Abstract
Phenanthrene-degrading bacteria were isolated from a 1-m2 intertidal sediment site in Boston Harbor. Samples were taken six times over 2 years. A total of 432 bacteria were isolated and characterized by biochemical testing. When clustered on the basis of phenotypic characteristics, the isolates could be separated into 68 groups at a similarity level of approximately 70%. Several groups (a total of 200 isolates) corresponded to well-characterized species belonging the genera Vibrio and Pseudomonas. Only 51 of the 437 isolates (<11.7% of the total) hybridized to a DNA probe that encodes the upper pathway of naphthalene and phenanthrene degradation in Pseudomonas putida NCIB 9816. A cluster analysis indicated that the species composition of the phenanthrene-degrading community changed significantly from sampling date to sampling date. At one sampling time, 12 6-mm-diameter core subsamples were taken within the 1-m2 site to determine the spatial variability of the degrading communities. An analysis of molecular variance, performed with the phenotypic characteristics, indicated that only 6% of the variation occurred among the 12 subsamples, suggesting that the subsamples were almost identical in composition. We concluded that the communities of phenanthrene-degrading bacteria in the sediments are very diverse, that the community structure undergoes significant change with time but does not vary significantly on a spatial scale of centimeters, and that the predominant genes that encode phenanthrene degradation in the communities are not well-characterized.
Polycyclic aromatic hydrocarbons (PAHs) are widespread pollutants in the marine environment. These hydrophobic compounds display a high affinity for organic matter and particles and accumulate in organic compound-rich marine sediments (21). An estimated 2.3 × 105 metric tons of PAHs enter aquatics systems every year (16). Urban estuaries in particular, such as Boston Harbor, contain elevated PAH concentrations in their sediments (26). High PAH levels are of public health concern because of the toxic, mutagenic, and carcinogenic properties of PAHs (7, 16). Therefore, bacteria present in contaminated marine sediments are of interest as agents of PAH bioremediation and as models of bacterial population dynamics.
Taxonomically diverse bacteria that are able to utilize low-molecular-weight PAHs, such as naphthalene, phenanthrene, and fluorene, as sources of carbon and energy have been isolated and characterized. For example, bacteria belonging to the genera Pseudomonas (6, 11, 22, 29), Alcaligenes (33), Vibrio (34), Mycobacterium (3, 4, 13), Comamonas (12), Rhodococcus (14), and Cycloclasticus (9) have been isolated from marine sediments and soils. Members of two other PAH-degrading genera (23) previously identified as members of the genera Pseudomonas, Burkholderia, and Sphingomonas are also likely to be isolated from marine waters. However, isolation of pure cultures, which is typically accomplished by enrichment methods, is not necessarily an indication of the importance of organisms as PAH degraders in situ. An understanding of the basic microbial ecology of PAH degraders is still lacking; one fundamental component is to characterize the spatial and temporal variability of the PAH-degrading communities.
We report here on the dynamics of communities or guilds of phenanthrene-degrading bacteria isolated from muddy intertidal sediments over a period of 2 years. One of the major objectives of this study was to determine the extent and scale of diversity of potential phenanthrene-degrading bacteria in moderately contaminated sediments. The isolates were phenotypically characterized and clustered to determine patterns of similarity. A second objective was to determine if the potential phenanthrene-degrading community changes significantly with time. Finally, we wished to determine what portion of the isolates contained the well-characterized genes encoding PAH catabolic pathways. These genes include a portion of the naphthalene dioxygenase gene, nahAaAb, isolated from Pseudomonas putida PpG7 (25, 37) and a gene cluster encoding the degradation of naphthalene, fluorene, and phenanthrene from P. putida NCIB 9816 (36).
MATERIALS AND METHODS
Study site.
Savin Hill Cove is a small extensively intertidal embayment of Boston Harbor (27). It receives PAHs from a storm drain and a combined sewer overflow, as well as from atmospheric deposition and nonpoint source runoff. Its sediments have a mean silt-clay content of 87.5%, a mean total organic carbon content of 34 mg/g, and a mean C/N ratio of 10.4. The site is moderately contaminated with PAHs, and PAH-degrading bacteria are abundant in the sediments (19).
Isolation of phenanthrene-degrading bacteria.
Surface sediment grab samples, ranging in weight from 0.5 to 2 g (wet weight), were taken from Savin Hill Cove over a period of 2 years. Only the top 0.5 cm of the sediment, the aerobic layer, was sampled. Single grab samples were taken by hand on 21 May 1992, 8 June 1992, 23 June 1993, and 18 March 1994. To determine spatial variability, two 6-mm-diameter core samples were taken approximately 15 cm apart on 13 May 1993, and on 11 June 1994 12 6-mm-diameter core samples were taken randomly over an area of 0.9 m2. The sediments were refrigerated at 4°C within 20 min of sampling and were processed within 1 h. Grab samples were placed in sterile 250-ml plastic containers with 50% headspace. The 6-mm-diameter core samples each had a 0.5-cm headspace.
Sediment samples were diluted in 1.5% Instant Ocean (Aquarium Systems, Mentor, Ohio) and were spread onto a modified medium of Anderson (1). This medium contained (per liter) 0.01 g of yeast extract, 0.01 g of peptone, 0.01 g of ferric chloride, 0.05 g of potassium phosphate, 15 g of agar, and either 500 ml of distilled water and 500 ml of filtered seawater or 1 liter of distilled water and 15 g of Instant Ocean. This method was used to enumerate bacteria that degraded phenanthrane as a sole carbon source or metabolized phenanthrene while they were growing on the peptone and yeast extract in the medium. The spread plates were incubated at 25°C for 2 to 3 days and then overlaid with phenanthrene (Aldrich Chemical Co., Milwaukee, Wis.) by spraying a 0.5% solution of phenanthrene in acetone with a chromatography sprayer onto the surfaces of the plates. The overlaid plates were incubated for 4 weeks, and colonies forming zones of clearing in the phenanthrene overlay were picked and streaked to determine purity. Isolates were stored at 4°C on dilute modified Luria-Bertani agar (containing [per liter of distilled water] 5 g of tryptone, 2.5 g of yeast extract, and 15 g of Instant Ocean, as well as 1.5% agar) and were preserved by freezing at −90°C in a solution containing 10% glycerol and 1.5% Instant Ocean.
Phenotypic testing.
The isolates were characterized by determining biochemical characteristics in 24-well tissue culture plates as described by Hansen and Sorheim (15). The methyl red, Voges-Proskauer, chitinase, amylase, β-galactosidase, and urease tests were not performed. Additional tests were performed in the same manner to determine the ability to utilize arabinose, citrate, gluconate, glucose, malate, maltose, mannitol, mannose, and N-acetylglucosamine. The assimilation broth contained (per liter) 0.01 g of yeast extract, 2.0 g of ammonium sulfate, 0.05 g of potassium phosphate, 15 g of Instant Ocean, and 1.0 g of agar. After autoclaving, the carbon sources were filter sterilized and added aseptically to a final concentration of 1% (wt/vol). Inoculations were performed by suspending colony material in 1.5% Instant Ocean in a multiwell plate and using a multipoint inoculator consisting of 24 stainless steel dowels imbedded in a polycarbonate block. In addition, the following morphological characteristics were determined: colony morphology, colony pigment, cell morphology, and the presence of cell inclusions. Selected organisms were also characterized with an API NFT kit (Analytab Products, Montalieu-Verciu, France). Several American Type Culture Collection cultures and one National Collection of Industrial Microorganisms culture were included in the battery of tests (Table 1).
TABLE 1.
Bacterial type culture strains used in this study
| Taxon | Straina | Source |
|---|---|---|
| Pseudomonas fluorescens biotype A | ATCC 13525 | Type strain |
| Pseudomonas fluorescens | ATCC 49838 | Quality control for Micro-Scan products |
| Pseudomonas putida biotype A | ATCC 12633 | Prefilter tanks, England |
| Pseudomonas putida | NCIMB 9816 | Soil |
| Vibrio alginolyticus | ATCC 17749 | Spoiled fish |
| Vibrio harveyi | ATCC 14126 | Dead luminescing amphipod |
| Vibrio parahaemolyticus | ATCC 27969 | Blue crab hemolymph |
| Vibrio parahaemolyticus | ATCC 17802 | Shirasu food poisoning |
| Vibrio proteolyticus | ATCC 15338 | Intestine of Limnoria tripunctata |
ATCC, American Type Culture Collection, Rockville, Md.; NCIMB, National Collections of Industrial and Marine Bacteria Ltd., Aberdeen, Great Britain.
Isolates were tentatively identified to either the genus or species level by comparing their phenotypic characteristics with those of American Type Culture Collection type cultures or by comparing biochemical test results, carbohydrate utilization patterns, and cell morphologies to those of species described in Bergey’s Manual of Systematic Bacteriology (17).
Colony hybridizations.
Two gene probes, both derived from P. putida NCIB 9816 (6), were used in this work. The 16-kb pY3-E16 probe contains the gene cluster encoding the upper pathway for the catabolism of phenanthrene, naphthalene, and fluorene (36). The initial genes of the 16-kb probe make up the smaller, 2.4-kb probe (pY3-2.4). The sequence of the 2.4-kb probe is nearly identical to the sequence of the nahAaAb genes of the NAH7 degradation pathway, which encode reductasenap and ferredoxinnap, respectively (28). Colony hybridizations were performed by using digoxigenin-labeled probes prepared with the Genius System (Boehringer Mannheim Biochemicals, Indianapolis, Ind.). Chemiluminescent detection was performed with a Lumi-Phos 530 apparatus (Boehringer Mannheim). The colonies were applied directly to the hybridization membrane (Magnagraph nylon transfer membrane; MSI, Westboro, Mass.) by using wooden applicator sticks; otherwise, all procedures were the procedures suggested by the manufacturer. P. putida NCIB 9816 and PpG7 were used as positive controls (8). Two pseudomonad strains that do not degrade phenanthrene were used as negative controls. All hybridizations were incubated at 65°C.
Data analysis.
The Levels of relatedness among the bacteria were determined from the phenotypic data by using Jaccard’s similarity index (30). The relationships among the degradative communities present in the 18 samples were analyzed by using the chord normalized expected species shared index (CNESS) (32). Phenograms were constructed by using unweighted pair group mean average (UPGMA) linkage. Indices and clustering were determined by using NTSYS-pc and COMPAH95 (24). COMPAH95 is available at http://www.es.umb.edu/edwebp.htm.
The variation in the composition of the phenanthrene-degrading community among samples was determined by applying the analysis of molecular variance (AMOVA) analytical model (10). This model delineates the extent of genetic or (in this case) phenotypic differentiation within and among populations. It was originally designed to study molecular variation in a single species. Information on DNA genotypes was incorporated into an analysis of variance format derived from a matrix of squared distances for all pairs of genotypes. Estimates of variance components at different levels of hierarchical subdivision were determined. The significance was tested by using a permutational approach. This approach was easily applied to ecological work (in this case using phenotypic characteristics rather than genotypic characteristics) in order to determine the variation within and among the degrading populations from each sampling date. The analysis was performed by using Winamova 1.04, a DOS-based Windows program available through anonymous ftp from acasun1.unige.ch.
RESULTS
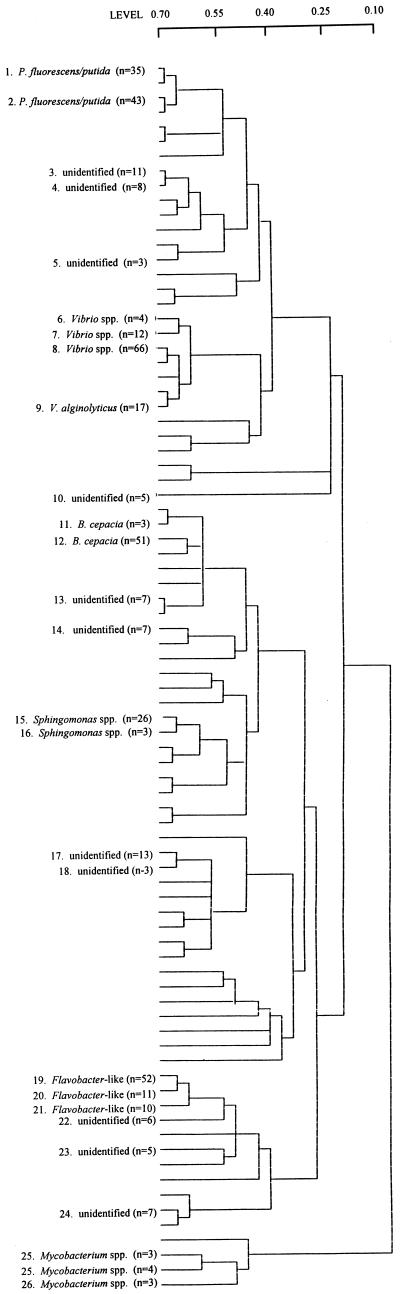
A total of 432 phenanthrene-degrading bacteria were isolated from 18 samples that were taken on six dates in May 1992, June 1992, May 1993, June 1993, March 1994, and June 1994. These isolates were characterized to determine the presence of 35 characteristics. Levels of phenotypic similarity between strains were calculated by using Jaccard’s index, and clustering was performed by using the UPGMA method. At a similarity level of 70%, 83 distinct taxonomic units were identified (Fig. 1). Twenty-seven of the taxa contained 3 to 66 isolates. The remaining 56 taxa contained only one or two members. Based on the placement of standard strains (Table 1), several taxa were tentatively identified. Taxa 1 and 2 consisted of aerobic gram-negative rods which were motile and produced a fluorescent pigment. Taxon 1 contained both Pseudomonas fluorescens and P. putida Savin Hill Cove isolates, and taxon 2 consisted of the P. fluorescens and P. putida type strains and one sediment isolate. Taxa 6 through 9 consisted of Vibrio spp. strains. These isolates were gram-negative, fermentative, motile rods, and at least 63% of them were capable of swarming behavior on agar plates, were arginine dehydrogenase negative, were lysine and ornithine decarboxylase and gelatinase positive, and were capable of utilizing sucrose. These bacteria were identified as Vibrio alginolyticus strains. All of the American Type Culture Collection Vibrio cultures belonged to taxon 8. Taxon 10 contained five unusual, pleomorphic degraders. These isolates were gram variable and formed star-shaped clusters when they were grown in cultures that were shaken and dense tangled mats when they were grown statically. Taxa 11 and 12 resembled Burkholderia cepacia. Taxa 15 and 16 were made up of Sphingomonas spp. Taxa 19 through 21 consisted of Flavobacter-like gram-negative, nonmotile, yellow-pigmented bacteria. These bacteria were oxidase positive and negative for all of the rest of the tests except the nitrite reduction and mannitol utilization tests. Finally, taxa 25 through 27 consisted of pigmented acid-fast bacteria identified as Mycobacterium spp.
FIG. 1.
Phenotypic similarities between isolates. Levels of similarity were calculated by using Jaccard’s index, and clustering was by the UPGMA method. At a similarity level of 70%, 83 taxonomic units were present. Taxonomic units containing three or more strains are numbered, and the presumptive identity of each, if known, is indicated.
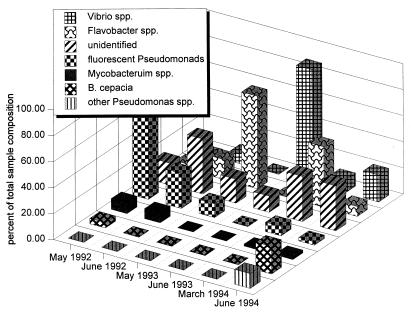
Figure 2 shows the change in the composition of the phenanthrene-degrading community with sampling date. The predominant group of phenanthrene degraders varied with time. Fluorescent pseudomonads (P. putida and P. fluorescens) predominated in May 1992, comprising 64% of all degraders that were isolated. In June 1992, fluorescent pseudomonads and Flavobacter-like spp. together accounted for 42% of the total, but unidentified bacteria comprised another 42%. In May 1993, 71% of the sample consisted of Flavobacter-like species. In June 1993, Vibrio spp. were the most numerous organisms (88% of the degraders isolated). In the March 1994 sample, 46% of the isolates were Flavobacter-like species, while the rest were mostly unidentified. On the last sample date, in June 1994, all of the groups were present, and no single group predominated.
FIG. 2.
Relative changes in species compositions of degrader communities.
None of the colonies hybridized strongly to either gene probe, compared to the control reactions. However, 11 of the March and June 1994 isolates (2.5% of the total isolates) hybridized weakly to the nahAaAb gene probe, which encodes the naphthalene dioxygenase. A greater proportion (11.7%) hybridized weakly to the 16-kb gene cluster, indicating that there was some homology with the upper-pathway genes other than the dioxygenase gene. In all, only 55 of 437 isolates (12.8%) hybridized to the nahAaAb probe or the 16-kb probe encoding the upper pathway of phenanthrene degradation (Table 2).
TABLE 2.
Colony hybridization of phenanthrene-degrading isolates to DNA probes constructed from genes encoding PAH-catabolic pathways
| Sample date | Total no. of isolates | No. of isolates weakly hybridizing to:
|
|
|---|---|---|---|
| nahAaAb probeb | pY3-E16b | ||
| May 1992 | 25 | 0 | 3 (12)c |
| June 1992 | 26 | 0 | 6 (23) |
| May 1993 | 38 | 0 | 10 (26) |
| June 1993 | 41 | 0 | 0 |
| March 1994 | 70 | 8 (11) | 4 (6) |
| June 1994 | 237 | 3 (1) | 28 (12) |
The nahAaAb probe is a 2.5-kb DNA probe encoding reductasenap and ferredoxinnap of the naphthalene dioxygenase gene.
pY3-E16 is a 16-kb DNA probe encoding the upper pathway for phenanthrene, naphthalene, and fluorene degradation and includes nahAaAb.
The values in parentheses are percentages.
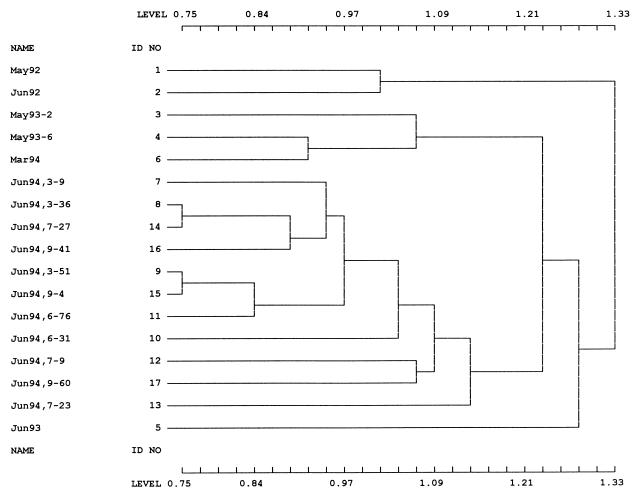
A cluster analysis was performed to determine the relatedness of samples with respect to the taxonomic structure. The resulting dendrogram, based on the CNESS index, reveals that all 12 samples taken at one time in June 1994 formed one distinct group, that the two samples taken at the same time in May 1993 also formed a distinct group, and that the two samples taken a month apart in 1992 formed a third group (Fig. 3). The organisms in the June 1993 sample, which consisted mostly of Vibrio spp., were not closely related to the other groups.
FIG. 3.
Dendrogram showing the relationships among the phenanthrene-degrading communities. The analysis was performed by determining how many isolates belonging to each of the 83 taxa were present in each community. Communities are identified by sampling date and subsample number.
An AMOVA analysis was performed by using the binary data matrix based on phenotypic characteristics. The analysis required a lower triangular distance-squared matrix; therefore, (1 − Jaccard’s coefficient)2 was used. The analysis was based on the data for the six sampling dates, which formed six test groups. Four of the groups consisted of one PAH-degrading community, one group (May 1993) consisted of two communities, and the June 1994 group consisted of 12 communities (total number of communities, 18). The design of the analysis is consistent with the design of a nested analysis of variance. The results of the AMOVA analysis indicate that 20.91% of the phenotypic variation was temporal and only 4.7% of the variation was spatial. The preponderance of the variation (74.3%) was attributed to differences among individuals in each community (Table 3). The phenotypic differences are significant (P < 0.002).
TABLE 3.
Analysis of variance of phenotypic distances for all populations
| Source of variation | Degrees of freedom | Variance | % of total variance | P value |
|---|---|---|---|---|
| Among sample dates | 5 | 0.0498 | 21.06 | <0.001 |
| Among populations, within dates | 12 | 0.0110 | 4.67 | <0.001 |
| Within populations | 417 | 0.1755 | 74.27 | <0.001 |
DISCUSSION
Our results indicate that the natural communities of phenanthrene-degrading bacteria in an intertidal sediment site are taxonomically diverse. This paradigm supports the emerging model of high microbial diversity in aquatic environments (2) and soils (31). We tentatively identified members of several genera, including the genera, Pseudomonas, Burkholderia, Sphingomonas, Flavobacter, Vibrio, and Mycobacterium. Many of these taxa occurred concurrently as a community of PAH-degrading bacteria in a single 0.5-g (wet weight) sediment sample. In addition, 140 isolates, or 32% of the total isolates, remained unidentified. While the ability to degrade phenanthrene is known to be spread widely across genera, this is the first time that such diversity has been reported from a single site. This high diversity is most likely a result of picking the phenanthrene degraders from primary spread plates. An alternative method, enrichment culturing, is typically used to isolate PAH-degrading bacteria, but enrichment selects for only the most rapidly growing strains under laboratory conditions. Even in this study, the diversity of PAH degraders was probably underestimated, since our methods were dependent on cell growth.
The spatial variation in the intertidal site among the communities of PAH degraders was low compared to the temporal variation. A separate AMOVA addressing the phenotypic variation between and among the isolates from 12 replicate core samples taken simultaneously showed that only 5% of the phenotypic variation was among the communities (Table 3). This indicates that there was little spatial variation in the site, whose area was approximately 1 m2. In contrast to the small spatial variation, the structure of the PAH-degrading communities changed significantly with time. In some communities, one taxon predominated. For example, 88% of the June 1993 sample was comprised of Vibrio spp., and 71% of the March 1994 sample contained Flavobacter-like spp. The pooled AMOVA results indicate that the majority (74.27%) of the phenotypic variation was found within each of the 18 communities examined. Only 4.67% of the total variation was present among the 12 June 1994 and 2 May 1993 communities. A substantial portion (21%) of the change in phenotypes was temporal. Thus, while each community itself was very diverse, there was a distinguishable change in the taxonomic composition of the communities at each sampling time.
All of the identified genera detected in Savin Hill Cove sediments have been previously isolated as phenanthrene degraders from aquatic sediments (11, 34). García-Valdés et al. (11) meticulously identified the naphthalene degraders Pseudomonas aeruginosa and P. putida from coastal sediments. Many of the phenanthrene-degrading bacteria isolated in our study were identified as pseudomonads or pseudomonad-like bacteria. Thus, pseudomonads appear to be as important in coastal ecosystems as they are in soils (5). We also isolated typical coastal bacteria (i.e., Vibrio spp.) that are capable of phenanthrene degradation. West et al. (34) have previously described Vibrio isolates as phenanthrene degraders in coastal sediments.
The DNA-DNA hybridization results indicate that naphthalene dioxygenase and the P. putida NCIB 9816 phenanthrene- and naphthalene-degradative genes play only a minor role in the Savin Hill Cove intertidal site. This confirms the results of a previous survey of naphthalene-degrading isolates from soils, freshwater, and marine sediments (20). In that report, only 28.8% of the marine naphthalene degraders hybridized to a nahABCD probe. It is not unexpected that these genes are not predominant in marine isolates, since they were isolated from a soil pseudomonad. Some of the isolates did hybridize weakly with the 16-kb probe but not with the nahAaAb probe, which suggests that there is some homology with at least a portion of the upper-pathway genes. The upper pathway includes genes that encode dehydrogenases, oxygenases, and a ring fission dioxygenase. These genes are not homologous with the archetypical dioxygenase genes of P. putida NCIB 9816.
While none of the isolates in this study displayed strong homology to the nahAaAb and 16-kb probes, it was possible to isolate strongly hybridizing phenanthrene-degrading bacteria from Savin Hill Cove intertidal sediments with naphthalene enrichment cultures (unpublished results). This suggests that the genes which we examined are present only as a minor fraction of the PAH-degradative genes in the community. Ultimately, the relative roles of various PAH-degrading bacteria and their pathways in nature should be examined with nonculture methods, but such an analysis must await isolation and genetic study of the predominant PAH degraders. Recently, Goyal and Zylstra (12) have cloned novel genes for phenanthrene oxidation in Comamonas testosteroni.
Our results are a major step in understanding the ecology of PAH-degrading bacteria in estuarine sediments. The work presented here indicates that the phenanthrene-degrading bacterial community in Savin Hill Cove intertidal sediments is a dynamic system. One source of diversity is the microadaptation of bacteria to the microhabitats on sediment particles (35). Environmental factors, such as temperature, salinity, predation, and organic loading, can also affect the composition of a microbial community (18). These are significant considerations in understanding the fate of PAHs in the system and for developing bioremediation protocols. Our results also show that a single snapshot of a natural community of degraders is not sufficient to characterize a degrading community.
ACKNOWLEDGMENTS
We thank Tom Goodkind for designing and making a multipoint inoculator. We also thank Pam DiBona, Jeff Plate, and John Walsh for reviewing the manuscript.
This work was supported in part by grant 8906397 from the National Science Foundation.
REFERENCES
- 1.Anderson J I W. Studies on micrococci isolated from the North Sea. J Appl Bacteriol. 1962;25:362–368. [Google Scholar]
- 2.Barns S M, Fundyga R E, Jeffries M W, Pace N R. Remarkable archaeal diversity detected in a Yellowstone National Park hot spring environment. Proc Natl Acad Sci USA. 1994;91:1609–1613. doi: 10.1073/pnas.91.5.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boldrin B, Tiehm A, Fritzsche C. Degradation of phenanthrene, fluorene, fluoranthene, and pyrene by a Mycobacterium sp. Appl Environ Microbiol. 1993;59:1927–1930. doi: 10.1128/aem.59.6.1927-1930.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burback B L, Perry J J. Biodegradation and biotransformation of groundwater pollutant mixtures by Mycobacterium vaccae. Appl Environ Microbiol. 1993;59:1025–1029. doi: 10.1128/aem.59.4.1025-1029.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cerniglia C E, Heitkamp M A. Microbial degradation of polycyclic aromatic hydrocarbons (PAH) in the aquatic environment. In: Varanasi U, editor. Metabolism of polycyclic aromatic hydrocarbons (PAH) in the aquatic environment. Boca Raton, Fla: CRC Press; 1989. pp. 41–68. [Google Scholar]
- 6.Davies J I, Evans W C. Oxidative metabolism of naphthalene by soil pseudomonads. Biochem J. 1964;91:251–261. doi: 10.1042/bj0910251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Djomo J E, Ferrier V, Gauthier L, Zollmoreux C, Marty J. Amphibian micronucleus test in vivo: evaluation of the genotoxicity of some major polycyclic aromatic hydrocarbons found in a crude oil. Mutagenesis. 1995;10:223–226. doi: 10.1093/mutage/10.3.223. [DOI] [PubMed] [Google Scholar]
- 8.Dunn N W, Gunsalus I C. Transmissible plasmid coding early enzymes of naphthalene oxidation in Pseudomonas putida. J Bacteriol. 1973;114:974–979. doi: 10.1128/jb.114.3.974-979.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dyksterhouse S E, Gray J P, Herwig R P, Lara J C, Staley J T. Cycloclasticus pugetii gen. nov., sp. nov., an aromatic hydrocarbon-degrading bacterium from marine sediments. Int J Sys Bacteriol. 1995;45:116–123. doi: 10.1099/00207713-45-1-116. [DOI] [PubMed] [Google Scholar]
- 10.Excoffier L, Smouse P E, Quattro J M. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.García-Valdés E, Cózar E, Rotger R, LaLucat J, Ursing J. New naphthalene-degrading marine Pseudomonas strains. Appl Environ Microbiol. 1988;54:2478–2485. doi: 10.1128/aem.54.10.2478-2485.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goyal A K, Zylstra G J. Molecular cloning of novel genes for polycyclic aromatic hydrocarbon degradation from Comamonas testosteroni GZ39. Appl Environ Microbiol. 1996;62:230–236. doi: 10.1128/aem.62.1.230-236.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grifoll M, Casellas M, Bayona J M, Solanas A M. Isolation and characterization of a fluorene-degrading bacterium—identification of ring oxidation and ring fission products. Appl Environ Microbiol. 1992;58:2910–2917. doi: 10.1128/aem.58.9.2910-2917.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grund E, Denecke B, Eichenlaub R. Rhodococcus sp. strain B4, isolated from a soil sample contaminated with polycyclic aromatic hydrocarbons, grows with naphthalene as the sole source of carbon and energy. Appl Environ Microbiol. 1992;58:1874–1877. doi: 10.1128/aem.58.6.1874-1877.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hansen G H, Sorheim R. Improved method for phenotypical characterization of marine bacteria. J Microbiol Methods. 1991;13:231–241. [Google Scholar]
- 16.Kennish M J. Ecology of estuaries: anthropogenic effects. Boca Raton, Fla: CRC Press, Inc.; 1992. [Google Scholar]
- 17.Krieg N R, Holt J G, editors. Bergey’s manual of systematic bacteriology. Vol. 1. Baltimore, Md: Williams & Wilkins; 1984. [Google Scholar]
- 18.Lee S, Fuhrman J A. Spatial and temporal variation of natural bacterioplankton assemblages studied by total genomic DNA cross-hybridization. Limnol Oceanogr. 1991;36:1277–1287. [Google Scholar]
- 19.MacGillivray A R, Shiaris M P. Biotransformation of polycyclic aromatic hydrocarbons by yeasts isolated from coastal sediments. Appl Environ Microbiol. 1993;59:1613–1618. doi: 10.1128/aem.59.5.1613-1618.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.MacGillivray A R, Shiaris M P. Microbial ecology of polycyclic aromatic hydrocarbon (PAH) degradation in coastal sediments. In: Chaudhry G R, editor. Biological degradation and bioremediation of toxic chemicals. Portland, Oreg: Timber Press/Discorides Press; 1995. pp. 125–147. [Google Scholar]
- 21.Means J C, Hassett J J, Wood S G, Banwart W L. Sorption properties of polynuclear aromatic hydrocarbons by sediments and soils. Environ Sci Technol. 1980;14:1524–1528. doi: 10.1021/es60172a005. [DOI] [PubMed] [Google Scholar]
- 22.Mueller J G, Chapman P J, Blattmann B O, Pritchard P H. Isolation and characterization of a fluoranthrene-utilizing strain of Pseudomonas paucimobilis. Appl Environ Microbiol. 1990;56:1079–1086. doi: 10.1128/aem.56.4.1079-1086.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mueller J G, Devereux R, Santavy D L, Lantz S E, Willis S G, Pritchard P H. Phylogenetic and physiological comparisons of PAH-degrading bacteria from geographically diverse soils. Antonie Leeuwenhoek In J Gen Mol Microbiol. 1997;71:329–343. doi: 10.1023/a:1000277008064. [DOI] [PubMed] [Google Scholar]
- 24.Rohlf F J. NTSYS-pc: Numerical taxonomy and multivariate analysis system, 1.8 ed. Stony Brook, N.Y: Exeter Software; 1993. [Google Scholar]
- 25.Schell M A. Cloning and expression in Escherichia coli of the naphthalene degradation genes from plasmid NAH7. J Bacteriol. 1983;153:822–829. doi: 10.1128/jb.153.2.822-829.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shiaris M P, Jambard-Sweet D. Distribution of polycyclic aromatic hydrocarbons in surficial sediments of Boston Harbor, Massachusetts, USA. Mar Pollut Bull. 1986;17:469–472. [Google Scholar]
- 27.Shiaris M P, Rex A C, Pettibone G W, Keay K, McManus P, Rex M A, Ebersole J, Gallagher E. Distribution of indicator bacteria and Vibrio parahaemolyticus in sewage-polluted intertidal sediments. Appl Environ Microbiol. 1987;53:1756–1761. doi: 10.1128/aem.53.8.1756-1761.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simon M J, Osslund T D, Saunders R, Ensley B D, Suggs S, Harcourt A, Suen W C, Cruden D L, Gibson D T, Zylstra G J. Sequences of genes encoding naphthalene dioxygenase in Pseudomonas putida strains PpG7 and NCIB-9816-4. Gene. 1993;127:31–37. doi: 10.1016/0378-1119(93)90613-8. [DOI] [PubMed] [Google Scholar]
- 29.Sisler F D, ZoBell D E. Microbial utilization of carcinogenic hydrocarbons. Science. 1947;106:521–522. doi: 10.1126/science.106.2761.521. [DOI] [PubMed] [Google Scholar]
- 30.Sneath P H A, Sokal R. Numerical taxonomy: the principles and practice of numerical classification. W. H. San Francisco, Calif: Freeman; 1973. [Google Scholar]
- 31.Torsvik V, Salte K, Sorheim R, Goksoyr J. Comparison of phenotypic diversity and DNA hgeterogeneity in a population of soil bacteria. Appl Environ Microbiol. 1990;56:776–781. doi: 10.1128/aem.56.3.776-781.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trueblood D D, Gallagher E D, Gould D M. 3 Stages of seasonal succession on the Savin Hill Cove mudflat, Boston Harbor. Limnol Oceanogr. 1994;39:1440–1454. [Google Scholar]
- 33.Weissenfels W D, Beyer M, Klein J. Degradation of phenanthrene, fluorene, and fluoranthrene by pure bacterial cultures. Appl Microbiol Biotechnol. 1990;32:479–484. doi: 10.1007/BF00903787. [DOI] [PubMed] [Google Scholar]
- 34.West P A, Okpokwasili G C, Brayton P R, Grimes D J, Colwell R R. Numerical taxonomy of phenanthrene-degrading bacteria isolated from the Chesapeake Bay. Appl Microbiol Biotechnol. 1984;48:988–993. doi: 10.1128/aem.48.5.988-993.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wise M G, Shimkets L J, Mcarthur J V. Genetic structure of a lotic population of Burkholderia (Pseudomonas) cepacia. Appl Environ Microbiol. 1995;61:1791–1798. doi: 10.1128/aem.61.5.1791-1798.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang Y J, Chen R F, Shiaris M P. Metabolism of naphthalene, fluorene, and phenanthrene—preliminary characterization of a cloned gene cluster from Pseudomonas putida NCIB 9816. J Bacteriol. 1994;176:2158–2164. doi: 10.1128/jb.176.8.2158-2164.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yen K-M, Serdar C M. Genetics of naphthalene metabolism in pseudomonads. Crit Rev Microbiol. 1988;15:247–268. doi: 10.3109/10408418809104459. [DOI] [PubMed] [Google Scholar]