Abstract
A complete nucleic-acid-based assay which consists of sample preparation, DNA amplification, and chromogenic detection was developed for quantifying potential toxin-producing cyanobacteria of interest to the public. The sample preparation strategy involves the same solid phase for cell concentration and DNA purification. For the detection step, we used a combination of competitive PCR amplification, sequence-specific labeling of oligonucleotide probes, hybridization of the labeled oligonucleotides to immobilized complements and, finally, chromogenic detection. The complete assay was tested with water containing toxin-producing cyanobacteria belonging to the genus Microcystis. A detection limit of 100 cells/ml and a quantitative range of more than 3 orders of magnitude were obtained. This approach can easily be adapted to a wide range of bacterial species and has the potential for simultaneous detection and quantitation of several different target organisms by a single assay.
Water blooms formed by cyanobacteria, particularly those belonging to the genus Microcystis, have a relatively high frequency of toxicity (between 25 and 70%) and constitute a potential health hazard for livestock and humans worldwide. Species of this genus can produce several toxins, with the hepatotoxic microcystins being the most potent (4, 21).
With the development of methods for detection and characterization of nucleic acids, such as hybridization (23), in vitro amplification (17), and DNA sequencing (18), novel approaches for environmental monitoring with nucleic acids are emerging (1, 2). Although nucleic acid techniques provide high sensitivity and specificity, there are some limitations for the routine use of these techniques. The methods for sample preparation are often labor-intensive, and the molecular results can be difficult to interpret, e.g., complex gel electrophoresis banding patterns. In addition, due to large variations in the sources and quality of environmental samples, problems with the preparation of cells and nucleic acids can be encountered (24). For these reasons, complete and reproducible assays—from water sampling to quantification of target organisms—are required for routine environmental monitoring. In this study, we present such a system for the environmental monitoring of potential toxin-producing cyanobacteria belonging to the genus Microcystis (26).
Previously, we have developed a method for preparing PCR-ready DNA from cyanobacteria in water by using the same solid phase for both cell concentration and DNA purification (15). Here, we have employed this approach for sample preparation in combination with a chromogenic detection method. To obtain high sensitivity, the detection method was based on coamplification of target and competitor DNA (competitive PCR) (12, 20). Thereafter, to obtain better specificity and dynamic range, one primer complementary to an internal segment of the amplified target and one primer complementary to an internal segment of the competitor were single-base extended (8, 13, 25) by thermocycling. The extended oligonucleotides were then hybridized to their immobilized complements and quantified by chromogenic detection, enabling both the detection of several targets and the simple interpretation of the results.
By combining the sample preparation and the detection steps in a complete assay on water samples, we obtained a detection limit of 100 cells/ml and a quantitative range of more than 3 orders of magnitude. These results show that both the sample preparation and the detection steps are quantitative. Furthermore, the methods used in this study are suitable for automation, providing a means for the development of high throughput systems for routine environmental monitoring.
MATERIALS AND METHODS
Organisms and sample preparation.
The organisms used are from the Norwegian Institute for Water Research. Cultivation was performed in medium Z8 (22). Illumination was provided by fluorescent lamps exposing the strains with 30 microeinsteins m−2s−1. Two different Microcystis aeruginosa strains (NIVA-CYA 228/1 and 43) were used as templates in the development of the assay. The system was also tested on experimentally modified water samples collected from Lake Akersvatnet, County of Vestfold, Norway. The cells were counted by microscopy in a Fuchst-Rosenthal counting chamber (Carl Hecht, Sondheim, Germany).
DNA was purified either by a standard phenol-chloroform protocol from cell pellets of unialgal cultures (14, 15) or by a solid-phase cell concentration and DNA purification protocol previously developed by Rudi et al. (15). In the solid-phase protocol, cells of cyanobacteria from 1 ml of aqueous solution were adsorbed for 20 min onto paramagnetic beads (final volume, 2 ml) in a buffer containing 50% isopropanol, 0.75 M ammonium acetate, and 1 U (the amount of beads in 200 μl of lysis buffer) of Dynabeads DNA DIRECT (Dynal A/S, Oslo, Norway). The magnetic beads and the adsorbed bacteria were attracted to the side of a 2-ml centrifuge tube by a MPC-Q magnet (Dynal A/S). Then, 20 μl of 4 M guanidine thiocyanate–1% Sarkosyl was added, and the incubation was continued at 65°C for 10 min. The DNA was precipitated onto the beads by the addition of 40 μl of 96% ethanol, with subsequent incubation at room temperature for 5 min. Finally, the DNA-and-bead complex was washed twice with 500 μl of 70% ethanol, with the magnet used between each washing. To remove residual ethanol, the complex was dried at 65°C for 5 min. The complete bead-and-DNA complex was then used in the amplification reactions.
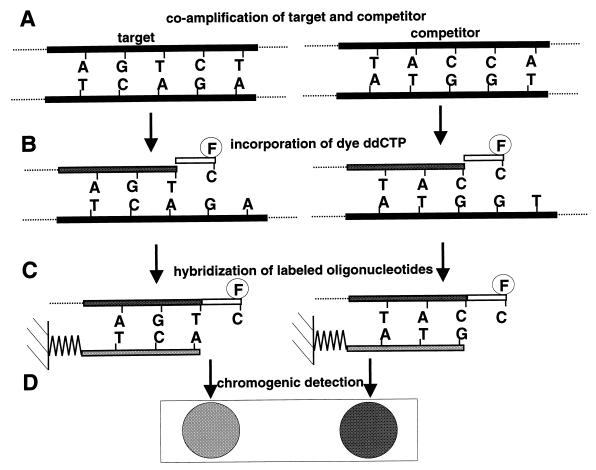
Competitive PCR (Fig. 1A).
FIG. 1.
Schematic representation of the quantitative labeling assay. (A) A known concentration of competitor DNA was added to the purified target and coamplified with the same primer pair. (B) Two oligonucleotides, one complementary to an internal segment of the competitor and one complementary to an internal segment of the target, were sequence specifically extended by a fluorescein-labeled dideoxycytosine by thermocycling. (C) The labeled primers were then hybridized to their immobilized complements. (D) A chromogenic detection of the label was performed, and the relative signal intensities were determined.
For selective amplification of genomic DNA from Microcystis, we used the 16S rDNA primers 5′-AGCCAAGTCTGCCGTCAAATCA-3′ (CH) and 5′-ACCGCTACACTGGGAATTCCTG-3′ (CI) developed by Rudi et al. (16). The competitor 5′-AGCCAAGTCTGCCGTCAAATCAAGCTGCCTCACTGCGGAGCTCGGACCAGGAATTCCCAGTGTAGCGGT-3′ is an oligonucleotide with sequences complementary to those of the PCR primers CH and CI and to that of the primer DK (see below) used in the cyclic labeling reaction. Amplification reactions with the GeneAmp 2400 PCR thermocycler (Perkin-Elmer, Norwalk, Conn.) contained 10 pmol of primers, 6 × 10−9 pmol of competitor, 200 μM (each) deoxynucleotide triphosphate, 10 mM Tris-HCl (pH 8.8), 1.5 mM MgCl2, 50 mM KCl, 0.1% Triton X-100, 1 U of DynaZyme DNA polymerase (Finnzymes Oy, Espoo, Finland), and purified DNA in a final volume of 50 μl. Prior to amplification, the DNA was denatured for 4 min at 94°C, and after amplification, an extension step for 7 min at 72°C was included. The cycling was done for 40 cycles with the following parameters: 94°C for 30 s, 58°C for 30 s, and 72°C for 30 s.
Cyclic labeling (Fig. 1B).
Five microliters of the PCR products from the competitive reaction was used in the cyclic labeling reaction. The deoxynucleotide triphosphates were dephosphorylated by the addition of 100 nmol of Tris-HCl (pH 8.0), 50 nmol of MgCl2, and 1 U of shrimp alkaline phosphatase (U.S. Biochemicals, Cleveland, Ohio), with subsequent incubation at 37°C for 1 h. Finally, the phosphatase was inactivated by heating at 96°C for 10 min.
The cyclic labeling reactions were carried out in 20-μl volumes containing 3 pmol of primer 5′-GTCCGAGCTCCGCAGTGAGGCAG-3′ (DK) complementary to the competitor, 3 pmol of primer 5′-TCTGCCAGTTTCCACCGCCTTTAGGT-3′ (DB) complementary to the Microcystis amplicon, 10 pmol of dideoxyATP, 10 pmol of dideoxyGTP, 10 pmol of dideoxyTTP (Boehringer GmbH, Mannheim, Germany), 7 pmol of fluorescein-12-dideoxyCTP (NEN, Boston, Mass.), 1.25 μl of Thermo Sequenase reaction buffer, 1.1 μl of enzyme dilution buffer, 0.15 μl of Thermo Sequenase (Amersham International plc, Buckinghamshire, England), and 6 μl of phosphatase-treated PCR product. The labeling was done for 25 cycles with the following parameters: 95°C for 30 s and 50°C for 4 min.
Hybridization and chromogenic detection (Fig. 1C and D).
One microliter (100 pmol/μl) of each of the primers 5′-ACCTAAAGGCGGTGGAAACTGGCAGA-3′ (DA) and 5′-CTGCCTCACTGCGGAGCTCGGAC-3′ (DJ) was spotted onto membrane strips (0.4 by 2 cm) GeneScreen (NEN) and then UV cross-linked with 5,000 J/cm2. An excess of the complementary primers was used to enable quantitative capture of the labeled probes. Primer DA is complementary to primer DB, and primer DJ is complementary to primer DK. The strips were prehybridized for 2 h at 37°C in a prehybridization solution containing 0.7× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 1× SPEP, 5× Denhardt’s solution, and 100 μg of heterologous DNA per ml (7). The products from the cyclic labeling reactions were added to 0.5 ml of hybridization solution (0.7× SSC, 1× SPEP, 1× Denhardt’s solution, 10% dextran sulfate, and 100 μg of heterologous DNA per ml) in a 2-ml centrifuge tube and denatured at 95°C for 5 min. The strips were added, and the incubation was continued with gentle inversion for 2 h at 37°C. The membrane strips were washed in 50 ml of 1× SSC–1% sodium dodecyl sulfate, then in 50 ml of 0.1× SSC–0.1% sodium dodecyl sulfate, and finally twice in 50 ml of 0.10 M Tris-HCl (pH 7.5)–0.15 M NaCl. Each washing was performed by brief vortexing at room temperature.
For antibody detection, the membrane strips were blocked with 20 ml of 0.10 M Tris-HCl (pH 7.5)–0.15 M NaCl–0.5% skimmed milk for 1 h and incubated in 10 ml of the same buffer containing 1/1,000 of antifluorescein-horseradish peroxidase conjugate (NEN) for 1 additional h. The membrane strips were washed three times by brief vortexing in 50 ml of 0.10 M Tris-HCl (pH 7.5)–0.15 M NaCl. The chromogenic reaction was done with the RENAISSANCE 4CN Plus for chromogenic detection of horseradish peroxidase for 5 min, according to the manufacturer’s recommendations (NEN).
The relative signal strengths were measured with a CCD video camera (Cohu high-performance CCD camera; San Diego, Calif.) and analyzed with Gel-Pro ANALYZER software (Media Cybernetics, Silver Spring, Md.).
RESULTS
The present results are based on the novel detection assay developed in this study and on the combination of this detection assay and the previously reported sample preparation approach with the same solid phase for cell concentration and DNA purification (15).
Detection assay of defined samples containing purified DNA.
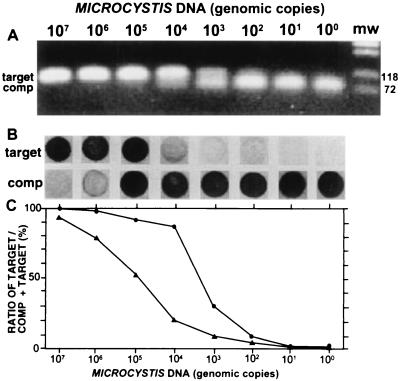
By titration experiments, the optimal amount of competitor (both for obtaining low detection limits and for reproducible amplifications) was determined to be 6 × 10−9 pmol (i.e., 3,600 molecules) per sample test (results not shown). Accordingly, 6 × 10−9 pmol of competitor was used in the testing of the assay of purified DNA from M. aeruginosa NIVA-CYA 43. Dilution series of Microcystis DNA from approximately 107 to 100 genomic copies (assuming a genome size of 5 ± 3 Mb [9]) were used in both the competitive PCR assay (Fig. 2A) and the subsequent labeling assay (Fig. 2B).
FIG. 2.
(A) Competitive PCR with dilution series of DNA isolated from M. aeruginosa NIVA-CYA 43; (B) labeling assay; (C) intensities of the target signals relative to the total signal intensities. The assays were performed with dilution series of purified DNA. The amount of DNA is given as genomic copies (assuming a genomic copy weight of 5 fg). (A) Ten microliters of the products from the competitive PCR was loaded in each lane on a 1.5% agarose gel (containing 30 μg of ethidium bromide per ml) and electrophoresed with 1× TBE at 100 V for 1 h. The products were visualized by UV transillumination. (B) The labeling assay was done as described in Materials and Methods. (C) Signal intensities (measured as the difference in the average pixel value on an 8-bit grayscale, between the signal and the background) for the target in panel A measured relative to the total signal intensities of both the target and the competitor (•). Respective values for the labeling assay in panel B are also shown (▴). Pictures were taken with a Cohu high-performance CCD camera and printed on a digital color printer (Mavigraph UP-D1500CNE; Sony, Tokyo, Japan).
Measurements of the ratio between target and competitor products, as determined by agarose gel electrophoresis, gave a quantitative range from 105 to 102 genomic copies (Fig. 2C). In contrast, the labeling assay gave a quantitative range from more than 107 to as few as 102 genomic copies. This dynamic range represents an increase of approximately 100-fold compared to that obtained by agarose gel electrophoresis detection assays (Fig. 2C). As few as 10 copies could be detected for the labeling assay by increasing the incubation time of the chromogenic detection reaction from 5 to 30 min. These detections, however, could not be done quantitatively with the detection systems used in this study, due to color density saturation for the competitor spot.
Effect of number of cycles in the labeling reaction on quantitative range.
The cyclic noncompetitive labeling reaction increased the quantitative range of the assay compared to that obtained by direct detection of the amplified DNA. For competitive PCR, with a logarithmic scale for the target concentration, the ratio of competitor and target signals resulted in a sigmoid curve with a relatively narrow quantitative range (Fig. 2C). A sigmoid curve was also obtained by performing the cyclic labeling assay with a few labeling cycles (data not shown). However, an increase in the number of labeling cycles resulted in label saturation of the competitor or the target oligonucleotides (all the probes were labeled) at each of the dilution series endpoints, leading to a curve with a wider quantitative range (Fig. 2C). Further increasing the cycle number resulted in a curve which was flatter at the middle because of label saturation of both oligonucleotides at this location (results not shown).
Quantification of Microcystis in water samples.
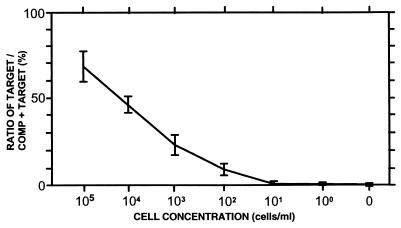
The complete quantitative assay, which includes the solid-phase cell concentration and DNA purification method, was carried out with a dilution series (105 to 100 cells/ml) of M. aeruginosa NIVA-CYA 43 and 228/1. Planktothrix agardhii NIVA-CYA 29 (filamentous) and Anabaena lemmermanii NIVA-CYA 83/1 (filamentous and heterocyst forming) were used as controls for reaction specificity. Microcystis cultures were diluted in pure water, water containing 105 cells of P. agardhii NIVA-CYA 29 per ml, water containing 105 cells of A. lemmermannii NIVA-CYA 83/1 per ml, and water sampled from Lake Akersvatnet.
There were no significant differences in either the specificity or the sensitivity of the assay for the different conditions tested. With 6 × 10−9 pmol of competitor, we obtained a quantitative range from more than 105 to as few as 102 cells/ml in all cases (results for M. aeruginosa NIVA-CYA 43 are shown in Fig. 3). By lengthening the incubation time for the chromogenic detection reaction from 5 to 30 min, we could detect as few as 10 cells/ml, but the determinations for the labeling assay with purified samples (see results above) were not quantitative. The detection curve for the complete detection assay (including solid-phase cell concentration and DNA purification) has about the same slope as the curve obtained for the dilution series of purified DNA (compare Fig. 2C and 3), indicating that method of cell concentration and DNA purification (sample preparation) is not affected by sample composition. Furthermore, these results show that the solid-phase cell concentration and DNA purification method can be used for quantitative sample preparations directly from water samples.
FIG. 3.
Complete assay with 6 × 10−9 pmol of competitor on dilution series of M. aeruginosa NIVA-CYA 43 in different aqueous environments. Cells were diluted in sterile water, water containing A. lemmermannii NIVA-CYA 83/1 (105 cells/ml), water containing P. agardhii NIVA-CYA 29 (105 cells/ml), and water from Lake Akersvatnet (sampled 30 May 1996). The percentages of the signal intensities for the target spots relative to the total signal intensities are shown with mean values for all the experiments. Error bars indicate the standard deviations, with 3 degrees of freedom for the combined analyses (the variance in the replication of each separate experimental condition was also in the same range). The complete assay, including solid-phase cell concentration and DNA purification, was performed as described in Materials and Methods.
The detection limit was dependent on the amount of competitor used, with a lower limit of 6 × 10−9 pmol of competitor. Increasing the amount of competitor 10-fold (6 × 10−8 pmol) also resulted in an increase in detection limit of about 10-fold (Fig. 4). However, lowering the amount of competitor to 6 × 10−10 pmol gave irreproducible results (data not shown). Thus, we conclude that 6 × 10−9 pmol was the optimal amount of competitor for obtaining both low detection limits and reproducible detections for the complete assay with water samples.
FIG. 4.
Complete assay with 10-fold-increased concentration of competitor (6 × 10−8 pmol) on dilution series of M. aeruginosa NIVA-CYA 43 in water from Lake Akersvatnet (samples 30 May 1996). (A) The complete assay, which includes the solid-phase cell concentration and DNA purification method, was performed as described in Materials and Methods. (B) Percentages of the signal intensities for the target spots relative to the total signal intensities. Pictures were taken with a Cohu high-performance CCD camera and printed on a digital color printer (Mavigraph UP-D1500CNE).
DISCUSSION
Our goal in this study was to develop a general method for sample preparation and then to employ a highly specific detection assay for quantifying the organisms of interest.
An assay consisting of a general sample preparation step and two specific detection steps.
The sample preparation did not discriminate between different species of the organisms tested. Thus, samples could be prepared from several different organisms without modification of the protocol (15). The specificity of the assay was obtained in the subsequent PCR amplification step and by the labeling of the oligonucleotide probes. Selective amplification of targets in a background of homologous nontargets can be difficult to achieve. We have used two specific steps in our detection assay to avoid this problem. First, the target was coamplified with the competitor with specific PCR primers. A low level of nontarget molecules may have been amplified in this reaction. However, the nontarget amplicons were excluded in the second step by selective labeling of the oligonucleotide probes based on signature sequences in the target and competitor amplicons. Finally, quantification was based on the signal ratios between the target and the competitor probes alone (independent of the nontarget molecules amplified in the first reaction). In our example, the two specific steps gave a high specificity in the detection reactions, e.g., as few as 100 cells of M. aeruginosa per ml could be detected and quantified in a background of 105 cells of other cyanobacteria per ml (Fig. 3).
Comparison of the complete assay developed in this work with standard quantitative assays.
The cell concentration step from environmental samples commonly involves specific antibody capture, centrifugation, or filtration. Then the DNA is purified by additional protocols. The major advantages of the combined solid-phase cell concentration and DNA purification method are the integration of these two steps and the simplicity of the method (15).
There are three main strategies for quantification of amplified DNA: size separation by electrophoresis, hybridization to capture probes, and real-time detection. The problems inherent in the gel electrophoresis method are detection of multiple targets in a single reaction and interpretation of the results. Size separation detection of multiplex amplifications is also difficult to achieve because the amplification ratios of amplicons with different sizes are dependent on DNA quality (5). In the present system, the different amplicons can have equal sizes, enabling more-accurate detections despite variable DNA quality. Furthermore, a single competitive reaction may be used for multiplex quantification, as discussed below.
The capture probe assay (11) is based on hybridization of the entire amplified fragments. Evidently, this assay is not suitable for separation and quantification of homologous amplicons, e.g., products of competitive amplifications. The different amplicons will form sandwich hybridizations at the homologous sites, leading to the capture of both target and nontarget fragments, even if the capture site is discriminating. Our detection assay, on the other hand, is based on the hybridization of labeled oligonucleotides and, as demonstrated in this work, is suitable for separation and quantification of homologous amplicons.
The ABI PRISM 7700 sequence detection system (Perkin-Elmer) provides real-time quantitative PCR amplification (10). According to the manufacturer, this system is accurate and fast and has a good dynamic range. However, multiplex assays are limited by the number of fluorochromes available and their overlapping fluorescent spectra. In the system described in this study, the hybridization step enables quantitative determinations of several targets by a single assay.
Complete assay for quantification of cyanobacteria in water.
Cyanobacteria belonging to the genus Microcystis can produce several different types of toxins, with the hepatotoxic microcystins being the most potent (4, 21). This toxin causes acute poisoning by liver damage and can promote carcinogenic tumors with long-term exposure to low doses (6). Thus, a continuous monitoring system to screen for the presence of the organisms producing this toxin is important.
Health authorities in Australia (New South Wales Blue-Green Algae Task Force, 1992) have already adopted a three-level alert system, as follows, based on cyanobacterial cell counts in water. At level 1 (500 to 2,000 cells/ml) water authorities are alerted, and water sampling for monitoring is increased. At level 2 (2,000 to 15,000 cells/ml), toxicity testing is carried out. At level 3, (over 15,000 cells/ml) water may be declared unsafe for human consumption if activated carbon is not available (3). With a detection limit for Microcystis of 100 cells/ml and a quantitative range of more than 3 orders of magnitude, our system seems suited for monitoring low Microcystis concentrations and for detection of potential toxic water blooms in drinking water. However, although we have tested several different conditions in this work, the versatility of the method has to be verified empirically by systematic screenings of natural water. Thus, we are currently developing a high-throughput automated system suitable for this purpose.
Development of multiplex assays.
The general sample preparation with the solid-phase cell concentration and DNA purification method, combined with the specificity in the detection method, makes the complete approach promising for multiplex determinations. By competitive PCR, multiple targets may be quantified with, for example, universal 16S rDNA primers in the competitive reaction. Then, different oligonucleotide probes can be labeled based on signature sequences for distinct bacterial groups, and finally, the ratios of the competitor signal can be compared to those for each bacterial group. A large number of simultaneous detections can also be achieved by hybridization of the labeled probes to high-density oligonucleotide arrays immobilized on glass chips, with subsequent direct detection of the fluorescein label (19). For practical purposes, multiplex quantitative assays are important both for environmental monitoring and for detection of toxic or pathogenic bacteria.
ACKNOWLEDGMENTS
This work has been supported by a grant from the Norwegian Research Council (NFR) to K.S.J. (grant no. 107622/420).
We especially thank Randi Skulberg for excellent work on preparing and cultivating the cyanobacterial species used in this work. Furthermore, we thank John E. Stacy and Heidi Rudi for critical reading of the manuscript.
REFERENCES
- 1.Bej A K, Mahbubani M H. Applications of the polymerase chain reaction (PCR) in vitro DNA-amplification method in environmental microbiology. In: Griffin H G, Griffin A M, editors. PCR technology: current innovations. Boca Raton, Fla: CRC Press, Inc.; 1994. pp. 327–339. [Google Scholar]
- 2.Bowman J P, Sayler G S. Nucleic acid techniques in the environmental detection of microorganisms and their activities. In: Picup R W, Saunders J R, editors. Molecular approaches to environmental microbiology. Chichester, United Kingdom: Ellis Horwood Limited; 1996. pp. 63–97. [Google Scholar]
- 3.Carmichael W W. Cyanobacterial toxins. In: Hallegraeff G M, Anderson D M, Cembella A D, editors. Manual on harmful marine microalgae. Paris, France: UNESCO; 1995. pp. 163–175. [Google Scholar]
- 4.Codd G A. Cyanobacterial toxins: occurrence, properties and biological significance. Water Sci Technol. 1995;32:149–156. [Google Scholar]
- 5.Deng G, Yu M, Smith H S. An improved method of competitive PCR for quantitation of gene copy number. Nucleic Acids Res. 1993;21:4848–4849. doi: 10.1093/nar/21.20.4848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Falconer, I. R. 1996. Potential impact on human health of toxic cyanobacteria. Phycologia 35(Suppl. 6):6–11.
- 7.Galau G A, Huges D W, Dure L., III Abscisic acid induction of cloned cotton late embryogenesis-aboundant (Lea) mRNAs. Plant Mol Biol. 1986;7:155–170. doi: 10.1007/BF00021327. [DOI] [PubMed] [Google Scholar]
- 8.Goldsborough A S, Kornberg T B. Allele-specific quantification of Drosophila engrailed and invected transcripts. Proc Natl Acad Sci USA. 1994;91:12696–12700. doi: 10.1073/pnas.91.26.12696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herdman M. Evolution and genetic properties of cyanobacterial genomes. In: Carr N G, Whitton B A, editors. The biology of cyanobacteria. Oxford, United Kingdom: Blackwell Scientific Publications; 1982. pp. 263–305. [Google Scholar]
- 10.Lee L G, Connell C R, Bloch W. Allelic discrimination by nic-translation PCR with fluorogenic probes. Nucleic Acids Res. 1993;21:3761–3766. doi: 10.1093/nar/21.16.3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mansy F, Hoyois B, DeVos M J, VanElsen A, Bollen A, Godfroid E. Colorimetric solid-phase capture hybridization assay for detection of amplified Borrelia burgdorferi. BioTechniques. 1996;21:122–125. doi: 10.2144/96211rr03. [DOI] [PubMed] [Google Scholar]
- 12.Möller A, Jansson J K. Quantification of genetically tagged cyanobacteria in Baltic Sea sediment by competitive PCR. BioTechniques. 1997;22:512–518. doi: 10.2144/97223rr02. [DOI] [PubMed] [Google Scholar]
- 13.Pastinen T, Partanen J, Syvänen A C. Multiplex, fluorescent solid-phase minisequencing for efficient screening of DNA sequence variation. Clin Chem. 1996;42:1391–1397. [PubMed] [Google Scholar]
- 14.Rudi K, Kroken M, Dahlberg O J, Deggerdal A, Jakobsen K S, Larsen F. Rapid, universal method to isolate PCR-ready DNA using magnetic beads. BioTechniques. 1997;22:506–511. doi: 10.2144/97223rr01. [DOI] [PubMed] [Google Scholar]
- 15.Rudi K, Larsen F, Jakobsen K S. Detection of toxin-producing cyanobacteria by use of paramagnetic beads for cell concentration and DNA purification. Appl Environ Microbiol. 1998;64:34–37. doi: 10.1128/aem.64.1.34-37.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rudi K, Skulberg O M, Larsen F, Jakobsen K S. Strain characterization and classification of oxyphotobacteria in clone cultures on the basis of 16S rRNA sequences from the variable regions V6, V7, and V8. Appl Environ Microbiol. 1997;63:2593–2599. doi: 10.1128/aem.63.7.2593-2599.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saiki R K, Scharf S, Faloona F, Mullis K B, Horn G T, Erlich H A, Arnheim N. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science. 1985;230:1350–1354. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- 18.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schena M, Shalon D, Davis R W, Brown P O. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1996;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 20.Siebert P D, Larrick J W. Competitive PCR. Nature. 1992;359:557–558. doi: 10.1038/359557a0. [DOI] [PubMed] [Google Scholar]
- 21.Sivonen, K. 1996. Cyanobacterial toxins and toxin production. Phycologia 35(Suppl. 6):12–24.
- 22.Skulberg R, Skulberg O M. Forskning med algekulturer.—NIVAs kultursamling av alger. [Research with algal cultures.—NIVA’s Culture collection of algae.] Oslo, Norway: Norsk Institutt for Vannforskning; 1990. [Google Scholar]
- 23.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 24.Steffan R J, Atlas R M. Polymerase chain reaction: applications in environmental microbiology. Annu Rev Microbiol. 1991;45:137–161. doi: 10.1146/annurev.mi.45.100191.001033. [DOI] [PubMed] [Google Scholar]
- 25.Syvänen A C, Aalto-Setälä K, Harju L, Kontula K, Söderlund H. A primer-guided nucleotide incorporation assay in the genotyping of apolipoprotein E. Genomics. 1990;8:684–692. doi: 10.1016/0888-7543(90)90255-s. [DOI] [PubMed] [Google Scholar]
- 26.Utkilen, H., O. M. Skulberg, B. Underdal, N. Gjølme, R. Skulberg, and J. Kotai. 1996. The rise and fall of a toxic population of Microcystis aeruginosa (Cyanophyceae/Cyanobacteria)—a decade of observations in Lake Akersvatnet, Norway. Phycologia 35(Suppl. 6):189–197.