Keywords: differential isoform usage, gene expression, hibernation, insulin resistance, PPAR signaling
Abstract
Hibernation in bears involves a suite of metabolical and physiological changes, including the onset of insulin resistance, that are driven in part by sweeping changes in gene expression in multiple tissues. Feeding bears glucose during hibernation partially restores active season physiological phenotypes, including partial resensitization to insulin, but the molecular mechanisms underlying this transition remain poorly understood. Here, we analyze tissue-level gene expression in adipose, liver, and muscle to identify genes that respond to midhibernation glucose feeding and thus potentially drive postfeeding metabolical and physiological shifts. We show that midhibernation feeding stimulates differential expression in all analyzed tissues of hibernating bears and that a subset of these genes responds specifically by shifting expression toward levels typical of the active season. Inferences of upstream regulatory molecules potentially driving these postfeeding responses implicate peroxisome proliferator-activated receptor gamma (PPARG) and other known regulators of insulin sensitivity, providing new insight into high-level regulatory mechanisms involved in shifting metabolic phenotypes between hibernation and active states.
INTRODUCTION
Hibernation in bears involves annual shifts in metabolism and physiology that enable bears to survive food-scarce winters (1–4). Notably, multiple tissues become insulin resistant during hibernation, only to have insulin sensitivity restored at the completion of the hibernation season (5–7). Recent studies of gene expression (8), differential isoform usage (9, 10), and cell culture experiments (7, 11) have made progress in identifying cellular and molecular mechanisms that drive key features of hibernation. However, these studies have relied primarily on broad comparisons between hibernating and nonhibernating bears, and questions remain regarding the mechanisms involved in the transitions into and out of hibernation.
To begin to dissect the role that feeding plays in the physiological transition between hibernation and the active season, we recently tested whether the feeding of a single macronutrient, glucose, is sufficient to “reverse” hibernation phenotypes. Glucose was chosen as the first macronutrient to explore given its relevance to insulin sensitivity (12). In this experiment, hibernating bears were fed glucose for a 10-day period and resulting changes in metabolical and physiological parameters including blood serum content, metabolic rate, body temperature, general activity, and insulin sensitivity were quantified (12). Following feeding, many of the measured phenotypes were restored to roughly 50% of their typical active season levels, suggesting that glucose consumption plays an important role in, but is not the sole driver of, the shift from hibernation active season physiology. Whether feeding elicits changes in gene expression in hibernating bears, and whether these changes shift toward active season levels, was not explored.
In a follow-up study, a series of adipocyte cell culture experiments were used to test whether blood serum from active season and glucose-fed hibernating bears stimulates changes in gene expression in hibernating bears (11, 13). Serum from active or fed bears was shown to elicit sweeping changes in gene expression in adipocytes, including expression of many genes involved in insulin signaling and other pathways previously implicated in driving hibernation phenotypes. Most notably, the expression profiles from hibernation adipocytes stimulated with either active or glucose-fed serum were nearly indistinguishable from those of active season cells stimulated with active season serum. Interestingly, levels of serum glucose and insulin did not differ significantly between pre- and post-fed bears (12), suggesting that other circulating factors in the serum are responsible for stimulating these shifts in gene expression. Proteomic comparisons of serum types revealed a set of eight proteins with differential abundance between hibernation and both active and glucose-fed serums, which may play important roles in stimulating changes in gene expression in adipocytes, and potentially other cell and tissue types, between active and hibernation seasons (13).
Together, these two studies indicate that feeding glucose (referred to simply as “feeding” hereafter) plays a key role in stimulating the physiological shift from hibernation to the active season, yet a discrepancy remains between the near complete reversal of gene expression at a cellular level and the partial reversal of metabolical and physiological phenotypes following midhibernation feeding. Here, we analyze tissue-level differential expression following midhibernation feeding in three key metabolic tissues–adipose, liver, and muscle—to better understand the molecular mechanisms involved in the physiological shift from hibernation to the active season and to test the hypothesis that midhibernation feeding causes genes that were previously differentially expressed (DE) during hibernation compared with the active season (i.e., genes with “hibernation-specific” expression) to shift back toward active season-like levels of expression. Given that midhibernation feeding partially restores insulin sensitivity in bears, we further hypothesize that the set of genes with reversed expression after feeding will include a disproportionate number of genes involved in insulin signaling. To investigate higher-level signaling controlling postfeeding expression, we also predict networks of upstream regulatory molecules (e.g., transcription factors) that likely 1) drive observed gene expression across multiple tissues and 2) are involved in regulating the candidate serum proteins previously implicated in modulating hibernation gene expression in adipocytes.
MATERIALS AND METHODS
Animal Care
Brown bears (Ursus arctos horribilis) used in this study were housed at the Washington State University Bear Research, Education, and Conservation Center (WSU Bear Center) in accordance with the Bear Care and Colony Health Standard Operating Procedures and under care protocols approved by the Washington State Institutional Animal Care and Use Committee (Protocols #6546, #6468). This study involved 11 bears (7 females, 4 males) ranging from 3 to 15 yr old at the time of sampling. See Refs. 7 and 8 for additional information about bear care procedures.
Glucose Feeding and Tissue Collection
Tissues used in this study were collected during a prior experiment described in detail in Ref. 12. In brief, six hibernating bears (4 females, 2 males) were fed a glucose solution (dextrose, Sigma-Aldrich, St. Louis, MO) at a daily volume calculated to replace 100% of the estimated daily energy expenditure of each animal (12). Although bears typically do not eat or drink during hibernation in nature (1), bears in the WSU Bear Center will readily accept a diluted honey and water solution during hibernation as a result of year-round positive-reinforcement training (14); for this experiment, the honey water solution was replaced with the glucose solution, as described previously (12). This feeding trial was conducted over the course of 10 consecutive days. Four bears (2 females, 2 males) were excluded from the feeding trial to serve as unfed controls.
Between 5 and 7 days before the feeding trial, all 11 bears were anesthetized using a combination of tiletamine HCL/zolazepam HCl (Telazol, Zoetis, Florham Park, NJ) and dexmedetomidine (Zoetis) as described in Refs. 7 and 12. During anesthesia, subcutaneous adipose biopsies were collected from the gluteal region using a 6-mm biopsy punch and immediately snap-frozen with liquid nitrogen (12). Liver and skeletal muscle (gastrocnemius) biopsies were collected using 14G tru-cut biopsy needles (Progressive Medicinal International, Vista, CA) and snap-frozen with liquid nitrogen. These samples comprise the “pre-experiment hibernation” treatment. Following the completion of the feeding trial, the 10 bears involved in the trial (6 fed, 4 not-fed; 1 bear was excluded from the trial entirely) were fasted for 5–7 days, at which point they were again anesthetized and adipose, muscle, and liver biopsies were collected following the same sampling protocol as the pre-experiment hibernation treatment, but sampling from a different part of the tissue (liver and muscle) or opposite glute (adipose) to minimize the impact of localized wound-healing responses in our gene expression data. These samples collected after the feeding trial comprise the “fed” and “not-fed” treatments. All samples were stored at −80°C before RNA extraction.
RNA Extraction, Library Preparation, and Sequencing
Frozen samples were placed into 600 μL QIAzol (Qiagen, Redwood City, CA) and homogenized using a TissueLyser LT (Qiagen). Following homogenization, 120 μL of chloroform was added to each sample. Samples were vortexed for 15 s to mix, centrifuged for 15 min at 4°C and 14,000 rpm, and incubated at room temperature for 1 min. The resulting aqueous phase was pipetted to a new tube and loaded into a QIAcube (Qiagen) for extraction using an RNeasy Mini kit (adipose and liver; Qiagen) or RNeasy Fibrous Tissue Mini Kit (muscle; Qiagen). Extracted RNA yield was quantified with a Qubit 2.0 (Invitrogen, Carlsbad, CA) and RNA quality was assessed with a BioAnalyzer 2100 [average RNA integrity number (RIN) = 6.94; Agilent Technologies, Santa Clara, CA]. Extracted RNA was stored at −80°C. We note that several RNA extractions were unsuccessful, and thus RNA-seq data were not generated for these samples (Supplemental Table S1).
RNA-seq libraries were prepared using the TruSeq Stranded Total RNA Prep Kit with Ribo-Zero Gold (Part #15031048 Rev. E, Illumina, San Diego, CA). Library concentration and quality were assessed with a Qubit 2.0 and BioAnalyzer 2100, respectively. Samples were pooled at equimolar concentrations and sequenced on one lane of an Illumina HiSeq 2500 (v4 reagents) with 100 base-pair (bp) paired-end reads at the Washington State University Genomics Core in Spokane, WA.
RNA-seq Data Processing and Quantification
Previously generated adipose RNA-seq data for hibernating and active brown bears (8) and from bear adipocyte cell culture experiments (13) were downloaded from NCBI (BioProject PRJNA413091) and processed alongside newly generated data as described below.
Raw RNA-seq reads were trimmed using TrimGalore v0.4.2 [Krueger (15); run with Cutadapt v2.7], removing reads less than 50bp in length and with quality scores lower than 20, and trimmed read quality was assessed using FastQC v0.11.9 (16). Trimmed reads were mapped to the brown bear genome [UrsArc1.0; NCBI: GCF_023065955.1; Armstrong et al. (17)] using STAR v2.7.6a (18) in two-pass mode and outputting only uniquely mapping reads with the flag –outFilterMultiMapNmax 1. Five samples were mapped poorly (less than 3 million uniquely mapped reads) and were therefore excluded from subsequent analyses (Supplement Table S1). Gene-level expression counts were quantified using featureCounts in the Subread package v1.6.3 (19). Separately, transcript-level expression counts were quantified using Kallisto v0.48.0 (20) and the brown bear transcriptome (UrsArc1.0; NCBI: GCF_023065955.1).
Differential Gene-Level Expression Analysis
Raw gene-level expression counts for data generated in this study and those generated by Jansen et al. (8) were imported into R (21) and differential expression analyses were conducted with DEseq2 v1.34.0 (22). For each tissue, pairwise comparisons were conducted for 1) not-fed versus fed, 2) pre-experiment hibernation versus fed, 3) pre-experiment hibernation versus not-fed, and 4) Jansen et al. (8) active season versus hibernation treatments (8). All pairwise comparison analyses were controlled for sex. Independent hypothesis weighting (IHW) was used to correct P values using baseMean values from DEseq2 as the covariate (23), and differentially expressed genes were defined as those with IHW-corrected P values < 0.05. Significantly differentially expressed genes are referred to hereafter as upregulated or downregulated if they are higher or lower, respectively, in the second treatment of each comparison compared with the first treatment; for example, upregulated genes in the not-fed versus fed comparison are more highly expressed in the fed treatment compared with the not-fed treatment. Gene counts were normalized using the variance-stabilizing transformation in DEseq2. Exploratory PCA plotting of the top 5,000 highly expressed genes in each tissue revealed one adipose sample (AF1N) and two liver samples (PL1N and CL1N) to be strong outliers; these samples were removed from the dataset and differential expression analysis was run again for adipose and liver. Final sample sizes for each condition are listed in Table 1. Raw and normalized gene counts are provided in Supplemental Table S2.
Table 1.
Summary of pairwise differential expression analyses
| Tissue | Data Source | Condition 1 | Condition 2 | No. of DE Genes |
|---|---|---|---|---|
| Adipose | Tissue biopsy, this study | Not fed (n = 4) | Fed (n = 5) | 160 (117 up, 43 down) |
| Tissue biopsy, this study | Pre-experiment hibernation (n = 9) | Fed (n = 5) | 1 (1 up, 0 down) | |
| Tissue biopsy, this study | Pre-experiment hibernation (n = 9) | Not fed (n = 4) | 0 | |
| Tissue biopsy, Jansen et al. (8) | Active season (n = 5) | Hibernation (n = 6) | 3,815 (1,791 up, 2,024 down) | |
| Adipocyte culture, Saxton et al. (13) | Hibernation cells, hibernation serum (n = 6) | Hibernation cells, postfeeding serum (n = 6) | 5,183 (2,604 up, 2,579 down) | |
| Liver | Tissue biopsy, this study | Not fed (n = 3) | Fed (n = 4) | 213 (190 up, 23 down) |
| Tissue biopsy, this study | Pre-experiment hibernation (n = 8) | Fed (n = 4) | 252 (201 up, 51 down) | |
| Tissue biopsy, this study | Pre-experiment hibernation (n = 8) | Not fed (n = 3) | 1 (0 up, 1 down) | |
| Tissue biopsy, Jansen et al. (8) | Active season (n = 5) | Hibernation (n = 6) | 2,743 (1,425 up, 1,318 down) | |
| Muscle | Tissue biopsy, this study | Not fed (n = 3) | Fed (n = 5) | 529 (264 up, 265 down) |
| Tissue biopsy, this study | Pre-experiment hibernation (n = 10) | Fed (n = 5) | 1,267 (589 up, 678 down) | |
| Tissue biopsy, this study | Pre-experiment hibernation (n = 10) | Not fed (n = 3) | 0 | |
| Tissue biopsy, Jansen et al. (8) | Active season (n = 6) | Hibernation (n = 6) | 602 (278 up, 324 down) |
Results of pairwise comparisons of gene-level expression. “Data source” indicates the sample type and source study of data underlying each pairwise comparison. Sample numbers are listed next to each condition.
Gene-level expression counts generated from adipocyte cell culture in a study by Saxton et al. (13) were also tested for differential expression following the approach described above; specifically, a pairwise comparison was conducted between hibernation cell/postfeeding serum (“HG”) and hibernation cell/hibernation serum (“HH”) treatments.
Significantly differentially expressed genes from different comparisons and experiments were intersected to assess overlap and plotted using the R packages ggplot2 v3.3.6 (24) and ggvenn v0.1.9 (25). Genes with differential gene expression were characterized for enrichment of Gene Ontology (GO) terms and KEGG pathways (26) using clusterProfiler v4.2.2 (27) in R.
Differential Isoform Usage Analysis
Raw transcript-level expression counts were imported into R and analyzed for differential isoform usage using the IsoformSwitchAnalyzeR package v1.16.0 (28). The DEXseq method (29) within IsoformSwitchAnalyzeR was used to identify differential isoform usage between treatments, controlling for sex. Differential isoform usage was assessed between active and hibernation season treatments from a study by Jansen et al. (8), between pre-experiment hibernation, fed, and not-fed samples generated in this study, and between HH and HG adipocyte/serum treatments from Saxton et al.’s study (13). Amino acid sequences for isoforms involved in switches were exported from R and protein domains were identified using pfam_scan.pl and the PFAM database version (30). Consequences of differential isoform usage were characterized using the analyzeSwitchConsequences tool within IsoformSwitchAnalyzeR to assess whether the involved isoforms differ substantially in intron retention, nonsense-mediated decay susceptibility, PFAM protein domains, open-reading frame sequence similarity, transcription start site (TSS) or transcription termination site (TTS), or length of the 3′ and/or 5′ untranslated region with default parameters. This analysis identifies changes in isoform usage that are most likely to have a true biologically relevant impact on a downstream phenotype while minimizing changes that are likely to be biologically inconsequential [i.e., a very small change in untranslated region (UTR) length or shift in TSS position; (28)]. Genes with differential isoform usage were characterized for enrichment of Gene Ontology (GO) terms and KEGG pathways using clusterProfiler v4.2.2 in R. The three samples identified as outliers in gene-level expression analyses (described above) were excluded from all isoform-level analyses.
Upstream Regulatory Molecule Inferences
To infer upstream regulatory molecules (i.e., transcription factors) that are responsible for regulating observed changes in gene expression after midhibernation feeding, we used CHEA3 [accessed March 2023; (31)] to perform transcription factor enrichment analysis on sets of differentially expressed genes. For each tissue, we ran CHEA3 on genes with reversed, upregulated expression after feeding (i.e., genes that are downregulated in hibernation compared with the active season, but subsequently upregulated after feeding) and separately genes with reversed, downregulated expression after feeding. All enrichment analyses with CHEA3 were conducted in R using the CHEA3 REST API. For each analysis, the top 25 predicted transcription factors based on mean rank scores were selected for subsequent characterization and analysis. Overlap of predicted regulatory molecules between tissues was assessed and plotted in R using ggvenn. Many predicted regulatory molecules of DE genes are also known to regulate other predicted regulatory molecules; we used the ggnetwork package v0.5.10 (32) in R to plot networks of coregulatory interactions between predicted regulatory molecules.
Separately, CHEA3 was also used to predict regulators of a set of candidate proteins shown previously to be differentially abundant in serum of hibernating versus fed or active bears (13) and therefore inferred to be involved in regulating, or potentially result from, differential gene regulation between seasons and after midhibernation feeding. A network of these proteins and their top inferred regulatory molecules was created using Cytoscape v3.9.1 (33).
RESULTS
Differential Expression and Isoform Usage following Glucose Feeding
Muscle tissues showed the greatest gene expression response to midhibernation feeding, with 529 significantly differentially expressed (DE) genes detected between fed and not-fed treatments; 213 and 160 DE genes were detected in liver and adipose, respectively (Table 1; Supplemental Table S3). In adipose and liver, the majority of DE genes were upregulated following feeding (117 out of 160 in adipose, 190 out of 213 in liver), whereas muscle exhibited similar numbers of up- and downregulated genes after feeding (264 up, 265 down; Table 1; Supplemental Table S3). In all tissues, the pre-experiment hibernation treatment did not differ substantially from the not-fed treatments, with a single DE gene detected between these treatments in liver [Serine peptidase inhibitor kazal type 1 (SPINK1); downregulated in not-fed] and no DE genes in adipose or muscle (Table 1; Supplemental Table S3).
Gene ontology (GO) overrepresentation analysis of genes upregulated after feeding in adipose revealed enrichment of a single term: collagen-containing extracellular matrix (Supplemental Fig. S1). No GO terms were significantly enriched in genes downregulated after feeding in adipose. Genes upregulated in liver after feeding were enriched for multiple GO terms and KEGG pathways relating to transmembrane transporter activity and metabolism, including peroxisome proliferator-activated receptor (PPAR) signaling, whereas downregulated genes were enriched for GO terms related to sulfur compound binding activity (Supplemental Fig. S1). In muscle, terms related to metabolism and the extracellular matrix were enriched among upregulated genes following feeding, whereas multiple terms related to the regulation of translation were enriched among downregulated genes (Supplemental Fig. S1). Enriched KEGG pathways for genes upregulated after feeding in muscle included diabetic cardiomyopathy, protein digestion and absorption, and multiple metabolism pathways, among others (Supplemental Fig. S1).
In addition to gene-level differential expression, changes in the expression levels individual isoforms of a given gene can impact downstream phenotypes by modifying the resulting gene product (34–36). Notably, changes in isoform usage (i.e., the relative expression levels of two or more isoforms of a gene) can occur with or without a change to overall gene-level abundance. Differential isoform usage (DIU) was previously documented between active, hyperphagia, and hibernation seasons in adipose, liver, and muscle tissue of brown bears, and may play a role in changes to insulin signaling and other aspects of hibernation physiology (9, 10). In the present study, analyses of DIU between fed and non-fed treatments detected 80 genes with changes in isoform usage in muscle tissue, 64 of which were predicted to result in a biologically significant functional consequence (i.e., a change in intron retention, nonsense mediated decay susceptibility, protein domains, TSS, TTS, and/or length of the 3′ or 5′ UTR between isoforms that change in usage; Table 2, Supplemental Table S4). The other two tissues showed a relatively muted response, with 20 (18 consequential) and 13 (10 consequential) genes with DIU detected in adipose and liver, respectively (Table 2; Supplemental Table S4). Genes with DIU after feeding were not significantly enriched for any GO terms or KEGG pathways in any tissues. In muscle, 10 genes with DIU were also found to be DE after feeding in gene-level expression analyses above; no genes with DIU in adipose or liver were also found to be DE after feeding.
Table 2.
Summary of differential isoform usage analyses
| No. of Genes with DIU |
|||||
|---|---|---|---|---|---|
| Tissue | Data source | Condition 1 | Condition 2 | All | Of Consequence |
| Adipose | Tissue biopsy, this study | Not fed (n = 4) | Fed (n = 5) | 20 | 18 |
| Tissue biopsy, this study | Pre-experiment hibernation (n = 9) | Fed (n = 5) | 9 | 6 | |
| Tissue biopsy, this study | Pre-experiment hibernation (n = 9) | Not fed (n = 4) | 7 | 6 | |
| Tissue biopsy, Jansen et al. (8) | Active season (n = 5) | Hibernation (n = 6) | 219 | 184 | |
| Adipocyte culture, Saxton et al. (13) | Hibernation cells, hibernation serum (n = 6) | Hibernation cells, postfeeding serum (n = 6) | 136 | 114 | |
| Liver | Tissue biopsy, this study | Not fed (n = 3) | Fed (n = 4) | 13 | 10 |
| Tissue biopsy, this study | Pre-experiment hibernation (n = 8) | Fed (n = 4) | 8 | 4 | |
| Tissue biopsy, this study | Pre-experiment hibernation (n = 8) | Not fed (n = 3) | 5 | 4 | |
| Tissue biopsy, Jansen et al. (8) | Active season (n = 5) | Hibernation (n = 6) | 30 | 25 | |
| Muscle | Tissue biopsy, this study | Not fed (n = 3) | Fed (n = 5) | 80 | 64 |
| Tissue biopsy, this study | Pre-experiment hibernation (n = 10) | Fed (n = 5) | 14 | 11 | |
| Tissue biopsy, this study | Pre-experiment hibernation (n = 10) | Not fed (n = 3) | 8 | 7 | |
| Tissue biopsy, Jansen et al. (8) | Active season (n = 6) | Hibernation (n = 6) | 65 | 44 | |
Results of pairwise comparisons of gene-level expression. “Data source” indicates the sample type and source study of data underlying each pairwise comparison. “Of consequence” indicates the number of genes with differential isoform usage (DIU) in which the involved isoforms differ in intron retention, nonsense-mediated decay susceptibility, presence of protein domains, the transcription start site and/or transcription termination site location, or the length of the 3′ and/or 5′ untranslated region.
Midhibernation Feeding Partially Reverses Hibernation-Specific Gene Expression
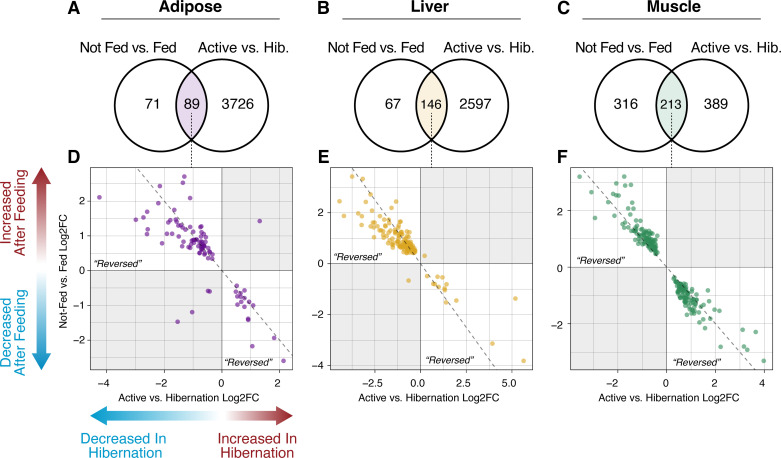
To assess whether feeding during hibernation shifts tissue-level gene expression toward active-season levels, we reanalyzed active season and hibernation RNA-seq data from study by Jansen et al. (8) and compared it to patterns of differential expression between fed and not-fed treatments (Fig. 1). For adipose and liver, the majority of differentially expressed genes after feeding are also differentially expressed between active and hibernation seasons (56% and 69%, respectively; Fig. 1, A and B). Muscle shared the largest number of DE genes with the active versus hibernation comparison (213 genes; Fig. 1C); muscle was also the only tissue that exhibited a greater number of DE genes unique to fed versus not-fed comparisons (316 genes; Fig. 1C) than were shared between both comparisons. In all three tissues, the majority of DE genes shared between fed versus not-fed and active versus hibernation comparisons showed “reversed” directions of expression (Fig. 1, D–F); for example, genes downregulated during hibernation were subsequently upregulated following midhibernation feeding, and vice versa. Specifically, 84 out of 89 (94%) shared DE genes in adipose showed reversed expression after feeding, whereas 144 out of 146 (99%) were reversed in liver (Fig. 1, D and E). All 213 shared DE genes in muscle showed reversed expression after feeding (Fig. 1F). All genes with “reversed” expression after feeding are provided in Supplemental Table S5.
Figure 1.
Reversal of hibernation-specific gene expression following feeding. Venn diagrams indicate overlapping differentially expressed (DE; determined using DEseq2; IHW P value < 0.05) genes after midhibernation feeding (left) and between active and hibernation seasons (right) in adipose (A), liver (B), and muscle tissue (C). The number of replicates (n) per treatment is provided in Table 1. Dot plots of log2-fold change values for overlapping DE genes in adipose (D), liver (E), and muscle (F). Positive values on the x-axis indicate increased expression during hibernation relative to the active season, whereas positive values on the y-axis indicate increased expression following feeding. The top-left and bottom-right quadrants correspond to genes with “reversed” expression following feeding (i.e., downregulation during hibernation and subsequent upregulation after feeding).
To characterize the function of genes with reversed expression after feeding, we conducted GO term and KEGG pathway overrepresentation analysis for each tissue using the set of DE genes between active and hibernation seasons in that tissue as the background. Genes with reversed and upregulated expression in post-fed adipose were enriched for four redundant terms related to extracellular organelles and vesicles (Supplemental Fig. S2). These same four terms were also enriched in reversed and post-fed upregulated genes in liver (Supplemental Fig. S2), in addition to multiple metabolism related terms, among others. No GO terms were enriched in reversed and downregulated genes in either adipose or liver. In muscle, reversed and upregulated genes were enriched for involvement in the mitochondrion, whereas reversed and downregulated genes were enriched for multiple terms relating to gene expression, the ribosome, and translation (Supplemental Fig. S2).
Comparisons of differential isoform usage after feeding and between active versus hibernation found no overlap in genes within adipose or liver. In muscle, three genes were found be DIU in both comparisons: zinc finger CCCH-type containing 11A (ZC3H11A), LIM domain binding 3 (LDB3), and plastin 3 (PLS3) (Supplemental Fig. S3 and Supplemental Table S4). Of these three, only LDB3 showed a “reversed” pattern of isoform usage between comparisons: isoform XM_044386246.2 decreases in isoform usage during hibernation but increased following feeding (Supplemental Fig. S3).
Shared Postfeeding Responses in Cell Culture and Tissue-Level Gene Expression
Serum collected following midhibernation feeding has been shown to stimulate changes in gene expression in cultured hibernating bear adipocytes, shifting expression to resemble patterns observed during the active season (13). To assess the degree to which changes in expression in adipocyte cell culture reflect changes in complex tissue, we reanalyzed RNA-seq data from cell culture experiments and compared it with differential expression and isoform usage in post-fed adipose tissue. We focused on the pairwise comparison between two cell culture treatments: hibernation cells stimulated with blood serum collected during hibernation (treatment HH; analogous to not-fed, hibernation adipose tissue) and hibernation cells stimulated with blood serum collected after the feeding trial (treatment HG; analogous to post-fed adipose tissue). A considerably larger gene expression response was observed in cell culture comparisons compared with post-fed tissue comparisons, with 5,183 significantly DE genes detected in cell culture compared with the 160 DE genes detected in post-fed adipose tissue (Supplemental Fig. S4). A total of 70 DE genes were detected in both cell culture and tissue-level comparisons, of which 48 (∼69%) shared the same direction of differential regulation in the analogous postfeeding treatments (Supplemental Fig. S4). Of these 48 genes, 34 overlap with the set of genes found to be “reversed” after feeding in the analyses described in the previous section. DE genes shared between cell culture and tissue responses were not enriched for any GO terms or KEGG pathways. Analysis of isoform switching between cell culture conditions identified 136 genes with an isoform switch, 114 of which are of consequence (Supplemental Table S4). However, none of these genes are shared with the 13 genes found to have an isoform switch in adipose tissue after feeding.
No Insulin Signaling Genes Exhibit Reversed Expression after Feeding
A small number of genes involved in insulin signaling were found to be differentially expressed after feeding (Supplemental Fig. S5). A single gene involved in insulin signaling, ATPase H+ transporting V0 subunit e1 (ATP6V0E1), was upregulated in adipose after feeding. Two genes, Sorbin and SH3 domain containing 1 (SORBS1) and eukaryotic translation initiation factor 4E binding protein 2 (EIF4EBP2), were upregulated in liver after feeding (Supplemental Fig. S5). Two additional genes, Insulin degrading enzyme (IDE) and ATPase H+ transporting V1 subunit D (ATP6V1D), were upregulated after feeding in muscle (Supplemental Fig. S5). All five of these genes exhibited weak upregulation (all log2 fold-changes < 1; Supplemental Fig. S5), and none show “reversed” expression (i.e., none were downregulated during hibernation before upregulation after feeding, and vice versa). A single insulin-related gene, glycogen synthase kinase 3 beta (GSK3B), exhibited a change in isoform usage after feeding (adipose tissue; Supplemental Fig. S6).
Upstream Regulators of Postfeeding Differential Gene Expression
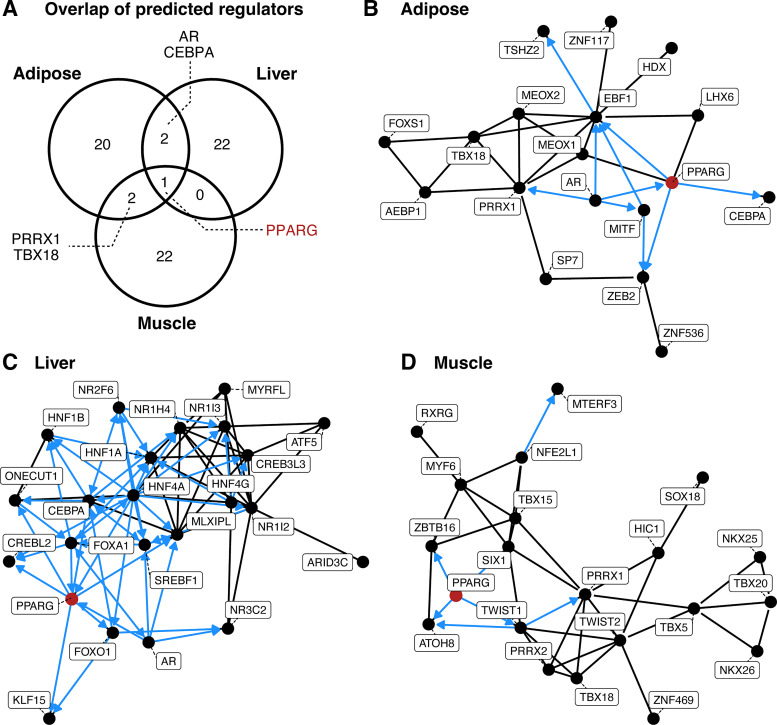
To identify higher-level regulatory molecules that may play a role in regulating observed changes in expression after feeding, we used CHEA3 to identify putative regulator molecules known to regulate genes with “reversed” gene expression after feeding (Supplemental Table S6). Many of the identified upstream regulatory molecules targeting DE genes also are known to play roles in regulating one another; coregulatory networks of the top 25 predicted regulatory molecules for reversed up- and downregulated genes in each tissue are shown in Fig. 2 and Supplemental Fig. S7.
Figure 2.
Predicted upstream regulatory molecules of genes with reversed, upregulated expression after midhibernation feeding. A: overlap of the top 25 CHEA3-predicted regulators of genes with reversed, upregulated expression following feeding in adipose, liver, and muscle. Coregulatory networks of predicted upstream regulators of reversed, upregulated genes in adipose (B), liver (C), and muscle (D). Edges represent known interactions between two regulators. Blue arrows indicate directional interactions that are supported by chromatin immunoprecipitation followed by sequencing (ChIP-seq) data in CHEA3 databases. PPARG, the only regulator predicted in all three tissues, is highlighted in red. PPARG, peroxisome proliferator-activated receptor gamma.
Two putative regulatory molecules were predicted to regulate reversed, upregulated genes in both adipose and liver [CCAAT enhancer binding protein alpha (CEBPA) and androgen receptor (AR); Fig. 2], whereas one regulator, SIX homeobox 5 (SIX5), was inferred to regulate reversed, downregulated genes in these two tissues (Supplemental Fig. S7). Two additional putative regulatory molecules were predicted to regulate reversed, upregulated genes in adipose and muscle [T-box transcription factor 18 (TBX18) and paired related homeobox 1 (PRRX1); Fig. 2]. A single regulatory molecule, peroxisome proliferator-activated receptor gamma (PPARG), was predicted to regulate reversed, upregulated genes in all three tissues (Fig. 2). PPARG was predicted to target 31 (46%), 20 (15%), and 40 (37%) reversed, upregulated genes in adipose, liver, and muscle, respectively (Supplemental Table S6). Of these genes, six were upregulated in two tissues following feeding, whereas a single PPARG-regulated gene, acyl-CoA synthetase family member 2 (ACSF2), was upregulated in all three tissues (Supplemental Table S6). The PPARG gene itself was found to be downregulated in adipose and upregulated in liver during hibernation (Supplemental Table S3), consistent with previous findings by Jansen et al. (8), however it did not exhibit reversed expression in any tissue following feeding. A coactivator of PPARG, PPARG coactivator 1 alpha (PPARGC1A or PGC1A), showed reversed, upregulated expression following feeding in liver tissue (Supplemental Table S5).
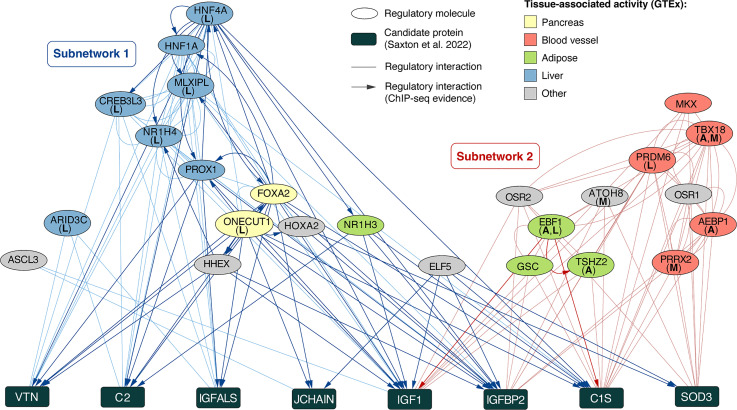
Upstream Regulators of Key Serum Proteins
Previously, eight proteins were identified with differential abundance in serum of hibernating bears compared with active and post-fed bears that may play a role in regulating, and/or result from, seasonal patterns of gene expression in bears (13). To better understand the regulation of these proteins, as well as their relationship to differentially expressed genes, we used CHEA3 to predict upstream regulatory molecules of these proteins (Supplemental Table S7). Upstream regulators of these candidate proteins form two distinct subnetworks of cointeracting regulators (Fig. 3). Members of “Subnetwork 1” interact with one another and collectively regulate all eight candidate proteins (Fig. 3). Members of “Subnetwork 2” only regulate four candidate proteins (Superoxide Dismutase 3 (SOD3), Complement C1s (C1S), Insulin Like Growth Factor Binding Protein 2 (IGFBP2), and Insulin Like Growth Factor 1 (IGF1); Fig. 3). No coregulatory interactions exist between members of these two subnetworks. Two upstream regulators in Subnetwork 1, One Cut Homeobox 1 (ONECUT1) and Forkhead Box A2 (FOXA2), were predicted to regulate seven out of eight candidate proteins (all proteins except SOD3; Fig. 3). Based on coexpression data available in CHEA3 [based on human gene expression data available in the GTEx database; GTeX Consortium (37)], these two molecules are enriched for regulatory activity in the pancreas (Fig. 3). ONECUT1 and FOXA2 further regulate, or are regulated by, multiple other upstream regulators identified by CHEA3 (Prospero Homeobox 1 (PROX1), Hepatocyte Nuclear Factor 4 Alpha (HNF4A), Hematopoietically Expressed Homeobox (HHEX), Homeobox A2 (HOXA2), MLX Interacting Protein Like (MLXIPL), and Hepatocyte Nuclear Factor 1-Alpha (HNF1A), many of which are enriched for regulatory activity in the liver according to CHEA3 (Fig. 3). Four upstream regulators enriched for activity in adipose tissue were also identified and inferred to regulate candidate proteins Complement 2 (C2), IGF1, IGFBP2, and C1S (Fig. 3). The remaining upstream regulators are associated with activity in blood or other body tissues (Fig. 3). Fourteen of the top upstream regulators for the candidate proteins were also inferred to regulate postfeeding expression of genes in one or more tissues (Fig. 3). This includes T-Box Transcription Factor 18 (TBX18), which was inferred to regulate reversed, upregulated genes after feeding in both muscle and adipose tissue (Figs. 2 and 3). AE Binding Protein 1 (AEBP1), a predicted regulator of candidate proteins IGFBP2, C1S, and SOD3 (Fig. 3), is also a predicted regulator of reversed, upregulated genes in adipose tissue after feeding (Fig. 2B). Interestingly, the gene encoding AEBP1 exhibits reversed and upregulated expression after feeding only in muscle (Supplemental Table S5).
Figure 3.
Predicted upstream regulators of candidate serum proteins previously implicated in hibernation signaling. Regulatory molecules are indicated with oval nodes, whereas candidate proteins are shown with rectangles. Edges represent known interactions between two regulators or a regulator and candidate protein. Arrows indicate directional interactions supported by ChIP-seq data in CHEA3 databases. Regulator nodes are colored based on enrichment for tissue-specific activity in CHEA3 databases (based on GTEx human gene expression data). Letters in parentheses indicate regulatory molecules that were also inferred to regulated reversed, upregulated genes (A= adipose, L = liver, M = muscle). Two discrete subnetworks of cointeracting regulatory molecules are indicated with blue (Subnetwork 1) and red (Subnetwork 2) edges.
DISCUSSION
Glucose feeding during hibernation has been shown to partially reverse insulin resistance and other characteristics of hibernation physiology in brown bears (12). In addition, it was recently shown that stimulating adipocytes from hibernating bears with serum collected from recently fed bears shifts adipocyte gene expression to resemble that of active, nonhibernating adipocytes (13). In this study, we analyzed the impact of midhibernation feeding on tissue-level gene expression patterns in adipose, liver, and muscle tissue, and complemented these prior studies by providing new insight into tissue-level signaling activity involved in hibernation and the transition from a hibernating to active season physiological state.
Overall Patterns of Postfeeding Gene Expression and Isoform Usage
We observed differential gene expression in all three tissues when comparing samples from fed to not-fed bears, indicating that feeding bears glucose during hibernation stimulates changes in gene regulation across multiple tissues. Despite exhibiting the largest change in gene expression between active and hibernation seasons [Table 1; (8)], adipose tissue showed the most muted response to feeding compared with liver and muscle. Furthermore, upregulated genes in adipose after feeding were only enriched for a single GO term relating to collagen and the extracellular matrix, potentially indicative of a lingering wound-healing response to the biopsy collected before the feeding trial. Consistent with previous findings supporting increased metabolism after midhibernation feeding (8), upregulated genes after feeding in liver and muscle were enriched for multiple GO terms related to metabolism. Upregulated genes in liver were also enriched for involvement in the PPAR signaling KEGG pathway; we discuss additional evidence for increased PPAR signaling activity following feeding below. In addition to metabolism, post-fed upregulated genes were enriched for protein digestion and absorption in muscle, whereas post-fed downregulated genes in muscle were enriched for GO terms and KEGG pathways related to translation. Previously, increased protein synthesis in muscle has been suggested as a means for preventing disuse atrophy in hibernating bears (8, 38); our findings suggest that feeding stimulates the reversal of, or at least decreases in, this elevated level of protein synthesis in muscle tissues. We note that the total number of RNA-seq reads per sample is relatively low in this study, which may result in genes with high variance between samples and/or fine-scale shifts in expression after feeding going undetected in these differential expression analyses. It is also possible that a small number of false positives are present; we therefore focus our interpretation and discussion below on the “reversed” genes that were selected based on 1) being significantly differentially expressed both in independent comparisons of not-fed versus fed samples and active versus hibernation samples and 2) showing opposing directions of expression in these comparisons (i.e., a gene that decreases expression during hibernation is subsequently upregulated, or reversed, after feeding), as we expect that few false positives would meet these criteria and are therefore less likely to influence these findings.
Feeding Reverses Hibernation-Specific Expression for a Subset of Genes
A key hypothesis in this study is that feeding “reverses” gene expression toward levels observed during the active state. To test this, we compared DE genes after feeding with DE genes observed between active and hibernation states using data from Jansen et al.’s study (8). In support of this hypothesis, nearly all genes found to be DE in both comparisons exhibited patterns of postfeeding gene-level expression consistent with a shift toward active season expression levels (i.e., gene expression was “reversed” after feeding). This supports the idea that feeding initiates a cascade of molecular, metabolical, and physiological changes that contribute to the transition of bears from a hibernating to active state. As previous studies have demonstrated that feeding results in partial reversal of several physiological aspects of hibernation, most notably increased insulin sensitivity (12), it is likely that this set of genes with reversed expression after feeding plays important roles in shifting cellular activity and metabolism as tissues respond to the reintroduction of external nutrition, as explored further below. We note that given the experimental differences between this study and study by Jansen et al. (8), we chose to define “reversed” genes as those with significant differential expression in opposite directions (i.e., downregulated in hibernation and upregulated after feeding) and did not assess or make conclusions about the degree of reversal of these genes (i.e., how closely a given gene returned to active season expression after feeding). In the future, experiments that investigate the effect of other macronutrients (i.e., fat or protein) and/or a complex diet will enable us to better understand the quantitative nature of expression reversal at a gene-by-gene level. Indeed, it is likely that the feeding of other nutrients, whether in isolation or in combination, stimulates differential expression and potentially the “reversal” of additional genes, and/or impacts the expression of genes identified herein to respond to glucose, in ways that we cannot detect in the current study.
Shared Cell- and Tissue-Level Gene Expression Responses to Midhibernation Feeding
To assess the degree to which adipocyte cell culture recapitulates tissue-level responses to feeding, we compared tissue-level gene expression for adipose to previously published cell culture RNA-seq data (13). Comparisons between hibernation cells stimulated with hibernation blood serum and hibernation cells stimulated with postfeeding blood serum detected a much greater number of DE genes than in adipose tissue postfeeding, the vast majority of which were not shared between cell- and tissue-level analyses. However, 34 out of 84 “reversed” genes in adipose were also DE in cell culture comparisons, indicating that despite the overall discrepancy between cell- and tissue-level analyses, an important core gene expression response to feeding/post-fed serum is indeed shared. Differences between DE responses detected in cell- and tissue-level analyses could be driven by several factors, including added noise in the tissue-level dataset from the inclusion of multiple cell types and the more complex environment of biological signals present in tissue compared with cell culture. These findings further argue for the incorporation of using single-cell resolution gene expression data in future studies so that we can simultaneously dissect cell-type specific patterns of expression in vivo and compare with expression patterns observed in vitro.
Differential Expression and Isoform Usage of Insulin Signaling Genes following Feeding
Many genes involved in insulin signaling and related metabolic pathways are differentially regulated during hibernation, when bears are insulin resistant (5, 7). Differential regulation of genes involved in insulin signaling was therefore proposed to play an important role in controlling insulin sensitivity during hibernation. Given that midhibernation feeding partially restored insulin sensitivity in hibernating bears (12), we hypothesized that insulin-related genes would be enriched for “reversed” gene expression following feeding. However, no insulin-related genes showed reversed expression after feeding in any tissue. Although not reversed, several insulin-related candidate genes did exhibit differential expression after feeding. Of note, insulin-degrading enzyme (IDE) was upregulated in muscle following feeding (Supplemental Fig. S5). IDE plays an important role in regulating circulating insulin levels by degrading insulin, and increased expression of IDE has been associated with improved insulin sensitivity in mice (39–41). Upregulation of IDE in muscle following feeding therefore may be an important step in returning to an insulin-sensitive state. A single insulin-related gene, GSK3B, exhibited changes in isoform usage in adipose tissue after feeding (Supplemental Fig. S6). The isoforms involved in this change in isoform usage exhibit different transcription termination sites and different length 3′ untranslated regions (UTRs). Previously, levels of phosphorylated GSK3B were shown to decrease during hibernation in adipose tissue, potentially playing a role in decreased insulin sensitivity (7). It is unclear whether this change in isoform usage relates to differential phosphorylation and/or downstream activity of GSK3B during hibernation.
Upstream Regulators of Genes with Reversed Expression Implicate PPARG as a Key Regulator of Shifts from Hibernation to Active Season Physiology
As we continue to dissect the molecular mechanisms of hibernation physiology in bears in different tissues and cell types, inferences of how these complex changes are controlled at a higher level and ultimately orchestrated across the entire body become increasingly valuable. In addition to revealing mechanisms regulating changes in gene expression, these inferences can also identify and implicate regulatory molecules that may not exhibit changes in gene-level regulation while having increased or decreased activity due to changes in phosphorylation, interactions with other proteins, or various other processes that are not detectable with RNA-sequencing alone. This experiment provides the opportunity to identify potential upstream regulatory molecules controlling a relatively small, yet likely critically important, set of differentially expressed genes involved in shifting hibernation physiology.
Predictions of upstream regulatory molecules identified multiple interacting transcription factors likely controlling the postfeeding up- and downregulation of reversed genes in adipose, liver, and muscle (Fig. 2, Supplemental Fig. S7). Although most of these regulators were predicted to target upregulated genes in a single tissue, five regulators were shared across two or more tissues. A single regulatory molecule, PPARG, was predicted to target upregulated genes in all three tissues (Fig. 2). PPARG is a transcription factor that forms a heterodimer with retinoid X receptor (RXRs) and, once activate via ligand binding, regulates many genes involved in adipocyte differentiation and maintenance (42). Notably, PPARG activation improves insulin sensitivity in patients with type II diabetes (42), and the activation of PPARG in adipose tissue alone may be sufficient to improve whole body insulin sensitivity (43). This suggests that postfeeding activation of PPARG may play a central role in body-wide shifts in metabolism and physiology, including the partial restoration of insulin sensitivity observed after midhibernation feeding (12). In each tissue, PPARG is coregulated by multiple other predicted regulatory molecules which may indicate tissue-specific nuances in PPARG action. Two key coregulators of PPARG, CCAAT enhancer binding protein alpha (CEBPA) and androgen receptor (AR), are predicted regulatory molecules for reversed, upregulated genes in both adipose and liver. CEBPA and PPARG cooperatively regulate numerous genes and processes in adipocytes, including insulin sensitivity (44, 45), whereas AR can impact insulin secretion in response to circulating hormone levels (46).
Midhibernation feeding of glucose was previously shown to result in a partial reduction of circulating fatty acids in brown bears (12). Fatty acids and PPARG are functionally interconnected in that some unsaturated fatty acids can bind to and activate PPARG, whereas PPARG is also capable of regulating fatty acid synthesis (47, 48). Links between fatty acids and PPARG function, along with the fact that we did not observe the reversed expression of insulin signaling genes after feeding, provide further support for an important role of fatty acids in the restoration of insulin sensitivity and modulation of metabolic processes following feeding that warrants further investigation in future studies. Furthermore, PGC1A, which was reversed and upregulated in liver after feeding, is permissive to PPARG activity, fatty acid metabolism, gluconeogenesis, and circadian rhythms (47–49). PGC1A may therefore represent another key node in the transition to the active season. The large number of reversed, upregulated genes in liver after feeding (Fig. 1B) is consistent with increased activity of PGC1A serving to elevate transcriptional activity at a broad, tissue-level scale (47–49).
Collectively, our findings implicate PPARG as a potential high-level regulator of shifts in insulin sensitivity and general metabolism in adipose, liver, and muscle tissues of hibernating bears, and suggests that increased regulatory activity of PPARG may play a particularly important role in shifting from a hibernation to an active season physiological state. We note, however, that experimental interrogation of PPARG is needed to functionally test these hypotheses, and such experiments should be prioritized in future studies of bear hibernation physiology.
Upstream Regulators of Candidate Serum Proteins Are Known Regulators of Insulin Sensitivity
Treating hibernating bear adipocytes with serum collected from either the active season or following midhibernation feeding induces changes in gene transcription to resemble that of active season adipocytes (13). Proteomic analysis of serum identified a set of candidate proteins that differ in abundance between hibernation versus active season or post-fed serum, and therefore may play a role in, or perhaps result from, changes in gene activity resulting from serum stimulation (13). However, none of these candidate proteins is known to act as transcription factors, and therefore their precise relevance to hibernation physiology and underlying gene expression remains somewhat unclear. To investigate potential connections between these proteins and differentially expressed genes, we identified known regulators of these proteins using CHEA3 and compared results with regulators inferred to target genes with reversed expression after feeding. The top predicted regulators form two distinct coregulatory networks, with one comprising interacting regulatory molecules known to be active in liver and pancreas (“Subnetwork 1,” Fig. 3) and the other comprising molecules enriched for activity primarily in blood vessels and adipose tissue (“Subnetwork 2,” Fig. 3).
Nearly all regulatory molecules in Subnetwork 1 have been previously implicated in the modulation of insulin sensitivity, often in studies of insulin resistance and/or type II diabetes. Two regulators in Subnetwork 1, ONECUT1 and FOXA2, were predicted to target seven out of the eight candidate proteins (Fig. 3). Both of these regulators are active in the pancreas, and FOXA2 has been shown to modulate insulin signaling and insulin sensitivity (50–52). ONECUT1 was also predicted to be a regulator of reversed, upregulated genes in the liver (Fig. 2C). Within Subnetwork 1, ONECUT1 and FOXA2 interact with multiple regulators of candidate proteins that are known to be enriched for involvement in lipid metabolism and/or insulin secretion, including HNF4A, HNF1A, NR1H2, PROX1, HHEX, and Acyl-CoA Synthetase Long Chain Family Member 3 (ACSL3) (53–58). Others, including MLXIPL and CAMP Responsive Element Binding Protein 3 Like 3 (CREB3L3), have been associated with changes in insulin sensitivity in multiple studies (59, 60). Six of the regulators in Subnetwork 1 were independently predicted to also regulate reversed gene expression exclusively in liver tissue (HNF4A, MLXIPL, CREB3L2, NR1H4, AT-Rich Interaction Domain 3C (ARID3C), and ONECUT1; Figs. 2 and 3).
Regulators in Subnetwork 2 were predicted to target only four candidate proteins: SOD3, C1S, IGFBP2, and IGF1 (Fig. 3). Regulators in this subnetwork exhibit a number of canonical functions, many of which are involved in development and cell differentiation (61–64). The functional relevance of this subnetwork of regulators is therefore less clear. However, one regulator in Subnetwork 2, EBF Transcription Factor 1 (EBF1), is known to play roles in adipogenesis and insulin signaling (65). EBF1 was enriched for activity in adipose tissue and was also a predicted regulator of reversed genes in muscle and adipose tissues (Figs. 2 and 3).
The involvement of numerous molecules with established associations to the regulation of insulin signaling and insulin sensitivity suggests that Subnetwork 1 is likely the most functionally relevant set of molecules involved in regulating both candidate proteins and genes with reversed expression. Regulators within Subnetwork 2 may play important but unclear roles in postfeeding shifts in physiology, but could also represent regulators of these molecules in other tissues, cell types, or contexts not relevant to the current study. Collectively, these findings provide valuable context for the involvement of these candidate serum proteins in hibernation and may suggest that these proteins represent important intermediate molecules in signaling cascades controlled by higher-level regulatory activity. Armed with this additional context and candidate regulators of these serum proteins, subsequent experimental studies that test the hypothesized roles of these regulatory molecules will provide a better understanding of the relevance of these candidate proteins in controlling metabolical and physiological shifts between hibernating and active states.
Conclusions
Here, we demonstrate that feeding glucose to hibernating bears initiates the partial reversal of hibernation-specific gene expression in adipose, liver, and muscle tissue, and implicate a number of higher-level regulatory molecules, most notably PPARG, as likely drivers of this reversal and postfeeding increases in insulin sensitivity. It is worth noting that the number of genes with reversed expression after feeding represents a relatively small portion of the total genes with differential expression between active and hibernation seasons. As has been suggested in previous studies (12), this provides further support that feeding alone is not sufficient to return hibernating bears to an active physiological state but instead represents a critical element of a more complex process by which bears emerge from hibernation. Moving forward, similar experiments that feed other key nutrients, such as fat or protein, or a nutritionally complex meal that is representative of the natural diet of emerging bears, will enable us to further dissect the role of feeding on the physiological transition between hibernation and an active state and the importance of particular nutrients in driving specific aspects of these physiological and metabolical shifts.
DATA AVAILABILITY
All newly generated RNA-seq data are available on NCBI under BioProject PRJNA988160. Previously generated data are available on NCBI under BioProject PRJNA413091. Code is available on Github at https://github.com/blairperry/midhib_feeding_uarctos.
SUPPLEMENTAL DATA
Supplemental Figs. S1–S7 and Supplemental Tables S1–S7: https://doi.org/10.6084/m9.figshare.22575124.
GRANTS
This work was supported by a National Science Foundation (NSF) Office of Polar Programs (OPP) grant to J.L.K. (Award No. 2312253) and an NSF OPP postdoctoral fellowship to B.W.P. (Award No. 2138649). Additional support was provided by the Cougar Cage program at Washington State University, the Interagency Grizzly Bear Committee, the US Department of Agriculture National Institute of Food and Agriculture (McIntire-Stennis project 1018967), Mazuri Exotic Animal Nutrition, the Raili Korkka Brown Bear Endowment, the Nutritional Ecology Endowment, and the Bear Research and Conservation Endowment at Washington State University. This research used resources from the Center for Institutional Research Computing at Washington State University.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
C.T.R., H.T.J., and J.L.K. conceived and designed research; S.T., M.W.S., B.D.E.H., C.T.R., H.T.J., and J.L.K. performed experiments; B.W.P., A.L.M., S.T., M.W.S., C.L., and O.E.C. analyzed data; B.W.P., E.P.V., H.T.J., and J.L.K. interpreted results of experiments; B.W.P. prepared figures; B.W.P. and J.L.K. drafted manuscript; B.W.P., S.T., M.W.S., E.P.V., C.L., B.D.E.H., O.E.C., C.T.R., H.T.J., and J.L.K. edited and revised manuscript; B.W.P., S.T., M.W.S., E.P.V., C.L., B.D.E.H., O.E.C., C.T.R., H.T.J., and J.L.K. approved final version of manuscript.
ACKNOWLEDGMENTS
We acknowledge and thank all the volunteers and staff at the Washington State University Bear Center.
Present address of M. W. Saxton: National Park Service, Katmai National Park and Preserve, King Salmon, AK 99613, USA.
REFERENCES
- 1. Nelson RA, Folk GE Jr, Pfeiffer EW, Craighead JJ, Jonkel CJ, Steiger DL. Behavior, biochemistry, and hibernation in black, grizzly, and polar bears. Bears their Biol Manag 5: 284–290, 1983. doi: 10.2307/3872551. [DOI] [Google Scholar]
- 2. Hissa R, Siekkinen J, Hohtola E, Saarela S, Hakala A, Pudas J. Seasonal patterns in the physiology of the European brown bear (Ursus arctos arctos) in Finland. Comp Biochem Physiol Part A Physiol 109: 781–791, 1994. doi: 10.1016/0300-9629(94)90222-4. [DOI] [PubMed] [Google Scholar]
- 3. Hellgren EC. Physiology of hibernation in bears. Ursus 10: 467–477, 1998. [jstor.org/stable/3873159] [Google Scholar]
- 4. Robbins CT, Lopez-Alfaro C, Rode KD, Tøien Ø, Nelson OL. Hibernation and seasonal fasting in bears: the energetic costs and consequences for polar bears. J Mammal 93: 1493–1503, 2012. doi: 10.1644/11-MAMM-A-406.1. [DOI] [Google Scholar]
- 5. McCain S, Ramsay E, Kirk C. The effects of hibernation and captivity on glucose metabolism and thyroid hormones in American black bear (Ursus americanus). J Zoo Wildl Med 44: 324–332, 2013. doi: 10.1638/2012-0146R1.1. [DOI] [PubMed] [Google Scholar]
- 6. Palumbo PJ, Wellik DL, Bagley NA, Nelson RA. Insulin and glucagon responses in the hibernating black bear. Bears Their Biol Manag 5: 291–296, 1980. doi: 10.2307/3872552. [DOI] [Google Scholar]
- 7. Rigano KS, Gehring JL, Hutzenbiler BDE, Chen AV, Nelson OL, Vella CA, Robbins CT, Jansen HT. Life in the fat lane: seasonal regulation of insulin sensitivity, food intake, and adipose biology in brown bears. J Comp Physiol B 187: 649–676, 2017. doi: 10.1007/s00360-016-1050-9. [DOI] [PubMed] [Google Scholar]
- 8. Jansen HT, Trojahn S, Saxton MW, Quackenbush CR, Hutzenbiler BDE, Nelson OL, Cornejo OE, Robbins CT, Kelley JL. Hibernation induces widespread transcriptional remodeling in metabolic tissues of the grizzly bear. Commun Biol 2: 336, 2019. doi: 10.1038/s42003-019-0574-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tseng E, Underwood JG, Evans Hutzenbiler BD, Trojahn S, Kingham B, Shevchenko O, Bernberg E, Vierra M, Robbins CT, Jansen HT. Long-read isoform sequencing reveals tissue-specific isoform expression between active and hibernating brown bears (Ursus arctos). G3 (Bethesda) 12: jkab422, 2022. doi: 10.1093/g3journal/jkab422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Perry BW, Armstrong EE, Robbins CT, Jansen HT, Kelley JL. Temporal analysis of gene expression and isoform switching in brown bears (Ursus arctos). Integr Comp Biol 62: 1802–1811, 2022. doi: 10.1093/icb/icac093. [DOI] [PubMed] [Google Scholar]
- 11. Hogan HRH, Hutzenbiler BDE, Robbins CT, Jansen HT. Changing lanes: seasonal differences in cellular metabolism of adipocytes in grizzly bears (Ursus arctos horribilis). J Comp Physiol B 192: 397–410, 2022. doi: 10.1007/s00360-021-01428-z. [DOI] [PubMed] [Google Scholar]
- 12. Jansen HT, Hutzenbiler BE, Hapner HR, McPhee ML, Carnahan AM, Kelley JL, Saxton MW, Robbins CT. Can offsetting the energetic cost of hibernation restore an active season phenotype in grizzly bears (Ursus arctos horribilis)? J Exp Biol 224: jeb242560, 2021. doi: 10.1242/jeb.242560. [DOI] [PubMed] [Google Scholar]
- 13. Saxton MW, Perry BW, Hutzenbiler BDE, Trojahn S, Gee A, Brown AP, Merrihew GE, Park J, Cornejo OE, MacCoss MJ, Ribbins CT, Jansen HT, Kelley JL. Serum plays an important role in reprogramming the seasonal transcriptional profile of brown bear adipocytes. Iscience 25: 105084, 2022. doi: 10.1016/j.isci.2022.105084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Joyce-Zuniga NM, Newberry RC, Robbins CT, Ware JV, Jansen HT, Nelson OL. Positive reinforcement training for blood collection in grizzly bears (Ursus arctos horribilis) results in undetectable elevations in serum cortisol levels: a preliminary investigation. J Appl Anim Welf Sci 19: 210–215, 2016. doi: 10.1080/10888705.2015.1126523. [DOI] [PubMed] [Google Scholar]
- 15. Krueger F. Trim Galore!: A wrapper tool around Cutadapt and FastQC to consistently apply quality and adapter trimming to FastQ files. 2014. https://github.com/FelixKrueger/TrimGalore.
- 16. Andrews S. FastQC: a quality control tool for high throughput sequence data. 2010. https://github.com/s-andrews/FastQC.
- 17. Armstrong EE, Perry BW, Huang Y, Garimella KV, Jansen HT, Robbins CT, Tucker NR, Kelley JL. A beary good genome: haplotype-resolved, chromosome-level assembly of the brown bear (Ursus arctos). Genome Biol Evol 14: evac125, 2022. doi: 10.1093/gbe/evac125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21, 2013. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30: 923–930, 2013. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 20. Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 34: 525–527, 2016. [Erratum in Nat Biotechnol 34: 888, 2016]. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 21.R Core Team. R: A language and environment for statistical computing. 2014.
- 22. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550, 2014. doi: 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ignatiadis N, Klaus B, Zaugg JB, Huber W. Data-driven hypothesis weighting increases detection power in genome-scale multiple testing. Nat Methods 13: 577, 2016. doi: 10.1038/nmeth.3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wickham H. ggplot2. Wiley Interdiscip Rev Comput Stat 3: 180–185, 2011. doi: 10.1002/wics.147. [DOI] [Google Scholar]
- 25. Yan L. ggvenn: Draw Venn Diagram by ‘ggplot2’. 2022. R package version 0.1.9. https://cran.r-project.org/package=ggvenn.
- 26. Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res 32: D277–D280, 2004. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yu G, Wang L-G, Han Y, He Q-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16: 284–287, 2012. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vitting-Seerup K, Sandelin A. IsoformSwitchAnalyzeR: analysis of changes in genome-wide patterns of alternative splicing and its functional consequences. Bioinformatics 35: 4469–4471, 2019. doi: 10.1093/bioinformatics/btz247. [DOI] [PubMed] [Google Scholar]
- 29. Li Y, Rao X, Mattox WW, Amos CI, Liu B. RNA-seq analysis of differential splice junction usage and intron retentions by DEXSeq. PLoS One 10: e0136653, 2015. doi: 10.1371/journal.pone.0136653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths‐Jones S, Khanna A, Marshall M, Moxon S, Sonnhammer ELL, Studholme DJ, Yeats C, Eddy SR. The Pfam protein families database. Nucleic Acids Res 32: D138–D141, 2004. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Keenan AB, Torre D, Lachmann A, Leong AK, Wojciechowicz ML, Utti V, Jagodnik KM, Kropiwnicki E, Wang Z, Ma’ayan A. ChEA3: transcription factor enrichment analysis by orthogonal omics integration. Nucleic Acids Res 47: W212–W224, 2019. doi: 10.1093/nar/gkz446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Briatte F. ggnetwork: Geometries to Plot Networks with’ggplot2’. 2016. R package version 0.5.10. https://CRAN.R-project.org/package=ggnetwork. [Google Scholar]
- 33. Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504, 2003. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature 456: 470–476, 2008. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Khoury MP, Bourdon J-C. p53 isoforms: an intracellular microprocessor? Genes Cancer 2: 453–465, 2011. doi: 10.1177/1947601911408893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pal S, Gupta R, Kim H, Wickramasinghe P, Baubet V, Showe LC, Dahmane N, Davuluri RV. Alternative transcription exceeds alternative splicing in generating the transcriptome diversity of cerebellar development. Genome Res 21: 1260–1272, 2011. doi: 10.1101/gr.120535.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.GTEx Consortium. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 369: 1318–1330, 2020. doi: 10.1126/science.aaz1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fedorov VB, Goropashnaya AV, Stewart NC, Toien O, Chang C, Wang H, Yan J, Showe LC, Showe MK, Barnes BM. Comparative functional genomics of adaptation to muscular disuse in hibernating mammals. Mol Ecol 23: 5524–5537, 2014. doi: 10.1111/mec.12963.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Merino B, Fernández-Díaz CM, Parrado-Fernández C, González-Casimiro CM, Postigo-Casado T, Lobatón CD, Leissring MA, Cózar-Castellano I, Perdomo G. Hepatic insulin-degrading enzyme regulates glucose and insulin homeostasis in diet-induced obese mice. Metabolism 113: 154352, 2020. doi: 10.1016/j.metabol.2020.154352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Venturini PR, Thomazini BF, Oliveira CA, Alves AA, Camargo TF, Domingues CEC, Barbosa-Sampaio HCL, Do Amaral MEC. Vitamin E supplementation and caloric restriction promotes regulation of insulin secretion and glycemic homeostasis by different mechanisms in rats. Biochem Cell Biol 96: 777–785, 2018. doi: 10.1139/bcb-2018-0066. [DOI] [PubMed] [Google Scholar]
- 41. González-Casimiro CM, Merino B, Casanueva-Álvarez E, Postigo-Casado T, Cámara-Torres P, Fernández-Díaz CM, Leissring MA, Cózar-Castellano I, Perdomo G. Modulation of insulin sensitivity by insulin-degrading enzyme. Biomedicines 9: 86, 2021. doi: 10.3390/biomedicines9010086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Leonardini A, Laviola L, Perrini S, Natalicchio A, Giorgino F. Cross-talk between PPAR and insulin signaling and modulation of insulin sensitivity. PPAR Res 2009: 818945, 2009. doi: 10.1155/2009/818945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sugii S, Olson P, Sears DD, Saberi M, Atkins AR, Barish GD, Hong S-H, Castro GL, Yin Y-Q, Nelson MC, Hsiao G, Greaves DR, Downes MA, Yu RT, Olefsky JM, Evans RM. PPARγ activation in adipocytes is sufficient for systemic insulin sensitization. Proc Natl Acad Sci USA 106: 22504–22509, 2009. doi: 10.1073/pnas.0912487106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Madsen MS, Siersbæk R, Boergesen M, Nielsen R, Mandrup S. Peroxisome proliferator-activated receptor γ and C/EBPα synergistically activate key metabolic adipocyte genes by assisted loading. Mol Cell Biol 34: 939–954, 2014. doi: 10.1128/MCB.01344-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wu Z, Rosen ED, Brun R, Hauser S, Adelmant G, Troy AE, McKeon C, Darlington GJ, Spiegelman BM. Cross-regulation of C/EBPα and PPARγ controls the transcriptional pathway of adipogenesis and insulin sensitivity. Mol Cell 3: 151–158, 1999. doi: 10.1016/s1097-2765(00)80306-8. [DOI] [PubMed] [Google Scholar]
- 46. Yu I-C, Lin H-Y, Sparks JD, Yeh S, Chang C. Androgen receptor roles in insulin resistance and obesity in males: the linkage of androgen-deprivation therapy to metabolic syndrome. Diabetes 63: 3180–3188, 2014. doi: 10.2337/db13-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Varga T, Czimmerer Z, Nagy L. PPARs are a unique set of fatty acid regulated transcription factors controlling both lipid metabolism and inflammation. Biochim Biophys Acta 1812: 1007–1022, 2011. doi: 10.1016/j.bbadis.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ahmadian M, Suh JM, Hah N, Liddle C, Atkins AR, Downes M, Evans RM. PPARγ signaling and metabolism: the good, the bad and the future. Nat Med 19: 557–566, 2013. doi: 10.1038/nm.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Finck BN, Kelly DP. PGC-1 coactivators: inducible regulators of energy metabolism in health and disease. J Clin Invest 116: 615–622, 2006. doi: 10.1172/JCI27794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Heller S, Li Z, Lin Q, Geusz R, Breunig M, Hohwieler M, Zhang X, Nair GG, Seufferlein T, Hebrok M. Transcriptional changes and the role of ONECUT1 in hPSC pancreatic differentiation. Commun Biol 4: 1298, 2021. doi: 10.1038/s42003-021-02818-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Puigserver P, Rodgers JT. Foxa2, a novel transcriptional regulator of insulin sensitivity. Nat Med 12: 38–39, 2006. doi: 10.1038/nm0106-38. [DOI] [PubMed] [Google Scholar]
- 52. Lantz KA, Vatamaniuk MZ, Brestelli JE, Friedman JR, Matschinsky FM, Kaestner KH. Foxa2 regulates multiple pathways of insulin secretion. J Clin Invest 114: 512–520, 2004. doi: 10.1172/JCI21149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hansen SK, Párrizas M, Jensen ML, Pruhova S, Ek J, Boj SF, Johansen A, Maestro MA, Rivera F, Eiberg H, Andel M, Lebl J, Pedersen O, Ferrer J, Hansen T. Genetic evidence that HNF-1α–dependent transcriptional control of HNF-4α is essential for human pancreatic β cell function. J Clin Invest 110: 827–833, 2002. doi: 10.1172/JCI15085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Miyachi Y, Miyazawa T, Ogawa Y. HNF1A mutations and beta cell dysfunction in diabetes. Int J Mol Sci 23: 3222, 2022. doi: 10.3390/ijms23063222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ketterer C, Müssig K, Machicao F, Stefan N, Fritsche A, Häring H-U, Staiger H. Genetic variation within the NR1H2 gene encoding liver X receptor β associates with insulin secretion in subjects at increased risk for type 2 diabetes. J Mol Med (Berl) 89: 75–81, 2011. doi: 10.1007/s00109-010-0687-1. [DOI] [PubMed] [Google Scholar]
- 56. Lecompte S, Pasquetti G, Hermant X, Grenier-Boley B, Gonzalez-Gross M, De Henauw S, Molnar D, Stehle P, Béghin L, Moreno LA. Genetic and molecular insights into the role of PROX1 in glucose metabolism. Diabetes 62: 1738–1745, 2013. doi: 10.2337/db12-0864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zhang J, McKenna LB, Bogue CW, Kaestner KH. The diabetes gene Hhex maintains δ-cell differentiation and islet function. Genes Dev 28: 829–834, 2014. doi: 10.1101/gad.235499.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Israr-ul HA, Longacre MJ, Stoker SW, Kendrick MA, O’Neill LM, Zitur LJ, Fernandez LA, Ntambi JM, MacDonald MJ. Characterization of Acyl-CoA synthetase isoforms in pancreatic beta cells: gene silencing shows participation of ACSL3 and ACSL4 in insulin secretion. Arch Biochem Biophys 618: 32–43, 2017. doi: 10.1016/j.abb.2017.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Nakagawa Y, Satoh A, Yabe S, Furusawa M, Tokushige N, Tezuka H, Mikami M, Iwata W, Shingyouchi A, Matsuzaka T, Kiwata S, Fujimoto Y, Shimizu H, Danno H, Yamamoto T, Ishii K, Karasawa T, Takeuchi Y, Iwasaki H, Shimada M, Kawakami Y, Urayama O, Sone H, Takekoshi K, Kobayashi K, Yatoh S, Takahashi A, Yahagi N, Suzuki H, Yamada N, Shimano H. Hepatic CREB3L3 controls whole-body energy homeostasis and improves obesity and diabetes. Endocrinology 155: 4706–4719, 2014. doi: 10.1210/en.2014-1113. [DOI] [PubMed] [Google Scholar]
- 60. Kursawe R, Caprio S, Giannini C, Narayan D, Lin A, D’Adamo E, Shaw M, Pierpont B, Cushman SW, Shulman GI. Decreased transcription of ChREBP-α/β isoforms in abdominal subcutaneous adipose tissue of obese adolescents with prediabetes or early type 2 diabetes: associations with insulin resistance and hyperglycemia. Diabetes 62: 837–844, 2013. doi: 10.2337/db12-0889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zhang L, Reidy SP, Nicholson TE, Lee H-J, Majdalawieh A, Webber C, Stewart BR, Dolphin P, Ro H-S. The role of AEBP1 in sex-specific diet-induced obesity. Mol Med 11: 39–47, 2005. doi: 10.2119/2005-00021.Ro. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ito Y, Toriuchi N, Yoshitaka T, Ueno-Kudoh H, Sato T, Yokoyama S, Nishida K, Akimoto T, Takahashi M, Miyaki S, Asahara H. The Mohawk homeobox gene is a critical regulator of tendon differentiation. Proc Natl Acad Sci USA 107: 10538–10542, 2010. doi: 10.1073/pnas.1000525107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wu S-P, Dong X-R, Regan JN, Su C, Majesky MW. Tbx18 regulates development of the epicardium and coronary vessels. Dev Biol 383: 307–320, 2013. doi: 10.1016/j.ydbio.2013.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Gao Y, Lan Y, Ovitt CE, Jiang R. Functional equivalence of the zinc finger transcription factors Osr1 and Osr2 in mouse development. Dev Biol 328: 200–209, 2009. doi: 10.1016/j.ydbio.2009.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Griffin MJ, Zhou Y, Kang S, Zhang X, Mikkelsen TS, Rosen ED. Early B-cell factor-1 (EBF1) is a key regulator of metabolic and inflammatory signaling pathways in mature adipocytes. J Biol Chem 288: 35925–35939, 2013. doi: 10.1074/jbc.M113.491936. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figs. S1–S7 and Supplemental Tables S1–S7: https://doi.org/10.6084/m9.figshare.22575124.
Data Availability Statement
All newly generated RNA-seq data are available on NCBI under BioProject PRJNA988160. Previously generated data are available on NCBI under BioProject PRJNA413091. Code is available on Github at https://github.com/blairperry/midhib_feeding_uarctos.