Abstract
The accumulated knowledge about the structure of protein-DNA complexes allowed us to understand the mechanisms of protein-DNA recognition and searching for a specific site on DNA. Obviously, the mechanism of specific DNA recognition by a protein must satisfy two requirements. First, the probability of incorrect binding should be very small. Second, the time to find the “correct” binding site should not be too long. If we assume that protein recognition of a precise site on DNA occurs at some distance from DNA and calculate global minima, we can avoid local minima at short distances. The only long-range interaction is the interaction of charges. The location of charges on DNA in three-dimensional space depends on the local conformation of DNA and thus reflects the DNA sequence and sets the spatial pattern for recognition. Various factors such as counter ion concentration, ionic strength, and pH can affect protein recognition of DNA. Nowadays, the theory of long-range interactions makes it possible to calculate the best mutual spatial arrangement of protein and DNA molecules by charged groups and avoid misplaced binding.
Keywords: Protein-DNA recognition, Electrostatic potential, Long-range interactions, Specific site, Protein sliding on DNA
Introduction
DNA–protein interactions play the key role in the processing of genetic information, such as replication, translation, repair, recombination, and so on. Conventionally, all DNA-binding proteins are classified into three types—specific, recognizing only one DNA sequence, multispecific, recognizing a pattern or set of patterns, and nonspecific, interacting with DNA regardless of sequence. This diversity of specificity is consistent with the functions of proteins, requiring varying degrees of sequence selectivity in DNA recognition. For example, transcription factors or restriction enzymes typically exhibit high selectivity, and DNA replication or packaging proteins can bind each DNA nucleotide sequence.
Specific proteins, such as the DNA-binding domains of transcription factor proteins, determine the primary specificity of the interaction, i.e., the affinity of binding by a particular protein to a particular oligonucleotide (Luscombe et al. 2001; Rohs et al. 2010) and the core pattern of DNA-binding sites. Local features of the three-dimensional structure of macromolecules and their direct consequences, such as the optimal orientation of hydrogen bonds between protein amino acids and nucleotides or the geometric parameters of DNA grooves, are reflected in the preferred sequences of binding sites (Oshchepkov et al. 2004). That is, the degree of similarity of different binding sites directly (direct contact between DNA and protein) or indirectly (physical properties of the local DNA site) reflects the protein’s preference for the recognized DNA site (Stormo 2013) and determines the pattern recognized by the protein in regulatory sequences.
Multispecific proteins recognize a pattern or set of DNA patterns. In general, for both specific and multispecific proteins, position-weighted matrices (PWMs) have proven to be a simple and convenient tool for creating a basic motif model. PWMs are based on the idea of independence of neighboring nucleotides, in terms of both their probability of being in functional sites and their contribution to the protein-DNA interaction energy. The PWM not only describes a set of degenerate substring binding sites, but also correlates with promoter activity in E. coli (Mulligan et al. 1984) and allows quantification of DNA–protein interaction energies. Key works (Berg and von Hippel 1987, 1988) provided PWM with a biophysical foundation: using statistical mechanics methods, the authors showed that PWM estimation is proportional to the affinity of the sites.
Analysis of known binding sites also shows the presence of correlated positions (Tomovic and Oakeley 2007), but the reliability of correlated observations has long been unobvious for small sets of a few tens of sequences. In fact, it is the small amount of data that has long limited the application and spreading of extended models that take into account the physicochemical properties of DNA (Oshchepkov et al. 2004) and remote correlations (Levitskiĭ et al. 2006). Interestingly, distant correlated contacts are markedly less common but are also possible (Jen-Jacobson 1997).
In fact, the binding abilities of each protein on different DNA sequences form a continuum between specific and nonspecific DNA binding abilities. In the case of wide sequence specificity, substitutions of one or more base pairs in the optimal sequence have only a minor effect on affinity binding. In the case of narrow sequence specificity, the replacement of a single base pair leads to a significant decrease in binding affinity. In addition, natural regulatory elements often contain suboptimal recognition sequences, which makes it possible to regulate gene expression over a very wide range, ‘switching’ a particular gene when the concentration of a transcription factor changes.
Specific DNA-binding proteins can differ in their ability to recognize and bind to specific DNA sequences and non-specific sites (selectivity coefficient) by more than 100-fold. There is also no relationship between DNA-binding affinity and sequence selectivity. Interactions in specific DNA–protein complex can be weak, and interactions in nonspecific DNA–protein complex can be strong. However, there are no selective and non-selective interactions.
From a physical point of view, the molecular interactions occurring in the protein-DNA binding region in these three types of complexes are the same. There are only a few types of interactions: π-π interactions (stacking) of nucleic bases with each other and with aromatic amino acid residues, electrostatic interactions between charged groups, protein-DNA hydrogen bonds and hydrogen bonds mediated by bound water, hydrophobic interactions, and van der Waals forces.
Stacking
Stacking interactions are usually mentioned when considering interactions within DNA. However, such interactions have been found in a number of DNA–protein complexes when the electron fields of nucleic bases interact with the electron fields aromatic amino acid residues. Typically, these complexes involve the opening of the DNA double strand and the eversion of one or more bases. In contrast to stacking in DNA, which makes more than half of the stability of the double helix (Yakovchuk et al. 2006), in the structure of the protein-DNA complex, stacking interactions appear to make a minimal contribution to stability.
Hydrogen bonds
Proteins and DNA are saturated with numerous functional groups containing hydrogen bond donors and/or acceptors. The backbone of the polypeptide chain, most amino acid side radicals in proteins, phosphate, sugar groups, and nucleic acid bases can form dense networks of hydrogen bonds. Hydrogen bond networks are highly cooperative because the length and geometry of the hydrogen bond are limited, and rearrangement of a single bond entails a cascade of rearrangements. In addition, water molecules at the DNA–protein interface provide an additional contribution.
Biologically active compounds of different types can bind to DNA using different interacting patterns. However, there may also be “universal” sets of interaction centers used by ligands of the same nature. Gursky et al. (1976; Livshitz et al. 1979) formulated the principles for peptides and antibiotics containing peptide or amide groups to be recognized for certain DNA sequences.
The key feature of such recognition is the formation of hydrogen bonds between the “donors” of the hydrogen bond, the amide groups of the ligand, and the “acceptors” of this bond, the N3 atoms of adenine or the O2 atoms of thymine and cytosine of the DNA molecule. These thymine, adenine, and cytosine atoms occupy positions in the canonical B-form of the DNA molecule that are linked by helical symmetry (translation by 3.4 angstroms along the helix axis and rotation by 36° translates them into each other). These atoms form a regular lattice of interaction centers—hydrogen bond acceptors. Considering only these key atoms for binding, we can represent a double-helical DNA molecule as a lattice of interaction centers. Such a lattice is “double”; i.e., it has two parallel linear chains of interaction centers, and if a section of DNA contains only AT pairs, such atoms, as hydrogen bond acceptors, will be equivalent. Mikhail Livshits and George Gursky et al. demonstrated that this binding scheme is able to predict correct specific binding site among a random sequence of DNA nucleotide pairs (Livshitz et al. 1979).
There are three points of view on the hydrogen bonds role in protein-DNA recognition. First, that network of hydrogen bonds between protein atoms and DNA atoms makes a great impact in the total free energy change and responsible for specific sequence recognition. The second view is that the hydrogen bonding network is cooperative and can adopt different topologies with a small change in energy between different states. So, hydrogen bonds’ networks cannot be specific. The third point of view is that both specific and non-selective DNA bindings occur due to hydrogen bonds (Kerppola 2001). The geometry of the hydrogen bond, including the distance between the donor and acceptor, affects the strength of the hydrogen bond. It is the hydrogen bonds within DNA that can determine local structural differences in the conformation or flexibility of DNA. Thus, the nucleotide sequence affects both the local equilibrium conformation and the mobility of the DNA helix. Hydrogen bonds within DNA contribute to the sequence dependence of DNA binding. In addition, hydrogen bonding networks including nucleotide bases, deoxyriboses, and phosphates can increase the energetic contribution of hydrogen bonds to selective DNA binding.
Another point of view is that hydrogen bond donors and acceptors are widespread on the protein surface, and hydrogen bond network only fixes the complex without participating in the searching for the exact site. So, hydrogen bonds contribute to binding affinity. In favor of this version is the fact that hydrogen bonds are highly energetic, about 5–15 kcal/mol per bond, and breaking the network of hydrogen bonds requires considerable effort.
Hydrogen bonds make a significant contribution to both enthalpy and entropy changes. The difference between the enthalpy of the hydrogen bonds formed and the enthalpy of the hydrogen bonds that are broken during the formation of the complex accounts for the enthalpy contribution. Restrictions of the degrees of freedom of protein and DNA atoms, as well as the binding or release of water molecules, determine the entropy contribution. The enthalpy and entropy contributions are different for different protein-DNA complexes (Kerppola 2001).
Hydrophobic effect
Hydrophobic atoms are unable to form hydrogen bonds with water or other molecules. A rigid “clathrate” network of hydrogen bonds is formed around hydrophobic surfaces. Such water molecules in such a grid have a smaller number of hydrogen bonds per molecule. In the process of complex formation, hydrophobic surfaces converge and water molecules of the “clathrate” network are released into the bulk solution, which leads to an increase in entropy. The released water molecules form more hydrogen bonds, and van der Waals interactions occur between the hydrophobic surface atoms, which reduces enthalpy. The total free energy gain favors the convergence of hydrophobic groups and is called the hydrophobic effect.
In addition to changes in free energy, most specific protein-DNA complexes are characterized by large negative changes in heat capacity. Heat capacity is the dependence of the enthalpy of a system on temperature. Part of the change in heat capacity is due to a decrease in the hydrophobic surface area available to the solvent. Also, apparently, the reduction of vibrational and rotational degrees of freedom of water molecules and amino acid side radicals at the protein-DNA interface contributes to the change in heat capacity (Ladbury et al. 1994). Protein-DNA complex formation is thermodynamically similar to protein folding, since it is accompanied by a significant increase in entropy and decrease in heat capacity (Kerppola 2001). Theoretically, complementary patterns of hydrophobic patches on the surfaces of molecules may contribute to selective recognition of DNA sequences.
Van der Waals forces
Close proximity of atoms of any type, including uncharged atoms, causes correlation between the permanent, induced, and instantaneous dipole moments of their atoms, which leads to the van der Waals attraction force. Over short distances, if the atoms are too close together, their electron clouds will overlap, causing a strong repulsion. So, it is necessary not only to make favorable contacts but also to avoid unfavorable ones. In fact, such a requirement corresponds to the steric complementarity of the surface shape of interacting macromolecules. The total free energy change upon DNA binding depends on the balance between attraction and repulsion (Jen-Jacobson 1997). The van der Waals forces between proteins and DNA act over a small distance between atoms, make a relatively small contribution to enthalpy, and depend weakly on the types of interacting atoms, so the contribution is proportional to the area of interaction. Van der Waals forces affect DNA binding affinity, not selectivity.
Electrostatic interactions
Electrostatic interactions include attraction of charges of different signs and repulsion of charges of the same sign, as well as dipole interactions. The strength of electrostatic interactions is proportional to the product of the absolute values of the charges and inversely proportional to the square of the distance between charged groups, as well as to the dielectric constant of the medium. It is generally believed that electrostatic interactions make the main contribution to the nonspecific DNA-binding capacity of proteins. However, the position of phosphates in space may depend on the local equilibrium conformation of the DNA structure, and hence on the base sequence (Ramirez-Carrozzi and Kerppola 2001). Charged protein residues can affect the structure of DNA, and vice versa. Positively charged groups can bend DNA toward the protein and negatively charged groups can bend DNA away from the protein (Ramirez-Carrozzi and Kerppola 2001). DNA structure is sensitive to the presence of counterions near phosphate groups. Removal of a counterion from a single phosphate can distort the DNA structure. Thus, electrostatic interactions explain the interdependence between protein binding and DNA structure.
In addition to the charge interactions proper, there are interactions of the next order of magnitude—dipole and quadrupole moments, which are responsible for the orientation of the protein in the electromagnetic field. Net charge, net dipole moment, and quadrupole moment in hybrid predictors could distinguish binding and non-binding proteins with more than 80% accuracy (Ahmad and Sarai 2004).
Protein-DNA binding affinity
The affinity of protein-DNA complex is difference in the free energy of the components in the solvent separately and of the complex together. The change in enthalpy and entropy is a result of differences in intermolecular and intramolecular protein-DNA interactions. These differences change Gibbs free energy and the binding constant as a function of temperature. Enthalpy and entropy changes during DNA binding can have opposite effects on the free energy of complex formation. Some protein-DNA complexes is enthalpy-driven, whereas the most complexes is entropy-driven. Thus, protein-DNA complexes differ both in the specific set of molecular interactions and in the thermodynamic consequences of these interactions.
At present, an enormous amount of data has been accumulated on the structures of protein-DNA complexes, on the patterns of molecular interactions in individual families, and on the influence of amino acid and nucleotide substitutions on the affinity of the complex. Based on this data, the mechanism of protein-DNA recognition has become better understood. In this review, I would like to highlight such works and the proposed recognition mechanisms.
Specific interactions require nonspecific ones
Many DNA-binding proteins probably form preliminary nonspecific complexes with DNA during recognition, because nonspecific binding to DNA may facilitate the search for specific DNA recognition sites. Most likely, such nonspecific binding has a very low binding constant and it is very difficult to experimentally confirm the existence of nonspecific complexes, but there is indirect evidence in favor of this hypothesis. For example, some proteins can move rapidly between neighboring nonspecific binding sites and thus slide along the DNA helix. This sliding process, where some proteins slide along the DNA searching for a specific site and skip the landing site about 40 times, is described in Wunderlich and Mirny (2008). In addition, the protein is shown to rotate around the DNA during sliding. The average energy barrier for sliding is 1.1 ± 0.2 kT (where k is Boltzmann’s constant and T is temperature in degrees Kelvin). For comparison, the average kinetic energy of thermal motion of molecules is 1.5 kT. Consequently, the thermal motion of proteins is sufficient to overcome the energy barrier between different nonspecific sites on DNA, allowing the protein to glide and quickly find target binding sites (Blainey et al. 2009). This allows the protein to scan more binding sites at one time than if the protein had to dissociate from the DNA before moving to a new site.
Influence of physicochemical factors on recognition accuracy
Many factors affect the accuracy of DNA site recognition. For example, when DNA is recognized and cut by some specific enzymes under abnormal conditions, the cut may occur at sites other than canonical sites. This enzyme activity was first observed in a study of the substrate specificity of EcoRI restriction endonuclease (Polisky et al. 1975) and was termed “star” activity. EcoRI endonuclease can cut different from the canonical GAATTC by one substitution sites. Among the conditions that cause such non-standard enzyme activity are high pH values (> 8.0), low ionic strength, glycerol concentration > 5%, high enzyme concentration (> 100 U/ug of DNA), and the presence of organic solvents (ethanol, DMSO, etc.). However, the efficiency of restriction enzymes can also be influenced by the sequence context. For example, the efficiency of restriction enzymes to cut a recognized sequence located at different sites can differ by 10–50-fold. This appears to be due to the influence of sequences surrounding the restriction site, and such influence can enhance or generally block enzyme binding or activity. A similar situation occurs when the recognized sites are located close to the ends of a linear DNA fragment. Most enzymes require some minimum number of residues surrounding the recognized site. Therefore, enzymes usually have specified end requirements.
Charged particles influence on the electrostatic interactions between proteins and DNA in two ways. First, positive ions are bound to DNA that partially neutralize the phosphate charge (Manning 1978). Density of bounded counterions on DNA does not depend on their concentration in solution. Counterions can be released by interactions between proteins and DNA. The binding of counterions on DNA thus increases the entropy change and decreases the enthalpy change of the protein-DNA interaction. Since the number of bound counterions is independent of their concentration, the dependence of the binding energy of protein-DNA on salt concentration can be used to estimate the number of counterions displaced from DNA during complex formation. Secondly, the presence of ions in the medium reduces the Debye charge shielding radius and exponentially reduces the strength of interaction between charged groups according to the Debye-Hückel equation. However, the charge density in DNA is so high that electrostatic interactions can have a long-range effect on the interaction between protein and DNA and direct contact between charged groups is not required (Kerppola 2001).
The dramatic difference in dielectric constants within proteins (e = 4) and in aqueous solution (e = 80) influences the electrostatic potential near proteins. So, the distribution of electrostatic potential depends on the overall shape of the protein (Honig and Nicholls 1995). Similarly, the distribution of potential on DNA depends on the shape of the molecule. This results in a high electrostatic potential in the narrow gaps of the protein. The binding site on the surface of DNA-binding proteins is usually positively charged. The different sign of the charge on the protein and DNA allows the protein to unfold to the DNA. In addition, the dipole moment of DNA-binding proteins also orientates the protein relative to the DNA (Ahmad and Sarai 2004). Thus, electrostatic interactions are a major determinant of non-specific DNA binding (Kerppola 2001). How they can contribute to the recognition of specific binding sites will be discussed below.
The process of searching for binding sites on DNA is probabilistic and long-range
We believe that star activity is possible because recognition of the binding site on DNA is probabilistic, since the difference in energy magnitude from other sites is not very large. This assumption is supported by the fact that some proteins in the process of sliding along DNA often overshoot the landing site (Wunderlich and Mirny 2008). The probabilistic nature of binding site recognition is also reflected in the probabilistic nature of amino acid-nucleotide contacts—a “recognition code” governing protein-DNA interaction (Benos et al. 2002).
In general, the mechanism of specific DNA recognition by protein must satisfy two requirements. On the one hand, the probability of incorrect binding should be very small. On the other hand, the time to find the binding site should not be too long. In a sense, these two requirements contradict each other. If in order to check the “correctness” of binding it is necessary that the protein binds on a site, then before getting to the correct site, one will have to randomly try many incorrect variants. This already requires quite a lot of time, especially if we take into account that the energy of non-specific binding still significantly exceeds kT, so the time it takes for the protein to detach from DNA in case of incorrect binding is not so short.
Nevertheless, such a mechanism of DNA–protein interaction, in which both of the above requirements can be fulfilled simultaneously, was proposed in the works of Namiot V.A. Within this mechanism, the search for the “correct” binding site is carried out not at the direct contact of DNA and protein, but when there is a certain gap between them, and they interact with each other due to the so-called long-range interactions. The only interaction satisfying the requirements of long-range interactions is the interaction of charges. In Namiot’s works, the theory of recognition by means of long-range interactions was first developed for modeling protein folding (Namiot et al. 2011a, b), then for the interaction of DNA molecule sequence determination (Namiot et al. 2012, 2013), and later generalized to various interactions between biological macromolecules (Namiot et al. 2016). In fact, this theory allows us to calculate global energy minima of long-range interactions between extended molecules, considering only the positions of charges in space or approximating biological macromolecules by linear chains of charges. Interaction energy for two parallel charged lines with a distance R between them and charges distributions и , in a general form can be written as an integral of the product of two Fourier images of these distributions (Formula 1)
| 1 |
Using Formula (1), it is possible to calculate the best mutual arrangement of molecules without bringing them closer together, thus avoiding local minima occurring at short distances and molecules getting closer together avoiding misplaced positions (Fig. 1). To apply this theory, it is sufficient to calculate the electrostatic potential along DNA, for example, using DNA Electrostatic Potential Properties Database (DEPPDB) (Osypov et al. 2010, 2012), and approximate the potential along the binding site on the protein. The authors of this server have previously shown that transcription factor binding sites gravitate toward high-potential regions. Other elements of the genome, such as terminators, also exhibit interesting electrostatic features. Most intriguing are gene starts that exhibit taxonomic correlations.
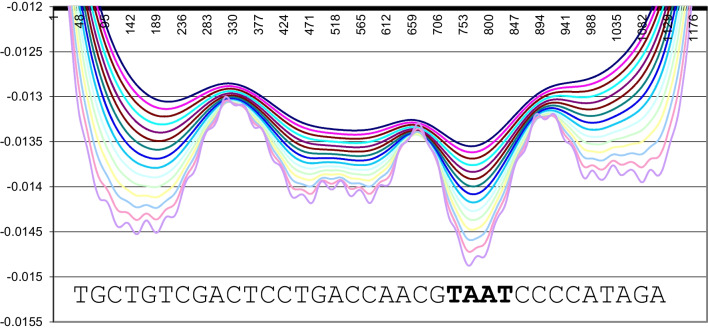
Fig. 1.
Interaction energy of bicoid protein (1zq3) and DNA (5′-TGCTGTCGACTCCTGACACCAACGTAATCCCCCCATAGAA-3′) with different distances between protein and DNA, from 4 to 60 Å with an increment 4.0 Å. Obtained by Formula (1). The nucleotide sequence of DNA is plotted on the X-axis; the energy of interaction in conventional units is plotted on the Y-axis. Figure taken from the report on RFBR grant 15–04-99605, 2017, with permission of the authors. The true site is shown in bold. Deeper lines correspond to shorter distances
In DEPPDB, the potential of DNA is calculated only on the basis of its nucleotide sequence (Osypov et al. 2012). However, it has now been established that the local conformation of DNA and its ability to bend (stiffness/flexibility) depend on the order of nucleic bases. The different overlapping area of neighboring nucleic bases leads to different stacking energy, which determines the local equilibrium conformation of nucleotides and the ability to bend a given section of DNA (El Hassan and Calladine 1996). The most charged region in the DNA structure is the phosphate groups, which form charged “rails” for protein sliding. In an ideal situation, the phosphate groups are equally spaced, with the same angle of rotation. However, a close examination reveals that this part of the structure, which is identical in all DNA nucleotides, forms an inhomogeneous pattern in space. This inhomogeneity arises precisely because of local structural inhomogeneity, creating non-uniformity in the distribution of negative charge. Thus, the different equilibrium conformation of local DNA sites leads to shifts in the position of charges of the sugar-phosphate backbone of DNA in three-dimensional space and distortion of the ideal pattern of charge arrangement. It is these distortions that create prerequisites for the possibility of recognizing specific sites on DNA, since the only field propagating over distances comparable to the size of proteins is electrostatic interactions.
If there are only positive charges in the binding region on the protein, there is no selectivity mechanism that ensures the recognition of negatively charged DNA. If the binding region contains both positive and negative charges, it becomes possible to provide selectivity through spatial complementarity of charges. Indeed, more than 80% of protein-DNA complex interfaces from PDB have negatively charged amino acids (Anashkina et al. 2008, 2018).
There remains one more type of interactions, which is not considered by any of the authors at the moment. It is about magnetic fields. A moving charge creates a magnetic field around itself, which acts on other moving charges. Consider two equally charged atoms uniformly rotating on a circle of radius r with frequency w. The magnetic field created by the moving charge depends on the plane of rotation of the charge and the direction of rotation, and the interaction of the two moving charges depends on the angle between the directions of the magnetic field created by these charges. Thus, unlike electrostatic interactions, two moving charges of the same sign can attract if they are placed in such a way that the axes of rotation of the charges lie on the same line and the magnetic field is directed in opposite directions. Protein and DNA atoms in the cell are not static; they are continuously moving due to thermal motion. Charged atoms of proteins and DNA make oscillations with various amplitude and frequency. Only the amplitude of the oscillation depends on the temperature, while the frequency is related to the geometric characteristics of the oscillating fragment. A function of enzymes is based on such thermal movements of fragments inside the protein (Hammes-Schiffer 2002). It is clear from general considerations that the magnetic interaction force of two moving point charges should be inversely proportional to the square of the distance between them, similar to the Coulomb interaction. In addition, the integral of the interaction of oscillating particles should turn to 0 for all interactions with non-matching frequency of oscillation. It would be interesting to see estimates for the magnitude of the magnetic interaction force of charged particles moving under the action of thermal fluctuations in the cell. This direction seems very promising, since magnetic interactions could explain the mechanism of attraction of proteins to certain sites in the cell through the occurrence of thermal fluctuations of charged groups with a certain frequency. Various posttranslational modifications of such charged groups can change their own frequency and regulate the interaction process.
Conclusions
The accumulated knowledge about the structure of protein-DNA complexes allowed us to understand the mechanisms of protein-DNA recognition and searching for a specific site on DNA. Obviously, the mechanism of specific DNA recognition by a protein must satisfy two requirements. First, the probability of incorrect binding should be very small. Second, the time to find the “correct” binding site should not be too long. If we assume that protein recognition of a precise site on DNA occurs at some distance from DNA and calculate global minima, we can avoid local minima occurring at short distances. There are only a few types of interactions: π-π interactions (stacking) of nucleic bases with each other and with aromatic amino acid residues, electrostatic interactions between charged groups, protein-DNA hydrogen bonds and hydrogen bonds mediated by bound water, hydrophobic interactions, and van der Waals forces. The only long-range interaction is the interaction of charges. The location of charges on DNA in three-dimensional space depends on the local conformation of DNA and thus reflects the DNA sequence and sets the spatial pattern for recognition. Various factors such as counter ion concentration, ionic strength, and pH can affect protein recognition of DNA. Nowadays, the theory of long-range interactions makes it possible to calculate the best mutual spatial arrangement of protein and DNA molecules by charged groups and avoid misplaced binding. We assume that many DNA-binding proteins probably form nonspecific preliminary complexes with DNA during recognition, because nonspecific binding to DNA may facilitate the search for specific DNA recognition sites. In the future, it would be interesting to study the contribution of thermal motion of charged groups and local magnetic fields to long-range protein-DNA interactions and recognition.
Author contribution
Anastasia A. Anashkina contributed to the study conception and design. Material preparation, data collection, and analysis were performed by Anastasia A. Anashkina. The first draft of the manuscript was written by Anastasia A. Anashkina and she commented on previous versions of the manuscript. She read and approved the final manuscript.
Data Availability
Not applicable.
Declarations
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent to publish
Not applicable.
Conflict of interest
The author declares no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Ahmad S, Sarai A. Moment-based prediction of DNA-binding proteins. J Mol Biol. 2004;341:65–71. doi: 10.1016/j.jmb.2004.05.058. [DOI] [PubMed] [Google Scholar]
- Anashkina A, Tumanyan V, Kuznetsov E, et al. Relative occurrence of amino acid-nucleotide contacts assessed by Voronoi-Delaunay tessellation of protein-DNA interfaces. Biophysics. 2008;53:199–201. doi: 10.1134/S0006350908030032. [DOI] [Google Scholar]
- Anashkina AA, Kuznetsov EN, Batyanovskii AV, et al. Protein-DNA interactions: statistical analysis of interatomic contacts in the major and minor grooves. Vavilov J Genet Breed. 2018;21:887–894. doi: 10.18699/VJ17.309. [DOI] [Google Scholar]
- Benos PV, Lapedes AS, Stormo GD. Is there a code for protein-DNA recognition? Probab(ilistical)ly. BioEssays News Rev Mol Cell Dev Biol. 2002;24:466–475. doi: 10.1002/bies.10073. [DOI] [PubMed] [Google Scholar]
- Berg OG, von Hippel PH. Selection of DNA binding sites by regulatory proteins: statistical-mechanical theory and application to operators and promoters. J Mol Biol. 1987;193:723–743. doi: 10.1016/0022-2836(87)90354-8. [DOI] [PubMed] [Google Scholar]
- Berg OG, von Hippel PH. Selection of DNA binding sites by regulatory proteins. Trends Biochem Sci. 1988;13:207–211. doi: 10.1016/0968-0004(88)90085-0. [DOI] [PubMed] [Google Scholar]
- Blainey PC, Luo G, Kou SC, et al. Nonspecifically bound proteins spin while diffusing along DNA. Nat Struct Mol Biol. 2009;16:1224–1229. doi: 10.1038/nsmb.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Hassan MA, Calladine CR. Propeller-twisting of base-pairs and the conformational mobility of dinucleotide steps in DNA. J Mol Biol. 1996;259:95–103. doi: 10.1006/jmbi.1996.0304. [DOI] [PubMed] [Google Scholar]
- Gursky AV, Tumanyan VG, Zasedatelev AS, et al. A code controlling specific binding of regulatory proteins to DNA. Mol Biol Rep. 1976;2:413–425. doi: 10.1007/BF00366264. [DOI] [PubMed] [Google Scholar]
- Hammes-Schiffer S. Impact of enzyme motion on activity. Biochemistry. 2002;41:13335–13343. doi: 10.1021/bi0267137. [DOI] [PubMed] [Google Scholar]
- Honig B, Nicholls A. Classical electrostatics in biology and chemistry. Science. 1995;268:1144–1149. doi: 10.1126/science.7761829. [DOI] [PubMed] [Google Scholar]
- Jen-Jacobson L. Protein-DNA recognition complexes: conservation of structure and binding energy in the transition state. Biopolymers. 1997;44:153–180. doi: 10.1002/(SICI)1097-0282(1997)44:2<153::AID-BIP4>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Kerppola TK (2001) Protein–DNA interactions: structure and energetics. In: Encyclopedia of Life Sciences. John Wiley & Sons, Ltd. 10.1038/npg.els.0001349
- Ladbury JE, Wright JG, Sturtevant JM, Sigler PB. A thermodynamic study of the trp repressor-operator interaction. J Mol Biol. 1994;238:669–681. doi: 10.1006/jmbi.1994.1328. [DOI] [PubMed] [Google Scholar]
- Levitskiĭ VG, Ignat’eva EV, Anan’ko EA, et al. Method SiteGA for the recognition of transcription factor binding sites. Biofizika. 2006;51:633–639. [PubMed] [Google Scholar]
- Livshitz MA, Gursky GV, Zasedatelev AS, Volkenstein MV. Equilibrium and kinetic aspects of protein-DNA recognition. Nucleic Acids Res. 1979;6:2217–2236. doi: 10.1093/nar/6.6.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luscombe NM, Laskowski RA, Thornton JM. Amino acid–base interactions: a three-dimensional analysis of protein–DNA interactions at an atomic level. Nucleic Acids Res. 2001;29:2860–2874. doi: 10.1093/nar/29.13.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning GS. The molecular theory of polyelectrolyte solutions with applications to the electrostatic properties of polynucleotides. Q Rev Biophys. 1978;11:179–246. doi: 10.1017/s0033583500002031. [DOI] [PubMed] [Google Scholar]
- Mulligan ME, Hawley DK, Entriken R, McClure WR. Escherichia coli promoter sequences predict in vitro RNA polymerase selectivity. Nucleic Acids Res. 1984;12:789–800. doi: 10.1093/nar/12.1Part2.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namiot VA, Batyanovskii AV, Filatov IV, et al. General theory of the long-range interactions in protein folding. Phys Lett A. 2011;375:2911–2915. doi: 10.1016/j.physleta.2011.06.030. [DOI] [Google Scholar]
- Namiot VA, Batyanovskii AV, Filatov IV, et al. On the optimal folding of protein molecules. Biophysics. 2011;56:596–601. doi: 10.1134/S0006350911040166. [DOI] [PubMed] [Google Scholar]
- Namiot VA, Anashkina AA, Filatov IV, et al. DNA sequencing using specific long-range interaction between macromolecules. Biophysics. 2012;57:716–721. doi: 10.1134/S0006350912060115. [DOI] [PubMed] [Google Scholar]
- Namiot VA, Anashkina AA, Filatov IV, et al. Long-range macromolecule interaction and “speed reading” long nucleotide sequences in DNA. Phys Lett A. 2013;377:323–328. doi: 10.1016/j.physleta.2012.11.029. [DOI] [Google Scholar]
- Namiot VA, Batyanovskii AV, Filatov IV, et al. Long-distance interactions and principles of molecular recognition at various biosystem organization levels. Biophysics. 2016;61:47–51. doi: 10.1134/S0006350916010188. [DOI] [Google Scholar]
- Oshchepkov DY, Vityaev EE, Grigorovich DA, et al. SITECON: a tool for detecting conservative conformational and physicochemical properties in transcription factor binding site alignments and for site recognition. Nucleic Acids Res. 2004;32:W208–W212. doi: 10.1093/nar/gkh474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osypov AA, Krutinin GG, Kamzolova SG. Deppdb–DNA electrostatic potential properties database: electrostatic properties of genome DNA. J Bioinform Comput Biol. 2010;8:413–425. doi: 10.1142/s0219720010004811. [DOI] [PubMed] [Google Scholar]
- Osypov AA, Krutinin GG, Krutinina EA, Kamzolova SG. DEPPDB - DNA electrostatic potential properties database. Electrostatic properties of genome DNA elements. J Bioinform Comput Biol. 2012;10:1241004. doi: 10.1142/S0219720012410041. [DOI] [PubMed] [Google Scholar]
- Polisky B, Greene P, Garfin DE, et al. Specificity of substrate recognition by the EcoRI restriction endonuclease. Proc Natl Acad Sci. 1975;72:3310–3314. doi: 10.1073/pnas.72.9.3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Carrozzi VR, Kerppola TK. Long-range electrostatic interactions influence the orientation of Fos-Jun binding at AP-1 sites. J Mol Biol. 2001;305:411–427. doi: 10.1006/jmbi.2000.4286. [DOI] [PubMed] [Google Scholar]
- Rohs R, Jin X, West SM, et al. Origins of specificity in protein-DNA recognition. Annu Rev Biochem. 2010;79:233–269. doi: 10.1146/annurev-biochem-060408-091030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stormo GD. Modeling the specificity of protein-DNA interactions. Quant Biol. 2013;1:115–130. doi: 10.1007/s40484-013-0012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomovic A, Oakeley EJ. Position dependencies in transcription factor binding sites. Bioinforma Oxf Engl. 2007;23:933–941. doi: 10.1093/bioinformatics/btm055. [DOI] [PubMed] [Google Scholar]
- Wunderlich Z, Mirny LA. Spatial effects on the speed and reliability of protein–DNA search. Nucleic Acids Res. 2008;36:3570–3578. doi: 10.1093/nar/gkn173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakovchuk P, Protozanova E, Frank-Kamenetskii MD. Base-stacking and base-pairing contributions into thermal stability of the DNA double helix. Nucleic Acids Res. 2006;34:564–574. doi: 10.1093/nar/gkj454. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.