Abstract
The microtubule-based spindle orchestrates chromosome segregation during cell division. Following more than a century of study, many components and pathways contributing to spindle assembly have been described, but how the spindle robustly assembles remains incompletely understood. This process involves the self-organization of a large number of molecular parts – up to hundreds of thousands in vertebrate cells – whose local interactions give rise to a cellular-scale structure with emergent architecture, mechanics and function. In this Review, we discuss key concepts in our understanding of spindle assembly, focusing on recent advances and the new approaches that enabled them. We describe the pathways that generate the microtubule framework of the spindle by driving microtubule nucleation in a spatially controlled fashion and present recent insights regarding the organization of individual microtubules into structural modules. Finally, we discuss the emergent properties of the spindle that enable robust chromosome segregation.
Introduction
The spindle, first described in the 1880s1 by Walther Flemming, is a bipolar, microtubule-based structure that positions chromosomes at its centre and segregates them into two daughter cells. Errors in spindle assembly and function can lead to aneuploidy, cancer and birth defects2,3. The process of spindle assembly, which occurs between prophase and metaphase of cell division (Fig. 1a), serves as an excellent system to study both fundamental concepts of cellular self-organization and mechanisms whose dysregulation leads to disease.
Fig. 1 ∣. Overview of spindle assembly, from molecular parts to cellular-scale properties.
a, Stages of spindle assembly. During prophase, centrosomes separate to opposite sides of the nucleus and chromosomes become condensed, followed by nuclear envelope breakdown at the end of prophase. In prometaphase, microtubules are nucleated from several sources (see also Fig. 2), and kinetochores begin to capture microtubules. Microtubules that interact with chromosome arms aid in chromosome congression to the middle of the cell. In metaphase, chromosomes align to form the metaphase plate, and bundles of microtubules attached to each kinetochore (termed k-fibres) mature. b, Spatial scales of spindle assembly. Spindle assembly requires formation of microtubules from αβ-tubulin dimers. This process relies on nucleation pathways that target the γ-tubulin ring complex (γ-TuRC) to promote microtubule nucleation in specified locations. Individual spindle microtubules assemble into structural modules through the action of crosslinkers and motor proteins. The spindle is characterized by emergent properties – that is, characteristics of the ensemble that are not exhibited by its component parts – such as its material properties and poleward microtubule flux.
During spindle assembly, the most basic building block of microtubules, tubulin, self-organizes from nanometre-scale dimers to micrometre-scale structural modules such as microtubule-based kinetochore fibres (k-fibres) (Fig. 1b). Although the identities of nearly all spindle parts (for example, microtubule nucleation factors, motors, crosslinkers) are known in many species, how these parts come together to give rise to the spindle’s robust architecture, mechanics and function remains far from clear. Indeed, both conceptually and technically, there is currently a gap between bottom-up approaches (reconstituting systems from their parts) and top-down approaches (breaking systems apart) to study spindle assembly.
We organize this Review into three parts, from the smaller to the larger spatial scales of spindle assembly (Fig. 1b). We first describe how the microtubule framework is built across space and time for efficient spindle assembly. Second, we introduce how motor proteins (such as dynein) and non-motor proteins (such as crosslinkers) organize this microtubule framework into key structural modules, including k-fibres, bridging fibres and astral microtubules, each of which are essential to spindle function. Third, we describe how these structural modules give rise to the metaphase spindle and discuss the emergent properties of the spindle, including spindle dynamics, mechanics and size, which ultimately enable accurate chromosome segregation. Although studying spindle assembly across diverse species will continue to reveal rich insights into the fundamental mechanisms of the cell division machinery, we focus our discussion on animal spindles. We highlight recent studies that have expanded classic concepts and describe how novel developments in molecular, physical and imaging approaches are improving our understanding of animal spindle assembly (Box 1).
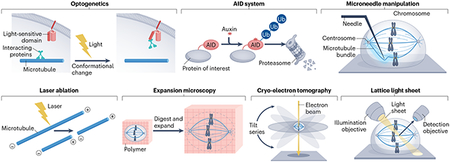
Box 1. Emerging technical approaches to study the spindle.
The process of spindle assembly is highly dynamic in space and time. Several emerging approaches, capable of manipulating and imaging the spindle with high spatiotemporal control and resolution, have complemented classical approaches to improve our understanding of spindle assembly.
Molecular approaches aim to define the mechanisms underlying spindle function. Molecular perturbations with improved spatiotemporal control include optogenetics and the auxin-inducible degron (AID) system. Optogenetic approaches entail tagging proteins of interest with engineered domains whose conformations are sensitive to specific wavelengths of light. This allows their localization or activity to be finely controlled via the spatial pattern, timing and/or intensity of illumination. This strategy has been employed to study the roles of the spindle proteins PRC1 (ref. 151) and EB1 (ref. 165), among others. The AID system takes advantage of a protein degradation system found in plants to deplete proteins with temporal but not spatial precision. This system can be applied to non-plant cells by genetically modifying proteins, such as NuMA, HURP and components of the RanGTP pathway15,176, to contain a small degron tag, which targets tagged proteins for rapid ubiquitylation (Ub) and degradation following induction with the plant hormone auxin. Both optogenetics and AID acutely affect protein behaviour, allowing for protein function at discrete stages of cell division to be studied. Acute perturbations can also reveal phenotypes previously obscured by cellular adaptation after long-term protein depletion. However, optogenetics and AID may not be possible for all proteins, including those with very slow turnover or those for which adding a tag disrupts protein function.
Physical approaches aim to measure and manipulate mechanical forces in the spindle. The spatiotemporal control that some physical approaches provide has been especially helpful in uncovering the specialized mechanics of the spindle in space and time, and probing their molecular basis. Microneedle manipulation, first developed half a century ago126 and recently adapted to mammalian cells212,217, exerts controllable forces of varying magnitude, direction and duration on specific spindle substructures such as kinetochore fibres. Microneedle manipulation in Xenopus egg extract spindles, where no surrounding membrane or cortex is present, can also provide quantitative measures of spindle material properties such as viscoelasticity238,239. Laser ablation acutely and locally severs spindle microtubules, allowing for the relaxation of spindle forces and breaks in spindle architecture. These approaches, while low throughput, provide insights to the material properties and robust structural maintenance of the spindle, and their more recent combination with molecular approaches is beginning to reveal their molecular basis.
Innovations in imaging approaches have improved the spatial and temporal resolution at which we can now observe spindle assembly. Expansion microscopy is a fixed-sample technique in which a sample is covalently bound to a swellable polymer, homogenized via enzymatic digestion, and linearly expanded to ~4.5-fold its original size. This expansion decreases crowdedness within the cell and allows higher spatial resolution of protein localization within the dense spindle architecture, observable through immunofluorescence and standard confocal microscopy271. Cryo-electron tomography utilizes thin frozen samples and acquisition of a series of data points at various (tilted) angles to provide three-dimensional reconstructions of spindle microtubules and structures, such as kinetochore fibres117,118, at angstrom resolution. Lattice light sheet microscopy scans samples using thin sheets of light, allowing high-resolution volumes of whole cells to be acquired with minimal phototoxicity at sub-second intervals60,272-274. With lattice light sheet microscopy, resolving the dynamics of all spindle microtubules is now achievable272,275. However, the slow speed associated with processing and analysing the large data sets generated from these experiments currently limit this technique.
Spindle microtubule nucleation
At the onset of cell division, interphase microtubule structures are disassembled into their basic αβ-tubulin subunits, supplying the cell with material to build the spindle framework. Efficient spindle assembly requires rapid formation of thousands of microtubules. However, spontaneous microtubule nucleation in cells is inefficient. The dividing cell uses several microtubule nucleation pathways to overcome this nucleation barrier at defined locations within the cell such as centrosomes, kinetochores and other microtubules. Together with proteins that organize microtubules and regulate their stability, these nucleation pathways give rise to the bipolar structure of the spindle, in which most microtubule plus ends (characterized by exposed β-tubulin subunits and usually fast growth) are oriented towards chromosomes and most microtubule minus ends (with exposed α-tubulin and slow growth) point towards spindle poles. Individual microtubules are dynamic, nucleating from their minus ends and constantly growing and shrinking, which occurs largely from their plus ends. The lifetimes of individual spindle microtubules (seconds to minutes) are much shorter than that of the spindle, which persists up to an hour to support chromosome segregation. Thus, microtubules must not only be nucleated to build the spindle but also to maintain it and allow it to remodel itself as cell division progresses. Here, we provide an overview of how the barrier to nucleation is overcome, how nucleation pathways regulate microtubule formation in space and time, and how these pathways cooperate to build the spindle.
Molecular basis of nucleation
Microtubules are cylindrical polymers containing 13 filaments composed of αβ-tubulin heterodimers, arranged in a head-to-tail configuration. For microtubules to form, individual tubulin dimers must interact laterally to create the cylindrical microtubule shape. This process requires the formation of a tubulin oligomer consisting of approximately eight tubulin dimers4, which is energetically unfavourable and makes spontaneous microtubule nucleation inefficient5 (Fig. 2a). In higher eukaryotes, efficient nucleation is promoted by a universal microtubule nucleation template, the γ-tubulin ring complex (γ-TuRC). This ~2.2-MDa cone-shaped complex is formed by γ-tubulin complex proteins 2–6 (GCP2–6), mitotic spindle organizing proteins 1 and 2 (MZT1 and MZT2), and actin6-9. Together, these proteins provide a scaffold to position 13 γ-tubulin monomers into a left-handed helix with 13-fold symmetry, roughly mimicking microtubule geometry6-9. This γ-TuRC geometry provides a microtubule minus end template upon which αβ-tubulin dimers assemble, facilitating the formation of critical lateral interactions between individual dimers necessary to overcome the kinetic nucleation barrier10. Recent studies revealed that γ-TuRC-facilitated nucleation requires the assembly of approximately four to seven tubulin dimers to nucleate a microtubule9,10, as opposed to the eight-dimer assembly needed for spontaneous nucleation10. Additionally, γ-TuRC requires the chTOG protein (also known as CKAP5, or Stu2 and XMAP215 in yeast and Xenopus, respectively), which recruits tubulin dimers to the template for assembly. Together, chTOG and γ-TuRC synergistically act to nucleate microtubules and form the universal microtubule nucleation module9-14 (Fig. 2a). Although mechanisms for γ-tubulin-independent nucleation have been reported in cells15,16 and studied in vitro17,18, bipolar spindle assembly requires γ-tubulin15,19 and relies mainly on γ-TuRC-dependent pathways, which are discussed in the next section.
Fig. 2 ∣. Microtubule nucleation during spindle assembly.
a, Spontaneous microtubule nucleation from αβ-tubulin dimers is energetically unfavourable and therefore an inefficient process. Nucleation is facilitated by the universal microtubule nucleation module that comprises the chTOG protein and the γ-tubulin ring complex (γ-TuRC), which mainly consists of γ-tubulin and γ-tubulin complex proteins 2–6 (GCP2–6). γ-TuRC provides a microtubule minus end template for nucleation whereas chTOG recruits αβ-tubulin dimers to speed up microtubule assembly. Microtubule nucleation is localized to specific spindle sites, such as centrosomes (b), existing microtubules (c), kinetochores (d) and centromeres via the chromosomal passenger complex (CPC) (e). Chromosomes play a key role in activating the small GTPase Ran to form RanGTP, which forms a gradient around chromosomes following nuclear envelope breakdown. This gradient spatially activates nucleation on existing microtubules (c) by creating a corresponding gradient of active spindle assembly factors (SAFs). Additionally, RanGTP activates nucleation at kinetochores (d). b, The pericentriolar matrix components pericentrin, centrosomal protein 192 (CEP192) and, possibly, CDK5 regulatory subunit-associated protein 2 (CDK5RAP2) localize γ-TuRC to the centrosome. c, In microtubule-mediated (branching) nucleation, the γ-TuRC-interacting complex augmin binds and localizes γ-TuRC and chTOG to the microtubule lattice. In Xenopus laevis egg extract, this mechanism also requires the RanGTP-activated SAF TPX2, which facilitates recruitment of the remaining factors to microtubules. This pathway results in microtubules of conserved polarity relative to the pre-existing microtubule, which are nucleated at a shallow angle (<30°) and often parallel to the mother microtubule. d, γ-TuRC is localized to kinetochores by the nucleoporin (Nup) complex Nup107–160 and Nup-interactor ELYS and activated in a RanGTP-dependent manner. e, Formation and localization of the chromosome passenger complex (CPC) to the inner centromere activates the CPC component Aurora B kinase. Aurora B phosphorylates and inactivates the microtubule destabilizing protein MCAK and the tubulin-sequestering protein stathmin to facilitate microtubule nucleation and polymerization towards kinetochores, independently of RanGTP.
Key to building a spindle is the spatial and temporal regulation of γ-TuRC nucleation activity. After nucleation, microtubule dynamics are largely modulated by proteins that tune growth and shrinkage, and regulate catastrophe and rescue rates20. It has been proposed that interactors of γ-TuRC, specific to each nucleation pathway, may serve as activators of its nucleation activity21-23 and we describe these in the following sections. How these activators function mechanistically is an open question. Proposed modes of activation include an induced conformational ‘ring closure’ of γ-TuRC to better mimic microtubule geometry, as observed in vitro with yeast γ-TuRC24. This conformational change may be achieved simply as a result of the lateral αβ-tubulin interactions formed between dimers upon assembly on γ-TuRC, which may promote a more microtubule-like configuration of the γ-TuRC structure10. Consequently, increased tubulin concentration alone would increase tubulin binding to γ-TuRC and bias γ-TuRC towards a conformationally activated state10. Recently, γ-TuRC regulation by local tubulin enrichment, driven by the formation of biomolecular condensates, was proposed to occur at centrosomes and on the microtubule lattice25-27. This enrichment may not only overcome the nucleation barrier9,10 but also provide the necessary raw material for spindle assembly exactly where it is needed.
γ-TuRC is targeted to defined locations to construct the spindle in a regulated manner. We next discuss these microtubule nucleation pathways, which are characterized by their means of specifically localizing and activating γ-TuRC at centrosomes and around chromosomes (Fig. 2b-e).
Centrosome-mediated nucleation
In centrosome-containing spindles, such as in animal somatic cells, centrosomes are located at the spindle poles. In preparation for cell division, centrosomes mature by expanding their surrounding pericentriolar material (PCM) through protein accumulation, which is largely controlled by PLK1 and Aurora A kinase activity28,29. Various PCM proteins then perform the key task of recruiting and anchoring γ-TuRC to centrosomes and recruiting microtubule nucleation effectors30-32 (Fig. 2b). The PCM protein CEP192 activates a cascade of protein phosphorylation through Aurora A and PLK1 kinases. While many details remain elusive, this cascade drives γ-TuRC and chTOG localization to centrosomes28,33 and activation of microtubule nucleation34, possibly through phosphorylation of the γ-TuRC interactor NEDD1 (ref. 35). CEP192 and NEDD1 localization is also facilitated by the PCM component pericentrin29, which can also directly anchor γ-TuRC36. Therefore, indirect effects caused by the interdependence of PCM protein accumulation make it difficult to disentangle the individual role of PCM proteins in γ-TuRC recruitment and can complicate functional studies. Additionally, the PCM component CDK5 regulatory subunit-associated protein 2 (CDK5RAP2) can activate γ-TuRC nucleation activity via its γ-TuRC nucleating activator (γ-TuNA) motif (also known as the CM1 motif)21,23,37, although conflicting reports exist on whether CDK5RAP2 can also directly localize γ-TuRC to centrosomes32,38.
In Caenorhabditis elegans, the CDK5RAP2 homologue SPD5 forms a protein-rich compartment, potentially through condensation, that is enhanced by PLK1 and the CEP192 homologue SPD2. This SPD5 compartment recruits the chTOG homologue ZYG9 and other regulatory factors (such as the C. elegans homologue of the Ran-regulated protein TPX2, discussed below), and enriches centrosomal tubulin concentrations tenfold25,26. These findings present a possible model of how tubulin and nucleation factors become enriched at the centrosome in the absence of a membrane, and thus may turn the centrosome into a potent microtubule organizing centre capable of facilitating spindle assembly in centrosome-containing cells.
Chromosome-mediated nucleation
Spindles in most plant cells, Xenopus egg extract and many other animal oocytes form in the absence of centrosomes39-42. Even in many centrosome-containing cells, centrosomes are dispensable for spindle assembly43-45. The ability of spindles to assemble without centrosomes is largely due to several other microtubule nucleation pathways that originate from chromosomes, many of which are regulated by the Ran pathway.
Ran-driven spindle assembly.
Ran is a small GTPase that is converted to its active RanGTP form by the chromatin-associated factor RCC1 (ref. 46). In interphase, RanGTP is concentrated in the nucleus and functions to release nuclear cargo proteins from the transport complex importin-α/β47, whose binding inhibits the function of many nuclear cargo proteins48-51. Many of these proteins regulate microtubule nucleation, organization and dynamics during cell division, and are termed spindle assembly factors (SAFs)48-51. After nuclear envelope breakdown, RanGTP diffuses into the surrounding cytoplasm, where GTP is hydrolysed to form inactive RanGDP, creating a chromatin-centred gradient of RanGTP (Fig. 2). This allows the release of SAFs from importin-α/β in a corresponding gradient52,53. This active SAF gradient facilitates spindle assembly in many systems53, although the mechanistic contributions of many individual SAFs to spindle assembly remain undetermined. Inhibition of the RanGTP pathway leads to severe spindle assembly defects in Xenopus egg extract and mild defects in mammalian cells50,53, which may indicate that chromosome-driven and centrosome-driven systems differentially utilize the Ran pathway during spindle assembly. Below, we describe the possible role of RanGTP in microtubule-mediated and kinetochore-mediated nucleation.
Microtubule-mediated nucleation.
Assembly of the spindle requires the formation of dense and uniformly polar microtubule arrays. The use of existing microtubules to form new microtubules with conserved microtubule polarity provides an elegant means of accelerating this process and has been observed in plants, Drosophila, Xenopus egg extract and human cells54-60. This process, also termed branching microtubule nucleation, nucleates microtubules at shallow angles along the surface of pre-existing microtubules, resulting in exponential microtubule self-amplification58. In vitro reconstitution of this process has been achieved using Xenopus, Drosophila and human proteins. In all cases, the minimal branching components consist of γ-TuRC and the octameric protein complex augmin61-63. The augmin complex is capable of both microtubule binding and γ-TuRC binding, allowing it to localize γ-TuRC to microtubules64,65 (Fig. 2c). Depletion of this universal branching factor leads to defects in k-fibre formation in Drosophila S2 and mammalian cells, and greatly reduces spindle microtubule mass in all spindles studied to date54,56,60,66-68. Thus, the functions of this pathway are conserved despite species-specific differences in its contribution to spindle assembly.
Additional factors are required in some species to drive efficient branching nucleation. In experiments based on Xenopus egg extract and in vitro assays using Xenopus proteins63 (but not in Drosophila S2 cells69), the branching pathway was found to require the SAF TPX2, which itself is regulated by the Ran pathway58. Although this process does not appear to be necessary for branching activity using human proteins in vitro62, the contributions of TPX2 and the Ran pathway in branching microtubule nucleation in human cells require further study.
In Xenopus egg extract, a branch site is initiated by the localization of RanGTP-released TPX2 to the microtubule lattice58,70. In vitro, this occurs by TPX2 condensates forming regularly spaced droplets on microtubules. These droplets confine the space along microtubules in which branching and nucleation factors must search to find each other, thus increasing the speed of branching complex formation in comparison to a uniform TPX2 coating27,71. In addition, TPX2 co-condenses with tubulin in vitro and thus may also supply the branch site with building material for more efficient nucleation27. Apart from mediating branching, TPX2 may stabilize growing microtubules, as seen in vitro72,73. However, further study is required to determine if any of these mechanisms are relevant to branching microtubule nucleation in vivo.
Besides TPX2, the microtubule-binding protein EML3 may facilitate binding of augmin to the microtubule lattice74. Additionally, augmin binding to γ-TuRC may be further supported by interaction with NEDD1 (ref. 56). Altogether, this nucleation pathway promotes spindle self-organization by increasing the number of spindle microtubules while preserving their orientation68,75, which may enable rapid kinetochore capture.
Kinetochore-mediated nucleation.
How spindle microtubules find kinetochores within large eukaryotic cells during the limited timescale of mitosis has been a major question of study. In principle, a solution is for kinetochores to nucleate their own microtubules. Initially proposed to be a safeguard for timely kinetochore attachment to the spindle in case other pathways fail43,76,77, recent studies suggest that microtubule nucleation from kinetochores begins at early prometaphase and is critical for timely kinetochore biorientation77-79. In HeLa cells and Xenopus egg extract, the nucleoporin (Nup) proteins Nup107–160 can localize γ-TuRC to unattached kinetochores via the Nup-interactor ELYS80-82. In Xenopus egg extract, microtubules nucleate from this complex in a RanGTP-dependent manner81 (Fig. 2d). Whereas the Nup107–160 complex is required for bipolar spindle assembly in Xenopus egg extract80, it is not necessary in HeLa cells83. Therefore, the relevance of this mechanism may be species dependent. Interestingly, the formation of kinetochore-associated microtubules requires TPX2 in mammalian cells84. This suggests that branching microtubule nucleation or TPX2-mediated microtubule stabilization at kinetochores may contribute to the formation of these kinetochore-associated microtubules.
Although microtubule nucleation from kinetochores would, in principle, result in incorrectly oriented microtubules with minus ends oriented towards kinetochores, electron microscopy reveals that these microtubules have the correct orientation with plus ends pointing to kinetochores85. The proper orientation could possibly be achieved if the kinetochore rapidly released newly nucleated microtubules, interacted with microtubule walls via the motor CENP-E and the depolymerase MCAK (also known as KIF2C), and subsequently recaptured plus ends78,86. However, a recent study visualizing the minus-end-localized protein ASPM during kinetochore-associated microtubule regrowth in Drosophila S2 cells argues against polarity inversion of kinetochore-mediated microtubules and favours a model in which kinetochores bind short microtubules nucleated in close proximity to the kinetochore by other mechanisms such as chromosome-mediated nucleation pathways87. Thus, visualization methods with better spatial and temporal resolution are required to establish exactly how microtubules form at kinetochores and how prevalent this mechanism is in comparison to other mechanisms active near chromosomes such as nucleation mediated by the chromosomal passenger complex (CPC).
CPC-mediated nucleation.
In the absence of a RanGTP gradient, Xenopus egg extract spindles still form around sperm chromatin, whereas no microtubules form around chromatin beads, which lack both kinetochores and centromeres88. This implies the existence of another spatially regulated pathway at either kinetochores or centromeres that acts complementary to and independent of RanGTP-dependent pathways. This pathway, which is able to compensate for loss of RanGTP-dependent microtubule formation, is mediated by the CPC88. In mammalian cells, this mechanism is required for kinetochore-associated microtubule formation84, reflecting a conserved role in generating spindle microtubules. The CPC consists of four proteins localized at the inner centromere: an inner centromere protein (INCENP) scaffold, Aurora B kinase, and the non-enzymatic subunits Borealin and Survivin (also known as BIRC5)89. Aurora B activity, promoted by CPC formation, phosphorylates and inactivates microtubule-destabilizing proteins such as MCAK and stathmin (Fig. 2e). This effectively promotes microtubule nucleation near kinetochores to facilitate kinetochore capture90-92. Passive diffusion of these phosphorylated Aurora B substrates, such as stathmin, away from the centromere creates a gradient that stabilizes spindle microtubules globally93,94, including microtubules nucleated by other pathways. Interestingly, the CPC appears to form a biomolecular condensate that can concentrate tubulin and directly nucleate microtubules in vitro95. In vivo, the ability of the CPC to form a condensate was proposed to be correlated to its localization to the centromere95. However, the formation of these condensates and their functional significance require validation96.
Regulation and synergy of nucleation pathways
How are these microtubule nucleation pathways coordinated in space and time to give rise to a unified spindle? Centrosomes and chromosomes both begin to nucleate microtubules within a tightly controlled time window, between 9 and 13 minutes following nuclear envelope breakdown in Drosophila spermatocytes97. In larger spindles (such as those in human cells and Xenopus egg extract), branching microtubule nucleation generates the majority of spindle microtubules55,56,67. In human cells, the dominance of the nucleation pathways undergoes a temporal shift, whereby the rate of centrosomal microtubule nucleation is initially higher (directly following nuclear envelope breakdown) than branching nucleation, which becomes the dominant pathway later60. A clear hierarchy of microtubule nucleation also exists in acentrosomal spindles where acentrosomal poles need to be established before acting as a nucleation centre; thus, microtubule nucleation from poles is delayed compared to chromosomal microtubule formation67,98. However, exactly when and where individual chromosome-mediated pathways are activated remains to be defined.
The presence of multiple nucleation pathways may safeguard spindle assembly by providing redundancy. For example, lack of centrosomal nucleation can be compensated for by Ran-mediated microtubule formation and vice versa43,59. Similarly, branching microtubule nucleation can be partially compensated for by the centrosome-mediated pathway in Drosophila54, and the CPC-mediated and Ran-mediated microtubule nucleation pathways can compensate for each other88,99. Despite these redundancies, nucleation pathways largely act in parallel and may work synergistically to speed up spindle assembly53,59. Interestingly, initial microtubule formation during meiosis I occurs at the nuclear envelope during breakdown, suggesting an additional Ran-independent and centriole-independent mechanism that contributes microtubules during spindle assembly100,101.
While microtubule nucleation pathways are regulated in space and time, microtubule nucleators and their activation factors are also transported throughout the spindle. For example, TPX2, although released in a RanGTP-dependent manner surrounding chromatin, is actively and passively transported to poles by motor proteins and poleward flux, respectively102, and thus may distribute branching microtubule nucleation further away from chromosomes. Similarly, γ-TuRC itself is, after binding the spindle, also transported to poles by motor proteins103,104, where it can continue nucleating microtubules. Thus, microtubule nucleation gives rise to spindle architecture and transport of nucleating factors reinforces this architecture by guiding where microtubule-based nucleation takes place.
Structural modules of the spindle
The complex task of chromosome segregation requires that, once nucleated, spindle microtubules are organized into structural modules that fulfil distinct roles such as generating force at kinetochores (k-fibres), providing structural stability (bridging fibres and other non-kinetochore microtubules) and positioning the spindle (astral microtubules) (Fig. 3). Microtubules must also be assembled into a bipolar structure with two focused poles to guide chromosome segregation into two daughter cells.
Fig. 3 ∣. Structural modules in the spindle.
Microtubule-associated proteins, including motors, crosslinkers and regulators of microtubule dynamics, organize microtubules into distinct structural modules. a, Kinetochore fibres (k-fibres) are bundles of parallel microtubules with their plus ends bound to the outer kinetochore protein NDC80. K-fibres are crosslinked by microtubule-associated proteins, including the kinesin KIF15, the transforming acidic coiled-coil-containing protein 3 (TACC3)–chTOG–clathrin complex, and HURP. b, Bridging fibres are bundles of antiparallel microtubules that maintain tension on k-fibres and align chromosomes. PRC1 and the kinesin-5 Eg5 bind and crosslink antiparallel microtubules, and PRC1 recruits the plus-end-directed motor MKLP1 to bridging fibres. c, Other non-kinetochore microtubules play important mechanical roles in stabilizing the spindle, anchoring k-fibres, and mediating polar ejection forces via the chromokinesins Kid (KIF22) and KIF4A. d, Astral microtubules extend from spindle poles towards the cell cortex, where they interact with the Gɑi–LGN–NuMA complex to position the spindle. Their lengths and numbers are regulated by the depolymerases MCAK and KIF18B, that are recruited to plus ends by plus-end-tracking protein EB1. e, Pole focusing is mediated by motors such as dynein that, together with its regulators (NuMA, dynactin), dwells at minus ends and carries these minus end cargos towards neighbouring minus ends. Black arrows indicate directions of motor stepping; blue arrows labelled ‘F’ indicate direction of force on microtubules.
This modular organization is mediated by a plethora of motor and non-motor microtubule-associated proteins (MAPs), which bundle, slide and focus microtubules. Mitotic motors include cytoplasmic dynein and members of several kinesin families (see table 1 in ref. 105). Many of these motors bind to microtubules through both a motor domain and a second microtubule-binding domain, allowing them to bridge two microtubules and thereby crosslink and slide them in an ATP-dependent manner. Non-motor MAPs include passive microtubule crosslinkers, which consume no energy but function in an ATP-independent fashion to regulate bundling, elasticity and friction within the spindle106. Microtubule polymerases and depolymerases also localize to specific structural modules, fine-tuning the dynamics of these modules as they arise during spindle assembly.
Some structural modules have been reconstituted in vitro, including microtubule asters that resemble spindle poles107-110 and anaphase midzone-like antiparallel microtubule bundles111-113. However, other modules, such as k-fibres and bridging fibres, have not yet been reconstituted outside the cell, limiting our understanding of how they form. In the following sub-sections, we discuss recent work, both in vitro and in cells, that has improved our understanding of the ultrastructure, regulation and function of the structural modules that together comprise the spindle and of their variation between species.
K-fibres
Microtubules with their plus ends embedded in kinetochores and their minus ends at or pointing towards spindle poles are known as kinetochore microtubules (KMTs). KMTs attached to the same kinetochore interact with each other and with non-KMTs in a parallel fashion to form k-fibres114. K-fibres generate pulling forces to move chromosomes, and their attachment to kinetochores satisfies the spindle assembly checkpoint115, allowing anaphase entry. Much of what we know about k-fibres comes from mammalian cells, where k-fibres are clearly identifiable by light and electron microscopy. The number of KMTs in a k-fibre varies between kinetochores within a cell and between species. For example, k-fibres vary from 4–40 KMTs in human RPE1 and HeLa cells116-118 to 25–60 KMTs in Indian muntjac skin fibroblasts119. KMTs are tightly bundled near kinetochores and gradually splay out to interact with other KMTs and non-KMTs towards poles117,118. Individual KMTs contact spindle poles either directly or indirectly through interactions with other pole-bound microtubules114,117,118.
How plus end polymerization and depolymerization of KMTs are coordinated within a k-fibre bundle to move chromosomes remains an open question. The coordinated regulation of k-fibre plus end and minus end dynamics is also poorly understood, and factors that accumulate on KMTs in a length-dependent manner may have important roles in this process. For example, the processive motor KIF18A accumulates more strongly at the plus ends of longer KMTs due to the higher number of motors that can bind (the ‘antenna model’ of length regulation)120,121. Upon arriving at plus ends, KIF18A suppresses microtubule dynamics122,123, serving as a feedback mechanism to maintain KMT length and promote chromosome alignment. In contrast, the microtubule stabilizer HURP (also known as DAP5) accumulates near the plus ends of k-fibres in a manner inversely proportional to their length through a mechanism that remains unclear124.
KMTs are thought to be the longest-lived microtubules in the spindle, with a half-life of ~7 min (10–20% of mitosis) in mammalian PtK1 cells125. Their stability stems from the tension they are under following plus end capture by the kinetochore-localized NDC80 complex126, and can be regulated by the phosphorylation state of NDC80 (ref. 127). The long lifetime of KMTs has both mechanical and biochemical implications. Mechanically, k-fibres can propagate and respond to forces on longer timescales than other microtubule populations in the spindle. Biochemically, KMTs can recruit distinct, slower-binding factors compared to other microtubules123. For accurate chromosome segregation, KMTs must be stable enough to preserve correct attachments between chromosomes and the spindle, and yet dynamic enough to allow for error correction when attachments are incorrect128. Many microtubule crosslinking proteins, such as the TACC3–chTOG–clathrin complex116, HURP129,130 and KIF15 (ref. 131), specifically localize to k-fibres (Fig. 3a), possibly due to a preference for binding bundled parallel microtubules. These proteins are thought to reinforce the bundling of KMTs and maintain k-fibre integrity132 and, upon physical manipulation with a microneedle (Box 1), k-fibres appear to behave as a single entity133.
How k-fibres form varies between species and remains an area of active research. In centrosome-containing cells, the search-and-capture model for k-fibre formation posits that dynamic centrosome-originated pioneer microtubules search the cytoplasm until they eventually capture kinetochores and become stabilized134 (Fig. 4a). This process could indeed be recapitulated via computational modelling135 considering: (1) RanGTP-dependent microtubule stabilization136; (2) pre-alignment of kinetochores towards poles through initial lateral microtubule–kinetochore interactions137; and (3) the expansion of the outer kinetochore during early mitosis138,139, which increases the surface area of kinetochores and their probability of capture135,139.
Fig. 4 ∣. K-fibre formation mechanisms.
a, Kinetochore fibre (k-fibre) formation can occur through two pathways, search-and-capture and chromosome-mediated assembly. In search-and-capture, dynamic centrosome-nucleated microtubules ‘search’ the surrounding cytoplasm for kinetochores as they grow and become stabilized upon kinetochore attachment. In chromosome-mediated assembly, microtubules nucleate via chromosome-mediated and kinetochore-mediated pathways described in Fig. 2. b, A nascent k-fibre typically begins to form when a kinetochore binds to the lateral surface of a microtubule. This laterally bound microtubule undergoes reorientation driven by the plus-end-directed kinesin CENP-E and the microtubule depolymerase MCAK, resulting in stable attachment of microtubule plus ends to the outer kinetochore NDC80 complex. The minus ends of these kinetochore-bound microtubules (KMTs) become bound by the minus-end-binding motor complex NuMA–dynein–dynactin. This minus-end-directed motor complex transports KMTs towards spindle poles (arrows). These nascent k-fibres additionally undergo branching microtubule nucleation, which serves to quickly amplify KMT numbers. c, The mature k-fibre forms both direct and indirect connections with the spindle poles, whereby KMTs of a single k-fibre that are bundled together near the kinetochore gradually splay out towards their minus ends to interact with other microtubules. Ongoing microtubule nucleation, mainly through branching nucleation, maintains the density of the k-fibre.
In addition to this search-and-capture mechanism, and in acentrosomal systems, k-fibres can originate through chromosome-mediated microtubule nucleation (Fig. 4a), followed by efficient lateral binding of these microtubules by kinetochores78,137,140,141 and subsequent conversion of kinetochore-bound microtubules to end-on attachments78,86 (Fig. 4b). The microtubule-binding protein NuMA can then bind the minus ends of KMTs, recruiting dynein and dynactin142. This NuMA–dynein–dynactin complex walks these minus ends as cargo towards the minus ends of nearby microtubules and thereby towards spindle poles76-78,84,143,144 (Fig. 4b). Upon formation of an initial KMT, amplification mechanisms ensure the establishment of k-fibres rather than relying on capture of additional KMTs (Fig. 4b). Branching microtubule nucleation biases microtubule amplification of the k-fibre towards kinetochores60,68. In vitro, TPX2 accumulates on more stable, long-lived microtubule lattice regions70, which could provide a mechanism to target branching microtubule nucleation to k-fibres. Altogether, the cooperation of multiple mechanisms ensures the robust and efficient formation of mature k-fibres (Fig. 4c). After formation, k-fibres are stabilized in part by bridging fibres, another structural module discussed in the next sub-section.
Bridging fibres
Many non-KMTs that cross the metaphase plate are crosslinked in antiparallel bundles known as bridging fibres118,145,146 (Fig. 3b), a structural module with important mechanical functions in the mammalian spindle. Near chromosomes, these bundles form one-to-one associations with pairs of sister k-fibres, that is, k-fibres that form on each of two sister chromatids147. Electron tomography has revealed that like k-fibres, bridging fibres splay apart closer to poles and their microtubules intercalate with multiple k-fibres118. Their minus ends form junctions with KMTs along the length of the k-fibre, although the identities of the factors mediating these junctions remain unknown. Some of these junctions are established as close as 2 μm from kinetochores in RPE1 cells118, suggesting that k-fibre anchorage within the spindle microtubule network differs outside this region. The number of microtubules within bridging fibres varies between cell types (6 ± 1 in PtK1 versus 14 ± 2 in HeLa cells, ~;25% and ~82% of the average number of microtubules in k-fibres, respectively)146. The higher number of bridging fibre microtubules in HeLa cells may reflect a more important mechanical role for bridging fibres in more curved spindles.
The crosslinker PRC1, which is specifically enriched on antiparallel microtubules111,148-150, has a central role in maintaining and organizing bridging fibres146,147,151. Optogenetic removal of PRC1 results in a ~2.5-fold reduction of bridging fibre microtubule density and reduces the levels of other molecular regulators localized to bridging fibres, including the plus-end-directed motors KIF4A and MKLP1 (also known as KIF23)151. Reduced levels ofthese factors after PRC1 removal have been proposed to result in increased antiparallel overlap, reduced outward sliding of the bridging fibre and disrupted chromosome alignment151.
Bridging fibres play an important mechanical role, acting to support k-fibre tension and curvature, which are required to maintain correct kinetochore–microtubule attachments and chromosome alignment. Optogenetic removal of PRC1 causes spindles to become more diamond shaped151. Laser severing of k-fibres in HeLa cells close to the kinetochore (and thus disconnecting the remaining k-fibre stub from the bridging fibre) has been shown to result in greater relaxation between sister kinetochores, further corroborating the role of bridging fibres in maintaining tension between kinetochores146. Bridging fibres may also accelerate chromosome biorientation by providing antiparallel microtubules in the middle of the spindle upon which dynein can transport nascent KMTs towards each pole79. Together, ultrastructural, optogenetic and biophysical approaches (Box 1) have revealed that the bridging fibre is a key structural module that aids in the assembly and mechanical stabilization of k-fibres.
Other non-KMTs
Much of the microtubule density of spindles is composed of non-KMTs that are not bundled into bridging fibres145. This microtubule population plays important roles in stabilizing spindle structure. The proportion of non-KMTs in the spindle ranges from ~50% of total microtubules in PtK1 cells152 to ~95% in Xenopus laevis extract spindles153 and ~86% in HeLa cells117. These non-KMTs are shorter on average than KMTs117,154,155 and arise mainly via branching microtubule nucleation55,60. Non-KMTs undergo rapid turnover (polymerizing and depolymerizing within seconds)156, which may allow spindles to quickly remodel as chromosomes move during mitosis.
During spindle assembly, non-KMTs aid in chromosome alignment via interactions with the chromokinesins Kid (also known as KIF22) and KIF4A. These plus-end-directed motors bind chromatin and generate polar ejection forces by walking along non-KMTs, thereby pushing chromosomes away from poles and towards the spindle equator157-159 (Fig. 3c). Non-KMTs are densest near poles owing to their continuous poleward transport104, generating higher polar ejection forces there and thus contributing to a self-organized chromosome-centring mechanism160-162.
Non-KMTs also interact with k-fibres and bridging fibres114,117,118, mechanically linking the various structural components of the spindle. As discussed below, these interactions are important to many emergent properties of the spindle such as k-fibre load bearing, poleward flux and elasticity. In sum, the dense network of non-KMTs helps align chromosomes and structurally integrate the spindle, making it robust against mechanical force.
Astral microtubules
Astral microtubules are dynamic, non-bundled microtubules that are nucleated at the centrosome and extend towards the cell cortex, with important roles in positioning the spindle (Fig. 3d). Their numbers and length are largely controlled by EB1 (also known as MAPRE1), a MAP that binds growing microtubule plus ends. EB1 recruits the depolymerases KIF18B and MCAK to mediate continuous astral microtubule pruning163,164, required to maintain spindle length as shown by optogenetic disruption of EB1 (ref. 165). Astral microtubules are molecularly distinguished by relatively low levels of tubulin detyrosination, whereas microtubules pointing towards the metaphase plate are detyrosinated. Tyrosinated tubulin inhibits the motility of the kinetochore motor CENP-E, biasing chromosome congression towards the metaphase plate and preventing chromosome accumulation near the cell cortex166.
The role of astral microtubules in positioning the spindle has been summarized in recent reviews167,168. Correctly positioning the spindle is important in both symmetric cell divisions, where astral microtubules enable the spindle to respond to cell shape cues169-171, and in asymmetric divisions, where daughter cells take on different fates. Throughout mitosis, astral microtubules are linked to the cell cortex via the evolutionary conserved ternary complex of Gɑi–LGN–NuMA172-174. This complex recruits dynein and dynactin to generate pulling forces that translate the spindle. Optogenetics (Box 1) has been valuable in studying spindle positioning, revealing that cortical recruitment of NuMA but not dynein itself is sufficient to orient the spindle in C. elegans and human HCT116 cells175,176. Although astral microtubules are not required to assemble a bipolar spindle, they fulfil a critical function in linking the spindle to cues from its tissue environment.
Spindle poles
Focused spindle poles incorporate microtubules belonging to several of the classes described above (KMTs, astral microtubules and other non-KMTs), and we here consider them as an independent architectural module that serves as a regulatory hub for microtubule nucleation and dynamics. Although spindle poles contain centrosomes in most cells, the formation of focused poles in acentrosomal oocytes39-41 and somatic cells after centrosome ablation43,45,177-180 has established that centrosomes are not necessary for the formation of focused spindle poles. Instead, pole focusing is driven by minus-end-directed motors that are able to dwell at microtubule minus ends and transport these minus end cargos along neighbouring microtubules – the same mechanism that forms microtubule asters in vitro109.
Dynein and its cofactors dynactin and NuMA are key players in pole focusing (Fig. 3e), and their inhibition leads to unfocused barrel-shaped or turbulent spindles39,107,108,181-184. NuMA localizes to minus ends, where it recruits dynactin and dynein to walk along neighbouring microtubules, facilitating the end-dwelling behaviour142. The large and multifunctional NuMA likely contributes to pole focusing in many ways: in addition to localizing dynein, it is a putative activating adaptor of dynein motility176,185 and may have auxiliary roles in organizing microtubules via passive crosslinking186 and/or phase separating at spindle poles187,188. The kinesin-14 family of motors, including Xenopus XCTK2, Drosophila Ncd and human HSET (also known as KIFC1), also contributes to pole focusing, although the exact mechanisms by which these motors organize spindle poles remain unclear. The relative importance of dynein and kinesin-14 varies between species, with Ncd playing a dominant role in Drosophila182, whereas HSET depletion has only mild phenotypes in human mitotic spindles189,190.
Functionally, poles and the motors that focus them are important in concentrating minus end regulatory factors like γ-tubulin. As γ-tubulin accumulates during pole maturation, it enhances microtubule nucleation and stabilizes the focused pole, even in acentrosomal spindles that initially lack nucleation at poles67,104. Pole focusing also functions to maintain a steady-state spindle shape, thereby ensuring that growing and shrinking microtubules are confined to the spindle region without interacting with organelles or generating cytoplasmic flows183. Interestingly, many plant spindles lack focused poles, suggesting that other geometric cues like the plant cell preprophase band (a ring of microtubules below the plasma membrane that marks the future division plane) can facilitate bipolar divisions while circumventing the need for pole focusing42.
Spindle bipolarity
The assembly of a spindle with two poles is critical to segregate chromosomes into two daughter cells. Monopolar, aster-shaped spindles are often the result of failed spindle assembly, and multipolar spindles are associated with genomic instability and cancer191. The bipolar homotetrameric kinesin-5 motors (Eg5 (also known as KIF11) in humans) play a central role in establishing spindle bipolarity in both acentrosomal and centrosome-containing spindles192,193 due to their ability to crosslink and slide antiparallel microtubules apart194,195.
In prophase, spindle bipolarity is templated by the sliding apart of duplicated centrosomes to opposite sides of the nucleus. This movement is driven by the pulling forces of dynein attached to the nuclear envelope196 and by Eg5-mediated antiparallel sliding of centrosomal microtubules. After nuclear envelope breakdown, microtubules from opposite centrosomes form additional antiparallel overlaps on which Eg5 can localize. Nascent k-fibres also promote bipolarity, potentially due to an outward pushing force on centrosomes from k-fibre polymerization at kinetochores197,198. In metaphase, spindles in many mammalian cell types remain bipolar even if Eg5 is inhibited due to the partially redundant activity of the kinesin-12 KIF15 (refs. 199-202), which allows maintenance though not initial generation of spindle bipolarity.
Branching microtubule nucleation also contributes to spindle bipolarity. Because new microtubules are formed at shallow branch angles from pre-existing microtubules58, this pathway both maintains microtubule polarity and generates closely juxtaposed arrays to which motors and crosslinkers can rapidly bind. Indeed, augmin depletion leads to multipolar spindles in both acentrosomal67 and centrosome-containing55 spindles. Thus, the cooperation of multiple motor-based and nucleation pathways between prophase and metaphase promotes the robust assembly of a bipolar spindle, setting the stage for chromosome biorientation and equal segregation into two daughter cells.
Emergent properties of the spindle
The spindle exhibits many cellular-scale emergent properties that are qualitatively different from those of its individual components and enable the spindle to segregate chromosomes accurately. These include dynamic features such as microtubule flux and continuous self-repair, mechanical features such as spatially varying viscoelasticity, and architectural features such as helical twist and the scaling of spindle size with cell size. While the full complexity of the emergent properties of the spindle cannot yet be reconstituted in vitro, recent studies have begun to elucidate the mechanistic basis for some of these features.
Robustness
The spindle is subject to mechanical and biochemical noise such as variations in gene expression and forces from neighbouring cells. However, even a single segregation error can give rise to disease203. Several failsafe mechanisms have been identified that allow the spindle to maintain its architecture and accurate function in the face of such variations.
Redundancy.
Redundancy exists in many spindle processes, including microtubule nucleation and motor activity. As discussed above, the spindle robustly generates the microtubule mass required for bipolar assembly. The Ran pathway can compensate for the absence of centrosomes43,59 and vice versa53. Several motors with partially redundant function also cooperate to promote correct spindle architecture (Fig. 5a). KIF15 maintains bipolarity when Eg5 is inhibited in metaphase199,200 and, when overexpressed, can fully compensate for loss of Eg5 in bipolar spindle assembly198,199. Further, kinesin-14 motors focus poles in a manner partially redundant with dynein, and cells can tolerate loss of the human kinesin-14 HSET during mitosis189,190 unless dynein is simultaneously inhibited204,205.
Fig. 5 ∣. Mechanisms underlying spindle robustness: redundancy, anchorage and dynamics.
a, Partial redundancy between motor proteins makes spindle assembly robust to perturbations and variations in motor activity. KIF15 can compensate for Eg5 inhibition to maintain spindle bipolarity. Similarly, NuMA–dynein–dynactin complexes have dominant roles in pole focusing in human spindles but the pole-focusing activity of HSET can compensate for reduced dynein activity. b, Plus-end-specific and minus-end-specific mechanisms reinforce key connections between the kinetochore fibre (k-fibre), pole, and kinetochore and these can be experimentally dissected by microneedle manipulation. On short (<20 s) timescales, PRC1-mediated crosslinking maintains k-fibre orientation, preventing k-fibres from pivoting around kinetochores. On longer (minutes) timescales, this reinforcement near kinetochores relaxes, allowing k-fibres to pivot around kinetochores. Plus ends undergo persistent polymerization while depolymerization at minus ends ceases, allowing the k-fibre to lengthen in response to external manipulation. These responses allow the k-fibre to locally dissipate force (F), remodelling at the individual k-fibre level to preserve global spindle architecture.
Motor redundancy has important implications for the therapeutic targeting of cell division such as in anticancer therapy. For example, KIF15 is essential for cells to acquire resistance to Eg5 inhibitors206. An interesting case of redundancy involves the opposing motors Eg5 and dynein. Inhibiting either motor alone leads to monopolar or unfocused spindles, respectively, but inhibiting both together results in bipolar spindles of normal size as the redundant motors discussed above can compensate for their functions205,207-209. However, these Eg5-inhibited and dynein-inhibited spindles are more mechanically fragile and more error-prone in chromosome segregation205, potentially as a consequence of locally altered microtubule organization205,210. This indicates that Eg5 and dynein make the spindle more robust to perturbations although their combined inhibition does not disrupt overall spindle architecture.
Local k-fibre remodelling to preserve global architecture.
Specialized mechanisms reinforce and repair key spindle connections at both the plus and minus ends of k-fibres. The NuMA–dynein–dynactin complex acts at minus ends to anchor k-fibres in the spindle. Not only does this complex transport KMT minus ends poleward during k-fibre formation, as discussed above, but it is also sufficient to re-establish k-fibre connections to the poles after acute perturbations such as laser severing143,144 (Box 1). Recent studies have also identified mechanisms that anchor the first few microns near k-fibre plus ends within the spindle microtubule network, protecting kinetochore–microtubule attachments from acute perturbation77,146,211,212. PRC1 has been shown to provide short-lived reinforcement in this region146,212, presumably by crosslinking KMTs with nearby antiparallel non-KMTs, thus preventing microtubule pivoting near kinetochores in response to external force supplied by a microneedle212 (Fig. 5b). NuMA has also been shown to support k-fibre anchorage near the plus end211, an effect that may reflect crosslinking between KMTs and the minus ends of short microtubules in the centre of the spindle. On the longer (approximately minutes) timescale, these reinforcements are relaxed212, allowing spindle remodelling and chromosome movement (Fig. 5b). Together, these mechanisms ensure that k-fibres remain robustly attached at kinetochores and anchored at poles.
Tension regulates microtubule dynamics at both plus and minus ends, enhancing spindle robustness to mechanical perturbation. Tension on microtubules decreases the catastrophe rate and increases the rescue frequency of microtubules at purified yeast kinetochores, stabilizing kinetochore–microtubule attachments under force213,214. In microneedle manipulation experiments in newt epithelial cells215, grasshopper spermatocytes216 and, more recently, mammalian PtK2 cells217, individual k-fibres elongated under applied force, demonstrating that similar principles apply to bundled k-fibres composed of many microtubules. At minus ends, depolymerization is inhibited when tension is applied to k-fibres either globally via cell compression218 or individually via a microneedle217 (Fig. 5b). The molecular mechanisms underlying these responses remain unknown, and they present an exciting opportunity to identify how the spindle physically and biochemically remodels under force. Collectively, these mechanisms allow the spindle to accommodate local deformations while limiting force propagation in space and time, suggesting a strategy to preserve global architecture and connectivity.
Microtubule flux
Spindle microtubules continuously treadmill towards poles, undergoing net polymerization at plus ends and depolymerization at minus ends in a phenomenon known as poleward flux219. Individual motor and non-motor MAPs with microtubule transport, polymerization or depolymerization activities must be spatially coordinated to allow spindle microtubules to maintain a steady-state length distribution while undergoing poleward flux. This is a complex process that has only recently been reconstituted for single microtubules, but not yet for bundles, in vitro220.
Eg5 was the first driver of flux identified in Xenopus egg extract221, and Eg5-mediated outward sliding of antiparallel microtubules is also important for flux in Drosophila222. However, Eg5 has a minor role in mammalian cells as flux is only reduced by around 25% in its absence223,224. Stronger effects on mammalian and Drosophila poleward flux were found by inhibiting the minus end destabilizing activity of the kinesin KIF2A (refs. 225-227) or kinetochore-localized CLASPs227-230. A recent study in human U2OS spindles proposed a model where instead of being the primary drivers of flux, KIF2A and CLASPs regulate plus and minus end dynamics in response to a driving force generated by multiple mitotic kinesins acting in concert224. According to this model, flux is driven by Eg5 and KIF15 acting in coordination with CENP-E in prometaphase and KIF4A in metaphase157,224. These antiparallel sliding and chromokinesin motors exert poleward forces on non-KMTs, which are only partially transmitted to k-fibres through crosslinkers such as NuMA and HSET224,227, explaining why non-KMTs flux faster than k-fibres104,224,231. Reliance on multiple motors would thus explain the difficulty in finding a principal driver of flux in mammalian cells.
Various functions have been proposed for flux, including maintaining and/or equalizing tension at kinetochores227,231, correcting erroneous kinetochore–microtubule attachments225,227, and regulating spindle length224. However, these functions remain controversial despite the high conservation of flux across all metazoan cell types studied to date. Multiple studies using complementary approaches have now shown that, in conditions where flux is reduced but spindle length is unchanged, chromosome segregation errors frequently occur in anaphase205,225,227. These studies support the view that poleward flux is an emergent phenomenon resulting from the coordinated activities of several motor and non-motor MAPs that is required to maintain accurate and dynamic kinetochore–microtubule attachments.
Material properties
Although the biophysical properties of microtubules and many individual MAPs are well understood in vitro, far less is known about the material properties of the spindle – its deformation, remodelling and structural failure under force. The spindle must respond to forces generated both internally (by motors and dynamic microtubules) and externally (by cortical motors and by neighbouring cells). Mechanical forces regulate many aspects of cell division, including chromosome movement232, spindle positioning233 and error correction at kinetochores126,234,235, and understanding each of these processes requires knowledge of the emergent mechanics of the spindle.
Several elegant studies using concepts of rheology have begun measuring the material properties of the spindle as a continuum, such as elasticity and viscosity, that exists in the fully assembled spindle but not in its component parts. These studies shed light on the heterogeneity of the spindle as a material as well as the mechanical principles that make it well suited to the timescales and force regimes of chromosome segregation. Experiments using microneedles, force-sensing cantilevers and cell confinement have demonstrated that invertebrate, mammalian and Xenopus egg extract spindles are stiffer in their long axes compared to their short axes218,236,237. This property may be functionally important for chromosome movement: for example, it may permit efficient longitudinal movements during chromosome congression, oscillation and segregation, while allowing chromosomes to deform their surroundings laterally. The dynamic and complex microstructure of the spindle also mediates distinct responses at different timescales of force application. In Xenopus egg extract spindles, the short axis deforms elastically on short (<10 s) or long (>100 s) timescales but is more viscous on intermediate timescales238. On the long axis, stiffness is highest at the pole and equator but the spindle is softer between them239 (Fig. 6a). These behaviours allow maximal structural plasticity on the timescale at which chromosomes move through the spindle, with minimal deformability near microtubule connections to chromosomes and poles. Microneedle manipulation of mammalian PtK2 cells has revealed that similar principles of spatially heterogeneous mechanics apply to spindles with prominent k-fibres212,217. Finally, recent studies of molecular diffusivity in mammalian spindles240 and mass density in Xenopus egg extract spindles241 have explored how particles move between closely packed spindle microtubules, which might be expected to impede diffusion. Surprisingly, diffusion and density do not differ between the spindle and the surrounding cytoplasm; instead, dynamic microtubules appear to fluidize the spindle region240, which may be important in enhancing the diffusion of large molecular complexes, such as the CPC or γ-TuRC, throughout the spindle. Together, these rheological studies illustrate mechanical features of the spindle across both axes and at different timescales that are optimized for its role in moving chromosomes.
Fig. 6 ∣. Spindle material properties.
a, The responses of the spindle to force are spatially and temporally heterogeneous. Along the short axis, Xenopus extract spindles behave elastically on short (<10 s) and long (>100 s) timescales. On intermediate timescales, corresponding to the time required for chromosome movement, the short axis of the spindle exhibits more viscous behaviour, dissipating energy as it deforms. The long axis of the spindle is also mechanically heterogeneous: it is stiffest near the poles and kinetochores but less stiff in the middle. The pole region exhibits solid-like behaviour, recovering its shape after deformation, whereas the middle and equator regions are more fluid-like. b, Dynein and its cofactors NuMA and dynactin generate contractile stress in vitro and in the spindle, clustering minus ends together. Eg5 mediates extensile stress, giving rise to nematic motifs of aligned, mixed-polarity microtubules. These opposing motor activities must be balanced to give rise to a bipolar spindle: an excess of contractile activity leads to monopolar spindles, and an excess of extensile sliding produces turbulent microtubule networks.
The molecular components of the spindle constantly dissipate energy in the form of GTP (microtubule dynamics) and ATP (motor proteins such as dynein), meaning that, although the spindle reaches a steady-state structure at metaphase, it does not reach thermodynamic equilibrium. Accordingly, the full complexity of the spindle cannot be fully captured by the rheological models described above. Additional insight into spindle material properties has been gained from active matter theories that model non-equilibrium systems. The microtubule architecture and dynamics of Xenopus egg extract spindles have been described by active liquid crystal theory, modelling the spindle as a droplet of microtubules that is shaped by microtubule turnover, mutual alignment, surface tension and motor-driven active stresses242. In this model, focused poles arise from a barrelling-type instability resulting from dynein activity243. Active matter theory has also been applied to the human spindle, where the cell membrane provides boundary conditions absent in the Xenopus egg extract system. In human cells, unchecked extensile sliding by Eg5 results in the bending and buckling of microtubules against the cell cortex, leading to chaotic, turbulent microtubule networks. This effect is opposed by dynein-mediated contractile stress that allows the spindle to maintain a steady-state shape183,244 (Fig. 6b).
Despite these quantitative physical measurements and theoretical models, many open questions remain regarding the molecular mechanisms that give rise to these material properties. Furthermore, many of the functional implications ascribed to these properties, such as their impact on chromosome motion, remain speculative and require more direct testing.
Spindle twist
The regulation and function of spindle twist is a new area of research sparked by a recent report that metaphase human U2OS and HeLa spindles are, on average, slightly left-handed245. Twist varies between cell types, with HeLa and U2OS spindles showing a stronger left-handed twist than RPE1 spindles205,245,246, and between mitotic stages, where twist peaks in early anaphase246. Interestingly, spindles of the amoeba Naegleria are, on average, right-handed247, suggesting that the left-handed bias may not be a highly conserved feature ofthe spindle.
Many mitotic motors have an off-axis (that is, they do not move exactly parallel to the microtubule) component in their stepping behaviour in vitro, resulting in helical motion around the microtubule track. Plus-end-directed motors studied thus far show a left-handed bias248-250 and minus-end-directed motors have a right-handed bias251-253, but how the spindle resists these additive torques to maintain its relatively untwisted global architecture is not known. The individual inhibition of several motors and MAPs, including Eg5, KIF18A and augmin, revealed that each contributes, albeit to a small degree, to left-handed twist245,246. Deletion of NuMA results in strong left-handed twist205, suggesting that NuMA–dynein motor activity or crosslinking may be important in counteracting left-handed twist.
Although the function, if any, of spindle twist remains unknown, a recent study proposed that twist allows the spindle to bear mechanical load along the pole–pole axis (for example, forces originating from neighbouring cells)246. Further research is required to determine whether maintaining spindle twist within a specific range is important for spindle function.
Spindle size and scaling
Spindle size scales roughly linearly with cell size in all species studied to date254,255, which allows the spindle to properly segregate chromosomes between daughter cells despite variation in cell size between developmental stages, tissues and species. Multiple models, involving both spindle-extrinsic and spindle-intrinsic mechanisms, have been proposed to explain spindle size scaling in different contexts. The limiting component model posits that there is a limiting component, or components, required for spindle assembly, such as the Xenopus protein XMAP215 (ref. 256), and spindles are larger in larger cells owing to greater amounts of this limiting component, assuming equal concentration between cells257 (Fig. 7a). Evidence for this mechanism was provided by encapsulating Xenopus egg extract in microfluidic droplets of defined sizes, where the total amount of cytoplasm could be carefully controlled while keeping cytoplasm composition unchanged258,259. Cellular surface area-to-volume ratio can also tune spindle size: as cells shrink and the surface area-to-volume ratio increases at later stages of X. laevis embryo development, the microtubule destabilizing activity of KIF2A is enhanced as more of its inhibitor becomes sequestered to the membrane260 (Fig. 7b). Similarly, studies in Xenopus and zebrafish suggest that membrane sequestration of a microtubule nucleation inhibitor may regulate spindle scaling261. Additionally, differing expression levels or activities of the MAPs KIF2A (ref. 262), the microtubule severing protein katanin263 and TPX2 (refs. 264,265) have been shown to regulate spindle size in a species-specific manner.
Fig. 7 ∣. Spindle scaling mechanisms.
The scale of the spindle is set by extrinsic and intrinsic mechanisms. a, Extrinsic mechanisms, whereby spindle size scales with cell size, include the limiting components model. This model posits that the size of the spindle is limited by the amount of spindle-building components available. Given that the concentration of the limiting component remains equal irrespective of cell size, a larger cell would contain more of this component and be able to build a larger spindle than a smaller cell. b, Extrinsic scaling can also result from changes in the cellular surface area-to-volume ratio. For example, this occurs if an inhibitor of a negative regulator of spindle size localizes to the cell membrane. As a result, in smaller cells with larger membrane-to-cytoplasm ratios, the inhibitor becomes sequestered at the membrane and enables the negative regulator to scale spindle size down. c, Intrinsic mechanisms for setting maximum spindle length have also been proposed. In very large cells such as early embryonic cells, spindle size can be determined by the scale of a gradient of microtubule nucleation activity such as a RanGTP gradient emanating from the chromosomes.
At very large cell diameters (≥140 μm), spindle size reaches a plateau254,255,258,261,266, revealing a second emergent phenomenon by which the spindle sets an intrinsic maximum length. This maximum is set, in part, by the spatial profile of microtubule nucleation activity, which peaks near chromosomes in large oocytes and large Xenopus egg extract spindles and is limited by diffusion at increasing distances from chromatin210,261,267 (Fig. 7c). Microtubule polymerization rates268, transport210,269 and lifetime269 also contribute to setting the intrinsic size scale of the spindle. Whether these models can account for the sizes of spindles with diverse architectures, such as mammalian spindles with prominent k-fibres, remains to be determined.
Future perspectives
Recent advances from in vitro reconstitution, Xenopus egg extract spindles and mammalian cells have provided a wealth of new insights into spindle assembly, allowing us to better link the structural and biochemical properties of individual molecules to the microtubule modules they build and the emergent properties they give rise to. Despite this progress, many open questions remain, and several new approaches promise to further unravel the mechanisms by which microtubules are nucleated and organized to build the spindle. Advances in imaging techniques, both the tracking of individual molecules as well as imaging whole spindle volumes at high spatiotemporal resolution, will enable us to identify individual events of interest within the dense spindle network and follow the dynamics of specific spindle substructures throughout their entire lifetimes. Improved approaches to apply spatiotemporal perturbations, such as optogenetics, auxin-inducible degrons and microneedle manipulation, now allow us to acutely modulate forces and molecular interactions in a specific region or mitotic phase (Box 1). Finally, new three-dimensional culture models provide exciting opportunities to understand how the tissue context can influence spindle assembly270 and how the spindle is rewired in disease. These approaches will help close the gap between in vitro and in vivo studies, reconstituting the spindle from the bottom up and dissecting its emergent properties from the top down.
Glossary
- Barrelling-type instability
A compression-induced deformation that is symmetric about the axis of compression such as a compression that transforms a cylinder into a barrel shape.
- Biomolecular condensates
Micro-scale compartments that lack surrounding membranes and are formed through weak multivalent interactions, concentrating proteins and nucleic acids.
- Biorientation
The state of a chromosome when both sister kinetochores are attached to microtubules linked to opposite spindle poles.
- Catastrophe
A parameter of microtubule dynamic instability that describes the stochastic switching of a microtubule into a rapidly shrinking, depolymerizing state.
- Chromokinesins
Microtubule plus-end-directed motors that bind to chromosome arms via specific DNA-binding motifs.
- CLASPs
TOG domain-containing proteins that recognize microtubule plus ends, where they suppress catastrophe and promote rescue.
- Elasticity
Ability of a material, by storing energy, to return to its original shape after being deformed. The stiffness (elastic modulus) determines the magnitude of a deformation.
- Emergent properties
Properties of an ensemble, produced by interactions between smaller components, that the smaller components in isolation do not exhibit.
- Metaphase plate
The plane formed by chromosomes after alignment at metaphase, located at the spindle equator.
- NDC80 complex
The core outer kinetochore protein complex that mediates direct binding to microtubules. The NDC80 complex consists of four subunits (Hec1/Ndc80, Nuf2, Spc24 and Spc25) and is present in multiple copies per kinetochore.
- Nematic
The liquid crystal phase in which filaments of mixed polarity are roughly aligned in parallel.
- Nucleoporin
(Nup). Broadly conserved family of proteins that comprise the nuclear pore complex, facilitating molecular transport between the cytoplasm and nucleus.
- Pericentriolar material
(PCM). The dense, structured protein matrix, composed mainly of scaffold proteins and microtubule nucleation factors, that accumulates around centrioles to create the centrosome.
- Polar ejection forces
Forces present during chromosome congression that push chromosome arms away from spindle poles.
- Self-organization
A stable order arising from local interactions between energy-consuming parts.
- Spindle assembly checkpoint
Signalling mechanism that monitors the attachment of kinetochores to spindle microtubules and inhibits progression of the cell cycle from metaphase to anaphase until sufficient attachments are made.
- Stathmin
A protein that regulates microtubule dynamics by sequestering tubulin dimers, which destabilizes microtubules and inhibits their growth.
- Rescue
A parameter of microtubule dynamic instability that describes the stochastic switching of a microtubule into a growing, polymerizing state.
- Rheology
The study of the deformation and flow of materials with both solid and fluid characteristics.
- Viscosity
Internal friction resisting deformation of a material, stemming from rearrangement of its components. The viscous drag coefficient determines the timescale of a deformation.
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.Flemming W. Zellsubstanz, kern undzelltheilung (Vogel, 1882). [Google Scholar]
- 2.Santaguida S & Amon A Short- and long-term effects of chromosome mis-segregation and aneuploidy. Nat. Rev. Mol. Cell Biol 16, 473–485 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Potapova T & Gorbsky GJ The consequences of chromosome segregation errors in mitosis and meiosis. Biology 10.3390/biology6010012 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thawani A & Petry S Molecular insight into how gamma-TuRC makes microtubules. J. Cell Sci 10.1242/jcs.245464 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Voter WA & Erickson HP The kinetics of microtubule assembly. Evidence for a two-stage nucleation mechanism. J. Biol. Chem 259, 10430–10438 (1984). [PubMed] [Google Scholar]
- 6.Zupa E, Liu P, Wurtz M, Schiebel E & Pfeffer S The structure of the gamma-TuRC: a 25-years-old molecular puzzle. Curr. Opin. Struct. Biol 66, 15–21 (2021). [DOI] [PubMed] [Google Scholar]
- 7.Liu P. et al. Insights into the assembly and activation of the microtubule nucleator gamma-TuRC. Nature 578, 467–471 (2020). [DOI] [PubMed] [Google Scholar]
- 8.Wieczorek M. et al. Asymmetric molecular architecture of the human gamma-tubulin ring complex. Cell 180, 165–175.e16 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Consolati T. et al. Microtubule nucleation properties of single human gammaTuRCs explained by their Cryo-EM structure. Dev. Cell 53, 603–617.e8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thawani A. et al. The transition state and regulation of gamma-TuRC-mediated microtubule nucleation revealed by single molecule microscopy. eLife 10.7554/eLife.54253 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thawani A, Kadzik RS & Petry S XMAP215 is a microtubule nucleation factor that functions synergistically with the gamma-tubulin ring complex. Nat. Cell Biol. 20, 575–585 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flor-Parra I, Iglesias-Romero AB & Chang F The XMAP215 ortholog Alp14 promotes microtubule nucleation in fission yeast. Curr. Biol 28, 1681–1691.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.King BR et al. XMAP215 and gamma-tubulin additively promote microtubule nucleation in purified solutions. Mol. Biol. Cell 31, 2187–2194 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gunzelmann J. et al. The microtubule polymerase Stu2 promotes oligomerization of the gamma-TuSC for cytoplasmic microtubule nucleation. eLife 10.7554/eLife.39932 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsuchiya K. et al. Ran-GTP is non-essential to activate NuMA for mitotic spindle-pole focusing but dynamically polarizes HURP near chromosomes. Curr. Biol 31, 115–127.e3 (2021). [DOI] [PubMed] [Google Scholar]
- 16.Sampaio P, Rebollo E, Varmark H, Sunkel CE & Gonzalez C Organized microtubule arrays in gamma-tubulin-depleted Drosophila spermatocytes. Curr. Biol 11, 1788–1793 (2001). [DOI] [PubMed] [Google Scholar]
- 17.Brunet S. et al. Characterization of the TPX2 domains involved in microtubule nucleation and spindle assembly in Xenopus egg extracts. Mol. Biol. Cell 15, 5318–5328 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Imasaki T. et al. CAMSAP2 organizes a gamma-tubulin-independent microtubule nucleation centre through phase separation. eLife 10.7554/eLife.77365 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McKinley KL & Cheeseman IM Large-scale analysis of CRISPR/Cas9 cell-cycle knockouts reveals the diversity of p53-dependent responses to cell-cycle defects. Dev. Cell 40, 405–420.e2 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gudimchuk NB & McIntosh JR Regulation of microtubule dynamics, mechanics and function through the growing tip. Nat. Rev. Mol. Cell Biol 22, 777–795 (2021). [DOI] [PubMed] [Google Scholar]
- 21.Choi YK, Liu P, Sze SK, Dai C & Qi RZ CDK5RAP2 stimulates microtubule nucleation by the gamma-tubulin ring complex. J. Cell Biol 191, 1089–1095 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu P, Choi YK & Qi RZ NME7 is a functional component of the gamma-tubulin ring complex. Mol. Biol. Cell 25, 2017–2025 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cota RR et al. MZT1 regulates microtubule nucleation by Unking gammaTuRC assembly to adapter-mediated targeting and activation. J. Cell Sci 130, 406–419 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Kollman JM et al. Ring closure activates yeast gammaTuRC for species-specific microtubule nucleation. Nat. Struct. Mol. Biol 22, 132–137 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Woodruff JB et al. The centrosome is a selective condensate that nucleates microtubules by concentrating tubulin. Cell 169, 1066–1077.e10 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Baumgart J. et al. Soluble tubulin is significantly enriched at mitotic centrosomes. J. Cell Biol 218, 3977–3985 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.King MR & Petry S Phase separation of TPX2 enhances and spatially coordinates microtubule nucleation. Nat. Commun 11, 270 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haren L, Stearns T & Luders J Plk1-dependent recruitment of gamma-tubulin complexes to mitotic centrosomes involves multiple PCM components. PLoS ONE 4, e5976 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee K & Rhee K PLK1 phosphorylation of pericentrin initiates centrosome maturation at the onset of mitosis. J. Cell Biol 195, 1093–1101 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haren L et al. NEDD1-dependent recruitment of the gamma-tubulin ring complex to the centrosome is necessary for centriole duplication and spindle assembly. J. Cell Biol 172, 505–515 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luders J, Patel UK & Stearns T GCP-WD is a gamma-tubulin targeting factor required for centrosomal and chromatin-mediated microtubule nucleation. Nat. Cell Biol 8, 137–147 (2006). [DOI] [PubMed] [Google Scholar]
- 32.Fong KW, Choi YK, Rattner JB & Qi RZ CDK5RAP2 is a pericentriolar protein that functions in centrosomal attachment of the gamma-tubulin ring complex. Mol. Biol. Cell 19, 115–125 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gomez-Ferreria MA et al. Human Cep192 is required for mitotic centrosome and spindle assembly. Curr. Biol 17, 1960–1966 (2007). [DOI] [PubMed] [Google Scholar]
- 34.Joukov V, Walter JC & De Nicolo A The Cep192-organized aurora A-Plk1 cascade is essential for centrosome cycle and bipolar spindle assembly. Mol. Cell 55, 578–591 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang X. et al. Sequential phosphorylation of Nedd1 by Cdk1 and Plk1 is required for targeting of the gammaTuRC to the centrosome. J. Cell Sci 122, 2240–2251 (2009). [DOI] [PubMed] [Google Scholar]
- 36.Zimmerman WC, Sillibourne J, Rosa J & Doxsey SJ Mitosis-specific anchoring of gamma tubulin complexes by pericentrin controls spindle organization and mitotic entry. Mol. Biol. Cell 15, 3642–3657 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rale MJ, Romer B, Mahon BP, Travis SM & Petry S The conserved centrosomin motif, γTuNA, forms a dimer that directly activates microtubule nucleation by the γ-tubulin ring complex (γTuRC). eLife 10.7554/eLife.80053 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tungadi EA, Ito A, Kiyomitsu T & Goshima G Human microcephaly ASPM protein is a spindle pole-focusing factor that functions redundantly with CDK5RAP2. J. Cell Sci 130, 3676–3684 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Heald R. et al. Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature 382, 420–425 (1996). [DOI] [PubMed] [Google Scholar]
- 40.Matthies HJ, McDonald HB, Goldstein LS & Theurkauf WE Anastral meiotic spindle morphogenesis: role of the non-claret disjunctional kinesin-like protein. J. Cell Biol 134, 455–464 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schuh M & Ellenberg J Self-organization of MTOCs replaces centrosome function during acentrosomal spindle assembly in live mouse oocytes. Cell 130, 484–498 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Zhang H & Dawe RK Mechanisms of plant spindle formation. Chromosome Res. 19, 335–344 (2011). [DOI] [PubMed] [Google Scholar]
- 43.Khodjakov A, Cole RW, Oakley BR & Rieder CL Centrosome-independent mitotic spindle formation in vertebrates. Curr. Biol 10, 59–67 (2000). [DOI] [PubMed] [Google Scholar]
- 44.Moutinho-Pereira S. et al. Genes involved in centrosome-independent mitotic spindle assembly in Drosophila S2 cells. Proc. Natl Acad. Sci. USA 110, 19808–19813 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Basto R. et al. Flies without centrioles. Cell 125, 1375–1386 (2006). [DOI] [PubMed] [Google Scholar]
- 46.Bischoff FR & Ponstingl H Catalysis of guanine nucleotide exchange on Ran by the mitotic regulator RCC1. Nature 354, 80–82 (1991). [DOI] [PubMed] [Google Scholar]
- 47.Gorlich D, Pante N, Kutay U, Aebi U & Bischoff FR Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO J. 15, 5584–5594 (1996). [PMC free article] [PubMed] [Google Scholar]
- 48.Cavazza T & Vernos I The RanGTP Pathway: from nucleo-cytoplasmic transport to spindle assembly and beyond. Front. Cell Dev. Biol. 3, 82 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gruss OJ et al. Ran induces spindle assembly by reversing the inhibitory effect of importin alpha on TPX2 activity. Cell 104, 83–93 (2001). [DOI] [PubMed] [Google Scholar]
- 50.Nachury MV et al. Importin beta is a mitotic target of the small GTPase Ran in spindle assembly. Cell 104, 95–106 (2001). [DOI] [PubMed] [Google Scholar]
- 51.Wiese C. et al. Role of importin-beta in coupling Ran to downstream targets in microtubule assembly. Science 291, 653–656 (2001). [DOI] [PubMed] [Google Scholar]
- 52.Kalab P, Weis K & Heald R Visualization of a Ran-GTP gradient in interphase and mitotic Xenopus egg extracts. Science 295, 2452–2456 (2002). [DOI] [PubMed] [Google Scholar]
- 53.Kalab P, Pralle A, Isacoff EY, Heald R & Weis K Analysis of a RanGTP-regulated gradient in mitotic somatic cells. Nature 440, 697–701 (2006). [DOI] [PubMed] [Google Scholar]
- 54.Goshima G, Mayer M, Zhang N, Stuurman N & Vale RD Augmin: a protein complex required for centrosome-independent microtubule generation within the spindle. J. Cell Biol 181, 421–429 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lawo S. et al. HAUS, the 8-subunit human Augmin complex, regulates centrosome and spindle integrity. Curr. Biol 19, 816–826 (2009). [DOI] [PubMed] [Google Scholar]
- 56.Uehara R. et al. The augmin complex plays a critical role in spindle microtubule generation for mitotic progression and cytokinesis in human cells. Proc. Natl Acad. Sci. USA 106, 6998–7003 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ho CM et al. Augmin plays a critical role in organizing the spindle and phragmoplast microtubule arrays in Arabidopsis. Plant. Cell 23, 2606–2618 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Petry S, Groen AC, Ishihara K, Mitchison TJ & Vale RD Branching microtubule nucleation in Xenopus egg extracts mediated by augmin and TPX2. Cell 152, 768–777 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hayward D, Metz J, Pellacani C & Wakefield JG Synergy between multiple microtubule-generating pathways confers robustness to centrosome-driven mitotic spindle formation. Dev. Cell 28, 81–93 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.David AF et al. Augmin accumulation on long-lived microtubules drives amplification and kinetochore-directed growth. J. Cell Biol 218, 2150–2168 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tariq A, Green L, Jeynes JCG, Soeller C & Wakefield JG In vitro reconstitution of branching microtubule nucleation. eLife 10.7554/eLife.49769 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang Y, Hong X, Hua S & Jiang K Reconstitution and mechanistic dissection of the human microtubule branching machinery. J. Cell Biol. 10.1083/jcb.202109053 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Alfaro-Aco R, Thawani A & Petry S Biochemical reconstitution of branching microtubule nucleation. eLife 10.7554/eLife.49797 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hsia KC et al. Reconstitution of the augmin complex provides insights into its architecture and function. Nat. Cell Biol 16, 852–863 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Song JG et al. Mechanism of how augmin directly targets the gamma-tubulin ring complex to microtubules. J. Cell Biol 217, 2417–2428 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhu H, Coppinger JA, Jang CY, Yates JR III & Fang G FAM29A promotes microtubule amplification via recruitment of the NEDD1-gamma-tubulin complex to the mitotic spindle. J. Cell Biol 183, 835–848 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Petry S, Pugieux C, Nedelec FJ & Vale RD Augmin promotes meiotic spindle formation and bipolarity in Xenopus egg extracts. Proc. Natl Acad. Sci. USA 108, 14473–14478 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Almeida AC et al. Augmin-dependent microtubule self-organization drives kinetochore fiber maturation in mammals. Cell Rep. 39, 110610 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Verma V & Maresca TJ Direct observation of branching MT nucleation in living animal cells. J. Cell Biol 218, 2829–2840 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Thawani A, Stone HA, Shaevitz JW & Petry S Spatiotemporal organization of branched microtubule networks. eLife 10.7554/eLife.43890 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Setru SU et al. A hydrodynamic instability drives protein droplet formation on microtubules to nucleate branches. Nat. Phys 17, 493–498 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Roostalu J, Cade NI & Surrey T Complementary activities of TPX2 and chTOG constitute an efficient importin-regulated microtubule nucleation module. Nat. Cell Biol 17, 1422–1434 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Reid TA et al. Suppression of microtubule assembly kinetics by the mitotic protein TPX2. J. Cell Sci 129, 1319–1328 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Luo J. et al. The microtubule-associated protein EML3 regulates mitotic spindle assembly by recruiting the Augmin complex to spindle microtubules. J. Biol. Chem 294, 5643–5656 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sanchez-Huertas C & Luders J The augmin connection in the geometry of microtubule networks. Curr. Biol 25, R294–R299 (2015). [DOI] [PubMed] [Google Scholar]
- 76.Khodjakov A, Copenagle L, Gordon MB, Compton DA & Kapoor TM Minus-end capture of preformed kinetochore fibers contributes to spindle morphogenesis. J. Cell Biol 160, 671–683 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Maiato H, Rieder CL & Khodjakov A Kinetochore-driven formation of kinetochore fibers contributes to spindle assembly during animal mitosis. J. Cell Biol 167, 831–840 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sikirzhytski V. et al. Microtubules assemble near most kinetochores during early prometaphase in human cells. J. Cell Biol 217, 2647–2659 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Renda F. et al. Non-centrosomal microtubules at kinetochores promote rapid chromosome biorientation during mitosis in human cells. Curr. Biol 32, 1049–1063.e4 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Orjalo AV et al. The Nup107-160 nucleoporin complex is required for correct bipolar spindle assembly. Mol. Biol. Cell 17, 3806–3818 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mishra RK, Chakraborty P, Arnaoutov A, Fontoura BM & Dasso M The Nup107-160 complex and γ-TuRC regulate microtubule polymerization at kinetochores. Nat. Cell Biol 12, 164–169 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yokoyama H. et al. The nucleoporin MEL-28 promotes RanGTP-dependent gamma-tubulin recruitment and microtubule nucleation in mitotic spindle formation. Nat. Commun 5, 3270 (2014). [DOI] [PubMed] [Google Scholar]
- 83.Rasala BA, Orjalo AV, Shen Z, Briggs S & Forbes DJ ELYS is a dual nucleoporin/kinetochore protein required for nuclear pore assembly and proper cell division. Proc. Natl Acad. Sci. USA 103, 17801–17806 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tulu US, Fagerstrom C, Ferenz NP & Wadsworth P Molecular requirements for kinetochore-associated microtubule formation in mammalian cells. Curr. Biol 16, 536–541 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Euteneuer U, Ris H & Borisy GG Polarity of kinetochore microtubules in Chinese hamster ovary cells after recovery from a colcemid block. J. Cell Biol 97, 202–208 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shrestha RL & Draviam VM Lateral to end-on conversion of chromosome-microtubule attachment requires kinesins CENP-E and MCAK. Curr. Biol 23, 1514–1526 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Popova JV et al. Genetic control of kinetochore-driven microtubule growth in Drosophila mitosis. Cells 10.3390/cells11142127 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Maresca TJ et al. Spindle assembly in the absence of a RanGTP gradient requires localized CPC activity. Curr. Biol 19, 1210–1215 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gassmann R. et al. Borealin: a novel chromosomal passenger required for stability of the bipolar mitotic spindle. J. Cell Biol 166, 179–191 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lan W. et al. Aurora B phosphorylates centromeric MCAK and regulates its localization and microtubule depolymerization activity. Curr. Biol 14, 273–286 (2004). [DOI] [PubMed] [Google Scholar]
- 91.Sampath SC et al. The chromosomal passenger complex is required for chromatin-induced microtubule stabilization and spindle assembly. Cell 118, 187–202 (2004). [DOI] [PubMed] [Google Scholar]
- 92.Ohi R, Sapra T, Howard J & Mitchison TJ Differentiation of cytoplasmic and meiotic spindle assembly MCAK functions by Aurora B-dependent phosphorylation. Mol. Biol. Cell 15, 2895–2906 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tan L & Kapoor TM Examining the dynamics of chromosomal passenger complex (CPC)-dependent phosphorylation during cell division. Proc. Natl Acad. Sci. USA 108, 16675–16680 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Niethammer P, Bastiaens P & Karsenti E Stathmin-tubulin interaction gradients in motile and mitotic cells. Science 303, 1862–1866 (2004). [DOI] [PubMed] [Google Scholar]
- 95.Trivedi P. et al. The inner centromere is a biomolecular condensate scaffolded by the chromosomal passenger complex. Nat. Cell Biol 21, 1127–1137 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Musacchio A. On the role of phase separation in the biogenesis of membraneless compartments. EMBO J. 41, e109952 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rebollo E, Llamazares S, Reina J & Gonzalez C Contribution of noncentrosomal microtubules to spindle assembly in Drosophila spermatocytes. PLoS Biol. 2, E8 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.So C. et al. A liquid-like spindle domain promotes acentrosomal spindle assembly in mammalian oocytes. Science 10.1126/science.aat9557 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Halpin D, Kalab P, Wang J, Weis K & Heald R Mitotic spindle assembly around RCC1-coated beads in Xenopus egg extracts. PLoS Biol. 9, e1001225 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wolff ID, Tran MV, Mullen TJ, Villeneuve AM & Wignall SM Assembly of Caenorhabditis elegans acentrosomal spindles occurs without evident microtubule-organizing centers and requires microtubule sorting by KLP-18/kinesin-12 and MESP-1. Mol. Biol. Cell 27, 3122–3131 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dumont J. et al. A centriole- and RanGTP-independent spindle assembly pathway in meiosis I of vertebrate oocytes. J. Cell Biol 176, 295–305 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ma N. et al. Poleward transport of TPX2 in the mammalian mitotic spindle requires dynein, Eg5, and microtubule flux. Mol. Biol. Cell 21, 979–988 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Heald R, Tournebize R, Habermann A, Karsenti E & Hyman A Spindle assembly in Xenopus egg extracts: respective roles of centrosomes and microtubule self-organization. J. Cell Biol 138, 615–628 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lecland N & Luders J The dynamics of microtubule minus ends in the human mitotic spindle. Nat. Cell Biol 16, 770–778 (2014). [DOI] [PubMed] [Google Scholar]
- 105.Prosser SL & Pelletier L Mitotic spindle assembly in animal cells: a fine balancing act. Nat. Rev. Mol. Cell Biol 18, 187–201 (2017). [DOI] [PubMed] [Google Scholar]
- 106.Gaska I, Armstrong ME, Alfieri A & Forth S The mitotic crosslinking protein PRC1 acts like a mechanical dashpot to resist microtubule sliding. Dev. Cell 54, 367–378.e5 (2020). [DOI] [PubMed] [Google Scholar]
- 107.Verde F, Berrez JM, Antony C & Karsenti E Taxol-induced microtubule asters in mitotic extracts of Xenopus eggs: requirement for phosphorylated factors and cytoplasmic dynein. J. Cell Biol 112, 1177–1187 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gaglio T, Saredi A & Compton DA NuMA is required for the organization of microtubules into aster-like mitotic arrays. J. Cell Biol 131, 693–708 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Surrey T, Nedelec F, Leibler S & Karsenti E Physical properties determining self-organization of motors and microtubules. Science 292, 1167–1171 (2001). [DOI] [PubMed] [Google Scholar]
- 110.Tan R, Foster PJ, Needleman DJ & McKenney RJ Cooperative accumulation of dynein-dynactin at microtubule minus-ends drives microtubule network reorganization. Dev. Cell 44, 233–247.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bieling P, Telley IA & Surrey T A minimal midzone protein module controls formation and length of antiparallel microtubule overlaps. Cell 142, 420–432 (2010). [DOI] [PubMed] [Google Scholar]
- 112.Wijeratne S & Subramanian R Geometry of antiparallel microtubule bundles regulates relative sliding and stalling by PRC1 and Kif4A. eLife 10.7554/eLife.32595 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hannabuss J. et al. Self-organization of minimal anaphase spindle midzone bundles. Curr. Biol 29, 2120–2130.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.McDonald KL, O’Toole ET, Mastronarde DN & McIntosh JR Kinetochore microtubules in PTK cells. J. Cell Biol 118, 369–383 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Long AF, Kuhn J & Dumont S The mammalian kinetochore-microtubule interface: robust mechanics and computation with many microtubules. Curr. Opin. Cell Biol 60, 60–67 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Booth DG, Hood FE, Prior IA & Royle SJ A TACC3/ch-TOG/clathrin complex stabilises kinetochore fibres by inter-microtubule bridging. EMBO J. 30, 906–919 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kiewisz R. et al. Three-dimensional structure of kinetochore-fibers in human mitotic spindles. eLife 10.7554/eLife.75459 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.O’Toole E, Morphew M & McIntosh JR Electron tomography reveals aspects of spindle structure important for mechanical stability at metaphase. Mol. Biol. Cell 31, 184–195 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Drpic D. et al. Chromosome segregation is biased by kinetochore size. Curr. Biol 28, 1344–1356.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Stumpff J, von Dassow G, Wagenbach M, Asbury C & Wordeman L The kinesin-8 motor Kif18A suppresses kinetochore movements to control mitotic chromosome alignment. Dev. Cell 14, 252–262 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Varga V. et al. Yeast kinesin-8 depolymerizes microtubules in a length-dependent manner. Nat. Cell Biol 8, 957–962 (2006). [DOI] [PubMed] [Google Scholar]
- 122.Du Y, English CA & Ohi R The kinesin-8 Kif18A dampens microtubule plus-end dynamics. Curr. Biol 20, 374–380 (2010). [DOI] [PubMed] [Google Scholar]
- 123.Stumpff J. et al. A tethering mechanism controls the processivity and kinetochore-microtubule plus-end enrichment of the kinesin-8 Kif18A. Mol. Cell 43, 764–775 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Dudka D, Castrogiovanni C, Liaudet N, Vassal H & Meraldi P Spindle-length-dependent HURP localization allows centrosomes to control kinetochore-fiber plus-end dynamics. Curr. Biol 29, 3563–3578.e6 (2019). [DOI] [PubMed] [Google Scholar]
- 125.Zhai Y, Kronebusch PJ & Borisy GG Kinetochore microtubule dynamics and the metaphase-anaphase transition. J. Cell Biol 131, 721–734 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Nicklas RB & Koch CA Chromosome micromanipulation. 3. Spindle fiber tension and the reorientation of mal-oriented chromosomes. J. Cell Biol 43, 40–50 (1969). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.DeLuca JG et al. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell 127, 969–982 (2006). [DOI] [PubMed] [Google Scholar]
- 128.Bakhoum SF, Genovese G & Compton DA Deviant kinetochore microtubule dynamics underlie chromosomal instability. Curr. Biol 19, 1937–1942 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Koffa MD et al. HURP is part of a Ran-dependent complex involved in spindle formation. Curr. Biol 16, 743–754 (2006). [DOI] [PubMed] [Google Scholar]
- 130.Sillje HH, Nagel S, Korner R & Nigg EA HURP is a Ran-importin beta-regulated protein that stabilizes kinetochore microtubules in the vicinity of chromosomes. Curr. Biol 16, 731–742 (2006). [DOI] [PubMed] [Google Scholar]
- 131.Begley MA et al. K-fiber bundles in the mitotic spindle are mechanically reinforced by Kif15. Mol. Biol. Cell 32, br11 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Nixon FM et al. The mesh is a network of microtubule connectors that stabilizes individual kinetochore fibers of the mitotic spindle. eLife 10.7554/eLife.07635 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Nicklas RB, Kubai DF & Hays TS Spindle microtubules and their mechanical associations after micromanipulation in anaphase. J. Cell Biol 95, 91–104 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kirschner M & Mitchison T Beyond self-assembly: from microtubules to morphogenesis. Cell 45, 329–342 (1986). [DOI] [PubMed] [Google Scholar]
- 135.Magidson V. et al. Adaptive changes in the kinetochore architecture facilitate proper spindle assembly. Nat. Cell Biol 17, 1134–1144 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wollman R. et al. Efficient chromosome capture requires a bias in the ‘search-and-capture’ process during mitotic-spindle assembly. Curr. Biol 15, 828–832 (2005). [DOI] [PubMed] [Google Scholar]
- 137.Magidson V. et al. The spatial arrangement of chromosomes during prometaphase facilitates spindle assembly. Cell 146, 555–567 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wynne DJ & Funabiki H Kinetochore function is controlled by a phospho-dependent coexpansion of inner and outer components. J. Cell Biol 210, 899–916 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Sacristan C. et al. Dynamic kinetochore size regulation promotes microtubule capture and chromosome biorientation in mitosis. Nat. Cell Biol 20, 800–810 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Mitchison T, Evans L, Schulze E & Kirschner M Sites of microtubule assembly and disassembly in the mitotic spindle. Cell 45, 515–527 (1986). [DOI] [PubMed] [Google Scholar]
- 141.Rieder CL & Alexander SP Kinetochores are transported poleward along a single astral microtubule during chromosome attachment to the spindle in newt lung cells. J. Cell Biol 110, 81–95 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Hueschen CL, Kenny SJ, Xu K & Dumont S NuMA recruits dynein activity to microtubule minus-ends at mitosis. eLife 10.7554/eLife.29328 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Elting MW, Hueschen CL, Udy DB & Dumont S Force on spindle microtubule minus ends moves chromosomes. J. Cell Biol 206, 245–256 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Sikirzhytski V. et al. Direct kinetochore-spindle pole connections are not required for chromosome segregation. J. Cell Biol 206, 231–243 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Mastronarde DN, McDonald KL, Ding R & McIntosh JR Interpolar spindle microtubules in PTK cells. J. Cell Biol 123, 1475–1489 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kajtez J. et al. Overlap microtubules link sister k-fibres and balance the forces on bi-oriented kinetochores. Nat. Commun 7, 10298 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Polak B, Risteski P, Lesjak S & Tolic IM PRC1-labeled microtubule bundles and kinetochore pairs show one-to-one association in metaphase. EMBO Rep. 18, 217–230 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Mollinari C. et al. PRC1 is a microtubule binding and bundling protein essential to maintain the mitotic spindle midzone. J. Cell Biol 157, 1175–1186 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kapitein LC et al. Microtubule-driven multimerization recruits ase1p onto overlapping microtubules. Curr. Biol 18, 1713–1717 (2008). [DOI] [PubMed] [Google Scholar]
- 150.Subramanian R. et al. Insights into antiparallel microtubule crosslinking by PRC1, a conserved nonmotor microtubule binding protein. Cell 142, 433–443 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Jagric M, Risteski P, Martincic J, Milas A & Tolic IM Optogenetic control of PRC1 reveals its role in chromosome alignment on the spindle by overlap length-dependent forces. eLife 10.7554/eLife.61170 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Brinkley BR & Cartwright J Jr. Cold-labile and cold-stable microtubules in the mitotic spindle of mammalian cells. Ann. N. Y. Acad. Sci 253, 428–439 (1975). [DOI] [PubMed] [Google Scholar]
- 153.Ohi R, Burbank K, Liu Q & Mitchison TJ Nonredundant functions of Kinesin-13s during meiotic spindle assembly. Curr. Biol 17, 953–959 (2007). [DOI] [PubMed] [Google Scholar]
- 154.Burbank KS, Groen AC, Perlman ZE, Fisher DS & Mitchison TJ A new method reveals microtubule minus ends throughout the meiotic spindle. J. Cell Biol 175, 369–375 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Yang G. et al. Architectural dynamics of the meiotic spindle revealed by single-fluorophore imaging. Nat. Cell Biol 9, 1233–1242 (2007). [DOI] [PubMed] [Google Scholar]
- 156.Saxton WM et al. Tubulin dynamics in cultured mammalian cells. J. Cell Biol 99, 2175–2186 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Wandke C. et al. Human chromokinesins promote chromosome congression and spindle microtubule dynamics during mitosis. J. Cell Biol 198, 847–863 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Antonio C. et al. Xkid, a chromokinesin required for chromosome alignment on the metaphase plate. Cell 102, 425–435 (2000). [DOI] [PubMed] [Google Scholar]
- 159.Funabiki H & Murray AW The Xenopus chromokinesin Xkid is essential for metaphase chromosome alignment and must be degraded to allow anaphase chromosome movement. Cell 102, 411–424 (2000). [DOI] [PubMed] [Google Scholar]
- 160.Brouhard GJ & Hunt AJ Microtubule movements on the arms of mitotic chromosomes: polar ejection forces quantified in vitro. Proc. Natl Acad. Sci. USA 102, 13903–13908 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Ke K, Cheng J & Hunt AJ The distribution of polar ejection forces determines the amplitude of chromosome directional instability. Curr. Biol 19, 807–815 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Rieder CL & Salmon ED Motile kinetochores and polar ejection forces dictate chromosome position on the vertebrate mitotic spindle. J. Cell Biol 124, 223–233 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Stout JR et al. Kif18B interacts with EB1 and controls astral microtubule length during mitosis. Mol. Biol. Cell 22, 3070–3080 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Tanenbaum ME et al. A complex of Kif18b and MCAK promotes microtubule depolymerization and is negatively regulated by Aurora kinases. Curr. Biol 21, 1356–1365 (2011). [DOI] [PubMed] [Google Scholar]
- 165.Dema A, van Haren J & Wittmann T Optogenetic EB1 inactivation shortens metaphase spindles by disrupting cortical force-producing interactions with astral microtubules. Curr. Biol 32, 1197–1205.e4 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Barisic M. et al. Mitosis. Microtubule detyrosination guides chromosomes during mitosis. Science 348, 799–803 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.di Pietro F, Echard A & Morin X Regulation of mitotic spindle orientation: an integrated view. EMBO Rep. 17, 1106–1130 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Lechler T & Mapelli M Spindle positioning and its impact on vertebrate tissue architecture and cell fate. Nat. Rev. Mol. Cell Biol 22, 691–708 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Thery M. et al. The extracellular matrix guides the orientation of the cell division axis. Nat. Cell Biol 7, 947–953 (2005). [DOI] [PubMed] [Google Scholar]
- 170.Fink J. et al. External forces control mitotic spindle positioning. Nat. Cell Biol 13, 771–778 (2011). [DOI] [PubMed] [Google Scholar]
- 171.Guild J, Ginzberg MB, Hueschen CL, Mitchison TJ & Dumont S Increased lateral microtubule contact at the cell cortex is sufficient to drive mammalian spindle elongation. Mol. Biol. Cell 28, 1975–1983 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Schaefer M, Shevchenko A, Shevchenko A & Knoblich JA A protein complex containing Inscuteable and the Galpha-binding protein Pins orients asymmetric cell divisions in Drosophila. Curr. Biol 10, 353–362 (2000). [DOI] [PubMed] [Google Scholar]
- 173.Gotta M, Dong Y, Peterson YK, Lanier SM & Ahringer J Asymmetrically distributed C. elegans homologs of AGS3/PINS control spindle position in the early embryo. Curr. Biol 13, 1029–1037 (2003). [DOI] [PubMed] [Google Scholar]
- 174.Du Q & Macara IG Mammalian Pins is a conformational switch that links NuMA to heterotrimeric G proteins. Cell 119, 503–516 (2004). [DOI] [PubMed] [Google Scholar]
- 175.Fielmich LE et al. Optogenetic dissection of mitotic spindle positioning in vivo. eLife 10.7554/eLife.38198 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Okumura M, Natsume T, Kanemaki MT & Kiyomitsu T Dynein-dynactin-NuMA clusters generate cortical spindle-pulling forces as a multi-arm ensemble. eLife 10.7554/eLife.36559 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Hornick JE et al. Amphiastral mitotic spindle assembly in vertebrate cells lacking centrosomes. Curr. Biol 21, 598–605 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Sir JH et al. Loss of centrioles causes chromosomal instability in vertebrate somatic cells. J. Cell Biol 203, 747–756 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Wong YL et al. Cell biology. Reversible centriole depletion with an inhibitor of Polo-like kinase 4. Science 348, 1155–1160 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Chinen T. et al. NuMA assemblies organize microtubule asters to establish spindle bipolarity in acentrosomal human cells. EMBO J. 39, e102378 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Merdes A, Heald R, Samejima K, Earnshaw WC & Cleveland DW Formation of spindle poles by dynein/dynactin-dependent transport of NuMA. J. Cell Biol 149, 851–862 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Goshima G, Nedelec F & Vale RD Mechanisms for focusing mitotic spindle poles by minus end-directed motor proteins. J. Cell Biol 171, 229–240 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Hueschen CL, Galstyan V, Amouzgar M, Phillips R & Dumont S Microtubule end-clustering maintains a steady-state spindle shape. Curr. Biol 29, 700–708.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Echeverri CJ, Paschal BM, Vaughan KT & Vallee RB Molecular characterization of the 50-kD subunit of dynactin reveals function for the complex in chromosome alignment and spindle organization during mitosis. J. Cell Biol 132, 617–633 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Renna C. et al. Organizational principles of the NuMA-Dynein interaction interface and implications for mitotic spindle functions. Structure 28, 820–829.e6 (2020). [DOI] [PubMed] [Google Scholar]
- 186.Harborth J, Wang J, Gueth-Hallonet C, Weber K & Osborn M Self assembly of NuMA: multiarm oligomers as structural units of a nuclear lattice. EMBO J. 18, 1689–1700 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Sun M. et al. NuMA regulates mitotic spindle assembly, structural dynamics and function via phase separation. Nat. Commun 12, 7157 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Ma H. et al. NuMA forms condensates through phase separation to drive spindle pole assembly. J. Mol. Cell Biol 10.1093/jmcb/mjab081 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Mountain V. et al. The kinesin-related protein, HSET, opposes the activity of Eg5 and cross-links microtubules in the mammalian mitotic spindle. J. Cell Biol 147, 351–366 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Cai S, Weaver LN, Ems-McClung SC & Walczak CE Kinesin-14 family proteins HSET/XCTK2 control spindle length by cross-linking and sliding microtubules. Mol. Biol. Cell 20, 1348–1359 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Ganem NJ, Godinho SA & Pellman D A mechanism linking extra centrosomes to chromosomal instability. Nature 460, 278–282 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Sawin KE, LeGuellec K, Philippe M & Mitchison TJ Mitotic spindle organization by a plus-end-directed microtubule motor. Nature 359, 540–543 (1992). [DOI] [PubMed] [Google Scholar]
- 193.Blangy A. et al. Phosphorylation by p34cdc2 regulates spindle association of human Eg5, a kinesin-related motor essential for bipolar spindle formation in vivo. Cell 83, 1159–1169 (1995). [DOI] [PubMed] [Google Scholar]
- 194.Kashina AS, Scholey JM, Leszyk JD & Saxton WM An essential bipolar mitotic motor. Nature 384, 225 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Kapitein LC et al. The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature 435, 114–118 (2005). [DOI] [PubMed] [Google Scholar]
- 196.Raaijmakers JA et al. Nuclear envelope-associated dynein drives prophase centrosome separation and enables Eg5-independent bipolar spindle formation. EMBO J. 31, 4179–4190 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Toso A. et al. Kinetochore-generated pushing forces separate centrosomes during bipolar spindle assembly. J. Cell Biol 184, 365–372 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Sturgill EG & Ohi R Kinesin-12 differentially affects spindle assembly depending on its microtubule substrate. Curr. Biol 23, 1280–1290 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Tanenbaum ME et al. Kif15 cooperates with eg5 to promote bipolar spindle assembly. Curr. Biol 19, 1703–1711 (2009). [DOI] [PubMed] [Google Scholar]
- 200.Vanneste D, Takagi M, Imamoto N & Vernos I The role of Hklp2 in the stabilization and maintenance of spindle bipolarity. Curr. Biol 19, 1712–1717 (2009). [DOI] [PubMed] [Google Scholar]
- 201.Drechsler H, McHugh T, Singleton MR, Carter NJ & McAinsh AD The Kinesin-12 Kif15 is a processive track-switching tetramer. eLife 3, e01724 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Reinemann DN et al. Collective force regulation in anti-parallel microtubule gliding by dimeric Kif15 kinesin motors. Curr. Biol 27, 2810–2820.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Umbreit NT et al. Mechanisms generating cancer genome complexity from a single cell division error. Science 10.1126/science.aba0712 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.van Heesbeen RG, Tanenbaum ME & Medema RH Balanced activity of three mitotic motors is required for bipolar spindle assembly and chromosome segregation. Cell Rep. 8, 948–956 (2014). [DOI] [PubMed] [Google Scholar]
- 205.Neahring L, Cho NH & Dumont S Opposing motors provide mechanical and functional robustness in the human spindle. Dev. Cell 56, 3006–3018.e5 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Sturgill EG, Norris SR, Guo Y & Ohi R Kinesin-5 inhibitor resistance is driven by kinesin-12. J. Cell Biol 213, 213–227 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Mitchison TJ et al. Roles of polymerization dynamics, opposed motors, and a tensile element in governing the length of Xenopus extract meiotic spindles. Mol. Biol. Cell 16, 3064–3076 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Tanenbaum ME, Macurek L, Galjart N & Medema RH Dynein, Lis1 and CLIP-170 counteract Eg5-dependent centrosome separation during bipolar spindle assembly. EMBO J. 27, 3235–3245 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Ferenz NP, Paul R, Fagerstrom C, Mogilner A & Wadsworth P Dynein antagonizes eg5 by crosslinking and sliding antiparallel microtubules. Curr. Biol 19, 1833–1838 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Brugues J, Nuzzo V, Mazur E & Needleman DJ Nucleation and transport organize microtubules in metaphase spindles. Cell 149, 554–564 (2012). [DOI] [PubMed] [Google Scholar]
- 211.Elting MW, Prakash M, Udy DB & Dumont S Mapping load-bearing in the mammalian spindle reveals local kinetochore fiber anchorage that provides mechanical isolation and redundancy. Curr. Biol 27, 2112–2122.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Suresh P, Long AF & Dumont S Microneedle manipulation of the mammalian spindle reveals specialized, short-lived reinforcement near chromosomes. eLife 10.7554/eLife.53807 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Franck AD et al. Tension applied through the Dam1 complex promotes microtubule elongation providing a direct mechanism for length control in mitosis. Nat. Cell Biol 9, 832–837 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Akiyoshi B. et al. Tension directly stabilizes reconstituted kinetochore-microtubule attachments. Nature 468, 576–579 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Skibbens RV & Salmon ED Micromanipulation of chromosomes in mitotic vertebrate tissue cells: tension controls the state of kinetochore movement. Exp. Cell Res 235, 314–324 (1997). [DOI] [PubMed] [Google Scholar]
- 216.Nicklas RB & Staehly CA Chromosome micromanipulation. I. The mechanics of chromosome attachment to the spindle. Chromosoma 21, 1–16 (1967). [DOI] [PubMed] [Google Scholar]
- 217.Long AF, Suresh P & Dumont S Individual kinetochore-fibers locally dissipate force to maintain robust mammalian spindle structure. J. Cell Biol 219, 10.1083/jcb.201911090 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Dumont S & Mitchison TJ Compression regulates mitotic spindle length by a mechanochemical switch at the poles. Curr. Biol 19, 1086–1095 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Mitchison TJ Polewards microtubule flux in the mitotic spindle: evidence from photoactivation of fluorescence. J. Cell Biol 109, 637–652 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Arpag G, Lawrence EJ, Farmer VJ, Hall SL & Zanic M Collective effects of XMAP215, EB1, CLASP2, and MCAK lead to robust microtubule treadmilling. Proc. Natl Acad. Sci. USA 117, 12847–12855 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Miyamoto DT, Perlman ZE, Burbank KS, Groen AC & Mitchison TJ The kinesin Eg5 drives poleward microtubule flux in Xenopus laevis egg extract spindles. J. Cell Biol 167, 813–818 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Brust-Mascher I, Sommi P, Cheerambathur DK & Scholey JM Kinesin-5-dependent poleward flux and spindle length control in Drosophila embryo mitosis. Mol. Biol. Cell 20, 1749–1762 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Cameron LA et al. Kinesin 5-independent poleward flux of kinetochore microtubules in PtK1 cells. J. Cell Biol 173, 173–179 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Steblyanko Y. et al. Microtubule poleward flux in human cells is driven by the coordinated action of four kinesins. EMBO J. 39, e105432 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Ganem NJ, Upton K & Compton DA Efficient mitosis in human cells lacking poleward microtubule flux. Curr. Biol 15, 1827–1832 (2005). [DOI] [PubMed] [Google Scholar]
- 226.Rogers GC et al. Two mitotic kinesins cooperate to drive sister chromatid separation during anaphase. Nature 427, 364–370 (2004). [DOI] [PubMed] [Google Scholar]
- 227.Matos I. et al. Synchronizing chromosome segregation by flux-dependent force equalization at kinetochores. J. Cell Biol 186, 11–26 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Maiato H, Khodjakov A & Rieder CL Drosophila CLASP is required for the incorporation of microtubule subunits into fluxing kinetochore fibres. Nat. Cell Biol 7, 42–47 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Maffini S. et al. Motor-independent targeting of CLASPs to kinetochores by CENP-E promotes microtubule turnover and poleward flux. Curr. Biol 19, 1566–1572 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Girao H. et al. CLASP2 binding to curved microtubule tips promotes flux and stabilizes kinetochore attachments. J. Cell Biol 10.1083/jcb.201905080 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Maddox P, Straight A, Coughlin P, Mitchison TJ & Salmon ED Direct observation of microtubule dynamics at kinetochores in Xenopus extract spindles: implications for spindle mechanics. J. Cell Biol 162, 377–382 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Nicklas RB Measurements of the force produced by the mitotic spindle in anaphase. J. Cell Biol 97, 542–548 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Garzon-Coral C, Fantana HA & Howard J A force-generating machinery maintains the spindle at the cell center during mitosis. Science 352, 1124–1127 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Miller MP, Asbury CL & Biggins S A TOG protein confers tension sensitivity to kinetochore-microtubule attachments. Cell 165, 1428–1439 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Chen GY et al. Tension promotes kinetochore-microtubule release by Aurora B kinase. J. Cell Biol 10.1083/jcb.202007030 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Begg DA & Ellis GW Micromanipulation studies of chromosome movement. I. Chromosome-spindle attachment and the mechanical properties of chromosomal spindle fibers. J. Cell Biol 82, 528–541 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Itabashi T. et al. Probing the mechanical architecture of the vertebrate meiotic spindle. Nat. Methods 6, 167–172 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Shimamoto Y, Maeda YT, Ishiwata S, Libchaber AJ & Kapoor TM Insights into the micromechanical properties of the metaphase spindle. Cell 145, 1062–1074 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Takagi J, Sakamoto R, Shiratsuchi G, Maeda YT & Shimamoto Y Mechanically distinct microtubule arrays determine the length and force response of the meiotic spindle. Dev. Cell 49, 267–278.e5 (2019). [DOI] [PubMed] [Google Scholar]
- 240.Carlini L, Brittingham GP, Holt LJ & Kapoor TM Microtubules enhance mesoscale effective diffusivity in the crowded metaphase cytoplasm. Dev. Cell 54, 574–582.e4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Biswas A, Kim K, Cojoc G, Guck J & Reber S The Xenopus spindle is as dense as the surrounding cytoplasm. Dev. Cell 56, 967–975.e5 (2021). [DOI] [PubMed] [Google Scholar]
- 242.Brugues J & Needleman D Physical basis of spindle self-organization. Proc. Natl Acad. Sci. USA 111, 18496–18500 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Oriola D, Julicher F & Brugues J Active forces shape the metaphase spindle through a mechanical instability. Proc. Natl Acad. Sci. USA 117, 16154–16159 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Roostalu J, Rickman J, Thomas C, Nedelec F & Surrey T Determinants of polar versus nematic organization in networks of dynamic microtubules and mitotic motors. Cell 175, 796–808.e14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Novak M. et al. The mitotic spindle is chiral due to torques within microtubule bundles. Nat. Commun 9, 3571 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Trupinic M. et al. The chirality of the mitotic spindle provides a mechanical response to forces and depends on microtubule motors and augmin. Curr. Biol 10.1016/j.cub.2022.04.035 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Velle KB et al. Naegleria’s mitotic spindles are built from unique tubulins and highlight core spindle features. Curr. Biol 32, 1247–1261.e6 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Yajima J, Mizutani K & Nishizaka T A torque component present in mitotic kinesin Eg5 revealed by three-dimensional tracking. Nat. Struct. Mol. Biol 15, 1119–1121 (2008). [DOI] [PubMed] [Google Scholar]
- 249.Maruyama Y. et al. CYK4 relaxes the bias in the off-axis motion by MKLP1 kinesin-6. Commun. Biol 4, 180 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Bormuth V. et al. The highly processive kinesin-8, Kip3, switches microtubule protofilaments with a bias toward the left. Biophys. J 103, L4–L6 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Elshenawy MM et al. Cargo adaptors regulate stepping and force generation of mammalian dynein-dynactin. Nat. Chem. Biol 15, 1093–1101 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Mitra A. et al. Kinesin-14 motors drive a right-handed helical motion of antiparallel microtubules around each other. Nat. Commun 11, 2565 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Nitzsche B. et al. Working stroke of the kinesin-14, ncd, comprises two substeps of different direction. Proc. Natl Acad. Sci. USA 113, E6582–E6589 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Wuhr M. et al. Evidence for an upper limit to mitotic spindle length. Curr. Biol 18, 1256–1261 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Crowder ME et al. A comparative analysis of spindle morphometrics across metazoans. Curr. Biol 25, 1542–1550 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Milunovic-Jevtic A, Jevtic P, Levy DL & Gatlin JC In vivo mitotic spindle scaling can be modulated by changing the levels of a single protein: the microtubule polymerase XMAP215. Mol. Biol. Cell 29, 1311–1317 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Goehring NW & Hyman AA Organelle growth control through limiting pools of cytoplasmic components. Curr. Biol 22, R330–R339 (2012). [DOI] [PubMed] [Google Scholar]
- 258.Good MC, Vahey MD, Skandarajah A, Fletcher DA & Heald R Cytoplasmic volume modulates spindle size during embryogenesis. Science 342, 856–860 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Hazel J. et al. Changes in cytoplasmic volume are sufficient to drive spindle scaling. Science 342, 853–856 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Wilbur JD & Heald R Mitotic spindle scaling during Xenopus development by kif2a and importin alpha. eLife 2, e00290 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Rieckhoff EM et al. Spindle scaling is governed by cell boundary regulation of microtubule nucleation. Curr. Biol 30, 4973–4983.e10 (2020). [DOI] [PubMed] [Google Scholar]
- 262.Miller KE, Session AM & Heald R Kif2a scales meiotic spindle size in Hymenochirus boettgeri. Curr. Biol 29, 3720–3727.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Loughlin R, Wilbur JD, McNally FJ, Nedelec FJ & Heald R Katanin contributes to interspecies spindle length scaling in Xenopus. Cell 147, 1397–1407 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Bird AW & Hyman AA Building a spindle of the correct length in human cells requires the interaction between TPX2 and Aurora A. J. Cell Biol 182, 289–300 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Helmke KJ & Heald R TPX2 levels modulate meiotic spindle size and architecture in Xenopus egg extracts. J. Cell Biol 206, 385–393 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Courtois A, Schuh M, Ellenberg J & Hiiragi T The transition from meiotic to mitotic spindle assembly is gradual during early mammalian development. J. Cell Biol 198, 357–370 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Decker F, Oriola D, Dalton B & Brugues J Autocatalytic microtubule nucleation determines the size and mass of Xenopus laevis egg extract spindles. eLife 10.7554/eLife.31149 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Reber SB et al. XMAP215 activity sets spindle length by controlling the total mass of spindle microtubules. Nat. Cell Biol 15, 1116–1122 (2013). [DOI] [PubMed] [Google Scholar]
- 269.Burbank KS, Mitchison TJ & Fisher DS Slide-and-cluster models for spindle assembly. Curr. Biol 17, 1373–1383 (2007). [DOI] [PubMed] [Google Scholar]
- 270.Knouse KA, Lopez KE, Bachofner M & Amon A Chromosome segregation fidelity in epithelia requires tissue architecture. Cell 175, 200–211.e13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Ponjavic I, Vukusic K & Tolic IM Expansion microscopy of the mitotic spindle. Methods Cell Biol. 161, 247–274 (2021). [DOI] [PubMed] [Google Scholar]
- 272.Chen BC et al. Lattice light-sheet microscopy: imaging molecules to embryos at high spatiotemporal resolution. Science 346, 1257998 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Sen O, Harrison JU, Burroughs NJ & McAinsh AD Kinetochore life histories reveal an Aurora-B-dependent error correction mechanism in anaphase. Dev. Cell 56, 3405 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Pamula MC et al. High-resolution imaging reveals how the spindle midzone impacts chromosome movement. J. Cell Biol 218, 2529–2544 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Yamashita N. et al. Three-dimensional tracking of plus-tips by lattice light-sheet microscopy permits the quantification of microtubule growth trajectories within the mitotic apparatus. J. Biomed. Opt 20, 101206 (2015). [DOI] [PubMed] [Google Scholar]









