.
FIG. 2.
Ps-PCR products and HaeIII RFLP patterns obtained from bulk DNA purified from an agricultural soil sample. PCR amplification (40 cycles) was performed with DNA from two soil subsamples (1 and 2) with annealing temperatures of either 62 or 65°C (62 or 65, respectively). (a) PCR products in a 1% agarose gel (only products of the 62°C annealing are shown). The molecular weight marker was λ HindIII. (b) HaeIII RFLP patterns in a 2% agarose gel. The molecular weight marker (MW) was ΦX174 HaeIII. Arrows, positions of RFLP pattern A shown in Fig. 1b2 (lanes 3, 4, 7 to 12, and 14 to 17).
Cloning of Ps-PCR products from agricultural soil DNA.
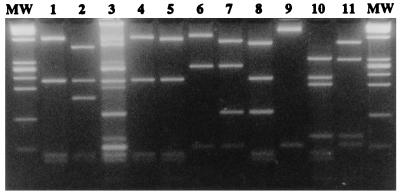
To identify the components present in the soil Ps-PCR-HaeIII RFLP fingerprint as illustrated in Fig. 2b, Ps-PCR products (62°C annealing) were cloned. The library was screened by the Ps-PCR protocol in combination with HaeIII RFLP analysis. Of 40 clones screened, 36 yielded positive Ps-PCR results (Ps-clones). RFLP analysis revealed eight different fragmentation paterns (Fig. 3, lanes 4 to 11). Pattern a (Fig. 3, lane 4) occurred 20 times (55.6%), was identical to pattern A observed for the majority of pure Pseudomonas cultures (Fig. 1b2, lanes 3, 4, 7 to 12, and 14 to 17), and is again shown in Fig. 3, lanes 1 and 4. Pattern w (Fig. 3, lane 7) occurred 10 times (27.7%). The other six patterns (d, e, j, x, y, and z; Fig. 3, lanes 5, 6, and 8 to 11) were all represented once (2.7% each) among the 36 Ps-clones. Gel electrophoresis through 4% MetaPhor revealed comigration of the eight identified patterns (Fig. 3, lanes 4 to 11) with the banding pattern observed in the complex soil DNA fingerprint (Fig. 3, lane 3). This finding, together with the observed frequency of the patterns, indicated that the 36 Ps-clones comprised the majority of Ps-PCR products amplified from the analyzed soil DNA.
FIG. 3.
HaeIII RFLP patterns of Ps-PCR products from pure Pseudomonas cultures, total soil DNA, and Ps-clones. Lanes 1 and 2, patterns A and C obtained from the field isolates P. fluorescens and P. marginalis, respectively; lane 3, high-resolution analysis of the soil DNA fingerprint shown in Fig. 2b (lane 1; 62°C annealing). The eight RFLP patterns (lanes 4 to 11) found among the 36 Ps-clones were pattern a (Ps-clone 3 [lane 4]), pattern d (Ps-clone 15 [lane 5]), pattern j (Ps-clone 1 [lane 6]), pattern w (Ps-clone 4 [lane 7]), pattern e (Ps-clone 14 [lane 8]), pattern x (Ps-clone 13 [lane 9]), pattern y (Ps-clone 2 [lane 10]), and pattern z (Ps-clone 6 [lane 11]). MetaPhor gel (4%) with molecular weight marker (MW) ΦX174 HaeIII was used.
Sequence analysis of Ps-PCR products from the agricultural soil DNA.
Complete sequences were determined for eight Ps-clones representing each of the eight RFLP patterns observed, followed by extensive sequence analyses. Similarity searches in the RDP and GenBank databases revealed that the sequences were of proteobacterial and dominantly Pseudomonas origin. CHIMERA_CHECK analysis in RDP suggested that Ps-clones 2 and 13 (patterns y and x) represented chimeric PCR products of sequences derived from the Oceanospirillum assemblage and Xanthomonas group or Nitrosomonas group and Pseudomonas subgroup genes, respectively (data not shown). SIMILARITY_RANK analyses indicated that Ps-clones 1, 3, and 15 (patterns j, a, and d, respectively) were Pseudomonas sequences and that Ps-clone 6 (pattern z) was derived from a microorganism closely related to the Nitrosomonas or Rhodocyclus group. The results obtained for Ps-clones 4 and 14 (patterns w and e, respectively) were not conclusive, since they indicated the presence of closely related sequences from the Oceanospirillum group and Pseudomonas subgroup or Legionella and Pseudomonas subgroups, respectively. The findings that HaeIII RFLP pattern w, represented by Ps-clone 4, occurred 10 times (27.7%) among the analyzed Ps-clones and that its banding pattern was found by independent PCR suggested that it did not represent a chimeric product. The phylogenetic analyses presented in the following section confirmed and further clarified these initial findings.
In order to evaluate a phylogenetic analysis approach, the 11 full-length Pseudomonas sequences and 4 selected closely related gamma proteobacterial outgroup sequences that encoded the entire Ps-PCR target region were obtained in the form of the 1,007-bp-long RDP alignment (Fig. 4, underlined names). Cluster analysis of this data set with 100 bootstrap samplings resulted in a phylogenetic tree consistent with the RDP phylogenetic data. The Pseudomonas sequences formed a distinct cluster that was joined by a branch of the closely related outgroup sequences (data not shown [but see Fig. 4]). This finding indicated that the Ps-PCR target area contained sufficient information to obtain phylogenetic results consistent with published data.
The 51 new Pseudomonas sequences and 4 new closely related outgroup sequences retrieved from GenBank and the 6 confirmed Ps-clone sequences were aligned with the 11 Pseudomonas and 4 non-Pseudomonas sequences, obtained as a 1,007-bp alignment from RDP. To align all 76 sequences, some adjustments of the RDP alignment were required, resulting in a final alignment length of 1,018 bp (data not shown). This data set was subjected to cluster analysis with 100 bootstrap samplings, resulting in the unrooted phylogenetic tree shown in Fig. 4. The 11 RDP Pseudomonas 16S rDNA sequences (9 from named Pseudomonas species plus those originally identified as Flavobacterium lutescens and Azospirillum isolates) did cluster with 52 related sequences, 47 of which were derived from named Pseudomonas strains. The 5 differently named sequences were from C. luteola (ATCC 43330 and IAM 13000), Flavimonas oryzihabitans (IAM 1568), and Ps-clones 3 and 15. This cluster of 63 sequences formed three main branches (I, II, and III in Fig. 4). Each of these branches was characterized by the specific occurrence of Pseudomonas (sensu stricto) species, i.e., P. putida and P. stutzeri in branch I, P. aeruginosa in branch II, and P. fluorescens and P. syringae in branch III. Ps-clones 3 and 15 localized in different subclusters of branch III. At the same branching point where these main branches originated, a P. oleovorans branch (one sequence) and a C. luteola branch (two sequences) diverged. Six sequences clustered in three branches between these 63 Pseudomonas 16S rDNA sequences and the sequence of SCB11, the outgroup 16S rDNA sequence most closely related to the genus Pseudomonas in RDP. These three branches were formed by Ps-clone 1 together with the undefined Pseudomonas isolate CRE 12; two Lake isolates, 45E3 and 52N3; and Ps-clones 4 and 14. All 69 sequences were located within the target area of the PS-PCR protocol (Fig. 4, open arrow) and formed a distinct cluster with a bootstrap value of 99%. Extensive cluster analysis of the Ps-clone 6 sequence and RDP proteobacterial sequences revealed that it clustered between the Gallionella assemblage (RDP group 2.13.2.5) and Nitrosomonas group (RDP group 2.13.2.6) of the beta proteobacterial subdivision (data not shown). Analyses of the same data set by the maximum-parsimony and maximum-likelihood methods revealed similar results, with two main differences compared to those for UPGMA analysis (data not shown). All methods identified the three clusters (Fig. 4); however, the parsimony and maximum-likelihood analyses identified branch III as a subgroup of branch I. The outgroup sequences also clustered, in agreement with the UPGMA analysis. The second difference involved the nine sequences within the target area of the Ps-PCR (Fig. 4, open arrow) but not located in branches I, II, and III. Parsimony analysis identified exactly the same target area as the UPGMA analysis but located Ps-clones 4 and 14 in branch III and Ps-clone 1 in branch I. Maximum-likelihood analysis located P. oleovorans (IAM 1508), the two lake isolates 45E3 and 52N3, and the Pseudomonas sp. isolate CRE 12 in the outgroup cluster. The two C. luteola (ATCC 43330 and IAM 13000) sequences clustered in branch I, and Ps-clone 1, 4, and 14 sequences clustered in branch III. In summary, all of these analyses revealed that the five Ps-clone sequences 1, 3, 4, 14, and 15, representing patterns j, a, w, e, and d, respectively, and 91.7% of the Ps-clones, clustered within the target area of the Ps-PCR protocol.
DISCUSSION
This study was conducted to develop a 16S rDNA-based PCR protocol for the detection and identification of members of the genus Pseudomonas (sensu stricto). The Ps-PCR primers were subjected to theoretical testing with database information and experimental testing on pure cultures and bulk DNA from an agricultural soil sample.
Database searches for 16S rDNA similarities revealed sequences related to Pseudomonas (sensu stricto) that derived from cultures originally identified by others as belonging to different genera. Pseudomonas classification has attracted much attention in the recent past, and extensive reclassification has been performed (5, 19, 34, 35). However, there still exist uncertainties (Table 1 and Fig. 4) (27). The Ps-PCR protocol may offer a rapid diagnostic tool for genus identification, which was strongly supported by its experimentally confirmed selectivity and the RFLP patterns obtained for Pseudomonas species (Fig. 1 and Table 3).
The 16S rRNA sequences retrieved from RDP and GenBank indicated that about 10% of the Pseudomonas sequences may display mismatches with the Ps-PCR primers (Table 1 and Fig. 4). Experimental evaluation of the PCR approach with pure cultures (Fig. 1 and Table 3) showed that 16S rRNA genes from a variety of Pseudomonas species were specifically amplified by the Ps-PCR. The test included P. aeruginosa (ATCC 10145), the published sequence of which displayed two mismatches with the reverse primer (44). This positive result under the high-stringency annealing condition of 68°C indicated a perfect match of the primers, since at the same temperature no product was obtained for B. pickettii (ATCC 27512; also referred to as Ralstonia pickettii [51]), which has only one mismatch in the forward primer (Table 1). At 72°C annealing, no amplification was observed for any of the samples, indicating the high stringency of the PCR conditions. Database information and available analysis tools (27) were very helpful in the process of primer design, but some caution should be used when interpreting the retrieved information. PCR or cloning artifacts and sequencing ambiguities may account for some of these sequence differences. It cannot be ruled out that some Pseudomonas isolates may have primer target sequences diverging from those described here; however, this was not observed among the 50 pure Pseudomonas cultures.
Stringency evaluation similar to that performed for pure cultures and discussed above was performed for the bulk soil DNA. The results revealed that for such samples, the annealing temperature needed to be lowered to 65°C (Fig. 3). This lower annealing temperature for the soil DNA may be explained by residual soil contaminants slightly altering the conditions for optimal PCR amplification (50). Analysis of B. pickettii indicated that an annealing temperature decrease by 3°C below the highest stringency may allow for amplification of sequences with single mismatches. Since the database analysis indicated that some Pseudomonas sp. strains may exhibit single mismatches and that no significant changes were observed between soil DNA fingerprints with annealing temperatures of 62 and 65°C (Fig. 2), the product of a PCR at 62°C annealing was cloned and analyzed in detail. Among 36 Ps-clones, eight different RFLP patterns were observed. Three patterns that represented only 8.3% of the clones were derived from nontarget sequences. Two were from chimeric PCR products which are a known possible artifact of PCR (26, 30), and one was a beta proteobacterial nontarget sequence. Thus, the reduced annealing temperature did not result in a high rate of nontarget sequence amplification. However, sequences with one mismatch may be more frequent in other samples, and it is recommended that the optimal annealing temperature be evaluated with preliminary DNA fingerprint analysis before cloning Ps-PCR amplification products.
Among the 50 Pseudomonas cultures that represented 21 species, 3 different HaeIII RFLP patterns were detected (Fig. 2; Table 3). Pattern A was found in 47 cultures (94%). The prominent occurrence of this pattern was confirmed by analysis of database information (Fig. 4). Among 63 Pseudomonas sequences in the databases, 52 (82.5%) displayed the HaeIII RFLP pattern A. Among the Pseudomonas cultures, pattern B was found once (2%) in P. agarici and was confirmed in GenBank with one occurrence (Table 3 and Fig. 4). Pattern C was derived from two of the tested cultures identified as P. marginalis with the Biolog system (Table 3). Six additional Pseudomonas sp. HaeIII RFLP patterns were identified in GenBank and were labelled patterns D through I (Fig. 4). The low variability of Pseudomonas 16S RFLP patterns has been reported earlier (25) and was in agreement with the database analysis shown in Fig. 4. The strong relatednes of the HaeIII RFLP patterns of Ps-PCR products may be used as a diagnostic criterion to assist with Pseudomonas identification.
Phylogenetic analyses revealed that the majority of Pseudomonas sequences clustered in three main branches (Fig. 4) characterized by P. putida and P. stutzeri (branch I), P. aeruginosa (branch II), and P. fluorescens and P. syringae (branch III). High similarity between the group of Pseudomonas and relatives in this study and the one of RDP was found, and a possible cluster of the genus Pseudomonas (sensu stricto) was identified (Fig. 4). This potential Pseudomonas branch formed the target area of the Ps-PCR protocol, included five of the six confirmed RFLP patterns found among the Ps-clones, and was largely supported by the three phylogenetic analyses performed in this study. In summary, five sequences (Ps-clones 1, 3, 4, 14, and 15) representing 91.7% of the Ps-clones clustered in the Pseudomonas target area and strongly supported the predicted high Pseudomonas selectivity of the Ps-PCR primer pair.
The Ps-PCR protocol in conjunction with RFLP analysis, sequence determination, and phylogenetic analyses may be used to compare Pseudomonas population structures from a variety of ecosystems and provide further insight about the occurrence, potential roles, and possible unidentified subgroups of this genus in different ecosystems. In addition, it may represent a rapid assay for confirmation of members of the genus Pseudomonas (sensu stricto).
ACKNOWLEDGMENTS
The information in this document has been funded in part by the U. S. Environmental Protection Agency. F.W. and G.D. were supported by postdoctoral research fellowships from the National Research Council, Washington, D.C., and acknowledge technical support received from the U.S. EPA. F.W. received additional support from the Ciba Geigy Jubiläumsstiftung, Basel, Switzerland.
We are grateful to B. Bochner, Biolog, Inc., for providing DNA from pure bacterial cultures; to L. A. Porteous, U.S. EPA, for providing purified agricultural soil DNA; and to S. J. Giovannoni, Oregon State University, for critically reviewing the manuscript.
REFERENCES
- 1.Alm E W, Oerther D B, Larsen N, Stahl D A, Raskin L. The oligonucleotide probe database. Appl Environ Microbiol. 1996;62:3557–3559. doi: 10.1128/aem.62.10.3557-3559.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Amann R, Ludwig W, Schulze R, Spring S, Moore E, Schleifer K-H. rRNA-targeted oligonucleotide probes for the identification of genuine and former pseudomonads. Syst Appl Microbiol. 1996;19:501–509. [Google Scholar]
- 4.Amann R I, Ludwig W, Schleifer K-H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brosch R, Lefèvre M, Grimont F, Grimont P A D. Taxonomic diversity of pseudomonads revealed by computer-interpretation of ribotyping data. Syst Appl Microbiol. 1996;19:541–555. [Google Scholar]
- 6.Christensen H, Poulsen L K. Detection of Pseudomonas in soil by rRNA targeted in situ hybridization. Vet Microbiol. 1994;26:1093–1096. [Google Scholar]
- 7.De Vos D, Lim A, Jr, De Vos P, Sarniguet A, Kersters K, Cornelis P. Detection of the outer membrane lipoprotein I and its gene in fluorescent and non-fluorescent pseudomonads: implications for taxonomy and diagnosis. J Gen Microbiol. 1993;139:2215–2223. doi: 10.1099/00221287-139-9-2215. [DOI] [PubMed] [Google Scholar]
- 8.De Vos P, Goor M, Gillis M, De Ley J. Ribosomal ribonucleic acid cistron similarities of phytopathogenic Pseudomonas species. Int J Syst Bact. 1985;35:169–184. [Google Scholar]
- 9.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 10.Gilligan P H. Microbiology of airway disease in patients with cystic fibrosis. Clin Microbiol Rev. 1991;4:35–51. doi: 10.1128/cmr.4.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giovannoni S J, Britschgi T B, Moyer C L, Field K G. Genetic diversity in Sargasso sea bacterioplancton. Nature. 1990;344:60–63. doi: 10.1038/345060a0. [DOI] [PubMed] [Google Scholar]
- 12.Grimberg S J, Stringfellow W T, Aitken M D. Quantifying the biodegradation of phenanthrene by Pseudomonas stutzeri P16 in the presence of a nonionic surfactant. Appl Environ Microbiol. 1996;62:2387–2392. doi: 10.1128/aem.62.7.2387-2392.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grimont P A D, Vancanneyt M, Lefèvre M, Vandemeulebroecke K, Vautrin L, Brosch R, Kersters K, Grimont F. Ability of Biolog and Biotype-100 systems to reveal the taxonomic diversity of the pseudomonads. Syst Appl Microbiol. 1996;19:510–527. [Google Scholar]
- 14.Guerin W F, Boyd S A. Maintenance and induction of naphthalene degradation activity in Pseudomonas putida and an Alcaligenes sp. under different culture conditions. Appl Environ Microbiol. 1995;61:4061–4068. doi: 10.1128/aem.61.11.4061-4068.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haigler B E, Pettigrew C A, Spain J C. Biodegradation of mixtures of substituted benzenes by Pseudomonas sp. strain JS150. Appl Environ Microbiol. 1992;58:2237–2244. doi: 10.1128/aem.58.7.2237-2244.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holloway B W. Pseudomonas genetics and taxonomy. In: Nakazawa T, Furukawa K, Haas D, Silver S, editors. Molecular biology of pseudomonads. Washington, D.C: ASM Press, American Society for Microbiology; 1996. pp. 22–32. [Google Scholar]
- 17.Johnsen K, Andersen S, Jacobsen C S. Phenotypic and genotypic characterization of phenanthrene-degrading fluorescent Pseudomonas biovars. Appl Environ Microbiol. 1996;62:3818–3825. doi: 10.1128/aem.62.10.3818-3825.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keel C, Weller D M, Natsch A, Defago G, Cook R J, Thomashow L S. Conservation of the 2,4-diacetylphloroglucinol biosynthesis locus among fluorescent Pseudomonas strains from diverse geographic locations. Appl Environ Microbiol. 1996;62:552–563. doi: 10.1128/aem.62.2.552-563.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kersters K, Ludwig W, Vancanneyt M, De Vos P, Gillis M, Schleifer K-H. Recent changes in classification of pseudomonads: an overview. Syst Appl Microbiol. 1996;19:465–477. [Google Scholar]
- 20.Kimura M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 21.Kingsford N M, Raadsma H W. Detection of Pseudomonas aeruginosa from ovine fleece washings by PCR amplification of 16S ribosomal RNA. Vet Microbiol. 1995;47:71–70. doi: 10.1016/0378-1135(95)00061-e. [DOI] [PubMed] [Google Scholar]
- 22.Klingler J M, Stowe S K, Obenhuber D C, Groves T O, Mishra S K, Pierson D L. Evaluation of the Biolog automated microbial identification system. Appl Environ Microbiol. 1992;58:2089–2092. doi: 10.1128/aem.58.6.2089-2092.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kragelund L, Leopold K, Nybroe O. Outer membrane protein heterogeneity within Pseudomonas fluorescens and P. putida and use of an OprF antibody as a probe for rRNA homology group I pseudomonads. Appl Environ Microbiol. 1996;62:480–485. doi: 10.1128/aem.62.2.480-485.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krieg N R, Holt J G, editors. Bergey’s manual of systematic bacteriology. Vol. 1. Baltimore, Md: Williams & Wilkins; 1984. [Google Scholar]
- 25.Laguerre G, Rigottier-Gros L, Lemanceau P. Fluorescent Pseudomonas species categorized by using polymerase chain reaction (PCR)/restriction fragment analysis of 16S rDNA. Mol Ecol. 1994;3:479–487. doi: 10.1111/j.1365-294x.1994.tb00126.x. [DOI] [PubMed] [Google Scholar]
- 26.Liesack W, Weyland H, Stackebrandt E. Potential risks of gene amplification by PCR as determined by 16S rDNA analysis of a mixed-culture of strict barophilic bacteria. Microb Ecol. 1991;21:191–198. doi: 10.1007/BF02539153. [DOI] [PubMed] [Google Scholar]
- 27.Maidack B L, Olsen G J, Larsen N, Overbeek R, McCaughey M J, Woese C R. The ribosomal database project (RDP) Nucleic Acids Res. 1996;24:82–85. doi: 10.1093/nar/24.1.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moore E R B, Mau M, Arnscheidt A, Böttger E C, Hutson R A, Collins M D, Van de Peer Y, De Wachter R, Timmis K N. The determination and comparison of the rRNA gene sequences of species of the genus Pseudomonas (sensu stricto) and estimation of natural intrageneric relationships. Syst Appl Microbiol. 1996;19:478–492. [Google Scholar]
- 29.Moyer C L, Dobbs F C, Karl D M. Phylogenetic diversity of the bacterial community from a microbial mat at an active, hydrothermal vent system, Loihi Seamount, Hawaii. Appl Environ Microbiol. 1995;61:1555–1562. doi: 10.1128/aem.61.4.1555-1562.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Odelberg S J, Weiss R B, Hata A, White R. Template-switching during DNA synthesis by Thermus aquaticus DNA polymerase. Nucleic Acids Res. 1995;23:2049–2057. doi: 10.1093/nar/23.11.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O’Sullivan D J, O’Gara F. Traits of fluorescent Pseudomonas spp. involved in suppression of plant root pathogens. Microbiol Rev. 1992;56:662–676. doi: 10.1128/mr.56.4.662-676.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Palleroni N J. Genus I. Pseudomonas (Migula 1894) In: Krieg N R, Holt J G, editors. Bergey’s manual of systematic bacteriology. Vol. 1. Baltimore, Md: Williams & Wilkins; 1984. pp. 141–199. [Google Scholar]
- 33.Palleroni N J. Human- and animal-pathogenic pseudomonads. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes. A handbook on the biology of bacteria: ecophysiology, isolation, identification, applications. New York, N.Y: Springer Verlag; 1992. pp. 3086–3103. [Google Scholar]
- 34.Palleroni N J. Present situation of the taxonomy of aerobic pseudomonads. In: Galli E, Silver S, Witholt B, editors. Pseudomonas: molecular biology and biotechnology. Washington, D.C: American Society for Microbiology; 1992. pp. 105–115. [Google Scholar]
- 35.Palleroni N J. Pseudomonas classification. A new case history in the taxonomy of gram-negative bacteria. Antonie Leeuwenhoek. 1993;64:231–251. doi: 10.1007/BF00873084. [DOI] [PubMed] [Google Scholar]
- 36.Palleroni N J, Kunisawa R, Contopoulou R, Doudoroff M. Nucleic acid homologies in the genus Pseudomonas. Int J Syst Bact. 1973;23:333–339. [Google Scholar]
- 37.Porteous L A, Seidler R J, Watrud L S. An improved method for purifying DNA from soil for polymerase chain reaction amplification and molecular ecology applications. Mol Ecol. 1997;6:787–791. [Google Scholar]
- 38.Sands D C, Schroth M N, Hildebrand D C. Pseudomonas. In: Schaad N W, editor. Laboratory guide for identification of plant pathogenic bacteria. St. Paul, Minn: American Phytopathological Society; 1980. pp. 36–44. [Google Scholar]
- 39.Sayler G S, Hooper S W, Layton A C, King J M H. Catabolic plasmids of environmental and ecological significance. Microb Ecol. 1990;19:1–20. doi: 10.1007/BF02015050. [DOI] [PubMed] [Google Scholar]
- 40.Schleifer K-H, Amann R, Ludwig W, Rothemund C, Springer N, Dorn S. Nucleic acid probes for the identification and in situ detection of pseudomonads. In: Galli E, Silver S, Withold B, editors. Pseudomonas. Molecular biology and biotechnology. Washington, D.C: American Society for Microbiology; 1992. pp. 127–134. [Google Scholar]
- 41.Spanggord R J, Spain J C, Nishino S F, Mortelmans K E. Biodegradation of 2,4-dinitrotoluene by a Pseudomonas sp. Appl Environ Microbiol. 1991;57:3200–3205. doi: 10.1128/aem.57.11.3200-3205.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stead D. Grouping of plant-pathogenic and some other Pseudomonas spp. by using cellular fatty acid profiles. Int J Syst Bacteriol. 1992;42:281–295. [Google Scholar]
- 43.Swofford D L. PAUP v.4.0.0d57: phylogenetic analysis using parsimony. Washington, D.C: Laboratory of Molecular Systematics, Smithsonian Institute; 1997. [Google Scholar]
- 44.Toschka H Y, Hoeptl P, Ludwig W, Schleifer K-H, Ulbrich N, Erdmann V A. Complete nucleotide sequence of a 16S ribosomal RNA gene from Pseudomonas aeruginosa. Nucleic Acids Res. 1988;16:2348–2348. doi: 10.1093/nar/16.5.2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tyler S D, Strathdee C A, Rozee K R, Johnson W M. Oligonucleotide primers designed to differentiate pathogenic pseudomonads on the basis of the sequencing of genes coding for 16S-23S rRNA internal transcribed spacer. Clin Diagn Lab Immun. 1995;2:448–453. doi: 10.1128/cdli.2.4.448-453.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vancanneyt M, Torck U, Dewettinck D, Vaerewijck M, Kersters K. Grouping of pseudomonads by SDS-PAGE of whole-cell proteins. Syst Appl Microbiol. 1996;19:556–568. [Google Scholar]
- 47.Vancanneyt M, Witt S, Abraham W-R, Kersters K, Fredrickson H L. Fatty acid content in whole-cell hydrolysates and phospholipid fractions of pseudomonads: a taxonomic evaluation. Syst Appl Microbiol. 1996;19:528–540. [Google Scholar]
- 48.Van de Peer Y, De Wachter R. TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput Appl Biosci. 1994;10:569–570. doi: 10.1093/bioinformatics/10.5.569. [DOI] [PubMed] [Google Scholar]
- 49.Weidner S, Arnold W, Pühler A. Diversity of uncultured microorganisms associated with the seagrass Halophila stipulacea estimated by restriction fragment length polymorphism analysis of PCR-amplified 16S rRNA genes. Appl Environ Microbiol. 1996;62:766–771. doi: 10.1128/aem.62.3.766-771.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Widmer F, Seidler R J, Watrud L S. Sensitive detection of transgenic plant marker gene persistence in soil microcosms. Mol Ecol. 1996;5:603–613. [Google Scholar]
- 51.Yabuuchi E, Kosako Y, Oyaizu H, Yano I, Hotta H, Hashimoto Y, Ezaki T, Arakawa M. Transfer of two Burkholderia and an Alcaligenes species to Ralstonia gen. nov.: proposal of Ralstonia pickettii (Ralston, Palleroni, and Doudoroff 1973) comb. nov., Ralstonia solanacearum (Smith 1896) comb. nov., Ralstonia eutropha (Davis 1969) comb. nov. Microbiol Immunol. 1995;39:897–904. doi: 10.1111/j.1348-0421.1995.tb03275.x. [DOI] [PubMed] [Google Scholar]