Abstract
Background:
Hepatitis E virus (HEV) is a leading cause of acute hepatitis and can cause chronic infections in immunocompromised patients. Although HEV infections can be treated with ribavirin, antiviral efficacy is hampered by resistance mutations, normally detected by virus sequencing.
Objectives:
High-throughput sequencing (HTS) allows for cost-effective complete viral genome sequencing. This enables the discovery and delineation of new subtypes, and revised the recognition of quasispecies and putative resistance mutations. However, HTS is challenged by factors including low viral load, sample degradation, high host background, and high viral diversity.
Study design:
We apply complete genome sequencing strategies for HEV, including a targeted enrichment approach. These approaches were used to investigate sequence diversity in HEV RNA-positive animal and human samples and intra-host diversity in a chronically infected patient.
Results:
Here, we describe the identification of potential novel subtypes in a blood donation (genotype 3) and in an ancient livestock sample (genotype 7). In a chronically infected patient, we successfully investigated intra-host virus diversity, including the presence of ribavirin resistance mutations. Furthermore, we found convincing evidence for HEV compartmentalization, including the central nervous system, in this patient.
Conclusions:
Targeted enrichment of viral sequences enables the generation of complete genome sequences from a variety of difficult sample materials. Moreover, it enables the generation of greater sequence coverage allowing more advanced analyses. This is key for a better understanding of virus diversity. Investigation of existing ribavirin resistance, in the context of minorities or compartmentalization, may be critical in treatment strategies of HEV patients.
Keywords: Hepatitis E Virus, high-throughput sequencing, targeted enrichment, virus diversity, ribavirin resistance, virus evolution
Introduction
Hepatitis E virus (HEV), species Paslahepevirus balayani, belongs to the genus Paslahepevirus of the family Hepeviridae (International Committee on the Taxonomy of Viruses). Eight genotypes have been defined, of which genotypes 1 and 2 only infect humans, whilst genotypes 3–6 infect mainly pigs and wild boars, with zoonotic spillover infections from genotypes 3 and 4 to humans [1–7]. Genotypes 7 and 8 are found in old world camelids [8,9].
The single-stranded, polyadenylated RNA genome of ~7.2 kb length encodes three open reading frames (ORF). ORF1 encodes non-structural proteins and is of variable length due to common host genome-derived insertions or deletions within a hypervariable region (HVR) [10–12]. ORF2 encodes the capsid protein, and ORF3 the multifunctional protein VP13 [13]. Genotype 1 encodes an additional ORF4 [14].
Annually, HEV infects ~20 million people, with ~56,000 fatalities [15,16]. Usually, the infection is self-limiting, however, chronic infections can occur among immunocompromised, such as transplant patients, and may lead to severe disease [17–19]. On rare occasions, HEV invades the central nervous system (CNS), leading to encephalitis, meningitis, and other neurological disorders [20,21]. In patients with chronic infections, ribavirin treatment is the standard of care [22]. However, treatment failure due to resistance mutations is common [23]. Prior to treatment, sequencing and analysis of HEV diversity in patients might allow predictions regarding treatment success/failure. For such predictions, partial genome sequencing e.g. by Sanger sequencing of RT-PCR amplicons is unlikely to be sufficient because it lacks the sensitivity to detect minor viral populations carrying resistance mutations. Here, high-throughput sequencing (HTS) approaches are of benefit. However, HTS, especially from clinical sample material, is challenging, given low viral loads, high host-background, or due to the presence of viral quasispecies.
In this study, we address these challenges for HEV sequencing, by using samples with a potential new subtype, degraded samples, and tissue samples from a fatal HEV infection.
Methods
Sample material.
In this proof of concept study, we sequenced HEV from RT-PCR positive samples that were residual samples from our diagnostic department, and which represent typical challenges as described in Table 1.
Table 1.
Overview of examined samples, patients, and research question
| Subject | Sample type | Viral load (IU/mL) | Description | Research question / challenge | Background described in |
|---|---|---|---|---|---|
| 1 | CSF | 6.15E+03 | patient had encephalitis of unknown origin with atypical symptoms, HEV infection retrospectively identified through HTS | Typing, Virus variant/mutation causative for CNS symptomatic? | this study (study previously mentioned in [39]) |
| 2 | FFPE sample from a liver biopsy | ~1E+04* | liver biopsy obtained after rejection of transplant, stored as FFPE mounted on a microscope slide for >5 years at room temperature | Typing, Sequencing of HEV from a degraded FFPE sample stored under improper conditions | Hillebrandt et al., unpublished |
| 3 | serum | 4.18E+04 | HEV detected in an Irish blood donation, subtyping through fragment sequencing failed | Typing, New variant/genotype that caused subtyping to fail? | [30] |
| 4 | serum | 6,96E+03 | HEV genotype 7 from a camel sampled in 1983 | Obtaining a genome of a serum sample as old as ~ 40 years with repeated freeze-thaw cycles | [40] |
| 5a | serum | 2.46E+06 | chronic HEV infection in a n immunocompromised patient with fatal outcome | Compartment-specific subpopulation that contributed to patients’ clinical outcome? (Subpopulation with) ribavirin resistance mutations? Intra-host viral evolution? High background of host nucleic acids. |
unpublished/this study |
| 5b | duodenum | 1.74E+06 | |||
| 5c | liver | 1.86E+06 | |||
| 5d | medulla oblongata | 5.62E+06 |
Due to the FFPE fixation this concentration is an estimate.
RNA extraction
Depending on sample type and accessible volume, we used different approaches to extract viral RNA: For cerebrospinal fluid (CSF) or serum up to 140 μL were extracted either in the Roche MagNAPure 96 system using the DNA and Viral NA Small Volume kit (Roche, Basel, Switzerland) or using the QIAamp Viral RNA Mini Kit (Qiagen, Hilden, Germany). For tissue extraction, a lentil-sized, frozen tissue sample was homogenised in 350 μL PBS using a steel bead and the TissueLyser II system (Qiagen). After centrifugation, 250 μL of supernatant were mixed with 250 μL DNA Tissue Lysis Buffer (Roche), extracted using the Roche MagNAPure 96 system, and eluted in 100 μL elution buffer. RNA extraction of the formalin-fixed, paraffin-embedded (FFPE) liver sample was performed using the RNeasy FFPE kit (Qiagen) according to the manufacturer’s instructions, but without DNase treatment.
HEV specific RT-qPCR
HEV RT-qPCR targeting an ORF2/ORF3 overlapping region was used and set up with the Qiagen OneStep RT-PCR kit [24]. For quantification of HEV RNA, a serial-diluted, in vitro transcribed RNA was used [25].
High-throughput sequencing and viral enrichment
HTS library preparation was performed using the KAPA RNA Hyper Prep Kit (Roche) according to the manufacturer’s instructions. Briefly, 5 μL RNA were used for fragmentation at 85°C for 5–6 min. RNA from the FFPE sample was incubated with the Fragment, Prime and Elute Buffer for 6 min at 4 °C. After ≤13 amplification cycles, the resulting libraries were quantified using the Qubit dsDNA HS Assay kit (Thermo Fisher Scientific™, Massachusetts, USA) and Agilent TapeStation using the HS D1000 Kit (Agilent, California, USA). Equimolar pooled libraries were paired-end sequenced on a NextSeq 550 (150/300 cycles, Illumina, California, USA).
As we previously described [26], a custom-made myBaits® set (ArborBioscience/BioCat, Heidelberg, Germany) was used for targeted enrichment following the manufacturer’s recommendations. Hybridization was performed for 18h at 65°C; washing steps were carried out at 65°C. The enriched libraries were amplified for 20–22 cycles using the KAPA Hifi HotStart Ready Mix and Library Amplification Primer Mix (Roche). Purified and quantified libraries were equimolar pooled and paired-end sequenced on a NextSeq 550 (150/300 cycles, Illumina).
The sequences of all baits can be found in the supplement and can also be ordered from BioCat (Heidelberg, Germany) using our initial reference number 181001–32.
Genome analysis and phylogenetic analysis
Reads were mapped using Geneious Prime v2022.0.1 (Biomatters, Auckland, New Zealand) and manually inspected. Consensus sequences were called with 75% highest quality setting of Geneious Prime. Checking for compartmentalization in case 5, we chose three consensus calling strategies: a strict one (90%), one that also considers the quality of the reads (75%), and a less strict one (50%).
Nucleotide (nt) and amino acid (aa) sequences were aligned using MAFFT (version 7.450). Pairwise identities of the HEV genome to reference sequences were calculated using Geneious Prime. A phylogenetic tree was constructed using the MrBayes plug-in in Geneious Prime, a GTR+I+G substitution model, and 500,000 replicates.
Results
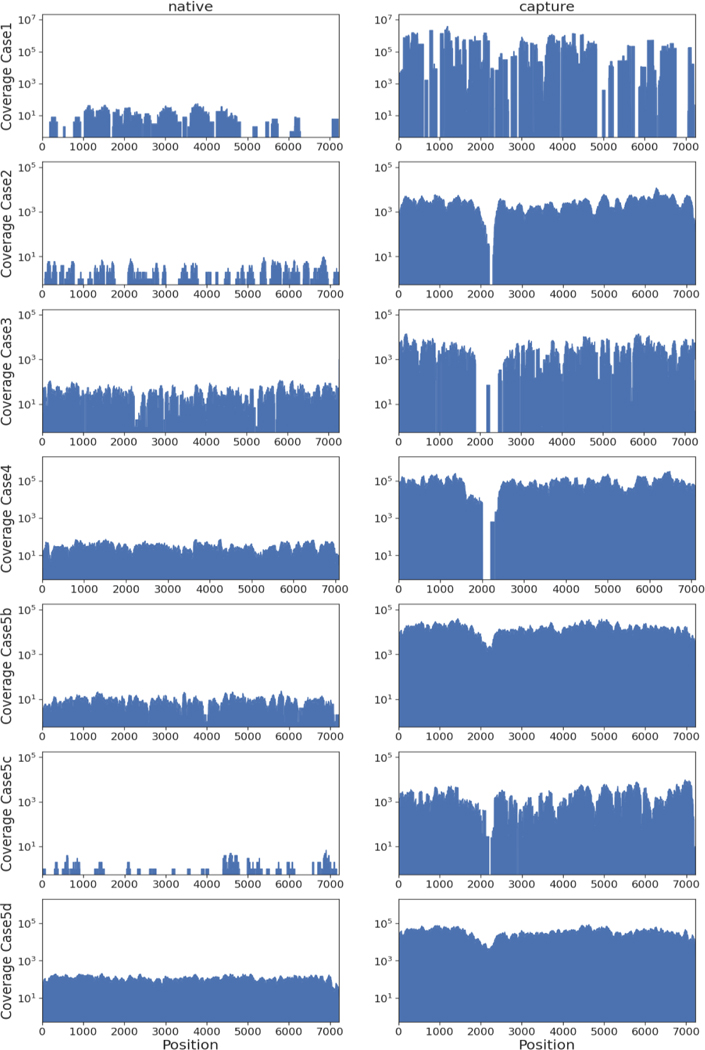
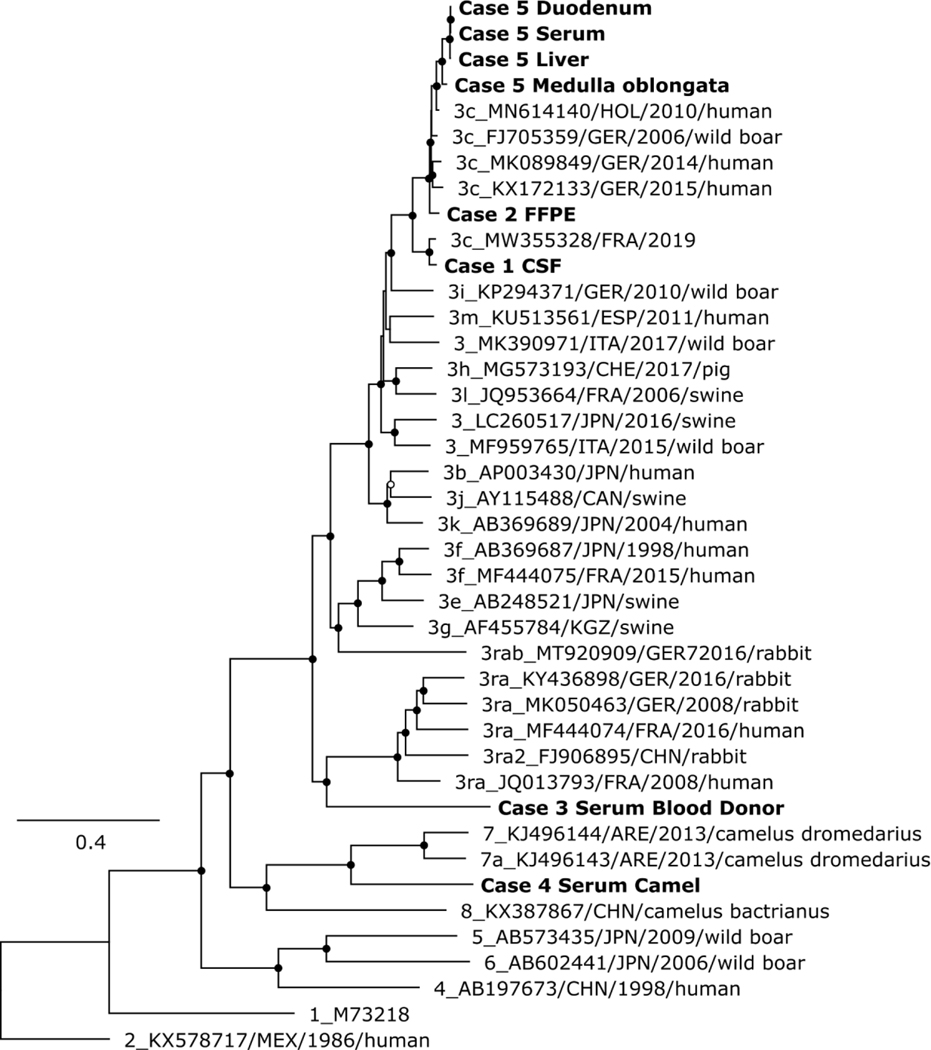
Using targeted viral enrichment and HTS, we generated (near) complete genomes for all samples. The viral loads of these samples ranged from 6.15×103 to 5.62×106 IU/mL (Table 1). As expected, we observed a lower or missing coverage for the HVR region. By our enrichment approach we increased the ratio of viral/total reads, up to 16,381-fold (Figure 1 and Table S1) compared to native HTS without targeted enrichment. The HEV sequences of cases 1, 2, and 5 could be determined as subtype 3c by phylogenetic analysis (Figure 2 and Table S2). The HEV sequences of case 3 and 4 were potentially new subtypes of genotype 3 and 7, respectively.
Figure 1.
Comparison of sequencing results for native HTS (left) and the same libraries after targeted enrichment (right) for case 1 (note that for the enriched results we combined the generated reads of two independent experiments), case 2, case 3, case 4, and the duodenum (b), liver (c), and medulla oblongata (d) sample of case 5.
Figure 2.
Phylogenetic analysis of the complete coding region of HEV. The analysis comprised all complete genome sequences generated in this study (given in bold), related 3c strains from humans, and reference Paslahepevirus balayani strains, as defined by Smith et al.[7]. Virus designations include GenBank accession number, country, collection year, and host, if available. Black circles at nodes indicate bootstrap support values of >90 % and white circles >75 %.
Recombination could not be detected using RDP4 [27] for any of the generated sequences (not shown). More specific results for individual cases are as follows:
Case 1: Low viral load
This case had encephalitis of unknown cause. HEV infection was retrospectively identified by untargeted HTS of a CSF sample. The presence of viral RNA was confirmed by RT-qPCR, revealing a low RNA concentration (6.15E+03 IU/mL). We enriched the original HTS library in two independent experiments. By combining the resulting reads, we obtained a genome coverage of 82.4% (Figure 1). To fill coverage gaps we used strain-specific primers (available upon request). Visualization of PCR products covering the HVR region showed presence of PCR amplicons with diverse size. Sanger sequencing confirmed the presence of two HVR variants (differing by 131 nt in length). In this case, the existence of multiple HVR variants hints towards a chronic HEV infection affecting the CNS. Unfortunately, neither clinical data nor longitudinal samples were available for analyses.
Case 2: Degraded sample material
Successful RNA extraction from a FFPE liver sample that had been stored for several years, mainly at room temperature, was confirmed by detection of beta-2 microglobulin mRNA (not shown). A near full-genome HEV sequence was obtained with an increase of the ratio of viral/total generated sequences of 1,344-fold. Sequencing of the entire HVR failed for the targeted enrichment approach, but was completed using results from the native HTS approach (Figure 1, Table S1). The successful complete genome sequencing from a FFPE sample using HTS and targeted enrichment emphasizes the benefit of these approaches for samples, where PCR amplicon-based approaches (e.g., with fragment sizes >500 bp) are likely to fail due to RNA fragmentation [28].
Case 3: A distinct HEV strain in an Irish blood donation
A full-genome HEV sequence was obtained through subsequent in silico iterations, resulting in a 7258 nt genome with a GC-content of 56.3 %, typical HEV genome organization, and highest similarity to MZ289095.1 with 80.78% nucleotide identity (Table S2). Sequence comparison and phylogenetic analysis confirmed this strain to be distinct, but most closely related to genotype 3 sequences (Figure 2). Of note, public genotyping tools (HEVnet, HEV-GLUE) failed to classify this sequence (https://www.rivm.nl/mpf/typingtool/hev/; http://hev.glue.cvr.ac.uk/, both accessed on 13.06.2023). Lacking defined ICTV genotype demarcation criteria, we suggest that this strain represents a previously undescribed subtype of genotype 3 based on the high nt distance in three genome regions recommended by the ICTV (≥17.9% methyltransferase, ≥20.4% polymerase, ≥18.3% ORF2 (aa121–413)) and its basal relationship to other genotype 3 sequences (Table 2, Figure 2).
Table 2.
Nt (upper) and aa (lower) identity (%) of HEV genomes obtained from the Irish blood donor (case 3) and the camel (case 4) with reference strains for selected genomic regions recommended by ICTV for species demarcation.
| ORF1 | ORF2 | |||||
|---|---|---|---|---|---|---|
| Genotype | Methyltransferase* | RNA-directed RNA polymerase* | Aa121–413* | |||
| Case 3 | Case 4 | Case 3 | Case 4 | Case 3 | Case 4 | |
| 1a | 75.9–77.9 | 76.2–77.5 | 73.6–74.3 | 74.8–75.6 | 78.6–79.5 | 78.6–79.9 |
| 85.6–86.7 | 87.3–89.0 | 85.6–86.4 | 86.9–87.9 | 95.2–95.8 | 93.8–94.6 | |
| 2b | 76.2 | 77 | 72.9 | 74.8 | 78.3 | 79.9 |
| 86.7 | 91.7 | 85.4 | 86 | 96.9 | 94.9 | |
| 3c | 77.5–82.1 | 74.8–79.2 | 76.0–79.6 | 75.5–78.2 | 78.2–81.7 | 77.4–80.6 |
| 91.2–95.0 | 91.2–96.1 | 89.3–92.4 | 88.9–92.2 | 96.9–98.3 | 93.2–94.6 | |
| 4d | 74.2–77.0 | 75.3–78.1 | 74.3–75.8 | 73.6–74.9 | 77.3–79.0 | 79.0–80.9 |
| 90.1–91.7 | 92.3–93.9 | 86.2–87.7 | 85.8–87.3 | 95.2–96.6 | 92.9–94.3 | |
| 5e | 74.8 | 75.9 | 75.2 | 74.7 | 79.1 | 79.3 |
| 89.0 | 91.2 | 86.4 | 85.6 | 96.0 | 94.1 | |
| 6f | 76.4–77.7 | 74.6–75.3 | 73.9–75.7 | 74.4–74.7 | 78.4–79.1 | 77.7–78.5 |
| 87.8–89.5 | 90.1–92.8 | 84.2–87.5 | 84.6–86.2 | 96.6 | 93.2 | |
| 7g | 75.9–76.1 | 79.4–81.2 | 74.4–75.7 | 80.9–81.2 | 78.6–79.1 | 81.4–81.9 |
| 91.2–91.7 | 96.7–97.2 | 88.1–88.9 | 94.3–94.9 | 96.0 | 95.8–96 | |
| 8h | 76.8–77.4 | 77.3–77.9 | 76.5–77.0 | 76.6–77.5 | 78.9–80.1 | 80.1–80.9 |
| 92.3–92.8 | 96.1–96.7 | 88.1–88.3 | 91.0–91.2 | 96.0 | 95.8 | |
Calculated with Geneious Prime 2022.0.1., including GenBank accession numbers:
AB248521, AB290312, AB290313, AB369687, AB369689, AF082843, AF455784, AP003430, AY115488, EU360977, FJ705359, FJ998008, JQ013793, JQ013794, JQ953664, KP294371, KU513561, KY436898, F444074, MF959764, MF959765, MK050463, MT920909, MW002523
as annotated for FJ705359.1
Case 4: A forty-year-old camel blood sample
Sequencing of a camel-derived HEV serum from 1983, resulted in a 7109 nt genome with a GC-content of 53.9%. The closest published sequence was obtained from a camel in Saudi-Arabia in 2017 (MW835253.1) with 81.81% nt identity. Targeted enrichment led to a nearly 3,000-fold increase of viral/total reads.
Similar to case 2, it was not possible to sequence the whole HVR (Figure 1, Table S2) using the enrichment approach. However, a combination of native and enrichment approaches led to a complete genome sequence.
Phylogenetic analysis and sequence comparisons showed the closest relationship of this sequence to sequences of genotype 7. As with case 3, established genotyping tools (HEVnet, HEV-GLUE) failed to classify this sequence. Because of the high nt distance (≥18.8% methyltransferase/polymerase, ≥18.1% ORF2 (aa121–413)), we suggest that the new strain define a distinct subtype of genotype 7 (Figure 2 and Table 2).
Case 5: Tissue samples and intra-host virus-diversity
We sequenced fresh-frozen duodenum, liver, and medulla oblongata samples obtained during autopsy of a patient with chronic HEV infection and CNS symptoms prior to death. HTS resulted in a full genome for the medulla oblongata sample, a near-full genome (99.9%) for the duodenum sample, but only a partial genome for the liver sample (43.0%). Through targeted viral enrichment, we obtained a full-genome for the duodenum and medulla oblongata sample and a near full-genome for the liver sample (99.7%). The resulting consensus sequences had a length of 7,162–7,227 nt with a GC-content of 53.4–54.8%. The genomes showed the typical HEV genome organization (Table S2).
We observed differences in the consensus sequences which were evenly distributed over the genome (Figure S1). We then undertook a more detailed analysis to test for potential compartmentalization. The HEV population of the medulla oblongata, the only CNS sample, was significantly different compared to the rest of the samples. Moreover, this sample showed a 3 nt insertion in the 3’ non-coding region by both sequencing approaches, not observed in the other samples/approaches. Sequencing of a serum sample taken 72 days prior to the autopsy revealed the greatest similarity to the duodenum sample and greatest distance to the CNS sample (Figure S1 and S2).
Analysis of mutations associated with ribavirin resistance [29] revealed the presence of several mutations, the frequency of which varied among sample types. Unexpectedly, fewer mutations were detected in the CNS compartment. Single nucleotide polymorphism abundance levels were comparable between sequencing approaches, indicating a low bias introduced through targeted enrichment (Figure 3). These results suggest the existence of distinct viral populations in the CNS.
Figure 3.
Abundance of mutations (variant A, black) that are associated with ribavirin resistance [29] per sample type and sequencing approach. Only genome positions with a coverage of at least 10 were analysed.
Discussion
In recent years, HTS has become increasingly available and cost-effective and has proven to be a useful tool for generating genome sequences and measuring intra-host diversity. However, HTS still has some limitations, especially the analysis of samples with (I) a very low viral load, (II) degraded nucleic acids, (III) novel pathogens or distinct viral strains/species, (IV) a high host-background, and (V) quasispecies/double-infection.
The first limitations can be circumvented by generating a large number of sequencing reads, however this incurs high costs and might not result in adequate depth of coverage. Targeted enrichment of viral sequences can help to overcome most limitations:
After detection of a slightly HEV-positive CSF sample (case 1), we used this approach to greatly increase the ratio of viral reads to total reads. Thereby, generating a near full-genome that was determined as subtype 3c.
Sample quality, influenced by e.g., storage, sample age, or fixation, can dramatically affect sequencing success [28]. Here, we extracted RNA from a FFPE liver biopsy (case 2) that had been sub-optimally stored for many years. After enrichment, we had a 1,280-fold increased ratio of viral to total generated reads and increased genome coverage from 67.6% to 98.8%. Moreover, we successfully enriched HEV sequences from a forty-year-old camel serum sample and increased the ratio of viral reads to sequenced reads by nearly 3,000-fold compared to the native HTS approach.
Most of the sequences generated in this study belonged to genotype 3c. We sequenced HEV from a blood donation (case 3) after genotyping failed [30]. Enrichment was again highly successful and enabled the generation of a full-genome sequence even though the closest-known reference sequence had a nt identity of only ~80 %, similar as for the viral strain obtained from the camel serum. Further analysis indicated that the sequences from the blood donor and the camel (case 4) belong to potentially new subtypes of genotypes 3 and 7, respectively. This demonstrates the effectiveness of the enrichment approach, even when dealing with diverse viral strains.
High host genome background in tissue samples poses another major challenge. Using targeted enrichment, we generated (near) full-genomes from different tissue types (case 5). Thereby, up to 91.5% of the generated sequencing reads (medulla oblongata) were of viral origin. We generated a (near) complete genome for the duodenum and medulla oblongata sample using a native sequencing approach, however, the depth of genome coverage did not allow us to make accurate conclusions about viral diversity in the duodenum sample. This could only be achieved by the targeted enrichment of viral sequences.
One of the patients (case 5) had a known chronic HEV infection and a fulminant phase with CNS symptoms shortly before death. Detection of mutations associated with ribavirin resistance was successful and, in the case of the liver sample, only possible due to the targeted enrichment approach. Analysis of longitudinal samples from such patients might allow conclusions regarding when the virus infiltrated the CNS. CNS involvement during HEV infection has been increasingly described in the past years [31–35]. We found CNS involvement in two of the five cases here. Importantly, HEV compartmentalization in the CNS may further challenge successful treatment of HEV, as antiviral drugs such as ribavirin may not reach sufficient levels in all compartments.
Limitations
Although HTS and our enrichment approach overcame sequencing challenges in many cases, there are still limitations. With a particularly low (barely detectable) amount of nucleic acid, a limiting factor is that enough molecules end up in the final library to cover the entire genome and subsequently be enriched.
Another limitation is that, in principle, it is possible to only enrich what is included in the capture bait design. However, as shown for the highly divergent and new subtypes of the Irish blood donor and camel-borne HEV, nucleotide divergence of ~20% to known sequences could successfully be enriched. Similar ratios have been achieved for other enrichment approaches [36,37].
The HVR remains a major challenge for HEV sequencing. Here, HEV sometimes integrates part of the host genome. As the insertion is likely random, it cannot be included in the bait design. As a consequence this reduces HEV genome sequence coverage in that region.
However, we observed that parts, and in some cases the entire HVR are covered, likely because baits bind to library molecules that are chimeric, consisting of HEV and host genome/HVR regions.
Bioinformatically challenging, and perhaps impossible with our approach, is the distinction between intra-host viral diversity and co-infection with closely-related, but, distinct viral strains. Long-read sequencing, for example enabled by nanopore sequencing, might be of use.
Conclusions:
Specific enrichment of viral sequences in combination with HTS is a useful approach, especially for samples with degraded sample material or high RNA/DNA background. It enables detection of viral diversity that might not otherwise be reliably measurable in the absence of exceedingly high sequencing depth. However, for generating a simple consensus sequence, especially for samples with reasonable viral load and low host nucleic acid background, HTS alone is often sufficient.
Outlook:
The availability of a robust whole genome sequencing approach at a relatively low (and decreasing) cost and rather short turnaround time leads us to advocate for its wider application in chronic or severe acute HEV infections. Whole genome sequencing could identify virus-intrinsic virulence factors (such as CNS affection). Furthermore, this will also provide a basis to assess and monitor both known and potentially emerging ribavirin resistance variants. This will also be critical when supplementing HEV treatment with other antiviral drugs, such as Sofosbuvir [38]. The timely availability of whole sequence data along with a comprehensive HEV sequence database will likely be of use for individualized therapy recommendations (discontinuation, dosing, or change of therapy).
The enrichment approach presented here can be readily applied to other pathogens and might facilitate sequencing of these. As for HEV deep sequencing of viruses prior to initiation of specific therapy can facilitate detection of resistance mutations initially present only as minorities.
Supplementary Material
Highlights.
Application of a combined HTS and specific enrichment approach for HEV.
Successfully generating complete genome sequences from challenging samples.
Identification of potential novel subtypes in a blood donor and in a camel.
Detection and analysis of ribavirin resistance mutations in virus subpopulations.
Evidence for HEV compartmentalization, including the central nervous system.
Funding
This research was funded by grants from the BMBF-ZooSeq (grant no. 01KI1905C) to C.D.and the German Federal Ministry of Health (BMG) (CHED-project grant No: ZMVI1-2518FSB705) To V.M.C and J.H. Parts of this work were supported by grants from NIAID-NIH CEIRS contract HHSN272201400008C to T.C.J. V.M.C. is a participant in the BIH-Charité Clinician Scientist Program funded by Charité-Universitätsmedizin Berlin and the Berlin Institute of Health.
Footnotes
Informed consent and patient details
No de-novo sampling was performed for this study. The ethical committee at Charité-Universitätsmedizin Berlin approved testing (EA2/194/21 and EA4/169/21).
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Declaration of Competing Interest
All authors declare no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Data Availability Statement
Sequence data is openly available in NCBI GenBank, Acc No. OQ567729-35. Further source data is available upon request.
References
- [1].Raj VS, Smits SL, Pas SD, Provacia LBV, Moorman-Roest H, Osterhaus ADME, Haagmans BL, Novel Hepatitis E Virus in Ferrets, the Netherlands, Emerg Infect Dis. 18 (2012) 1369–1370. 10.3201/eid1808.111659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Krog JS, Breum SØ, Jensen TH, Larsen LE, Hepatitis E Virus Variant in Farmed Mink, Denmark, Emerg Infect Dis. 19 (2013) 2028–2030. 10.3201/eid1912.130614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Xia J, Zeng H, Liu L, Zhang Y, Liu P, Geng J, Wang L, Wang L, Zhuang H, Swine and rabbits are the main reservoirs of hepatitis E virus in China: detection of HEV RNA in feces of farmed and wild animals, Arch Virol. 160 (2015) 2791–2798. 10.1007/s00705-015-2574-0. [DOI] [PubMed] [Google Scholar]
- [4].Spahr C, Knauf‐Witzens T, Vahlenkamp T, Ulrich RG, Johne R, Hepatitis E virus and related viruses in wild, domestic and zoo animals: A review, Zoonoses Public Hlth. 65 (2018) 11–29. 10.1111/zph.12405. [DOI] [PubMed] [Google Scholar]
- [5].Kamar N, Izopet J, Pavio N, Aggarwal R, Labrique A, Wedemeyer H, Dalton HR, Hepatitis E virus infection, Nat Rev Dis Primers. 3 (2017) 17086. 10.1038/nrdp.2017.86. [DOI] [PubMed] [Google Scholar]
- [6].Villalba MM, Martínez DC, Ahmad I, Lay LR, Corredor MB, March CG, Martínez LS, Martínez-Campo LS, Jameel S, Hepatitis E virus in bottlenose dolphins Tursiops truncatus, Dis Aquat Organ. 123 (2017) 13–18. 10.3354/dao03085. [DOI] [PubMed] [Google Scholar]
- [7].Smith DB, Izopet J, Nicot F, Simmonds P, Jameel S, Meng X-J, Norder H, Okamoto H, van der Poel WHM, Reuter G, Purdy MA, Update: proposed reference sequences for subtypes of hepatitis E virus (species Orthohepevirus A), J Gen Virol. 101 (2020) 692–698. 10.1099/jgv.0.001435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Woo PCY, Lau SKP, Teng JLL, Cao K-Y, Wernery U, Schountz T, Chiu TH, Tsang AKL, Wong P-C, Wong EYM, Yuen K-Y, New Hepatitis E Virus Genotype in Bactrian Camels, Xinjiang, China, 2013, Emerg Infect Dis. 22 (2016) 2219–2221. 10.3201/eid2212.160979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Woo PCY, Lau SKP, Teng JLL, Tsang AKL, Joseph M, Wong EYM, Tang Y, Sivakumar S, Xie J, Bai R, Wernery R, Wernery U, Yuen K-Y, New Hepatitis E Virus Genotype in Camels, the Middle East, Emerg Infect Dis. 20 (2014) 1044–1048. 10.3201/eid2006.140140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Tsarev SA, Emerson SU, Reyes GR, Tsareva TS, Legters LJ, Malik IA, Iqbal M, Purcell RH, Characterization of a prototype strain of hepatitis E virus., Proc National Acad Sci. 89 (1992) 559–563. 10.1073/pnas.89.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Ahmad I, Holla RP, Jameel S, Molecular virology of hepatitis E virus, Virus Res. 161 (2011) 47–58. 10.1016/j.virusres.2011.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Papp C-P, Biedermann P, Harms D, Wang B, Kebelmann M, Choi M, Helmuth J, Corman VM, Thürmer A, Altmann B, Klink P, Hofmann J, Bock C-T, Advanced sequencing approaches detected insertions of viral and human origin in the viral genome of chronic hepatitis E virus patients, Sci Rep-Uk. 12 (2022) 1720. 10.1038/s41598-022-05706-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Graff J, Torian U, Nguyen H, Emerson SU, A Bicistronic Subgenomic mRNA Encodes both the ORF2 and ORF3 Proteins of Hepatitis E Virus, J Virol. 80 (2006) 5919–5926. 10.1128/jvi.00046-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Nair VP, Anang S, Subramani C, Madhvi A, Bakshi K, Srivastava A, Shalimar, Nayak B, CT RK, Surjit M, Endoplasmic Reticulum Stress Induced Synthesis of a Novel Viral Factor Mediates Efficient Replication of Genotype-1 Hepatitis E Virus, Plos Pathog. 12 (2016) e1005521. 10.1371/journal.ppat.1005521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Rein DB, Stevens GA, Theaker J, Wittenborn JS, Wiersma ST, The global burden of hepatitis E virus genotypes 1 and 2 in 2005, Hepatology. 55 (2012) 988–997. 10.1002/hep.25505. [DOI] [PubMed] [Google Scholar]
- [16].WHO Hepatitis E, (n.d.). https://www.who.int/news-room/fact-sheets/detail/hepatitis-e.
- [17].Haagsma EB, van den Berg AP, Porte RJ, Benne CA, Vennema H, Reimerink JHJ, Koopmans MPG, Chronic hepatitis E virus infection in liver transplant recipients, Liver Transplant. 14 (2008) 547–553. 10.1002/lt.21480. [DOI] [PubMed] [Google Scholar]
- [18].Nassim K, Janick S, Jean-Michel M, Leila O, Jean-Marie P, Joëlle G, Olivier C, Laure E, Florence A, Marie D, Dominique D, Jean-Pierre V, Jacques I, Lionel R, Hepatitis E Virus and Chronic Hepatitis in Organ-Transplant Recipients, New Engl J Med. 358 (2008) 811–817. 10.1056/nejmoa0706992. [DOI] [PubMed] [Google Scholar]
- [19].Azman AS, Ciglenecki I, Oeser C, Said B, Tedder RS, Ijaz S, The incubation period of hepatitis E genotype 1: insights from pooled analyses of travellers, Epidemiol Infect. 146 (2018) 1533–1536. 10.1017/s0950268818001097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Zhou X, Huang F, Xu L, Lin Z, de Vrij FMS, Ayo-Martin AC, van der Kroeg M, Zhao M, Yin Y, Wang W, Cao W, Wang Y, Kushner SA, Peron JM, Alric L, de Man RA, Jacobs BC, van Eijk JJ, Aronica EMA, Sprengers D, Metselaar HJ, de Zeeuw CI, Dalton HR, Kamar N, Peppelenbosch MP, Pan Q, Hepatitis E Virus Infects Neurons and Brains, J Infect Dis. 215 (2017) 1197–1206. 10.1093/infdis/jix079. [DOI] [PubMed] [Google Scholar]
- [21].Abravanel F, Pique J, Couturier E, Nicot F, Dimeglio C, Lhomme S, Chiabrando J, Saune K, Péron J-M, Kamar N, Evrard S, de Valk H, Cintas P, Izopet J, study group H, Acute hepatitis E in French patients and neurological manifestations, J. Infect. 77 (2018) 220–226. 10.1016/j.jinf.2018.06.007. [DOI] [PubMed] [Google Scholar]
- [22].Kamar N, Abravanel F, Behrendt P, Hofmann J, Pageaux GP, Barbet C, Moal V, Couzi L, Horvatits T, Man RAD, Cassuto E, Elsharkawy AM, Riezebos-Brilman A, Scemla A, Hillaire S, Donnelly MC, Radenne S, Sayegh J, Garrouste C, Dumortier J, Glowaki F, Matignon M, Coilly A, Figueres L, Mousson C, Minello A, Dharancy S, Rerolle JP, Lebray P, Etienne I, Perrin P, Choi M, Marion O, Izopet J, Group HEVRS, Bellière J, Cointault O, Bello AD, Espostio L, Hebral AL, Lavayssière L, Lhomme S, Mansuy JM, Wedemeyer H, Nickel P, Bismuth M, Stefic K, Büchler M, D’Alteroche L, Colson P, Bufton S, Ramière C, Trimoulet P, Pischke S, Todesco E, Soussan RS, Legendre C, Mallet V, Johannessen I, Simpson K, Ribavirin for Hepatitis E Virus Infection After Organ Transplantation: A Large European Retrospective Multicenter Study, Clin Infect Dis. 71 (2019) 1204–1211. 10.1093/cid/ciz953. [DOI] [PubMed] [Google Scholar]
- [23].Todt D, Meister TL, Steinmann E, Hepatitis E virus treatment and ribavirin therapy: viral mechanisms of nonresponse, Curr Opin Virol. 32 (2018) 80–87. 10.1016/j.coviro.2018.10.001. [DOI] [PubMed] [Google Scholar]
- [24].Jothikumar N, Cromeans TL, Robertson BH, Meng XJ, Hill VR, A broadly reactive one-step real-time RT-PCR assay for rapid and sensitive detection of hepatitis E virus, J Virol Methods. 131 (2006) 65–71. 10.1016/j.jviromet.2005.07.004. [DOI] [PubMed] [Google Scholar]
- [25].Corman VM, Eickmann M, Landt O, Bleicker T, Brünink S, Eschbach-Bludau M, Matrosovich M, Becker S, Drosten C, Specific detection by real-time reverse-transcription PCR assays of a novel avian influenza A(H7N9) strain associated with human spillover infections in China, Eurosurveillance. 18 (2013). 10.2807/ese.18.16.20461-en. [DOI] [PubMed] [Google Scholar]
- [26].Schilling-Loeffler K, Viera-Segura O, Corman VM, Schneider J, Gadicherla AK, Schotte U, Johne R, Cell Culture Isolation and Whole Genome Characterization of Hepatitis E Virus Strains from Wild Boars in Germany, Microorg. 9 (2021) 2302. 10.3390/microorganisms9112302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Martin DP, Murrell B, Golden M, Khoosal A, Muhire B, RDP4: Detection and analysis of recombination patterns in virus genomes, Virus Evol. 1 (2015) vev003. 10.1093/ve/vev003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].von Ahlfen S, Missel A, Bendrat K, Schlumpberger M, Determinants of RNA Quality from FFPE Samples, PLoS ONE. 2 (2007) e1261. 10.1371/journal.pone.0001261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Todt D, Gisa A, Radonic A, Nitsche A, Behrendt P, Suneetha PV, Pischke S, Bremer B, Brown RJP, Manns MP, Cornberg M, Bock CT, Steinmann E, Wedemeyer H , In vivo evidence for ribavirin-induced mutagenesis of the hepatitis E virus genome, Gut. 65 (2016) 1733–1743. 10.1136/gutjnl-2015-311000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Baylis SA, O’Flaherty N, Burke L, Hogema B, Corman VM, Identification of rabbit hepatitis E virus (HEV) and novel HEV clade in Irish blood donors, J Hepatol. (2022). 10.1016/j.jhep.2022.04.015. [DOI] [PubMed]
- [31].Dalton HR, van Eijk JJJ, Cintas P, Madden RG, Jones C, Webb GW, Norton B, Pique J, Lutgens S, Devooght-Johnson N, Woolson K, Baker J, Saunders M, Househam L, Griffiths J, Abravanel F, Izopet J, Kamar N, van Alfen N, van Engelen BGM, Hunter JG, van der Eijk AA, Bendall RP, Mclean BN, Jacobs BC, Hepatitis E virus infection and acute non-traumatic neurological injury: A prospective multicentre study, J. Hepatol. 67 (2017) 925–932. 10.1016/j.jhep.2017.07.010. [DOI] [PubMed] [Google Scholar]
- [32].Wang Y, Wang S, Wu J, Jiang Y, Zhang H, Li S, Liu H, Yang C, Tang H, Guo N, Peppelenbosch MP, Wei L, Pan Q, Zhao J, Hepatitis E virus infection in acute non-traumatic neuropathy: A large prospective case-control study in China, EBioMedicine. 36 (2018) 122–130. 10.1016/j.ebiom.2018.08.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Deroux A, Brion JP, Hyerle L, Belbezier A, Vaillant M, Mosnier E, Larrat S, Morand P, Pavese P, Association between hepatitis E and neurological disorders: Two case studies and literature review, J Clin Virol. 60 (2014) 60–62. 10.1016/j.jcv.2014.01.026. [DOI] [PubMed] [Google Scholar]
- [34].Pasha SA, Pasha SA, Suhasini T, Rao DA, Hepatitis E Virus-Associated Acute Encephalitic Parkinsonism., J. Assoc. Physicians India. 66 (2018) 92–3. [PubMed] [Google Scholar]
- [35].Murkey JA, Chew KW, Carlson M, Shannon CL, Sirohi D, Sample HA, Wilson MR, Vespa P, Humphries RM, Miller S, Klausner JD, Chiu CY, Hepatitis E virus-associated meningoencephalitis in a lung transplant recipient diagnosed by clinical metagenomic sequencing, Open Forum Infect. Dis. 4 (2017) ofx121-. 10.1093/ofid/ofx121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Wylie TN, Wylie KM, Herter BN, Storch GA, Enhanced virome sequencing using targeted sequence capture, Genome Res. 25 (2015) 1910–1920. 10.1101/gr.191049.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Paskey AC, Frey KG, Schroth G, Gross S, Hamilton T, Bishop-Lilly KA, Enrichment post-library preparation enhances the sensitivity of high-throughput sequencing-based detection and characterization of viruses from complex samples, BMC Genom. 20 (2019) 155. 10.1186/s12864-019-5543-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Gömer A, Klöhn M, Jagst M, Nocke M, Pischke S, Horvatits T, zur Wiesch JS, Müller T, Hardtke S, Cornberg M, Wedemeyer H, Behrendt P, Steinmann E, Todt D, Emergence of resistance-associated variants during sofosbuvir treatment in chronically infected hepatitis E patients, Hepatology. Publish Ahead of Print (2023). 10.1097/hep.0000000000000514. [DOI] [PMC free article] [PubMed]
- [39].Schneider J, Bachmann F, Choi M, Kurvits L, Schmidt ML, Bergfeld L, Meier I, Zuchowski M, Werber D, Hofmann J, Ruprecht K, Eckardt K, Jones TC, Drosten C, Corman VM, Autochthonous West Nile virus infection in Germany: Increasing numbers and a rare encephalitis case in a kidney transplant recipient, Transbound Emerg Dis. 69 (2022) 221–226. 10.1111/tbed.14406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Rasche A, Saqib M, Liljander AM, Bornstein S, Zohaib A, Renneker S, Steinhagen K, Wernery R, Younan M, Gluecks I, Hilali M, Musa BE, Jores J, Wernery U, Drexer JF, Drosten C, Corman VM, Hepatitis E Virus Infection in Dromedaries, North and East Africa, United Arab Emirates, and Pakistan, 1983–2015, Emerg Infect Dis. 22 (2016) 1249–1252. 10.3201/eid2207.160168. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data is openly available in NCBI GenBank, Acc No. OQ567729-35. Further source data is available upon request.