Abstract
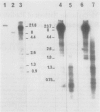
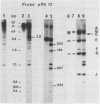
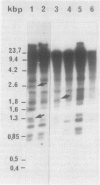
The methylation pattern of radish Raphanus sativus nuclear rDNA has been investigated using the Hpa II, Msp I, and Hha I restriction enzymes. The presence of numerous target sites for these enzymes has been shown using cloned rDNA fragments. A large fraction of the numerous rDNA units are heavily methylated, being completely resistant to Hpa II and Hpa I. However, specific sites are constantly available in another fraction of the units and are therefore unmethylated. The use of different probes allowed us to demonstrate that hypomethylated sites are present in different regions. Major hypomethylated Hha I sites have been mapped in the 5′ portion of 25S rRNA coding sequence. Among the hypomethylated fraction, different methylation patterns coexist. It has been possible to demonstrate that methylation patterns are specific for particular units. The Hha I pattern of rDNA in tissues of different developmental stages was analyzed. Evidence for possible tissue specific differences in the methylation pattern is reported.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bird A. P., Southern E. M. Use of restriction enzymes to study eukaryotic DNA methylation: I. The methylation pattern in ribosomal DNA from Xenopus laevis. J Mol Biol. 1978 Jan 5;118(1):27–47. doi: 10.1016/0022-2836(78)90242-5. [DOI] [PubMed] [Google Scholar]
- Bird A. P., Taggart M. H., Gehring C. A. Methylated and unmethylated ribosomal RNA genes in the mouse. J Mol Biol. 1981 Oct 15;152(1):1–17. doi: 10.1016/0022-2836(81)90092-9. [DOI] [PubMed] [Google Scholar]
- Bird A. P., Taggart M. H., Smith B. A. Methylated and unmethylated DNA compartments in the sea urchin genome. Cell. 1979 Aug;17(4):889–901. doi: 10.1016/0092-8674(79)90329-5. [DOI] [PubMed] [Google Scholar]
- Bird A. P., Taggart M. H. Variable patterns of total DNA and rDNA methylation in animals. Nucleic Acids Res. 1980 Apr 11;8(7):1485–1497. doi: 10.1093/nar/8.7.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A., Taggart M., Macleod D. Loss of rDNA methylation accompanies the onset of ribosomal gene activity in early development of X. laevis. Cell. 1981 Nov;26(3 Pt 1):381–390. doi: 10.1016/0092-8674(81)90207-5. [DOI] [PubMed] [Google Scholar]
- Busslinger M., deBoer E., Wright S., Grosveld F. G., Flavell R. A. The sequence GGCmCGG is resistant to MspI cleavage. Nucleic Acids Res. 1983 Jun 11;11(11):3559–3569. doi: 10.1093/nar/11.11.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deumling B. Sequence arrangement of a highly methylated satellite DNA of a plant, Scilla: A tandemly repeated inverted repeat. Proc Natl Acad Sci U S A. 1981 Jan;78(1):338–342. doi: 10.1073/pnas.78.1.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M., Wang R. Y. 5-Methylcytosine in eukaryotic DNA. Science. 1981 Jun 19;212(4501):1350–1357. doi: 10.1126/science.6262918. [DOI] [PubMed] [Google Scholar]
- Ellis T. H., Goldsbrough P. B., Castleton J. A. Transcription and methylation of flax rDNA. Nucleic Acids Res. 1983 May 25;11(10):3047–3064. doi: 10.1093/nar/11.10.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach W. L., Bedbrook J. R. Cloning and characterization of ribosomal RNA genes from wheat and barley. Nucleic Acids Res. 1979 Dec 11;7(7):1869–1885. doi: 10.1093/nar/7.7.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsbrough P. B., Cullis C. A. Characterisation of the genes for ribosomal RNA in flax. Nucleic Acids Res. 1981 Mar 25;9(6):1301–1309. doi: 10.1093/nar/9.6.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsbrough P. B., Ellis T. H., Cullis C. A. Organisation of the 5S RNA genes in flax. Nucleic Acids Res. 1981 Nov 25;9(22):5895–5904. doi: 10.1093/nar/9.22.5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenbaum Y., Naveh-Many T., Cedar H., Razin A. Sequence specificity of methylation in higher plant DNA. Nature. 1981 Aug 27;292(5826):860–862. doi: 10.1038/292860a0. [DOI] [PubMed] [Google Scholar]
- Julien R., Delseny M. Differential labelling of DNA in higher plants. Biochem Biophys Res Commun. 1972 Aug 7;48(3):578–584. doi: 10.1016/0006-291x(72)90387-7. [DOI] [PubMed] [Google Scholar]
- Keshet E., Cedar H. Effect of CpG methylation on Msp I. Nucleic Acids Res. 1983 Jun 11;11(11):3571–3580. doi: 10.1093/nar/11.11.3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunnath L., Locker J. Variable methylation of the ribosomal RNA genes of the rat. Nucleic Acids Res. 1982 Jul 10;10(13):3877–3892. doi: 10.1093/nar/10.13.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leder P., Tiemeier D., Enquist L. EK2 derivatives of bacteriophage lambda useful in the cloning of DNA from higher organisms: the lambdagtWES system. Science. 1977 Apr 8;196(4286):175–177. doi: 10.1126/science.322278. [DOI] [PubMed] [Google Scholar]
- Macleod D., Bird A. DNAase I sensitivity and methylation of active versus inactive rRNA genes in xenopus species hybrids. Cell. 1982 May;29(1):211–218. doi: 10.1016/0092-8674(82)90105-2. [DOI] [PubMed] [Google Scholar]
- McClelland M. The effect of site specific methylation on restriction endonuclease cleavage (update). Nucleic Acids Res. 1983 Jan 11;11(1):r169–r173. doi: 10.1093/nar/11.1.235-c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rae P. M., Steele R. E. Absence of cytosine methylation at C-C-G-G and G-C-G-C sites in the rDNA coding regions and intervening sequences of Drosophila and the rDNA of other insects. Nucleic Acids Res. 1979 Jul 11;6(9):2987–2995. doi: 10.1093/nar/6.9.2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin A., Riggs A. D. DNA methylation and gene function. Science. 1980 Nov 7;210(4470):604–610. doi: 10.1126/science.6254144. [DOI] [PubMed] [Google Scholar]
- Siegel A., Kolacz K. Heterogeneity of pumpkin ribosomal DNA. Plant Physiol. 1983 May;72(1):166–171. doi: 10.1104/pp.72.1.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spena A., Viotti A., Pirrotta V. Two adjacent genomic zein sequences: structure, organization and tissue-specific restriction pattern. J Mol Biol. 1983 Oct 5;169(4):799–811. doi: 10.1016/s0022-2836(83)80137-5. [DOI] [PubMed] [Google Scholar]
- Wigler M. H. The inheritance of methylation patterns in vertebrates. Cell. 1981 May;24(2):285–286. doi: 10.1016/0092-8674(81)90317-2. [DOI] [PubMed] [Google Scholar]