Abstract
N6-methyladenosine (m6A) methylation of human immunodeficiency virus type 1 (HIV-1) RNA regulates viral replication, and the m6A of host RNA is affected by HIV-1 infection, but its global pattern and function are still unclear. In this study, we report that the number and position of m6A peaks in huge genes of human microglial HMC3 cells were modulated by a single cycle HIV-1 pseudotyped with VSV-G envelope glycoprotein infection using methylated RNA immunoprecipitation sequencing (MeRIP-seq). A conjoint analysis of MeRIP-seq and high-throughput sequencing for mRNA (RNA-seq) explored four groups of clearly classified genes, including 45 hyper-up (m6A-mRNA), 45 hyper-down, 120 hypo-up, and 54 hypo-down genes, in HIV-1 infected cells compared to uninfected ones. KEGG pathway analysis showed that these genes were mainly enriched in the Wnt and TNF signaling pathway, and cytokine–cytokine receptor interaction, which might be related to the immune response in HMC3 cells. And some of these genes might be associated with the pathway of axon guidance and neuroactive ligan-receptor interaction, which affect the neuronal state. However, the cognitive disorders caused by HIV-1 is associated with inflammatory changes that have not yet been well clarified. Furthermore, we confirmed the expression and m6A levels of four genes using RT-PCR and MeRIP-qPCR. Similar to the sequencing results, the expressions of these genes were significantly upregulated by HIV-1 infection. And the m6A level of IL-6 was downregulated, and those of HLA-B, CFB, and OLR1 were upregulated. These results suggest that HIV-1-induced changes in gene expression may be achieved through the regulation of methylation. Our study revealed the global m6A methylation and gene expression patterns under HIV-1 infection in human microglia, which might provide clues for understanding the interaction between HIV-1 and host cells and the cognitive disorders caused by HIV-1.
Keywords: N6-methyladenosine, HIV-1, HMC3, MeRIP-seq, RNA-Seq
1. Introduction
N6-methyladenosine (m6A) is considered to be the most abundant modification of eukaryotic mRNA [1], which plays important roles in controlling cellular gene expression and physiological function by affecting the stability [[2], [3], [4], [5]] translation, splicing [2,6,7], export, and decay [[8], [9], [10]] of mRNAs. Viral infection is one of the most crucial factors to interfere with the host's physiological state, and m6A plays significant roles in the life cycle of viruses and the interaction between virus and host [11]. It was reported that the m6A methylation of human immunodeficiency virus type 1 (HIV-1) RNA regulates viral replication. Spot-blot detection showed that the m6A level of T-cell RNA is upregulated [12], but the global pattern and function of m6A methylation under HIV-1 infection are still unknown.
‘RRACH’ has been demonstrated to be a common sequence for m6A modification, where R is adenine/guanine, H is adenine/cytosine/uracil, and A is the m6A site [[13], [14], [15]]. It was also shown that m6A methylation tended to be enriched in the 3′ UTR, long intron, exon, and stop codon. Multiple proteins were discovered to have significant roles in the process of m6A methylation. METTL3/14/16 (methyltransferase-like 3/14/16), and its additional adaptor molecules (namely RBM15/15B, WTAP, HAKAI, KIAA1429, VIRMA, and ZC3H13) [[16], [17], [18], [19], [20]] have been identified as components of the methyltransferase complex and are called ‘writers’. ALKBH5 (alkB homolog 5) and FTO (fat mass and obesity-associated protein) have been identified as demethylases with ‘erasers’ functions [[21], [22], [23]]. YTHDF1/2/3 (YTH-family proteins 1/2/3), YTHDC1/2 (YTH domain-containing proteins 1/2), and IGF2BP1/2/3 (insulin-like growth factor 2 mRNA binding proteins 1/2/3) were identified as m6A ‘readers’ that mediate specific functions of methylated mRNA transcripts [1,[24], [25], [26]]. Numerous studies focusing on m6A methylation of RNA have demonstrated that the regulators (writers, erasers, and readers) of m6A RNA methylation are involved in various human cancers and metabolic diseases [27]. In particular, m6A modification has significant impacts on lipid metabolic regulation [28]. In summary, m6A-associated proteins have shown more evidence of the physiological effects of m6A methylation, and the regulation of these proteins may influence m6A levels of cellular mRNAs.
m6A modification has been reported in many viral RNA transcripts including the adenovirus, influenza virus, avian sarcoma virus, simian virus 40, Rous sarcoma virus, and HIV-1 [12,[29], [30], [31], [32], [33]]. It can regulate the replication of different viruses, causing either inhibition or promotion. Therefore, it is important to research the biological functions of m6A modification in different viruses to understand the pathogenesis of viruses and develop prevention methods. HIV-1 can regulate the modification of host nucleic acids and proteins, thereby inducing a cellular response to infection and causing pathological changes [[34], [35], [36]]. Studies have found that HIV-1 mRNA contains multiple m6A modification sequences and that m6A levels are upregulated in CD4+ T cells infected by HIV-1 [12,37]. METTLL3, METTL14, and WTAP, as small-molecule activators, can promote the m6A modification of HIV-1 RNA and generate virions in cells incubating copies of HIV-1 proviruses [38]. The m6A modification can also allow HIV-1 to evade host immune recognition and promote its replication [12,39]. These reports suggest that m6A modification plays a significant role in the interaction between HIV-1 and host cells, but how to influence cellular gene expression through the regulation of m6A modification remains unclear.
Macrophages and T lymphocytes are the main target cells of HIV-1 infection [40]. Microglia, as intracranial macrophages, play important roles in brain inflammation and are the foremost cells infected by HIV-1 in the central nervous system [41]. When microglia are activated, large amounts of cytokines, chemokines, and other neurotoxic proteins are released which may contribute to the pathological neuroinflammation observed in HIV-associated neurocognitive disorders (HAND) [42]. Another report suggested that neuroinflammation caused by HIV-1 infection is the main reason for neurodegeneration, which is the main pathology of HAND [43].
We detected m6A levels of RNAs in three target cell lines infected by pseudotyped HIV-1. Compared with uninfected cells, the m6A levels were significantly elevated in MT-4 T cells, THP-1 macrophages, and HMC3 microglial cells. In this study, our objective was to investigate the transcriptome-wide m6A profiles in HMC3 cells infected with HIV-1 using methylated RNA immunoprecipitation sequencing (MeRIP-seq) and mRNA high-throughput sequencing (RNA-seq). In addition, we verified the levels of m6A and the expression level of differentially expressed genes in the sequencing data using MeRIP-qPCR and RT-PCR.
2. Materials and methods
2.1. Cell culture and antibodies
Human microglial cells HMC3 and human renal epithelial cells HEK-293T were grown in Dulbecco's modified Eagle medium (DMEM, Gibco, UT, USA) with 10 % fetal bovine serum (FBS, Gibco, UT, USA). Human monocytic cells THP-1 and human leukemia cells MT-4 were grown in Roswell Park Memorial Institute 1640 medium (RPMI-1640, Gibco, UT, USA) with 10 % FBS. All cells were cultured at 37 °C with 5 % CO2 and purchased from the Cell Bank of the Chinese Academy of Science (Shanghai, China). THP-1 cells were stimulated with 100 mM phorbol-12-myristate-13-acetate (PMA, Thermo, CA, USA) for 48 h (THP-1/PMA) to enable macrophage differentiation.
An antibody against HIV-1 p24 (Santa Cruz Biotechnology, CA, USA) was diluted to 1:1000. An antibody against β-Actin (Proteintech, IL, USA) was diluted to 1:1000. An antibody against m6A (Cell Signaling Technology, MA, USA) was diluted to1:500. An antibody against IBA1 (Abcam, Cambridge, UK) was diluted to1:500.
2.2. Pseudotyped HIV-1 preparation and single-round infection
The HIV-1 proviral plasmid pNL4-3.Luc.R + E−was provided by Prof. K. Tokunaga (NIID, Japan), which lacks nef and env expression and contains the luciferase gene. We replaced the luciferase gene with the EGFP gene to produce the pNL4-3.EGFP.R + E−vector. Briefly, 293T were seeded in a six-well plate with appropriate density, and the confluence rate reached 80 % on the second day, then, 15 μg of pNL4-3.EGFP.R + E−plasmid and 5 μg of VSV-G expression plasmid were co-transfected using 40 μl Lipofectamine 2000 reagent (Invitrogen, CA, USA). After 4 h, the culture medium was changed, and the virus-containing supernatants were collected at 44 h. The supernatants were used as pseudotyped HIV-1 (HIV-1-EGFP), and a 0.45 μm filter was used to filter the supernatants. Then, they were divided into equal samples for the inoculation assay. A p24 ELISA kit (Advanced BioScience Laboratories, Inc, USA) was used to measure the viral concentration.
THP-1/PMA, HMC3, or MT-4 cells (1 × 106/well) were seeded into a six-well plate and inoculated with HIV-1-EGFP at 100 ng of p24 per mL. After 4 h of infection, the medium was renewed and further cultured for 44 h. Then, the cells were collected for further detection.
2.3. Western blot
Cell samples were rinsed gently with PBS and lysed using RIPA Lysis Buffer (Beyotime Biotechnology, Beijing, China) containing protease inhibitors (Roche, Shanghai, China) for 30 min at 4 °C. The lysate was centrifugated at 1000 g as a protein sample. The protein concentrations were quantified using a BCA kit (Beyotime Biotechnology, Beijing, China). Then, 5 × SDS sample buffer was added to a 20 μg protein sample and subjected to 10 % SDS-PAGE. The proteins were transferred to polyvinylidene fluoride (PVDF) membranes (Millipore, MA, USA). Then, 5 % non-fat milk in TBST (20 mM Tris, 0.9 % NaCl, and 0.05 % Tween 20) buffer was used to block the membrane for 1 h and using the primary antibody incubated the membrane overnight at 4 °C. After rinsing with, the membrane was incubated with the appropriate HRP-conjugated goat anti-mouse antibody for 1 h at room temperature, the membrane was washed with TBST for 3 times again. The Chemidoc XRS Gel Imaging System (Bio-Rad, CA, USA) was used to visualize protein bands with an ECL substrate (Millipore, MA, USA).
2.4. m6A dot blot detection
RNA samples were adjusted to 100 ng/μl for one assay. The diluted RNA was denatured at 95 °C for 3 min and was immediately chilled on ice. Then, 2 μl of RNA was dropped directly onto a Hybond-N+ membrane (GE Healthcare, PA, USA), and cross-linked for 20–50 s. After washing with 10 ml TBST for 5 min, the membrane was blocked in 10 ml TBST containing 5 % non-fat milk for 1 h at room temperature with gentle shaking and then incubated with the m6A antibody overnight at 4 °C. The following procedure was performed as described above for the Western blot. The densitometry of the relative m6A levels was standardized with methylene blue staining.
2.5. MeRIP-seq and RNA-seq
Poly (A) RNA was purified with Dynabeads Oligo (DT) for 2 cycles and cleaved using a Magnesium RNA Fragmentation Module (NEB, MA, USA) at 86 °C for 7 min then, stored at 4 °C for 2 h. Next, 10 % of the RNA was used as input RNA, and the remaining fragments were incubated with an m6A antibody for 2 h at 4 °C as IP RNA. The IP RNA was reverse-transcribed into cDNA using SuperScript™ II Reverse Transcriptase (Invitrogen, CA, USA). Then, a base A was added to the blunt end of each strand, ready to attach to the indexed adapters. The second-strand DNA labeled with U was treated with a heat-labile UDG enzyme (NEB, MA, USA). Finally, we performed paired-end sequencing (PE150) on an Illumina Novaseq™ 6000 platform (LC-Bio Technology, Hangzhou, China) following the vendor's recommended protocol.
2.6. Quantitative real-time PCR (qRT-PCR) and MeRIP-qPCR
RNA (2 μg) was reverse-transcribed using M-MLV Reverse Transcriptase (Takara, Tokyo, Japan). The produced cDNAs were used as templates to determine the mRNA expression levels using TB Green® Premix Ex Taq™ II (Takara, Tokyo, Japan). The housekeeping GAPDH gene was used for normalization, and the 2−ΔΔCt method was used to calculate the relative amounts of PCR products. The primers for qRT-PCR are shown in Table 1.
Table 1.
The primers for qRT-PCR.
| genes | Primer sequences |
|---|---|
| CFB-F | 5′- CCCGTTCTCGAAGTCGTGTG -3′ |
| CFB-R | 5′- GCACTGGAGTACGTGTGTCC -3′ |
| HLA-B-F | 5′- CAGTTCGTGAGGTTCGACAG -3′ |
| HLA-B-R | 5′- CAGCCGTACATGCTCTGGA -3′ |
| IL-6-F | 5′-ACTTCCATCCAGTTGCCTTCTTGG -3′ |
| IL-6-R | 5′- TTAAGCCTCCGACTGTGAAGTG -3′ |
| OLR1-F | 5′- TCCGAATGTTGATTATGCCTCG -3′ |
| OLR1-R | 5′- TGACAGCGGAATGTTCTTCCC -3′ |
| GAPDH-F | 5′- GAGCCCGCAGCCTCCCGCTT -3′ |
| GAPDH-R | 5′- CCCGCGGCCATCACGCCACAG -3′ |
Methylated m6A RNA immunoprecipitation (Me-RIP) was performed using an EpiQuikTM CUT&RUN m6A RNA Enrichment Kit (Epigentek, NY, USA). Total RNA was incubated with an anti-m6A antibody. Then, the Cleavage Enzyme Mix was used to separate the m6A-bound mRNA, and RNA-binding beads were used to purify the fragments. After 2 washes with 90 % ethanol, the RNA was released from the beads with an elution buffer. A small proportion of fragmented RNA was used as the input control. The m6A IP samples and input samples were collected for RT-qPCR as described above. The primers for MeRIP-qPCR are shown in Table 2.
Table 2.
The primers for MeRIP-qPCR.
| genes | Primer sequences |
|---|---|
| CFB-F | 5′- TCCTAACTCCCACTCCTCTC -3′ |
| CFB-R | 5′- CCAGTGGGCAGGACTGGCCTTC -3′ |
| HLA-B-F | 5′- GCACATAGAAATGCTATTGAT -3′ |
| HLA-B-R | 5′- TATCCTTGTTTGCACATCAT -3′ |
| IL-6-F | 5′- CCAGCCTTTCCAGAGGAAT -3′ |
| IL-6-R | 5′- ATAAGCATGCCAGTAGA -3′ |
| OLR1-F | 5′- GAAGTCTTAATATTGCGA -3′ |
| OLR1-R | 5′- TGTAAGAGTACATTATCTT -3′ |
2.7. Data analysis
For the MeRIP-seq analysis, exomePeak2 software was used to identify differentially methylated m6A peaks. For the RNA-seq analysis, the R package DESeq2 was used to identify differentially expressed genes. All the data satisfied the following criteria: log2 (fold change) ≥ 1 or log2 (fold change) ≤ −1 and p-value <0.05. HOMER (http://homer.ucsd.edu/homer/motif) was used for de novo and known motif finding, followed by localization of the motif with respect to peak summit. GO and KEGG pathway analyses were performed based on the differentially methylated protein-coding genes and differentially expressed genes (https://www.omicstudio.cn/index). The visualization of differentially methylated gene alignment was performed using the Integrative Genomics Viewer (IGV). GraphPad Prism 8 was used to perform the statistical analysis, and p values were calculated using two-tailed unpaired Student's t-tests (*p < 0.05, **p < 0.01, ***p < 0.001).
3. Results
3.1. HIV-1 infection upregulates m6A levels
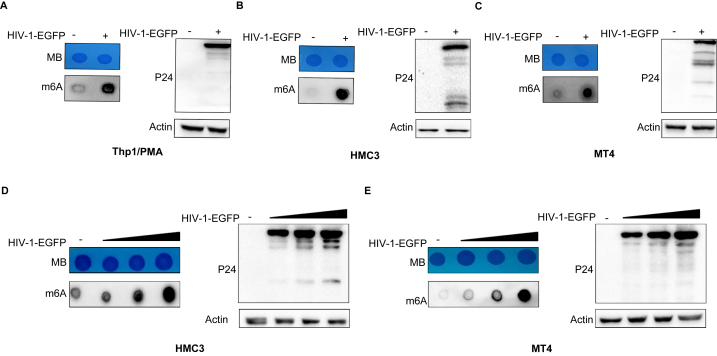
Since m6A modification is involved in the interaction between the virus and host cells, we detected the m6A modification levels in HIV-1-infected target cells. THP-1/PMA, HMC3, and MT-4 cells were infected with pseudotyped HIV-1-EGFP at 100 ng of p24/mL for 48 h, the m6A levels of cellular RNA were detected by a dot-blot assay, and total cellular RNA was stained with methylene blue (MB) as a loading control. The results showed that the cellular m6A levels increased in these cell lines (Fig. 1A, B, C). To confirm the increase was due to HIV-1 infection, we further performed infection experiments with virus concentration gradients with MT-4 and HMC3 cells. The m6A levels increased in a virus-dose-dependent manner (Fig. 1D and E). These results confirmed that HIV-1 infection can enhance the m6A methylation levels of target cells.
Fig. 1.
HIV-1 infection upregulates m6A levels of cellular RNA.
THP-1/PMA, HMC3, and MT4 cells were infected with HIV-1-EGFP at 100 ng of p24/ml for 48 h (A, B, C), HMC3 and MT4 cells were infected with HIV-1-EGFP at 0, 25, 50, and 100 ng of p24/ml for 48 h (D, E). Then, the m6A levels in total RNA (200 ng) from infected or uninfected cells were measured with a dot-blot assay using m6A-specific antibodies, and MB staining of RNA was used as a loading control. The gag levels of infected cells were quantified using Western blots. Quantification of relative m6A levels is shown as representative results from three (A, B, C) or one (D, E) independent experiments (n = 3 or 1).
3.2. Overview of the m6A methylation in HIV-1-infected HMC3 cells
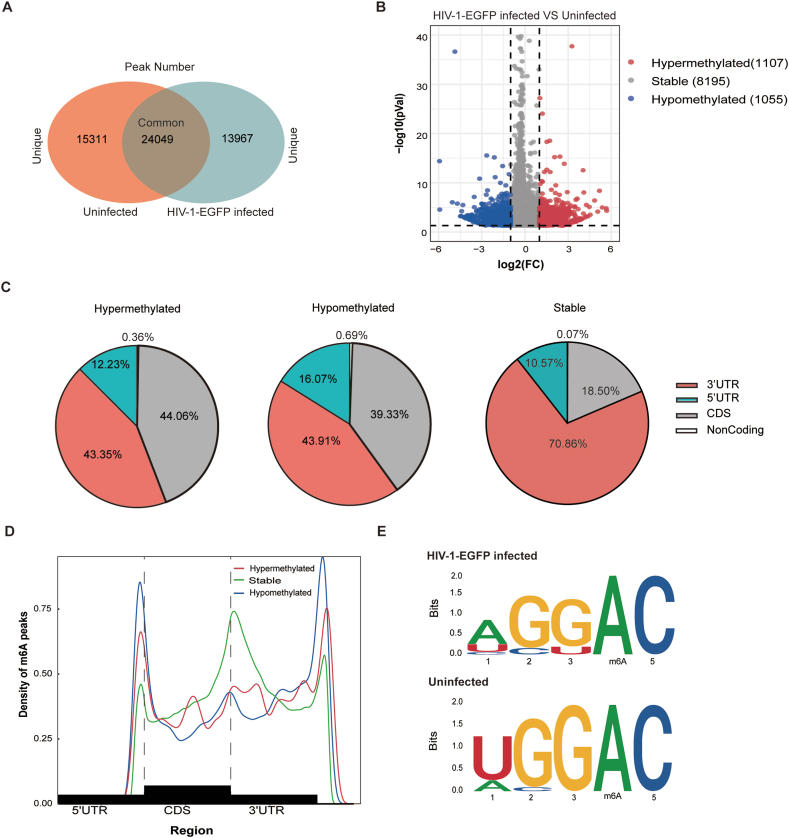
HMC3 cells are derived from human microglial cells, which are the main cell in the brain to cause neuroinflammation once activated. Hence, we systematically analyzed the effects of HIV-1 infection on the m6A methylation of cellular RNA in HMC3 cells after 48 h of HIV-1-EGFP infection. By MeRIP-seq analysis, the results showed that there were 24049 common peaks, including 15311 unique peaks in uninfected cells and 13967 unique peaks in HIV-1-EGFP-infected HMC3 cells (Fig. 2A). We further compared the m6A peak changes in the infected cells with the uninfected ones, and 1109 hyper-methylation and 1055 hypo-methylation peaks were identified (Fig. 2B).
Fig. 2.
Characteristics of m6A methylation in HIV-1-infected HMC3 cells. (A) A Venn diagram showing the common and unique m6A peaks in HIV-1-EGFP-infected and uninfected cells. (B) A volcano plot showing the significantly differential m6A peaks in HIV-1-EGFP-infected and uninfected cells. The percentage (C) and accumulation (D) of hyper-methylated, hypo-methylated, and stable m6A peaks among the four transcript segments. (E) A representative motif analysis of the HIV-1-EGFP infected and uninfected groups based on the data in C.
To analyze the distribution of m6A peaks in mRNAs, the peaks were divided into four transcribed fragments: the 5′UTR, the coding sequence (CDS), the noncoding sequence, and the 3′UTR. Compared to the uninfected cells, HIV-1 infection led to a significant increase in m6A peaks in the CDS (hyper, 44.06 % vs. 18.50 %; hypo, 39.33 % vs. 18.50 %), a significant decrease in the 3′UTR (hyper, 43.35 % vs. 70.86 %; hypo, 43.91 % vs. 70.86 %), and a relative increase in the 5′UTR (up, 12.23 % vs. 10.57 %; down, 16.07 % vs. 10.57 %) (Fig. 2C). Further analysis showed that the 3′ UTR and stop codon were the main enrichment regions of m6A peaks, their distributions and densities across the mRNA transcripts were similar (Fig. 2D). The typical motif was identified based on these data. There was a small difference in the m6A methylation point between the two groups (Fig. 2E). These results indicated that HIV-1 infection significantly affected the level and distribution of m6A methylation in human microglial cells.
3.3. Differentially methylated RNAs induced by HIV-1 are involved in important biological pathways
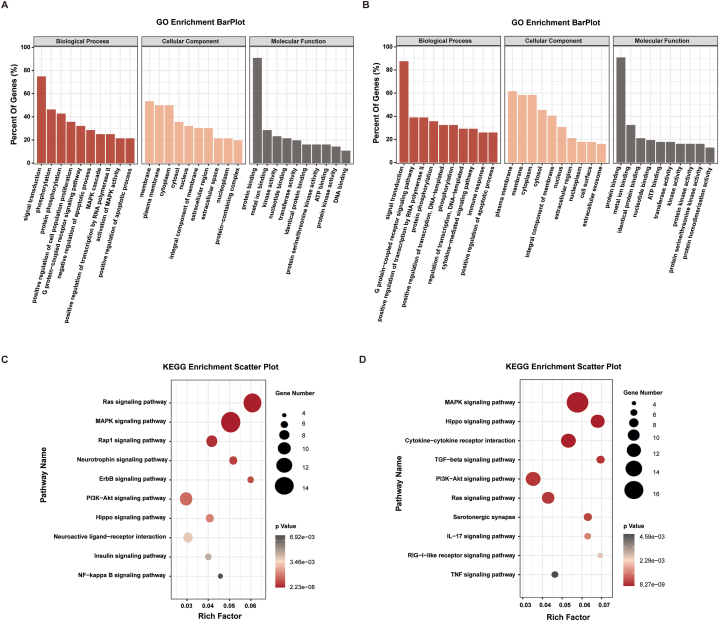
To detect the biological importance of m6A modification in HIV-1-EGFP-infected HMC3 cells, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were performed. The GO analysis categorized genes into three functional groups, including biological processes (BPs), cellular components (CCs), and molecular functions (MFs). The genes significantly enriched in BPs, CCs, and MFs with upregulated or downregulated m6A peaks are shown in Fig. 3A and B. For the BP category, we observed that up-regulated methylated m6A peaks were significantly enriched in the signal transduction, phosphorylation, and protein phosphorylation. Down-regulated methylated m6A peaks were significantly enriched in the signal transduction, G protein−coupled receptor signaling pathway and positive regulation of transcription by RNA polymerase II. The KEGG pathway analysis showed that upregulated m6A peaks were significantly associated with the Ras, MAPK, Rap1, and PI3K-Akt signaling pathway (Fig. 3C). And downregulated m6A peaks were significantly associated with the MAPK, Hippo, PI3K-Akt, and Ras signaling pathway, and Cytokine−cytokine receptor interaction (Fig. 3D). These results suggest that m6A modification has complicated roles in HIV-1 infection and various pathways related to the immune and nervous systems.
Fig. 3.
GO and KEGG pathway analyses of differentially methylated mRNA.
The GO terms of genes with upregulated (A) and downregulated (B) m6A peaks in the HIV-1-EGFP infected HMC3 cells are shown. The KEGG pathways of genes with upregulated (C) and downregulated (D) m6A peaks in the HIV-1-EGFP infected HMC3 cells are shown.
3.4. Conjoint analyses of MeRIP-Seq and RNA-seq data
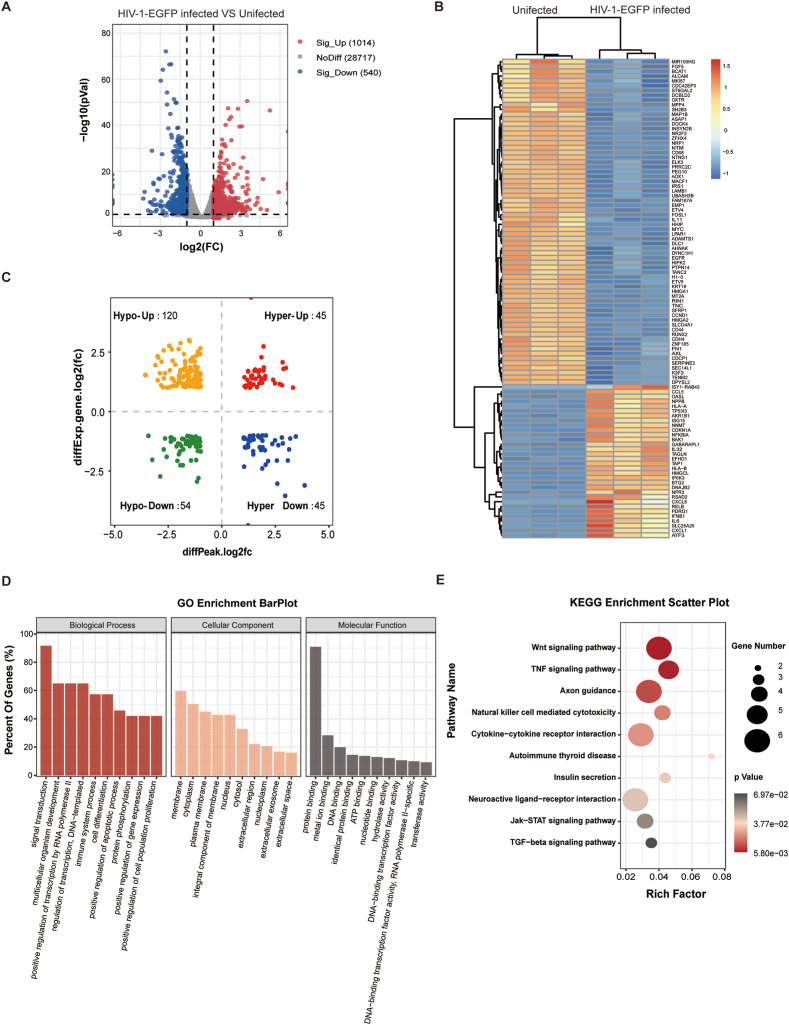
To further explore the functional significance of m6A methylation differences under HIV-1-EGFP infection, we analyzed the transcriptome profiles using RNA-seq data (MeRIP-Seq input library). A volcano plot shows that the numbers of significantly upregulated and significantly downregulated genes were 1014 and 540, respectively (Fig. 4 A), and the top 100 genes (up: 32, down: 68) with the smallest P-values are shown in Fig. 4B. Next, we performed a cross-analysis of the MeRIP-seq and RNA-seq data. All the genes with differential m6A peaks and differential mRNA levels (a total of 264 genes) were divided into four groups: differentially hyper-methylated m6A peaks with differentially upregulated genes (hyper-up, 45), differentially hyper-methylated m6A peaks with differentially downregulated genes (hyper-down, 45), differentially hypo-methylated m6A peaks with differentially upregulated genes (hypo-up, 120), and differentially hypo-methylated m6A peaks with differentially downregulated genes (hypo-down, 54) (Fig. 4C). The top 10 % of these genes are shown in sTable1. The biological significance of these genes was investigated using GO and KEGG pathway analyses. The GO analysis indicated that these genes are mainly involved in signal transduction, multicellular organism development, the positive regulation of transcription from the RNA polymerase II promoter, and the regulation of transcription involved in the cellular response to chemical stimuli (Fig. 4D). The KEGG pathway analysis revealed that 264 genes were primarily enriched in the Wnt and TNF signaling pathway, Axon guidance, and cytokine–cytokine receptor interaction (Fig. 4E). It is suggested that the m6A methylation modulated by HIV-1 might be one of the factors affecting the expression of these genes, and most of them are related to immune response and axon signaling pathways in HMC3 cells.
Fig. 4.
Identification and enrichment analysis of differentially expressed genes and their association with the m6A peak (A) Volcano plot showing the significantly differentially expressed genes in HIV-1-EGFP-infected and uninfected cells. (B) Heatmap plot showing the differentially expressed genes in HIV-1-EGFP-infected and uninfected cells. (C) Four-quadrant plot showing the distribution of genes with significant changes in both the m6A modification (x-axis) and mRNA levels (y-axis). GO analysis (D) and KEGG analysis (E) of four-quadrant plot genes in C.
3.5. Validation of the m6A methylation and expression data with MeRIP-qPCR and RT-PCR
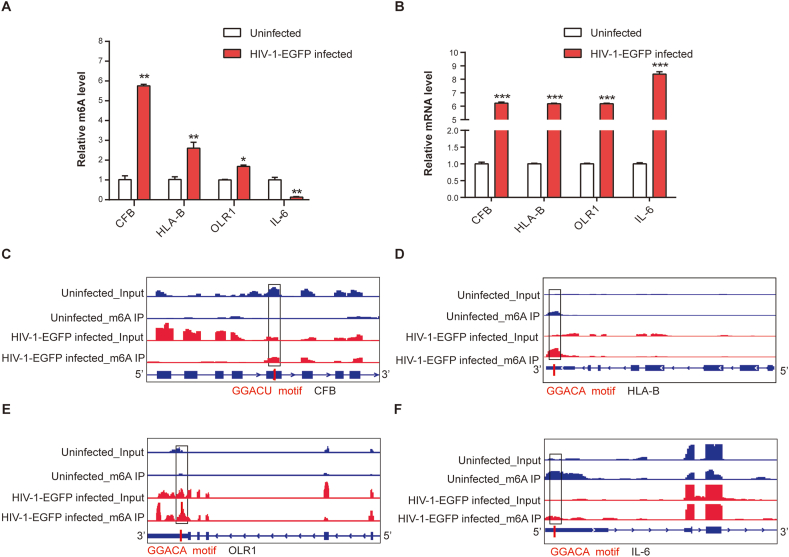
To further confirm the results of our MeRIP-seq data, we validated the m6A methylation and expression alterations of several of the top genes (sTable 1) using MeRIP-qPCR. The MeRIP-qPCR results showed significantly elevated m6A levels of HLA-B, CFB, and OLR1 and a significantly reduced m6A level of IL-6 RNA in HMC3 cells infected with HIV-1-EGFP (Fig. 5A). We further tested these gene expression levels using qRT-PCR. The results indicated that HIV-1-EGFP infection increased the expression levels of these four genes by more than six times (Fig. 5B). Furthermore, to clarify the altered intensity of the differentially methylated sites in these two groups, we analyzed the changes in m6A methylation of the four genes using Integrative Genomics Viewer (IGV). Compared to uninfected cells, the m6A levels of HLA-B, CFB, and OLR1 increased and that of IL-6 decreased in HIV-1-EGFP-infected cells, as shown in the small boxes in Fig. 5C–F. In addition, the m6A site distributions of the four target genes were analyzed. The m6A sites of HLA-B and CFB were in the CDS, the m6A site of OLR1 was in the 3′UTR, and the site of IL-6 was in the 5′UTR. These results suggest that HIV-1 infection might affect the expression of different genes by regulating the m6A methylation levels in different mRNA regions.
Fig. 5.
MeRIP-qPCR assays and Integrative Genomics Viewer (A) MeRIP-qPCR quantitative analysis of m6A level changes of HLA-B, IL-6, CFB, and OLR1 between HIV-1-EGFP infected and uninfected cells using immunoprecipitation with a specific m6A antibody. (B) qRT-PCR validation of expression of four genes between HIV-1-EGFP infected and uninfected cells. The m6A site distributions of the four genes were analyzed using IGV: CFB (C), HLA-B (D), OLR1 (E), and IL-6 (F). (*p < 0.05, **p < 0.01, ***p < 0.001, Student's t-test).
4. Discussion
Several studies have demonstrated that the mRNA of HIV-1 can be modified by m6A methylation to regulate the replication of the virus. For example, several m6A editing sites in HIV-1 3′UTRs can enhance the expression of the viral genes [44]. The suppression or enhancement of the m6A modification level by silencing METTL3 and METTL14 or ALKBH5 can decrease or increase HIV-1 replication [12]. On the other hand, HIV-1 infection can also regulate the m6A modification of cellular RNA, possibly through its viral gp120 protein [37]. However, the modification pattern and the effects on the expression of genes are still unknown. In our study, we confirmed that HIV-1 infection can enhance the m6A methylation of cellular RNA in three target cell lines. Further systematic analysis with MeRIP-seq revealed the global m6A methylation patterns of cellular RNA in HMC3 cells infected with HIV-1, and the results of some genes were validated using MeRIP-qPCR and qRT-PCR.
Our data showed that the m6A peak number and region of HIV-1-infected cells were distinct from those of uninfected cells. It is well known that m6A methylation is an invertible process involving several enzymes, including writers, erasers, and readers [16]. The global changes in m6A modification by HIV-1 infection could result from the abnormal expression of these enzymes. However, the expression of these enzymes remained unchanged in the HIV-1-infected HMC3 cells, according to our RNA-seq data. M6A methylation is a complex process, and other unknown factors could be involved. Recently, it was reported that exon junction complexes (EJCs) act as an m6A suppressor, protecting exon-junction-proximal RNA in coding sequences from methylation and regulating mRNA stability by m6A suppression [45]. Again, the limited sample size in this study may be the main reason. A larger sample size should be investigated in future studies.
Nagaraja et al. reported that HIV-1 infection enhanced the m6A modification through its envelope protein gp120; however, they found no significant changes in the m6A-related enzymes in gp120-expressing cells [37]. Another study found that enterovirus 71 (EV71) RNA undergoes m6A modification during viral infection, which alters the expression and localization of m6A methyltransferase, demethylase, and their binding proteins [46]. Although Zhang et al. found that m6A modification of cellular mRNA was not significantly altered by HIV-1 infection in T cells using LC-MS/MS method [47].In our study, even though we used the VSV-G pseudotyped virus without gp120 protein, a significant increase in the m6A modification of cellular RNA was also observed using a dot-blot assay. It may be because of the different detection methods used in different studies. It is necessary to perform further investigation. These results indicate that, in addition to gp120, other viral proteins might also affect the m6A process. These viral proteins need to be further identified, as does the mechanism by which they interfere with the m6A modification process in target cells.
The KEGG pathway analyses showed that the genes with upregulated m6A-modified sites were related to the Ras signaling pathway, which is one of the main pathways for immune responses [48]. The conjoint analyses of the MeRIP-Seq and RNA-Seq data found that 264 genes showed differential m6A peaks and expression induced by pseudotyped HIV-1, which were enriched in the Wnt and TNF signaling pathway, Axon guidance, and cytokine–cytokine receptor interaction, which is related to the immune responses and physiological functions of the nervous system. HMC3 is the target cell of HIV-1 in the brain, and it is also the immune cell that plays an important role in HAND [49]. It was reported that Ras signaling induces Ros generation via the downstream effector MEK/ERK [50]. We found that HIV-1 infection could lead to the m6A modification of lots of proteins that are involved in the above pathways, which indicates potential interactions between HIV-1 and the host cells, especially in the neuropathological change during HAND. Since HIV-1 infection can activate microglia through multiple pathways, we are trying to demonstrate that it can induce neuroinflammation through the regulation of m6A modification. we detected the m6A modification in the HIV-1 infected cells incubated with m6A inhibitor DAA. The results showed that the increase of methylation induced by HIV-1 infection was suppressed by the addition of DAA (sFigure1A, B), and the higher level of IBA1 induced by virus infection is decreased by DAA treatment (sFigure1C). These results implied that m6A methylation might be one of the reasons for the activation of inflammation under HIV-1 infection. HIV-1 transgenic rats study found neuronal cell death and degeneration through increased nitroxidative stress and immune response pathway [51]. Our results provide more information about the interaction between the virus and host, such as which pathway is targeted by the virus by regulating m6A modification.
Although our m6A detection results showed that the total m6A level significantly increased in HIV-1-infected cells (Fig. 1), almost half of the changed genes showed a decreased m6A methylation level, even though the expression level of their mRNA increased. In the HIV-1 infected cells, the interaction between virus and cells is complex and constantly changing, the m6A modification also is regulated in a variety of ways, and the region of m6A modification also made different affection on the gene expression. The results imply that the m6A modification might be the reason for the changes in gene expression and that these genes might play important roles in the pathogenesis of HIV-1 infection. HIV-1 infection enhances IL-6 expression through multiple pathways [52], the methylation might also be one way to affect the gene expression, and how it affects also need to be investigated more. And it was also reported that the activation of the immune reaction plays important roles in HAND development, including IL-6 upregulation [53]. These results suggest that the activation of an inflammatory signaling different than m6A methylation, and IL-6 might be related to TNF pathway. It is necessary to confirm the m6A modified region of IL-6 and to clarify the relation between that and the expression of IL-6.
Lipid metabolism exerts a profound impact on the maintenance of human physiology and health status. It is reported that m6A modification of RNA also plays important roles on lipid metabolic regulation [28], we further examined the MDA to confirm the levels of lipid peroxidation in the treated cells. MDA levels increased in HIV-1 infected cells, and DAA incubation prevent the increase of MDA (sFigure1D). These results suggested that the increase of lipid peroxidation level in HMC3 during HIV-1 infection might be partially due to the m6A methylation increasing. It has been reported that CFB promotes adipocyte differentiation, maturation, and lipid droplet formation [54]. OLR1 implements its role in lipid metabolism [55], also is has been shown that OLR1 promotes the release of inflammatory cytokines and is associated with metabolic diseases [56], suggesting that OLR1 might involve in inflammatory response by regulating lipid metabolism. The detailed mechanism is worth to further investigate. In addition, distinct HLA-B alleles, were associated with HIV-1 infection control [57]. However, the relationship between HLA-B methylation and viral infection needs further investigation.
In summary, via high-throughput sequencing and biological experiments, we confirmed that HIV-1 infection can enhance m6A methylation in T cells and macrophages and showed distinct global patterns in microglial HMC3 cells infected with HIV-1. The differentially expressed genes might be due to the regulation of m6A methylation by HIV-1 infection, and these genes are closely related to immune activation. Our study provides new clues for understanding the interaction between HIV-1 and microglia-mediated by m6A modifications on cellular RNA. In this study we performed the investigation just in vitro, the findings are very interesting, but the results are limited. The conclusions need to be confirmed with primary microglia or the virus-positive macrophages from HIV-1 infected individuals in future study, and the phenotypic and mechanism analysis is necessary to be performed with HIV-1 infected humanized mice.
Funding
This work was supported by the National Natural Science Foundation of China (grant number 31670716), and the Research Fund of Jianghan University (grants number 08190001 and 08190006).
Data availability statement
Data will be made available on request.
CRediT authorship contribution statement
Qian Peng: Data curation, Project administration, Writing – original draft, Methodology. Jialu Qiao: Data curation, Methodology, Writing – original draft. Weiling Li: Validation. Qiang You: Data curation. Song Hu: Formal analysis. Yuchen Liu: Data curation. Wei Liu: Investigation. Kanghong Hu: Writing – review & editing. Binlian Sun: Funding acquisition, Resources, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e21307.
Contributor Information
Kanghong Hu, Email: hukh@hbut.edu.cn.
Binlian Sun, Email: binlian17@jhun.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Yang Y., Hsu P.J., Chen Y.S., Yang Y.G. Dynamic transcriptomic m(6)A decoration: writers, erasers, readers and functions in RNA metabolism. Cell Res. 2018;28:616–624. doi: 10.1038/s41422-018-0040-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tang C., Klukovich R., Peng H., Wang Z., Yu T., Zhang Y., et al. ALKBH5-dependent m6A demethylation controls splicing and stability of long 3'-UTR mRNAs in male germ cells. Proc. Natl. Acad. Sci. U. S. A. 2018;115:E325–e333. doi: 10.1073/pnas.1717794115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zheng Q., Hou J., Zhou Y., Li Z., Cao X. The RNA helicase DDX46 inhibits innate immunity by entrapping m(6)A-demethylated antiviral transcripts in the nucleus. Nat. Immunol. 2017;18:1094–1103. doi: 10.1038/ni.3830. [DOI] [PubMed] [Google Scholar]
- 4.Vu L.P., Pickering B.F., Cheng Y., Zaccara S., Nguyen D., Minuesa G., et al. The N(6)-methyladenosine (m(6)A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat. Med. 2017;23:1369–1376. doi: 10.1038/nm.4416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bertero A., Brown S., Madrigal P., Osnato A., Ortmann D., Yiangou L., et al. The SMAD2/3 interactome reveals that TGFβ controls m(6)A mRNA methylation in pluripotency. Nature. 2018;555:256–259. doi: 10.1038/nature25784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartosovic M., Molares H.C., Gregorova P., Hrossova D., Kudla G., Vanacova S. N6-methyladenosine demethylase FTO targets pre-mRNAs and regulates alternative splicing and 3'-end processing. Nucleic Acids Res. 2017;45:11356–11370. doi: 10.1093/nar/gkx778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiao W., Adhikari S., Dahal U., Chen Y.S., Hao Y.J., Sun B.F., et al. Nuclear m(6)A reader YTHDC1 regulates mRNA splicing. Mol. Cell. 2016;61:507–519. doi: 10.1016/j.molcel.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 8.Wei W., Ji X., Guo X., Ji S. Regulatory role of N(6) -methyladenosine (m(6) A) methylation in RNA processing and human diseases. J. Cell. Biochem. 2017;118:2534–2543. doi: 10.1002/jcb.25967. [DOI] [PubMed] [Google Scholar]
- 9.Liu J., Yue Y., Han D., Wang X., Fu Y., Zhang L., et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 2014;10:93–95. doi: 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Y., Li Y., Toth J.I., Petroski M.D., Zhang Z., Zhao J.C. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat. Cell Biol. 2014;16:191–198. doi: 10.1038/ncb2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y., Zhou X. N(6)-methyladenosine and its implications in viruses. Dev. Reprod. Biol. 2022 doi: 10.1016/j.gpb.2022.04.009. S1672-0229(22).00083-3. [DOI] [PubMed] [Google Scholar]
- 12.Lichinchi G., Gao S., Saletore Y., Gonzalez G.M., Bansal V., Wang Y., et al. Dynamics of the human and viral m(6)A RNA methylomes during HIV-1 infection of T cells. Nat. Microbiol. 2016;1 doi: 10.1038/nmicrobiol.2016.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meyer K.D., Saletore Y., Zumbo P., Elemento O., Mason C.E., Jaffrey S.R. Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dominissini D., Moshitch-Moshkovitz S., Schwartz S., Salmon-Divon M., Ungar L., Osenberg S., et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 15.Csepany T., Lin A., Baldick C.J., Jr., Beemon K. Sequence specificity of mRNA N6-adenosine methyltransferase. J. Biol. Chem. 1990;265:20117–20122. [PubMed] [Google Scholar]
- 16.Shi H., Wei J., He C. Where, when, and how: context-dependent functions of RNA methylation writers, readers, and erasers. Mol. Cell. 2019;74:640–650. doi: 10.1016/j.molcel.2019.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ping X.L., Sun B.F., Wang L., Xiao W., Yang X., Wang W.J., et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24:177–189. doi: 10.1038/cr.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwartz S., Mumbach M.R., Jovanovic M., Wang T., Maciag K., Bushkin G.G., et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5' sites. Cell Rep. 2014;8:284–296. doi: 10.1016/j.celrep.2014.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horiuchi K., Kawamura T., Iwanari H., Ohashi R., Naito M., Kodama T., Hamakubo T. Identification of Wilms' tumor 1-associating protein complex and its role in alternative splicing and the cell cycle. J. Biol. Chem. 2013;288:33292–33302. doi: 10.1074/jbc.M113.500397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pendleton K.E., Chen B., Liu K., Hunter O.V., Xie Y., Tu B.P., Conrad N.K. The U6 snRNA m(6)A methyltransferase METTL16 regulates SAM synthetase intron retention. Cell. 2017;169:824–835.e814. doi: 10.1016/j.cell.2017.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng G., Dahl J.A., Niu Y., Fedorcsak P., Huang C.M., Li C.J., et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jia G., Fu Y., Zhao X., Dai Q., Zheng G., Yang Y., et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011;7:885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gerken T., Girard C.A., Tung Y.C., Webby C.J., Saudek V., Hewitson K.S., et al. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science. 2007;318:1469–1472. doi: 10.1126/science.1151710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bi Z., Liu Y., Zhao Y., Yao Y., Wu R., Liu Q., et al. A dynamic reversible RNA N(6) -methyladenosine modification: current status and perspectives. J. Cell. Physiol. 2019;234:7948–7956. doi: 10.1002/jcp.28014. [DOI] [PubMed] [Google Scholar]
- 25.Patil D.P., Pickering B.F., Jaffrey S.R. Reading m(6)A in the transcriptome: m(6)a-binding proteins. Trends Cell Biol. 2018;28:113–127. doi: 10.1016/j.tcb.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang H., Weng H., Sun W., Qin X., Shi H., Wu H., et al. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 2018;20:285–295. doi: 10.1038/s41556-018-0045-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang X., Liu B., Nie Z., Duan L., Xiong Q., Jin Z., et al. The role of m6A modification in the biological functions and diseases. Signal Transduct. Targeted Ther. 2021;6:74. doi: 10.1038/s41392-020-00450-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y., Wang Y., Gu J., Su T., Gu X., Feng Y. The role of RNA m6A methylation in lipid metabolism. Front. Endocrinol. 2022;13 doi: 10.3389/fendo.2022.866116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Price A.M., Hayer K.E., McIntyre A.B.R., Gokhale N.S., Abebe J.S., Della Fera A.N., et al. Direct RNA sequencing reveals m(6)A modifications on adenovirus RNA are necessary for efficient splicing. Nat. Commun. 2020;11:6016. doi: 10.1038/s41467-020-19787-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.To E.E., Luong R., Diao J., Jj O.L., Brooks D.A., Vlahos R., Selemidis S. Novel endosomal NOX2 oxidase inhibitor ameliorates pandemic influenza A virus-induced lung inflammation in mice. Respirology. 2019;24:1011–1017. doi: 10.1111/resp.13524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kane S.E., Beemon K. Precise localization of m6A in Rous sarcoma virus RNA reveals clustering of methylation sites: implications for RNA processing. Mol. Cell Biol. 1985;5:2298–2306. doi: 10.1128/mcb.5.9.2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Canaani D., Kahana C., Lavi S., Groner Y. Identification and mapping of N6-methyladenosine containing sequences in simian virus 40 RNA. Nucleic Acids Res. 1979;6:2879–2899. doi: 10.1093/nar/6.8.2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krug R.M., Morgan M.A., Shatkin A.J. Influenza viral mRNA contains internal N6-methyladenosine and 5'-terminal 7-methylguanosine in cap structures. J. Virol. 1976;20:45–53. doi: 10.1128/jvi.20.1.45-53.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Freed E.O. HIV-1 assembly, release and maturation. Nat. Rev. Microbiol. 2015;13:484–496. doi: 10.1038/nrmicro3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moir S., Chun T.W., Fauci A.S. Pathogenic mechanisms of HIV disease. Annu. Rev. Pathol. 2011;6:223–248. doi: 10.1146/annurev-pathol-011110-130254. [DOI] [PubMed] [Google Scholar]
- 36.Goff S.P. Host factors exploited by retroviruses. Nat. Rev. Microbiol. 2007;5:253–263. doi: 10.1038/nrmicro1541. [DOI] [PubMed] [Google Scholar]
- 37.Tirumuru N., Wu L. HIV-1 envelope proteins up-regulate N(6)-methyladenosine levels of cellular RNA independently of viral replication. J. Biol. Chem. 2019;294:3249–3260. doi: 10.1074/jbc.RA118.005608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Selberg S., Zusinaite E., Herodes K., Seli N., Kankuri E., Merits A., Karelson M. HIV replication is increased by RNA methylation METTL3/METTL14/WTAP complex activators. ACS Omega. 2021;6:15957–15963. doi: 10.1021/acsomega.1c01626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen S., Kumar S., Espada C.E., Tirumuru N., Cahill M.P., Hu L., et al. N6-methyladenosine modification of HIV-1 RNA suppresses type-I interferon induction in differentiated monocytic cells and primary macrophages. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ocwieja K.E., Sherrill-Mix S., Mukherjee R., Custers-Allen R., David P., Brown M., et al. Dynamic regulation of HIV-1 mRNA populations analyzed by single-molecule enrichment and long-read sequencing. Nucleic Acids Res. 2012;40:10345–10355. doi: 10.1093/nar/gks753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bilimoria P.M., Stevens B. Microglia function during brain development: new insights from animal models. Brain Res. 2015;1617:7–17. doi: 10.1016/j.brainres.2014.11.032. [DOI] [PubMed] [Google Scholar]
- 42.Hong S., Banks W.A. Role of the immune system in HIV-associated neuroinflammation and neurocognitive implications. Brain Behav. Immun. 2015;45:1–12. doi: 10.1016/j.bbi.2014.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ginsberg S.D., Alldred M.J., Gunnam S.M., Schiroli C., Lee S.H., Morgello S., Fischer T. Expression profiling suggests microglial impairment in human immunodeficiency virus neuropathogenesis. Ann. Neurol. 2018;83:406–417. doi: 10.1002/ana.25160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kennedy E.M., Bogerd H.P., Kornepati A.V., Kang D., Ghoshal D., Marshall J.B., et al. Posttranscriptional m(6)A editing of HIV-1 mRNAs enhances viral gene expression. Cell Host Microbe. 2016;19:675–685. doi: 10.1016/j.chom.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Uzonyi A., Dierks D., Nir R., Kwon O.S., Toth U., Barbosa I., et al. Exclusion of m6A from splice-site proximal regions by the exon junction complex dictates m6A topologies and mRNA stability. Mol. Cell. 2023;83:237–251.e237. doi: 10.1016/j.molcel.2022.12.026. [DOI] [PubMed] [Google Scholar]
- 46.Hao H., Hao S., Chen H., Chen Z., Zhang Y., Wang J., et al. N6-methyladenosine modification and METTL3 modulate enterovirus 71 replication. Nucleic Acids Res. 2019;47:362–374. doi: 10.1093/nar/gky1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Q., Kang Y., Wang S., Gonzalez G.M., Li W., Hui H., et al. HIV reprograms host m(6)Am RNA methylome by viral Vpr protein-mediated degradation of PCIF1. Nat. Commun. 2021;12:5543. doi: 10.1038/s41467-021-25683-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jha M.K., Sarode A.Y., Saha B. Ras isoforms selectively regulate antigen-specific immune response. Cytokine. 2020;126 doi: 10.1016/j.cyto.2019.154914. [DOI] [PubMed] [Google Scholar]
- 49.Agrawal L., Louboutin J.P., Reyes B.A., Van Bockstaele E.J., Strayer D.S. HIV-1 Tat neurotoxicity: a model of acute and chronic exposure, and neuroprotection by gene delivery of antioxidant enzymes. Neurobiol. Dis. 2012;45:657–670. doi: 10.1016/j.nbd.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 50.Mayes D.A., Rizvi T.A., Titus-Mitchell H., Oberst R., Ciraolo G.M., Vorhees C.V., et al. Nf1 loss and Ras hyperactivation in oligodendrocytes induce NOS-driven defects in myelin and vasculature. Cell Rep. 2013;4:1197–1212. doi: 10.1016/j.celrep.2013.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cho Y.E., Lee M.H., Song B.J. Neuronal cell death and degeneration through increased nitroxidative stress and tau phosphorylation in HIV-1 transgenic rats. PLoS One. 2017;12 doi: 10.1371/journal.pone.0169945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bhargavan B., Kanmogne G.D. Toll-like receptor-3 mediates HIV-1-Induced interleukin-6 expression in the human brain endothelium via TAK1 and JNK pathways: implications for viral neuropathogenesis. Mol. Neurobiol. 2018;55:5976–5992. doi: 10.1007/s12035-017-0816-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee Y.J., Yeo I.J., Choi D.Y., Yun J., Son D.J., Han S.B., Hong J.T. Amyloidogenic, neuroinflammatory and memory dysfunction effects of HIV-1 gp120. Arch Pharm. Res. (Seoul) 2021;44:689–701. doi: 10.1007/s12272-021-01340-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Matsunaga H., Iwashita M., Shinjo T., Yamashita A., Tsuruta M., Nagasaka S., et al. Adipose tissue complement factor B promotes adipocyte maturation. Biochem. Biophys. Res. Commun. 2018;495:740–748. doi: 10.1016/j.bbrc.2017.11.069. [DOI] [PubMed] [Google Scholar]
- 55.Ardicli S., Dincel D., Samli H., Senturk N., Karalar B., Unlu S., et al. Association of polymorphisms in lipid and energy metabolism-related genes with fattening performance in Simmental cattle. Anim. Biotechnol. 2022:1–13. doi: 10.1080/10495398.2022.2152557. [DOI] [PubMed] [Google Scholar]
- 56.Yang G., Xiong G., Feng M., Zhao F., Qiu J., Liu Y., et al. OLR1 promotes pancreatic cancer metastasis via increased c-myc expression and transcription of HMGA2. Mol. Cancer Res. 2020;18:685–697. doi: 10.1158/1541-7786.MCR-19-0718. [DOI] [PubMed] [Google Scholar]
- 57.Valenzuela-Fernández A., Cabrera-Rodríguez R., Casado C., Pérez-Yanes S., Pernas M., García-Luis J., et al. Contribution of the HIV-1 envelope glycoprotein to AIDS pathogenesis and clinical progression. Biomedicines. 2022;10 doi: 10.3390/biomedicines10092172. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.