Abstract
Caloric restriction (CR), which extends lifespan in rodents, leads to increased hepatic fatty acid β-oxidation and oxidative phosphorylation (OXPHOS), with parallel changes in proteins and their mRNAs. Genetic mutants that extend lifespan, including growth hormone receptor knockout (GHRKO) and Snell dwarf (SD) mice, have lower respiratory quotient, suggesting increased reliance on fatty acid oxidation, but the molecular mechanism(s) of this metabolic shift have not yet been worked out. Here we show that both GHRKO and SD mice have significantly higher mRNA and protein levels of enzymes involved in mitochondrial and peroxisomal fatty acid β-oxidation. In addition, multiple subunits of OXPHOS complexes I-IV are upregulated in GHRKO and SD livers, and Complex V subunit ATP5a is upregulated in liver of GHRKO mice. Expression of these genes is regulated by a group of nuclear receptors and transcription factors including peroxisome proliferator-activated receptors (PPARs) and estrogen-related receptors (ERRs). We found that levels of these nuclear receptors and their co-activator PGC-1α were unchanged or downregulated in liver of GHRKO and SD mice. In contrast, NCOR1, a co-repressor for the same receptors, was significantly downregulated in the two long-lived mouse models, suggesting a plausible mechanism for the changes in FAO and OXPHOS proteins. Hepatic levels of HDAC3, a co-factor for NCOR1 transcriptional repression, were also downregulated. The role of NCOR1 is well established in the contexts of cancer and metabolic disease, but may provide new mechanistic insights into metabolic control in long-lived mouse models.
Supplementary Information
The online version contains supplementary material available at 10.1007/s11357-023-00849-8.
Keywords: Longevity, β-oxidation, OXPHOS, Mitochondria, NCOR1, GHRKO
Introduction
Changes in cellular fuel utilization, including modifications in lipid synthesis, storage, and mobilization, have been linked to lifespan and age-dependent physiological status in invertebrates [1], mice [2, 3], and humans [4], and are likely to contribute to increased risks of metabolic syndrome, obesity, and diabetes in human populations [5, 6]. Many lifespan-extending interventions in mice have elevated fatty acid β-oxidation. Caloric restriction (CR) induces whole body fatty acid oxidation (FAO) and increases reliance on fatty acids for fuel [7]. CR also upregulates many enzymes involved in the β-oxidation pathway [8]. Transgenic overexpression of fibroblast growth factor 21 (FGF21) also extends murine lifespan and induces hepatic FAO [9, 10]. PTEN overexpression, another genetic intervention that extends murine lifespan, has been shown to induce FAO in calf hepatocyte cell cultures, and knockdown of PTEN inhibited FAO [11, 12]. Pharmacological interventions, like rapamycin, has also been shown to induce FAO in cell cultures of rat hepatocytes and skeletal muscle cells [13, 14]. Growth hormone receptor knockout (GHRKO) mice are more dependent on fat oxidation for fuel, more resistant to diet-induced obesity, and show higher levels of FAO [15–17].
Mitochondrial FAO is physically and functionally associated with oxidative phosphorylation (OXPHOS). For example, FAO provides reducing equivalents that enter the OXPHOS pathway at the level of complexes I and III. Additionally, FAO trifunctional protein (TFP) enzyme interacts with complex I of the OXPHOS pathway [18–20]. Using blue native polyacrylamide gel electrophoresis to separate OXPHOS complexes from liver mitochondrial extracts reveals co-migration of FAO enzymes with OXPHOS complexes [20]. Furthermore, many genetic disorders in OXPHOS proteins impair FAO, and conversely pharmacological inhibitors of FAO reduce OXPHOS capacity [21, 22]. In humans, fibroblasts from patients with loss of acyl-CoA dehydrogenase medium chain (ACADM) demonstrate defects in biogenesis, stability, and activity of OXPHOS complexes [23]. In terms of relationship to longevity, a recent study found that upregulation of OXPHOS is one of the major hepatic gene expression signatures associated with extended lifespan in multiple murine interventions including CR, growth hormone deficiency, and rapamycin treatment [24]. Another study found OXPHOS gene upregulation to be the top common transcriptional signature in cochlea, hippocampus, heart, liver, kidney, gastrocnemius, and white adipose tissues in CR mice. They also found this signature to be conserved in CR flies and in tissues from CR rats and rhesus monkeys [25].
Many OXPHOS and FAO genes are regulated by a group of nuclear receptors (NR) and transcription factors (TF) that have been implicated in longevity and metabolic reprogramming in many long-lived mouse models. Peroxisome proliferator-activated receptors (PPARs) α and δ, and estrogen-related receptors (ERRs), are the master transcriptional regulators for FAO and OXPHOS genes, respectively [26–31]. There is little information on ERRs in mouse longevity models, but PPARs have been studied in long-lived mice. Livers from CR mice had significantly higher protein levels of PPARα, significantly lower levels of PPARβ/δ, and no significant changes in levels of PPARγ. GHRKO mice had significantly higher levels of hepatic PPARα and γ, and significantly lower levels of PPARβ/δ. Skeletal muscle of both GHRKO and CR mice had significantly lower PPARα, β/δ, and γ levels [32, 33]. PPARs and ERRs, as well as other NRs/TFs, are co-activated by binding to peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α). Consistent with these findings, PGC-1α was reported in multiple studies to be upregulated in liver and adipose tissue of CR mice, and in liver of Snell Dwarf mice [34–38]. Many of the metabolic benefits of protein restriction (PR) diets, which also extend lifespan, have been linked to increased hepatic fibroblast growth factor 21 (FGF-21) and the subsequent increase in PGC-1α [10, 39–43]. PGC-1α in vitro overexpression in the 3T3‐L1 preadipocyte cell line led to significant enrichment of PPAR signaling, fatty acid metabolism, and OXPHOS KEGG pathways [44].
Nuclear receptor corepressor 1 (NCOR1) is also able to regulate PPARs and ERRs [45, 46]. NCOR1 has recently been identified as a conserved metabolic switch that regulates oxidative metabolism signaling [46]. It forms a repressive complex that inhibits the activity of both PPARs and ERRs and downregulates their transcription targets [45, 47–50]. While the role of NCOR1 in metabolic regulation is well established, and resembles metabolic phenotypes seen in long-lived murine models, levels of NCOR1 in those models have not been evaluated.
Here we report levels of FAO enzymes and OXPHOS subunits in males and females of long-lived endocrine mutant mice, and evaluate transcriptional regulators known to influence these enzymes.
Methods
Mice
GHRKO and SD mice were generated and maintained as previously described [51–53], and used at 5–6 months of age. Experiments were conducted on 5–6 male and 5–6 female mice for control and mutant groups, unless otherwise specified. Mice used in this study were fed ad libitum, unless otherwise specified.
All experimental protocols were reviewed and approved by the University of Michigan Institutional Animal Care and Use Committee (IACUC).
Western blotting
Liver samples were collected from mice at a specific time window (from 9:30 to 11:30 a.m.) to ensure the changes in protein levels were not influenced by circadian rhythms. Proteins from liver tissue were homogenized and extracted in freshly prepared Laemmli lysis buffer supplemented with Protease and Phosphatase Inhibitor Cocktail (Thermo, PI78440). Protein concentration was measured using a BCA assay (Thermo, 23227). The protein extracts were separated by SDS/PAGE on a 4–15% precast gel (BioRad, 4561096), and transferred to Immun-Blot PVDF Membrane (BioRad, 1620177). Membranes were then evaluated using EcoBright Femto HRP 100 (Innovative Solutions, EBFH100). Histone H3 was used as a protein loading control, and no significant changes were found in GHRKO or SD Histone H3 levels (data not shown). A full list of primary antibodies used can be found in Table S1. Quantification was performed using ImageJ software.
RNA isolation and cDNA synthesis
Murine liver tissue was homogenized, and RNA samples were extracted using the RNeasy Mini Kit (Qiagen, 74104). The concentration of total RNA was measured using Nanodrop One (Thermo). cDNA was reverse transcribed from 2 μg of total RNA using iScript cDNA reverse transcription kit (BioRad, 1708891).
Quantitative real-time PCR
qPCR was performed using TaqMan Fast Advanced Master Mix (Applied Biosystems, 4444557). RT-PCR was performed using quantitative PCR systems (Thermo) with corresponding TaqMan gene expression assay probes (Thermo, 4331182). 18S was simultaneously assayed as a loading control. A full list of TaqMan assays used in this paper can be found in Table S2. The expression levels of different mRNA were reported as CT values.
Statistical analysis
Statistical analyses and plotting were conducted in GraphPad Prism (version 9). All data are presented as mean ± SEM. Data from male and female mice were not pooled. 2-way ANOVAs were used for reporting sex effect, genotype effect, and their interaction. An unpaired Student t-test was performed when a significant interaction term was noted.
Results
Higher levels of mitochondrial fatty acid β-oxidation proteins in liver of SD and GHRKO mice
Multiple murine lifespan-extending interventions, including CR and FGF21 treatment, show higher levels of hepatic fatty acid β-oxidation [8–10]. CR mice were previously reported to have higher protein levels of multiple mitochondrial β-oxidation enzymes [8]. GHRKO mice were previously reported to have reduced respiratory quotient (RQ) indicating an increased reliance on fat oxidation rather than carbohydrate metabolism [17]. We therefore undertook a comprehensive survey of proteins involved in hepatic fatty acid oxidation in GHRKO and SD mice (Fig. S1).
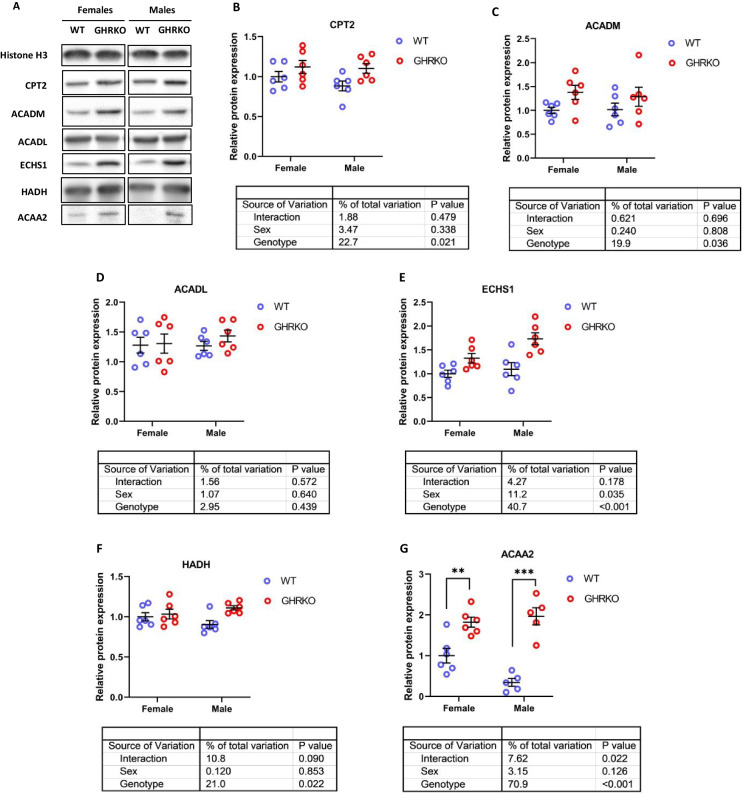
We found that levels of carnitine palmitoyltransferase 2 (CPT2), an enzyme that transports fatty acids across the mitochondrial membrane, were significantly upregulated in GHRKO and SD liver (Figs. 1A, B, S2A, and S2B). We measured protein levels of two acyl-CoA dehydrogenases, long chain (ACADL) and ACADM, which differ in specificity based on fatty acid chain length. We found that GHRKO and SD livers had significantly higher ACADM (Figs. 1C and S2C), but that ACADL levels were not significantly changed (Figs. 1D and S2D). This agrees with previously published data on ACADL in SD livers, and ACADM in CR livers [8, 54]. Enoyl-CoA Hydratase, Short Chain 1 (ECHS1), the enzyme responsible for the second step of mitochondrial fatty acid β-oxidation, was significantly upregulated in both GHRKO and SD livers (Figs. 1E and S2E). Hydroxyacyl-CoA Dehydrogenase (HADH) and acetyl-CoA acyltransferase 2 (ACAA2), the enzymes responsible for the final two steps of mitochondrial β-oxidation were both significantly upregulated in GHRKO and SD livers (Figs. 1F, G, S2F, and S2G).
Fig. 1.
Effect of growth hormone receptor knockout on mitochondrial fatty acid β-oxidation enzymes in liver. (A) Representative western blots for mitochondrial FAO enzymes in liver lysates from female and male wild type (WT) and GHRKO mice. (B-G) Scatter plots of CPT2 (B), ACADM (C), ACADL (D), ECHS1 (E), HADH (F), and ACAA2 (G). Data show mean ± S.E.M. Each symbol represents an individual mouse. n = 5–6 for each group. Two-way ANOVA was used for analysis of genotype effect, sex effect, and their interaction. Unpaired t-test was used for each sex separately when the interaction term was significant. ** P < 0.01, *** p < 0.001
GHRKO and SD mice have higher levels of peroxisomal fatty acid β-oxidation proteins
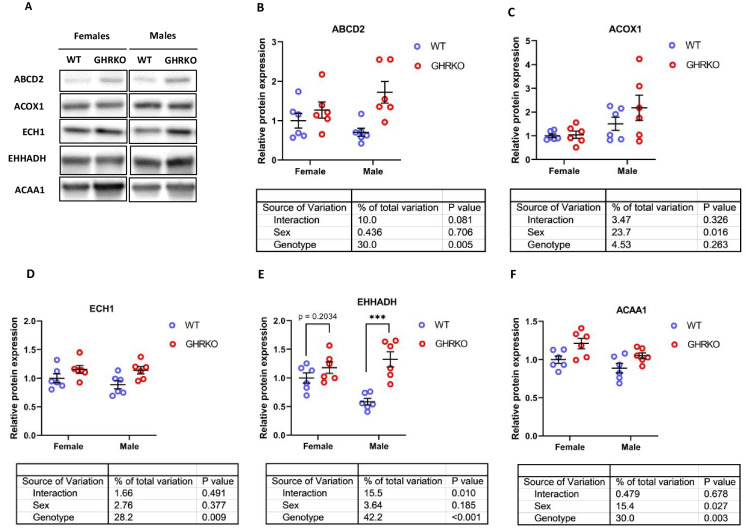
Peroxisomes are another cellular site for fatty acid β-oxidation (Fig. S1). Previous studies reported an increase in peroxisomal FAO proteins in liver of acarbose-treated and SD mice [55, 56]. In CR mice, acyl-CoA Oxidase 1 (ACOX1), which is responsible for the first of four peroxisomal β-oxidation reactions, was downregulated compared to wild type mice [8].
ATP Binding Cassette Subfamily D Member 2 (ABCD2), the protein responsible for transporting fatty acids into the peroxisome, was significantly upregulated in GHRKO and SD livers (Figs. 2A, B, S3A, and S3B). ACOX1 was elevated in SD liver (Fig. S3C), but did not show a significant effect in GHRKO liver (Fig. 2C). In both models, ACOX1 was significantly higher in males than in females. Enoyl-CoA Hydratase (ECH1), the peroxisomal counterpart of mitochondrial ECHS1, was significantly upregulated in both GHRKO and SD livers (Figs. 2D and S3D). Enoyl-CoA Hydratase And 3-Hydroxyacyl CoA Dehydrogenase (EHHADH), which catalyzes the third reaction in the peroxisomal fatty acid β-oxidation pathway, was significantly upregulated in male GHRKO mice and in both sexes of SD mice, with a non-significant effect (p = 0.2) in female GHRKO mice (Figs. 2E and S3E). Peroxisomal acetyl-CoA acyltransferase 1 (ACAA1) was significantly upregulated in GHRKO mice, but not in SD mice (p-value = 0.09 in SD) (Figs. 2F and S3F).
Fig. 2.
Hepatic levels of peroxisomal fatty acid β-oxidation enzymes in WT and GHRKO livers. (A) Representative images of western blot data for peroxisomal FAO enzymes in liver lysates from female and male WT and GHRKO mice. (B-F) Scatter plots of ABCD2 (B), ACOX1 (C), ECH1 (D), EHHADH (E), and ACAA1 (F). Data show mean ± S.E.M. Each symbol represents an individual mouse. n = 5–6 for each group. Two-way ANOVA was used for analysis of genotype effect, sex effect, and their interaction. Unpaired t-test was used when the interaction term was significant. *** p < 0.001
Decreased growth hormone signaling also elevates expression of oxidative phosphorylation proteins
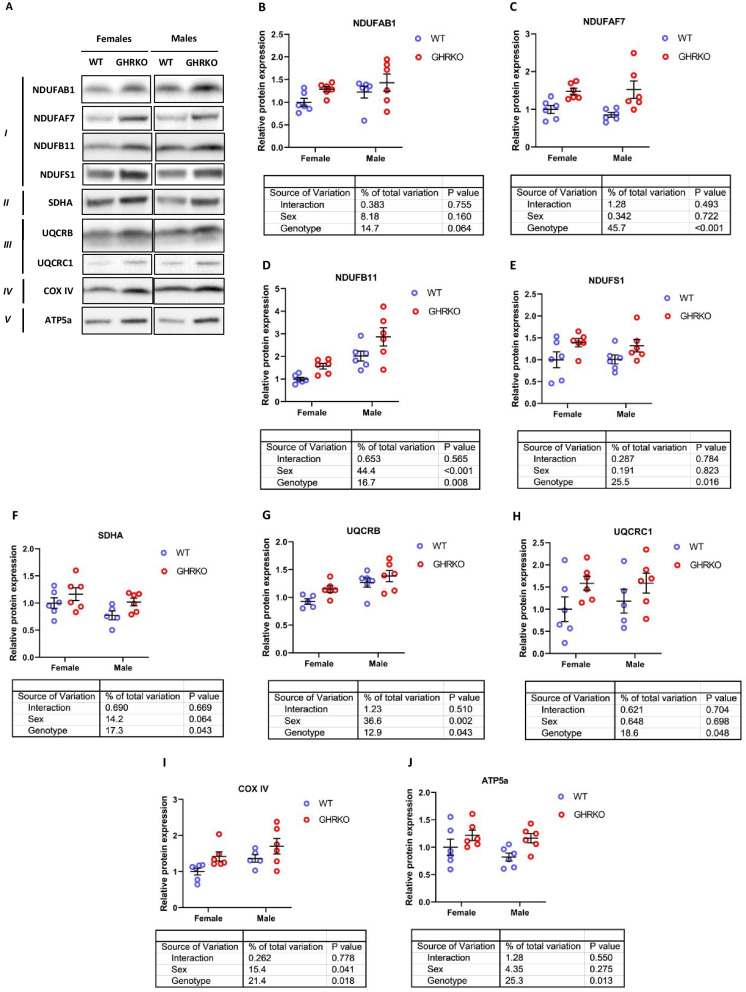
Mitochondrial fatty acid oxidation is physically and functionally linked to the oxidative phosphorylation (OXPHOS) pathway, with proteins in both pathways co-regulated on the transcriptional level by common nuclear receptors and shared transcription factors [18, 20, 35, 44, 45, 50]. Hence, our next aim was to measure the levels of different protein subunits that make up the five OXPHOS complexes.
First, we measured the levels of four subunits in complex I. NDUFAF7, NDUFB11, and NDUFS1 were all significantly upregulated in GHRKO livers, and NDUFAB1 showed a similar effect that approached conventional significance thresholds (p = 0.06) (Fig. 3A–E). In SD livers, NDUFAF7, NDUFB11, and NDUFAB1 were all significantly upregulated, while NDUFS1 showed a similar trend (p-value = 0.10) (Figures S4A-S4E). We also measured levels of SDHA (complex II), UQCRB and UQCRC1 (complex III), COX IV (complex IV), and ATP5a (complex V). The subunits from complexes II to V were all significantly upregulated in GHRKO livers (Fig. 3F–K). This is consistent with previous studies showing that upregulation of OXPHOS is a common transcriptional signature of long-lived models including CR and GHRKO [24, 25]. In Snell Dwarf livers, subunits from complexes II to IV (SDHA, UQCRB, UQCRC1, and COX IV) were significantly upregulated, while ATP5a showed a similar trend (p-value = 0.06) (Figure S4F-S4K).
Fig. 3.
Hepatic levels of OXPHOS subunit proteins in WT and GHRKO livers. (A) Representative western blot images for OXPHOS subunits in liver lysates from female and male WT and GHRKO mice. (B-J) Scatter plots of NDUFAB1 (B), NDUFAF7 (C), NDUFB11 (D), NDUFS1 (E), SDHA (F), UQCRB (G), UQCRC1 (H), COX IV (I), and ATP5a (J). Data show mean ± S.E.M. Each symbol represents an individual mouse. n = 5–6 for each group. Two-way ANOVA was used for analysis of genotype effect, sex effect, and their interaction
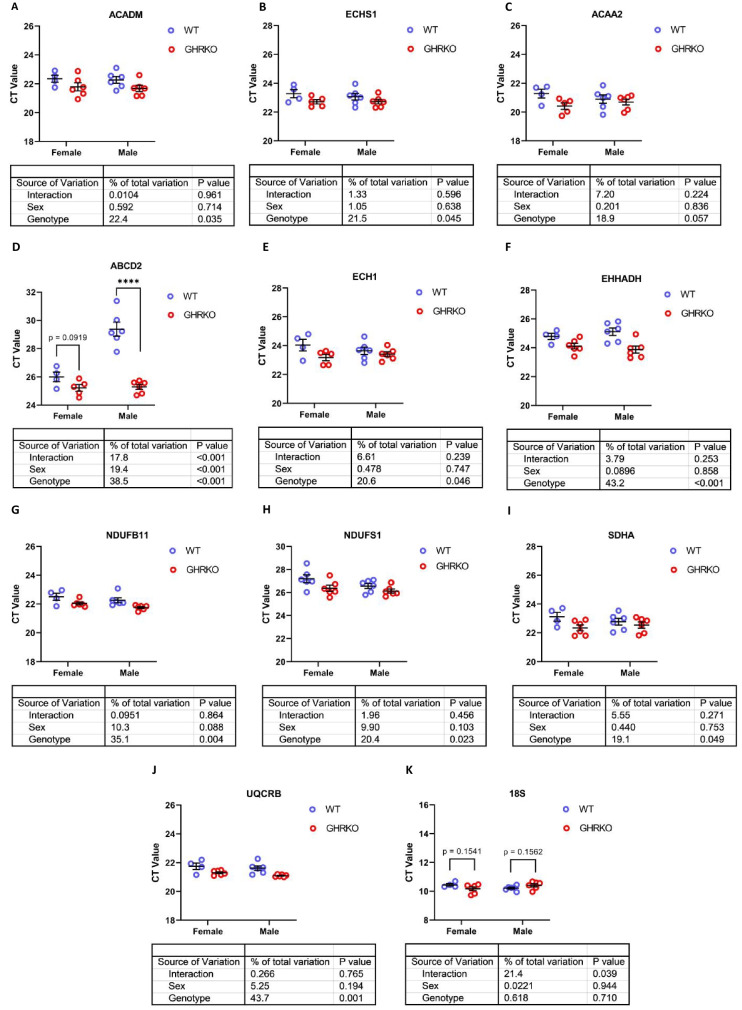
Upregulation of mRNA encoding proteins of fatty acid oxidation and oxidative phosphorylation
We then sought to determine if these changes in fatty acid oxidation enzymes and OXPHOS proteins reflected transcriptional changes. We found that mRNA levels of mitochondrial fatty acid β-oxidation enzymes ACADM, ECHS1, and ACAA2 were all significantly upregulated in GHRKO and SD livers, except for mRNA for ACAA2 in GHRKO livers, which approached our significance threshold (p = 0.06) (Figs. 4A–C and S5A-S5C). mRNA for peroxisomal enzymes ECH1 and EHHADH was also significantly upregulated in GHRKO mice. mRNA for ABCD2 showed a [Sex x Genotype] interaction in GHRKO mice, with significant elevation in males, and a similar trend (p = 0.09) in female mice (Fig. 4D–F). In SD mice, only ECH1 mRNA was significantly upregulated in both males and females, while ABCD2 and EHHADH were significantly upregulated in male mice, with female mice showing similar trends (p = 0.08 and p = 0.1, respectively) (S5D-S5F). Finally, OXPHOS genes NDUFB11, NDUFS1, SDHA, and UQCRB were all significantly increased at the mRNA level in both mouse models (Figs. 4G–J and S5G-S5J). The OXPHOS data are consistent with previous reports showing that OXPHOS genes are upregulated in CR, GHRKO, and SD livers [24]. The levels of 18S mRNA were used as a control and did not show any statistically significant changes in GHRKO or SD liver samples (Figs. 4K and S5K).
Fig. 4.
Hepatic mRNA levels of FAO and OXPHOS genes in WT and GHRKO livers. (A-K) Scatter plots of ACADM (A), ECHS1 (B), ACAA2 (C), ABCD2 (D), ECH1 (E), EHHADH (F), NDUFB11 (G), NDUFS1 (H), SDHA (I), UQCRB (J), and 18S (K). Data shows mean ± S.E.M. Each symbol represents an individual mouse. n = 4–6 for each group. Two-way ANOVA was used for analysis of genotype effect, sex effect, and their interaction. Unpaired t-test was used when interaction was significant. **** p < 0.0001
Transcriptional upregulation of oxidative metabolism proteins in GHRKO and SD livers is not explained by levels of corresponding transcription factors
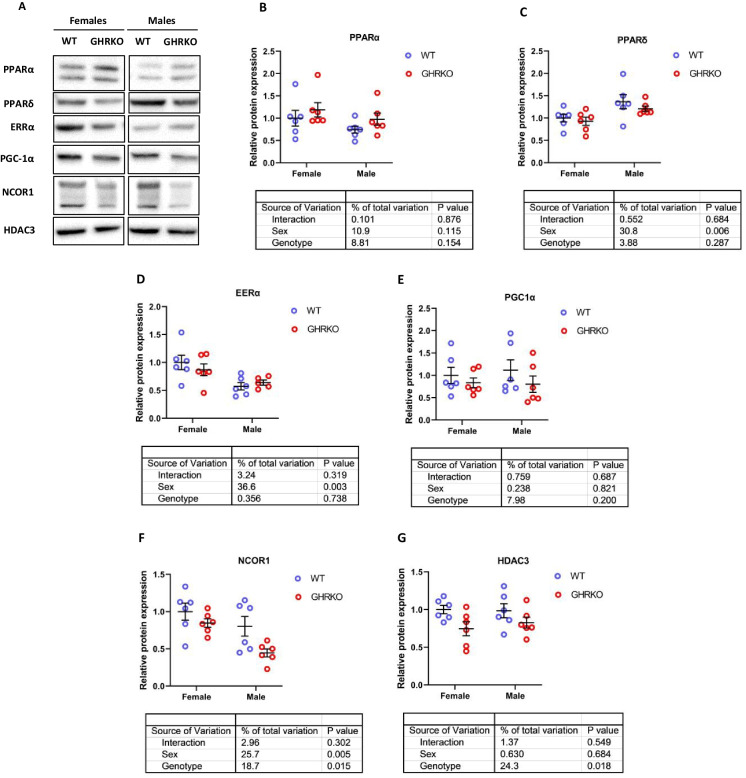
The proteins involved in fatty acid β-oxidation and OXPHOS are transcriptionally controlled via a group of transcription factors called nuclear receptors, which includes peroxisome proliferator-activated receptors (PPARs) and estrogen-related receptors (ERRs) [44, 45]. Protein levels and activity of different PPARs have long been hypothesized to play an important role in metabolic regulation in long-lived murine models [28, 31, 37, 44, 55, 57, 58]. A common co-activator for those receptors, peroxisome proliferator-activated receptor-γ coactivator-1α (PGC-1α), has also been positively implicated in multiple murine lifespan-extending pathways [21, 31, 33, 44].
Previous studies have reported an increase in PPARα protein in GHRKO and SD liver tissue [33, 55].While we saw an upward trend in GHRKO livers, the effect did not reach statistical significance (Fig. 5A and B). In SD livers, PPARα protein was significantly downregulated (Figures S6A and S6B), contradicting previous reports. The discrepancy could stem from the difference in genetic backgrounds or diets of SD mice [55]. PPARδ, another PPAR receptor that acts as a transcription factor for mitochondrial and peroxisomal β-oxidation enzymes, showed no significant change in GHRKO livers (Fig. 5C) and was not evaluated in SD livers. ERRα, a nuclear receptor and transcription factor responsible for OXPHOS proteins transcription, was also unchanged in GHRKO livers (Fig. 5D); both proteins did show sexual dimorphism, with PPARδ higher in males and ERRα higher in females. In SD livers, EERα showed a decline in female mice that approached statistical significance (p = 0.051) (Fig. S6C). Surprisingly, PGC-1α, a co-activator for PPARs and ERRs, was not significantly changed in GHRKO or SD livers (Figs. 5E and S6D). This disagrees with a previously published study that found PGC-1α to be significantly upregulated in SD livers [38]. This difference could be due to changes in feeding conditions of the mice or in time of the day when the samples were collected, since PGC-1α influences and is in turn influenced by circadian rhythms [59].
Fig. 5.
Hepatic levels of PPAR signaling network proteins in WT and GHRKO livers. (A) Representative images of western blot data for different nuclear receptors and their co-regulators in liver lysates from female and male WT and GHRKO mice. (B-G) Scatter plots of PPARα (B), PPARδ (C), ERRα (D), PGC-1α (E), NCOR1 (F), and HDAC3 (G). Data show mean ± S.E.M. Each symbol represents an individual mouse. n = 5–6 for each group. Two-way ANOVA was used for analysis of genotype effect, sex effect, and their interaction
Overall, the changes in transcription factors and co-activator levels did not explain the significant transcriptional upregulation observed in most of the downstream targets.
Diminished levels of nuclear receptor co-repressor NCOR1 in GHRKO and Snell dwarf mice
Nuclear receptor co-repressor 1 (NCOR1) is an inhibitor of PPARs and ERRs [45, 47, 48]. Tissue-specific knockdown of NCOR1 has been shown to increase oxidative phosphorylation and fatty acid oxidation in murine liver and muscle tissues [50, 60]. We tested the hypothesis that NCOR1 decline in GHRKO and SD livers could be responsible for increased transcription of genes regulated by PPARs and ERRs in mouse liver. We found that NCOR1 was indeed significantly downregulated in GHRKO and SD livers by at least 15% and 25%, respectively (Figs. 5F and S6E). NCOR1 forms a complex with Histone deacetylase 3 (HDAC3) that is necessary for metabolic functions of NCOR1 [61, 62]. HDAC3 levels were also significantly reduced in GHRKO and SD livers (Figs. 5G and S6F). These results suggest that reduced NCOR1 and HDAC3 may contribute to the coordinated increase of transcription of OXPHOS subunits and β-oxidation enzymes in GHRKO and Snell mice.
Discussion
Murine lifespan extension can be achieved via dietary changes (e.g., CR and methionine restriction), genetic mutations (e.g., GHRKO and SD), or pharmacologically (e.g., rapamycin and acarbose) [2, 56, 63–65]. Finding shared signatures and mechanisms for longevity among these models would provide important clues for developing new lifespan extending interventions and for a more detailed understanding of pathways that control aging rate and risks of late-life diseases. One such shared signature is the upregulation of mRNA for fatty acid β-oxidation and OXPHOS in long-lived mice [24, 25]. RNA-seq data show OXPHOS and FAO pathways to be upregulated in CR and GHRKO livers [7, 25]. Furthermore, male mice treated with rapamycin for the first 45 days of life were long-lived, had lower levels of enrichment of inflammatory pathway genes including interferon α and IL6-JAK-STAT3 signaling, and higher levels of enrichment of metabolic pathway genes including OXPHOS, FAO, gluconeogenesis, and adipogenesis [66]. Overexpression of phosphatase and tensin homolog (PTEN), which also extends lifespan in mice, increases FAO in calf hepatocytes [11]. Consistently, knocking down PTEN in calf hepatocytes inhibits FAO [12].
One weakness of inference based on mRNA data alone (i.e. on “genetic signatures”) is that there can be substantial discrepancies between mRNA levels and levels of the corresponding proteins [54, 63, 67–69]. Many studies of long-lived mice have highlighted the importance of differential translation of specific sets of mRNAs. For example, cap-independent translation of many mitochondrial proteins is upregulated in GHRKO, SD, rapamycin-treated, and acarbose-treated mice without increases in the mRNA levels of the corresponding genes [38, 63]. Additionally, proteolysis via ubiquitination, autophagy, or chaperone-mediated autophagy can also alter the levels of specific sets of proteins in long-lived mouse models [54, 67, 70–72]. Differences in genetic signatures based on mRNA levels are not sufficient to show that a specific pathway is causative for, linked to, or even present in models of lifespan extension. Our new data help to clarify these ambiguities by evaluation of proteins related to OXPHOS, FAO, and their transcriptional regulators. We found that FAO and OXPHOS proteins, as well as mRNA, are significantly upregulated in GHRKO and SD livers. This finding adds to the body of evidence that OXPHOS and FAO pathways are upregulated in long-lived mutant mice, as they are in CR animals [8]. Interestingly, we found that ACADM, one of the enzymes responsible for the first β-oxidation step in the mitochondria, was significantly upregulated in both GHRKO and SD livers. ACADL, which catalyzes the same reaction for longer fatty acids, was not significantly changed in either model. This could suggest that in both GHRKO and SD livers, different acetyl-CoA dehydrogenases could be differentially regulated based on hepatic use of fatty acids in specific length classes.
Our results are consistent with previous studies on metabolic phenotypes in Ames dwarf and GHRKO mice, which rely more on fatty acids than on carbohydrates. The previous data included measures of energy expenditure, respiratory quotient, oxygen consumption, and body temperature [15, 17].
Most of the changes we report were seen in both sexes, but sex-specific peroxisomal FAO enzymes were an exception to this generalization. ABCD2 mRNA levels in GHRKO and SD, EHHADH protein level in GHRKO livers, and its EHHADH mRNA level in SD livers all showed a significant interaction term, and in all these cases, male mice showed the more extreme changes.
Increased oxidative metabolism and β-oxidation could reflect increased abundance of these proteins, or perhaps increased enzymatic activity with minimal change in protein levels. Previous research provided some evidence to support each of these hypotheses. For example, in liver of CR mice, protein levels of CPT1, ACADM, ACADL, and HADHB were found to be significantly upregulated [8]. There are also multiple reports that post-translational modifications of those enzymes (e.g., SIRT3-mediated hyperacetylation of OXPHOS and FAO enzymes) contribute to their modulated activity and stability [8, 73–75]. In this study, we confirmed increased abundance of many proteins involved in those two pathways. More work will be needed to see if activity of these enzymes is also higher in the long-lived mice.
The transcription of FAO and OXPHOS genes is regulated by PPARs and ERRs, respectively [26, 27, 29, 30, 57]. There are some previous data suggesting PPARα upregulation in livers of CR, GHRKO, and SD mice [33, 55], and data on muscle showing that PPARα is unchanged in GHRKO mice and downregulated in CR mice [32]. Indeed, many studies have reported PGC-1α to be upregulated in liver, muscle, and adipose tissue of different longevity models [38, 44]. Since PGC-1α acts as a co-activator for many NRs and TFs, including PPARs and ERRs, it has attracted interest as a potential target for anti-aging interventions.
In this study, we did not find any significant upregulation in PPARs, ERRs, or PGC-1α that could explain FAO and OXPHOS transcriptional upregulation in GHRKO and SD mice. It is possible that this discrepancy may reflect circadian control of PPARs and PGC-1α [59, 76]. Indeed, the relationship between circadian clock and energy metabolism is strong and well-established. For example, it was previously shown that caloric restriction induces better rhythmicity in hepatic ketogenesis as well as higher levels of PPARα, FGF21, ACADL and ECHS1 [77]. Additionally, previous work has demonstrated that PGC-1α influences circadian clock genes, and liver PGC-1α is crucial for integrating mammalian clock with energy metabolism [59]. Similarly, and of relevance to the current study, disruption of HDAC3-NCOR1 interaction led to changes in circadian behavior, and generated mice that are leaner and have higher energy expenditure and insulin sensitivity. Levels of different FAO enzymes and PPARs in mice with disrupted HDAC3-NCOR1 appeared to have a circadian phase-shift that resulted in them being higher than or similar to WT levels depending on time of day. For example, PPARα level in that model was almost the same as WT mice at ZT5 (five hours from lights on), and was significantly higher than WT mice at ZT10, before falling back to lower than WT levels at ZT15 [76]. Other studies have also demonstrated that activity of PPARs can be modulated without alteration in PPAR protein levels [32, 50].
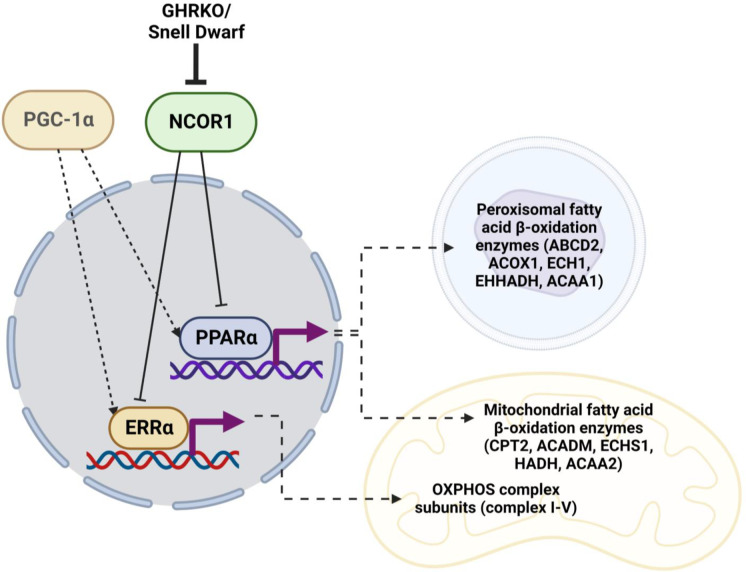
Transcription of OXPHOS and FAO mRNA is also influenced by other co-regulators, including receptor-interacting protein 140 (RIP140), silencing mediator for retinoid or thyroid-hormone receptors (SMRT), and NCOR1 [47, 48, 78]. Those regulators have not previously been evaluated in slow-aging mice.. NCOR1 antagonizes PGC-1α in skeletal muscle oxidative metabolism [45]. Deletion of NCOR1 in skeletal muscles or adipose tissue leads to increased insulin sensitivity [50, 79]. Additionally, mTORC1 controls fasting-induced ketogenesis through phosphorylation of S6K and subsequent activation of NCOR1 [80]. NCOR1 mediates this inhibition of NRs by forming a repressive complex with HDAC3 [61, 62]. Our results show downregulation of hepatic NCOR1 and HDAC3 in both GHRKO and SD mice, and suggest that the NCOR1-HDAC3 repressive complex may play an important role as a regulator of metabolic phenotypes linked to increased longevity (Fig. 6).
Fig. 6.
Postulated mechanism of NCOR1 regulation of PPARα- and ERRα-mediated transcription in GHRKO and SD. Created with BioRender.com
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary file1 (PDF 3416 KB) Supplementary Figure 1: Fatty acid β-oxidation pathways in mitochondria and peroxisome. Created with BioRender.com. Supplementary Figure 2: Effect of Snell dwarf mutation (Pit1dw) on hepatic levels of mitochondrial fatty acid β-oxidation enzymes. (A) Representative images of western blot data for different mitochondrial FAO enzymes in liver lysates from female and male wild type (WT) and SD mice. (B-G) Scatter plots of CPT2 (B), ACADM (C), ACADL (D), ECHS1 (E), HADH (F), and ACAA2 (G). Data show mean ± S.E.M. Each symbol represents an individual mouse. n = 5-6 for each group. Two-way ANOVA was used for analysis of genotype effect, sex effect, and their interaction. Unpaired t-test was used when interaction was significant. ** P < 0.01, *** p < 0.001. Supplementary Figure 3: Hepatic levels of peroxisomal fatty acid β-oxidation enzymes in WT and SD livers. (A) Representative images of western blot data for different peroxisomal FAO enzymes in liver lysates from female and male WT and SD mice. (B-F) Scatter plots of ABCD2 (B), ACOX1 (C), ECH1 (D), EHHADH (E), and ACAA1 (F). Data show mean ± S.E.M. Each symbol represents an individual mouse. n = 5-6 for each group. Two-way ANOVA was used for analysis of genotype effect, sex effect, and their interaction. Supplementary Figure 4: Hepatic levels of OXPHOS subunit proteins in WT and SD livers. (A) Representative images of western blot data for different OXPHOS complexes subunits in liver lysates from female and male WT and SD mice. (B-J) Scatter plots of NDUFAB1 (B), NDUFAF7 (C), NDUFB11 (D), NDUFS1 (E), SDHA (F), UQCRB (G), UQCRC1 (H), COX IV (I), and ATP5a (J). Data show mean ± S.E.M. Each symbol represents an individual mouse. n = 5-6 for each group. Two-way ANOVA was used for analysis of genotype effect, sex effect, and their interaction. Supplementary Figure 5: Hepatic mRNA levels of FAO and OXPHOS genes in WT and SD livers. (A-K) Scatter plots of ACADM (A), ECHS1 (B), ACAA2 (C), ABCD2 (D), ECH1 (E), EHHADH (F), NDUFB11 (G), NDUFS1 (H), SDHA (I), UQCRB (J), and 18S (K). Data show mean ± S.E.M. Each symbol represents an individual mouse. n = 5-6 for each group. Two-way ANOVA was used for analysis of genotype effect, sex effect, and their interaction. Unpaired t-test was used when interaction was significant. *** p < 0.001, **** p < 0.0001. Supplementary Figure 6: Hepatic levels of PPAR signaling network proteins in WT and SD livers. (A) Representative images of western blot data for different nuclear receptors and their co-regulators in liver lysates from female and male WT and SD mice. (B-F) Scatter plots of PPARα (B), ERRα (C), PGC-1α (D), NCOR1 (E), and HDAC3 (F). Data show mean ± S.E.M. Each symbol represents an individual mouse. n = 5-6 for each group. Two-way ANOVA was used for analysis of genotype effect, sex effect, and their interaction
Acknowledgements
This work was funded by National Institutes of Health (NIH) grants AG024824, AG023122, AG064706 and National Institute on Aging (NIA) grant T32-AG000114
Data availability
All data supporting the findings of this study are included within this paper and its Supplementary Information.
Declarations
Competing interests
The authors have no relevant financial or non-financial interests to disclose.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Rubio-Tomás T, Tavernarakis N. Lipid metabolism and ageing in Caenorhabditis elegans: a complex interplay. Biogerontology. 2022;23(5):541–557. doi: 10.1007/s10522-022-09989-4. [DOI] [PubMed] [Google Scholar]
- 2.Bartke A, Westbrook R. Metabolic characteristics of long-lived mice. Front Genet. 2012;3:288–288. doi: 10.3389/fgene.2012.00288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Araki S, Okazaki M, Goto S. Impaired lipid metabolism in aged mice as revealed by fasting-induced expression of apolipoprotein mRNAs in the liver and changes in serum lipids. Gerontology. 2004;50(4):206–215. doi: 10.1159/000078349. [DOI] [PubMed] [Google Scholar]
- 4.Johnson AA, Stolzing A. The role of lipid metabolism in aging, lifespan regulation, and age-related disease. Aging Cell. 2019;18(6):e13048–e13048. doi: 10.1111/acel.13048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dominguez LJ, Barbagallo M. The biology of the metabolic syndrome and aging. Curr Opin Clin Nutr Metab Care. 2016;19(1):5–11. doi: 10.1097/MCO.0000000000000243. [DOI] [PubMed] [Google Scholar]
- 6.Wilson PW, et al. Metabolic syndrome as a precursor of cardiovascular disease and type 2 diabetes mellitus. Circulation. 2005;112(20):3066–3072. doi: 10.1161/CIRCULATIONAHA.105.539528. [DOI] [PubMed] [Google Scholar]
- 7.Bruss MD, et al. Calorie restriction increases fatty acid synthesis and whole body fat oxidation rates. Am J Physiol Endocrinol Metab. 2010;298(1):E108-16. [DOI] [PMC free article] [PubMed]
- 8.Mezhnina V, et al. CR reprograms acetyl-CoA metabolism and induces long-chain acyl-CoA dehydrogenase and CrAT expression. Aging Cell. 2020;19(11):e13266–e13266. doi: 10.1111/acel.13266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fletcher JA, et al. Fibroblast growth factor 21 increases hepatic oxidative capacity but not physical activity or energy expenditure in hepatic peroxisome proliferator-activated receptor γ coactivator-1α-deficient mice. Exp Physiol. 2018;103(3):408–418. doi: 10.1113/EP086629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Potthoff MJ, et al. FGF21 induces PGC-1alpha and regulates carbohydrate and fatty acid metabolism during the adaptive starvation response. Proc Natl Acad Sci USA. 2009;106(26):10853–10858. doi: 10.1073/pnas.0904187106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fu S, et al. Increase of fatty acid oxidation and VLDL assembly and secretion overexpression of PTEN in cultured hepatocytes of newborn calf. Cell Physiol Biochem. 2012;30(4):1005–1013. doi: 10.1159/000341477. [DOI] [PubMed] [Google Scholar]
- 12.Zhao B, et al. Knockdown of phosphatase and tensin homolog (PTEN) inhibits fatty acid oxidation and reduces very low density lipoprotein assembly and secretion in calf hepatocytes. J Dairy Sci. 2020;103(11):10728–10741. doi: 10.3168/jds.2019-17920. [DOI] [PubMed] [Google Scholar]
- 13.Brown NF, et al. The mammalian target of rapamycin regulates lipid metabolism in primary cultures of rat hepatocytes. Metabolism. 2007;56(11):1500–1507. doi: 10.1016/j.metabol.2007.06.016. [DOI] [PubMed] [Google Scholar]
- 14.Sipula IJ, Brown NF, Perdomo G. Rapamycin-mediated inhibition of mammalian target of rapamycin in skeletal muscle cells reduces glucose utilization and increases fatty acid oxidation. Metabolism. 2006;55(12):1637–1644. doi: 10.1016/j.metabol.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 15.List EO, et al. Endocrine parameters and phenotypes of the growth hormone receptor gene disrupted (GHR-/-) mouse. Endocr Rev. 2011;32(3):356–386. doi: 10.1210/er.2010-0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berryman DE, et al. Effect of growth hormone on susceptibility to diet-induced obesity. Endocrinology. 2006;147(6):2801–2808. doi: 10.1210/en.2006-0086. [DOI] [PubMed] [Google Scholar]
- 17.Westbrook R, et al. Alterations in oxygen consumption, respiratory quotient, and heat production in long-lived GHRKO and Ames dwarf mice, and short-lived bGH transgenic mice. J Gerontol A Biol Sci Med Sci. 2009;64(4):443–451. doi: 10.1093/gerona/gln075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y, et al. Mitochondrial fatty acid oxidation and the electron transport chain comprise a multifunctional mitochondrial protein complex. J Biol Chem. 2019;294(33):12380–12391. doi: 10.1074/jbc.RA119.008680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frerman FE. Reaction of electron-transfer flavoprotein ubiquinone oxidoreductase with the mitochondrial respiratory chain. Biochimica et Biophysica Acta (BBA) - Bioenergetics. 1987;893(2):161–169. doi: 10.1016/0005-2728(87)90035-1. [DOI] [PubMed] [Google Scholar]
- 20.Wang⁎ Y, et al. Evidence for the physical association of mitochondrial fatty acid oxidation and oxidative phosphorylation complexes. Mitochondrion. 2011;11(4):644. [DOI] [PMC free article] [PubMed]
- 21.Bjørndal B, et al. Associations between fatty acid oxidation, hepatic mitochondrial function, and plasma acylcarnitine levels in mice. Nutr Metab. 2018;15:10–10. doi: 10.1186/s12986-018-0241-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Venizelos N, von Döbeln U, Hagenfeldt L. Fatty acid oxidation in fibroblasts from patients with defects in β-oxidation and in the respiratory chain. J Inherit Metab Dis. 1998;21(4):409–415. doi: 10.1023/A:1005310809714. [DOI] [PubMed] [Google Scholar]
- 23.Lim SC, et al. Loss of the mitochondrial fatty Acid β-Oxidation protein Medium-Chain Acyl-Coenzyme A dehydrogenase disrupts oxidative phosphorylation protein complex stability and function. Sci Rep. 2018;8(1):153–153. doi: 10.1038/s41598-017-18530-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tyshkovskiy A, et al. Identification and application of gene expression signatures associated with lifespan extension. Cell Metab. 2019;30(3):573–593.e8. doi: 10.1016/j.cmet.2019.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barger JL, et al. A conserved transcriptional signature of delayed aging and reduced disease vulnerability is partially mediated by SIRT3. PLoS ONE. 2015;10(4):e0120738–e0120738. doi: 10.1371/journal.pone.0120738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Staels B. PPARS: Fatty acid-activated receptors controlling lipid metabolism and inflammation. Atherosclerosis. 2000;151(1):86. doi: 10.1016/S0021-9150(00)80388-3. [DOI] [Google Scholar]
- 27.Burkart EM, et al. Nuclear receptors PPARbeta/delta and PPARalpha direct distinct metabolic regulatory programs in the mouse heart. J Clin Investig. 2007;117(12):3930–3939. doi: 10.1172/JCI32578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barberá MJ, et al. Peroxisome proliferator-activated receptor α activates transcription of the brown fat uncoupling Protein-1 gene. J Biol Chem. 2001;276(2):1486–1493. doi: 10.1074/jbc.M006246200. [DOI] [PubMed] [Google Scholar]
- 29.Alaynick WA, et al. ERRγ directs and maintains the transition to oxidative metabolism in the postnatal heart. Cell Metab. 2007;6(1):13–24. doi: 10.1016/j.cmet.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 30.Dufour CR, et al. Genome-wide orchestration of cardiac functions by the orphan nuclear receptors ERRα and γ. Cell Metab. 2007;5(5):345–356. doi: 10.1016/j.cmet.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 31.Wang Y-X, et al. Peroxisome-Proliferator-Activated receptor δ activates fat metabolism to prevent obesity. Cell. 2003;113(2):159–170. doi: 10.1016/S0092-8674(03)00269-1. [DOI] [PubMed] [Google Scholar]
- 32.Masternak MM, et al. Caloric restriction results in decreased expression of peroxisome proliferator-activated receptor superfamily in muscle of normal and long-lived growth hormone receptor/binding protein knockout mice. J Gerontol A Biol Sci Med Sci. 2005;60(10):1238–1245. doi: 10.1093/gerona/60.10.1238. [DOI] [PubMed] [Google Scholar]
- 33.Masternak MM, et al. Effects of caloric restriction and growth hormone resistance on the expression level of peroxisome proliferator-activated receptors superfamily in liver of normal and long-lived growth hormone receptor/binding protein knockout mice. J Gerontol A Biol Sci Med Sci. 2005;60(11):1394–1398. doi: 10.1093/gerona/60.11.1394. [DOI] [PubMed] [Google Scholar]
- 34.Nisoli E, et al. Calorie restriction promotes mitochondrial biogenesis by inducing the expression of eNOS. Science. 2005;310(5746):314–317. doi: 10.1126/science.1117728. [DOI] [PubMed] [Google Scholar]
- 35.Anderson RM, et al. Dynamic regulation of PGC-1alpha localization and turnover implicates mitochondrial adaptation in calorie restriction and the stress response. Aging Cell. 2008;7(1):101–111. doi: 10.1111/j.1474-9726.2007.00357.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Corton JC, et al. Mimetics of caloric restriction include agonists of lipid-activated nuclear receptors. J Biol Chem. 2004;279(44):46204–46212. doi: 10.1074/jbc.M406739200. [DOI] [PubMed] [Google Scholar]
- 37.Fujii N, et al. Sterol regulatory element-binding protein-1c orchestrates metabolic remodeling of white adipose tissue by caloric restriction. Aging Cell. 2017;16(3):508–517. doi: 10.1111/acel.12576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ozkurede U, et al. Cap-independent mRNA translation is upregulated in long-lived endocrine mutant mice. J Mol Endocrinol. 2019;63(2):123–138. doi: 10.1530/JME-19-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Green CL, Lamming DW. Regulation of metabolic health by essential dietary amino acids. Mech Ageing Dev. 2019;177:186–200. doi: 10.1016/j.mad.2018.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hill CM, et al. Low protein-induced increases in FGF21 drive UCP1-dependent metabolic but not thermoregulatory endpoints. Sci Rep. 2017;7(1):8209–8209. doi: 10.1038/s41598-017-07498-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Laeger T, et al. FGF21 is an endocrine signal of protein restriction. J Clin Investig. 2014;124(9):3913–3922. doi: 10.1172/JCI74915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maida A, et al. A liver stress-endocrine nexus promotes metabolic integrity during dietary protein dilution. J Clin Investig. 2016;126(9):3263–3278. doi: 10.1172/JCI85946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pezeshki A, et al. Low protein diets produce divergent effects on energy balance. Sci Rep. 2016;6:25145–25145. doi: 10.1038/srep25145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miller KN, et al. PGC-1a integrates a metabolism and growth network linked to caloric restriction. Aging Cell. 2019;18(5):e12999–e12999. doi: 10.1111/acel.12999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pérez-Schindler J, et al. The corepressor NCoR1 antagonizes PGC-1α and estrogen-related receptor α in the regulation of skeletal muscle function and oxidative metabolism. Mol Cell Biol. 2012;32(24):4913–4924. doi: 10.1128/MCB.00877-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lima TI, et al. Role of NCoR1 in mitochondrial function and energy metabolism. Cell Biol Int. 2018;42(6):734–741. doi: 10.1002/cbin.10973. [DOI] [PubMed] [Google Scholar]
- 47.Mottis A, Mouchiroud L, Auwerx J. Emerging roles of the corepressors NCoR1 and SMRT in homeostasis. Genes Dev. 2013;27(8):819–835. doi: 10.1101/gad.214023.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ritter MJ, et al. Nuclear receptor CoRepressors, NCOR1 and SMRT, are required for maintaining systemic metabolic homeostasis. Mol Metab. 2021;53:101315–101315. doi: 10.1016/j.molmet.2021.101315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sen K, et al. NCoR1 controls immune tolerance in conventional dendritic cells by fine-tuning glycolysis and fatty acid oxidation. Redox Biol. 2023;59:102575–102575. doi: 10.1016/j.redox.2022.102575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamamoto H, et al. NCoR1 is a conserved physiological modulator of muscle mass and oxidative function. Cell. 2011;147(4):827–839. doi: 10.1016/j.cell.2011.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou Y, et al. A mammalian model for Laron syndrome produced by targeted disruption of the mouse growth hormone receptor/binding protein gene (the Laron mouse) Proc Natl Acad Sci U S A. 1997;94(24):13215–13220. doi: 10.1073/pnas.94.24.13215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Coschigano KT, et al. Assessment of growth parameters and life span of GHR/BP gene-disrupted mice. Endocrinology. 2000;141(7):2608–2613. doi: 10.1210/endo.141.7.7586. [DOI] [PubMed] [Google Scholar]
- 53.Flurkey K, et al. Lifespan extension and delayed immune and collagen aging in mutant mice with defects in growth hormone production. Proc Natl Acad Sci U S A. 2001;98(12):6736–6741. doi: 10.1073/pnas.111158898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Endicott SJ, et al. Lysosomal targetomics of ghr KO mice shows chaperone-mediated autophagy degrades nucleocytosolic acetyl-coA enzymes. Autophagy. 2022;18(7):1551–1571. doi: 10.1080/15548627.2021.1990670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stauber AJ, et al. Constitutive expression of peroxisome proliferator-activated receptor alpha-regulated genes in dwarf mice. Mol Pharmacol. 2005;67(3):681–694. doi: 10.1124/mol.104.007278. [DOI] [PubMed] [Google Scholar]
- 56.Herrera JJ, et al. Acarbose has sex-dependent and -independent effects on age-related physical function, cardiac health, and lipid biology. JCI insight. 2020;5(21):e137474. doi: 10.1172/jci.insight.137474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vernia S, et al. The PPARα-FGF21 hormone axis contributes to metabolic regulation by the hepatic JNK signaling pathway. Cell Metab. 2014;20(3):512–525. doi: 10.1016/j.cmet.2014.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim K, Pyo S, Um SH. S6 kinase 2 deficiency enhances ketone body production and increases peroxisome proliferator-activated receptor alpha activity in the liver. Hepatology. 2012;55(6):1727–1737. doi: 10.1002/hep.25537. [DOI] [PubMed] [Google Scholar]
- 59.Liu C, et al. Transcriptional coactivator PGC-1α integrates the mammalian clock and energy metabolism. Nature. 2007;447(7143):477–481. doi: 10.1038/nature05767. [DOI] [PubMed] [Google Scholar]
- 60.Ou-Yang Q, et al. Distinct role of nuclear receptor corepressor 1 regulated de novo fatty acids synthesis in liver regeneration and hepatocarcinogenesis in mice. Hepatology (Baltimore, Md.) 2018;67(3):1071–1087. doi: 10.1002/hep.29562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sun Z, et al. Deacetylase-independent function of HDAC3 in transcription and metabolism requires nuclear receptor corepressor. Mol Cell. 2013;52(6):769–782. doi: 10.1016/j.molcel.2013.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Armour SM, et al. An HDAC3-PROX1 corepressor module acts on HNF4α to control hepatic triglycerides. Nat Commun. 2017;8(1):549–549. doi: 10.1038/s41467-017-00772-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shen Z, et al. Cap-independent translation: A shared mechanism for lifespan extension by rapamycin, acarbose, and 17α-estradiol. Aging Cell. 2021;20(5):e13345. doi: 10.1111/acel.13345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sun L, et al. Life-span extension in mice by preweaning food restriction and by methionine restriction in middle age. J Gerontol A Biol Sci Med Sci. 2009;64(7):711–722. doi: 10.1093/gerona/glp051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Miller RA, et al. Methionine-deficient diet extends mouse lifespan, slows immune and lens aging, alters glucose, T4, IGF-I and insulin levels, and increases hepatocyte MIF levels and stress resistance. Aging Cell. 2005;4(3):119–125. doi: 10.1111/j.1474-9726.2005.00152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shindyapina AV, et al. Rapamycin treatment during development extends life span and health span of male mice and Daphnia magna. Sci Adv. 2022;8(37):eabo5482–eabo5482. doi: 10.1126/sciadv.abo5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Endicott SJ, et al. Long-lived mice with reduced growth hormone signaling have a constitutive upregulation of hepatic chaperone-mediated autophagy. Autophagy. 2021;17(3):612–625. doi: 10.1080/15548627.2020.1725378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gerdes Gyuricza I, et al. Genome-wide transcript and protein analysis highlights the role of protein homeostasis in the aging mouse heart. Genome Res. 2022;32(5):838–852. doi: 10.1101/gr.275672.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Takemon Y, et al. Proteomic and transcriptomic profiling reveal different aspects of aging in the kidney. Elife. 2021;10:e62585. [DOI] [PMC free article] [PubMed]
- 70.Park SH, Choi WH, Lee MJ. Effects of mTORC1 inhibition on proteasome activity and levels. BMB Rep. 2022;55(4):161–165. doi: 10.5483/BMBRep.2022.55.4.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang L, et al. Ubiquitylome study identifies increased histone 2A ubiquitylation as an evolutionarily conserved aging biomarker. Nat Commun. 2019;10(1):2191. doi: 10.1038/s41467-019-10136-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hansen M, Rubinsztein DC, Walker DW. Autophagy as a promoter of longevity: insights from model organisms. Nat Rev Mol Cell Biol. 2018;19(9):579–593. doi: 10.1038/s41580-018-0033-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hebert AS, et al. Calorie restriction and SIRT3 trigger global reprogramming of the mitochondrial protein acetylome. Mol Cell. 2013;49(1):186–199. doi: 10.1016/j.molcel.2012.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pougovkina O, et al. Mitochondrial protein acetylation is driven by acetyl-CoA from fatty acid oxidation. Hum Mol Genet. 2014;23(13):3513–3522. doi: 10.1093/hmg/ddu059. [DOI] [PubMed] [Google Scholar]
- 75.Schwer B, et al. Calorie restriction alters mitochondrial protein acetylation. Aging Cell. 2009;8(5):604–606. doi: 10.1111/j.1474-9726.2009.00503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Alenghat T, et al. Nuclear receptor corepressor and histone deacetylase 3 govern circadian metabolic physiology. Nature. 2008;456(7224):997–1000. doi: 10.1038/nature07541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mezhnina V, et al. Circadian clock controls rhythms in ketogenesis by interfering with PPARα transcriptional network. Proc Natl Acad Sci U S A. 2022;119(40):e2205755119. doi: 10.1073/pnas.2205755119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nautiyal J, Christian M, Parker MG. Distinct functions for RIP140 in development, inflammation, and metabolism. Trends Endocrinol Metab. 2013;24(9):451–459. doi: 10.1016/j.tem.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 79.Li P, et al. Adipocyte NCoR knockout decreases PPARγ phosphorylation and enhances PPARγ activity and insulin sensitivity. Cell. 2011;147(4):815–826. doi: 10.1016/j.cell.2011.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sengupta S, et al. mTORC1 controls fasting-induced ketogenesis and its modulation by ageing. Nature. 2010;468(7327):1100–1104. doi: 10.1038/nature09584. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary file1 (PDF 3416 KB) Supplementary Figure 1: Fatty acid β-oxidation pathways in mitochondria and peroxisome. Created with BioRender.com. Supplementary Figure 2: Effect of Snell dwarf mutation (Pit1dw) on hepatic levels of mitochondrial fatty acid β-oxidation enzymes. (A) Representative images of western blot data for different mitochondrial FAO enzymes in liver lysates from female and male wild type (WT) and SD mice. (B-G) Scatter plots of CPT2 (B), ACADM (C), ACADL (D), ECHS1 (E), HADH (F), and ACAA2 (G). Data show mean ± S.E.M. Each symbol represents an individual mouse. n = 5-6 for each group. Two-way ANOVA was used for analysis of genotype effect, sex effect, and their interaction. Unpaired t-test was used when interaction was significant. ** P < 0.01, *** p < 0.001. Supplementary Figure 3: Hepatic levels of peroxisomal fatty acid β-oxidation enzymes in WT and SD livers. (A) Representative images of western blot data for different peroxisomal FAO enzymes in liver lysates from female and male WT and SD mice. (B-F) Scatter plots of ABCD2 (B), ACOX1 (C), ECH1 (D), EHHADH (E), and ACAA1 (F). Data show mean ± S.E.M. Each symbol represents an individual mouse. n = 5-6 for each group. Two-way ANOVA was used for analysis of genotype effect, sex effect, and their interaction. Supplementary Figure 4: Hepatic levels of OXPHOS subunit proteins in WT and SD livers. (A) Representative images of western blot data for different OXPHOS complexes subunits in liver lysates from female and male WT and SD mice. (B-J) Scatter plots of NDUFAB1 (B), NDUFAF7 (C), NDUFB11 (D), NDUFS1 (E), SDHA (F), UQCRB (G), UQCRC1 (H), COX IV (I), and ATP5a (J). Data show mean ± S.E.M. Each symbol represents an individual mouse. n = 5-6 for each group. Two-way ANOVA was used for analysis of genotype effect, sex effect, and their interaction. Supplementary Figure 5: Hepatic mRNA levels of FAO and OXPHOS genes in WT and SD livers. (A-K) Scatter plots of ACADM (A), ECHS1 (B), ACAA2 (C), ABCD2 (D), ECH1 (E), EHHADH (F), NDUFB11 (G), NDUFS1 (H), SDHA (I), UQCRB (J), and 18S (K). Data show mean ± S.E.M. Each symbol represents an individual mouse. n = 5-6 for each group. Two-way ANOVA was used for analysis of genotype effect, sex effect, and their interaction. Unpaired t-test was used when interaction was significant. *** p < 0.001, **** p < 0.0001. Supplementary Figure 6: Hepatic levels of PPAR signaling network proteins in WT and SD livers. (A) Representative images of western blot data for different nuclear receptors and their co-regulators in liver lysates from female and male WT and SD mice. (B-F) Scatter plots of PPARα (B), ERRα (C), PGC-1α (D), NCOR1 (E), and HDAC3 (F). Data show mean ± S.E.M. Each symbol represents an individual mouse. n = 5-6 for each group. Two-way ANOVA was used for analysis of genotype effect, sex effect, and their interaction
Data Availability Statement
All data supporting the findings of this study are included within this paper and its Supplementary Information.