Abstract
We have previously demonstrated that Slc6a13-deficient (Slc6a13−/−; KO) mice are resistant to P. multocida infection, which might be in connection with macrophage-mediated inflammation; however, the specific metabolic mechanism is still enigmatic. Here we reproduce the less sensitive to P. multocida infection in overall survival assays as well as reduced bacterial loads, tissue lesions, and inflammation of lungs in KO mice. The transcriptome sequencing analysis of wild-type (WT) and KO mice shows a large number of differentially expressed genes that are enriched in amino acid metabolism by functional analysis. Of note, glycine levels are substantially increased in the lungs of KO mice with or without P. multocida infection in comparison to the WT controls. Interestingly, exogenous glycine supplementation alleviates P. multocida infection-induced inflammation. Mechanistically, glycine reduces the production of inflammatory cytokines in macrophages by blocking the activation of inflammasome (NALP1, NLRP3, NLRC4, AIM2, and Caspase-1). Together, Slc6a13 deficiency attenuates P. multocida infection through lessening the excessive inflammatory responses of macrophages involving glycine-inflammasome signaling.
Keywords: Slc6a13, Pasteurella multocida, Glycine, Macrophage, Inflammasome
Introduction
Bacterial pathogens have been shown to induce metabolic alterations in hosts, including fatty acid, glucose, and even amino acids [1, 2, 3, 4]. For example, enterotoxigenic Escherichia coli change the abundance of amino acids (isoleucine, glutamine, asparagine, glycine, and γ-amino butyric acid [GABA]) in the serum and the jejunum of piglets [5, 6]. Moreover, the levels of amino acids (serine, glycine, threonine, arginine, proline, tyrosine, methionine, leucine, and lysine) in the mouse lung show large differences after P. multocida infection [7]. Increasing investigations have found that amino acid metabolism in the host affects the process of pathogen infection [8]. For instance, glycine, as a nonessential amino acid, could inhibit the inflammatory responses in multiple organs (e.g., lung, liver, and intestines) induced by pathogenic bacteria and/or LPS [9, 10], which may be involved in the inactivation of NF-κB, mTOR, and NLRP3 [11, 12, 13]. However, the mechanisms underlying the amino acid metabolism in host that resists pathogen infections remain poorly understood.
The GABAergic system includes GABA, GABA receptors, glutamate decarboxylase, vesicular inhibitory amino acid transporter, GABA transaminase, and GABA transporters (GATs) [14]. It has been reported that the GABAergic system has inhibitory effect on the central nervous system of vertebrates [15, 16]. However, some studies have also found that the GABAergic system enhances immune responses. For example, GABA promotes intestinal Th17 cell differentiation and interleukin (IL)-17 expressions during enterotoxigenic Escherichia coli infection in piglets and mice [6]. Furthermore, GAT2 (encoding by Slc6a13) deficiency promotes Th17 cell responses through the activation of GABA-mTOR signaling [17]. Additionally, our recent research demonstrates that Slc6a13 deficiency alleviates P. multocida capsular type A CQ2 (PmCQ2) infection in mice [18]. Nevertheless, the amino acid metabolism-associated anti-infection mechanism of Slc6a13 deficiency is still unknown.
In this study, we demonstrate that Slc6a13 deficiency improves mouse survival through reducing excessive inflammation induced by P. multocida infection. Furthermore, Slc6a13 deficiency remarkably shapes glycine metabolism in the mouse lung. Exogenous glycine administration lowers bacterial colonization and curtails macrophage-mediated hyperinflammation, while enhancing the survival rate in mice during P. multocida infection. Mechanistically, glycine reduces the production of inflammatory cytokines in macrophages by the inactivation of inflammasomes. Our findings reveal an unidentified role of glycine in preventing and/or treating macrophage-associated diseases (e.g., pathogenic infection).
Materials and Methods
Bacterial Strains and Growth Conditions
The highly virulent bovine P. multocida (PmCQ2) was isolated from a lung of a calf with pneumonia in Chongqing, China [19]. PmCQ2 was streaked on a Martin agar plate and incubated for 24 h at 37°C, and then a single colony was inoculated into 5 mL Martin broth liquid medium for 12 h (37°C, 220 r/min).
Experimental Animals and Ethics Statement
Female C57BL/6 and KM mice (7–8 weeks old, weight 18–22 g) were purchased from the Hunan SJA Laboratory Animal Co., Ltd (Changsha, China). Female Slc6a13−/− mice (7–8 weeks old, weight 18–22 g) with a C57BL/6 background (online suppl. Fig. 1a; see www.karger.com/doi/10.1159/000525089 for all online suppl. material) were kindly provided by Prof. Wenkai Ren (South China Agricultural University). The mice were housed in individually ventilated pathogen-free cages (temperature at 20–30°C, relative humidity at 50–60%, and lighting cycle at 12 h/day) with free access to feed (Lab Mice Diet, SWS9102, Jiangsu Synergy Medical Bioengineering Co., Ltd., China) and water, and acclimated for 4 days before starting the experiments. This study was carried out in accordance with the principles of the Basel Declaration and Recommendations of the Laboratory Animal Ethical Commission of the Southwest University (Permit No. IACUC-20190510-01), Chongqing, China.
Glycine in P. multocida Infection
Wild-type (WT) and Slc6a13−/− mice were randomly divided into four groups (n = 10 per group), respectively: WT + P. multocida + saline, WT + P. multocida + glycine (2 mg/kg; intranasally, i.n.), Slc6a13−/− + P. multocida + saline, and Slc6a13−/− + P. multocida + glycine (2 mg/kg; intranasally, i.n.). Glycine (A110167, Sandon Biotech Shanghai, China) was administered to mice for 7 consecutive days by intranasal before P. multocida infection, whereas solvent control mice received equal amounts of saline. Then all mice were challenged with P. multocida (2.2 × 105 CFUs) by intraperitoneal injection. The survival rate of mice during 7 days was monitored and recorded; those showing severe clinical signs were considered moribund and were humanely euthanized by intraperitoneal injection of 100 μL pentobarbital sodium (1.5%). After treated as described above, the lung tissues and serum samples of mice were collected at 8, 16, and 24 h postinfection for later analysis.
Bacterial Colonization
The bacterial loads were measured as previously described [7]. Briefly, the tissues (n = 10 in each group) were homogenized aseptically and bacterial colonization were quantified by 10-fold serial dilution in saline. These different dilutions were plated in triplicate on Martin's broth agar and were incubated at 37°C up to 24 h to count CFUs.
Histopathological Examination Staining
The histopathological examination experiments were performed as described in previous study [20]. Briefly, the lung tissues (n = 6 in each group) were immediately fixed in the 4% paraformaldehyde (PFA) for 24 h, dehydrated in graded ethanol, and then embedded in paraffin wax. The tissues were sliced at 3 μm thick and then stained with hematoxylin and eosin.
Fluorescence in situ Hybridization
The fluorescence in situ hybridization (FISH) experiments were performed as described in previous study [21]. Briefly, the tissues (n = 6 in each group) were fixed in the 4% PFA for 1 h, dehydrated in graded ethanol, and then embedded in paraffin wax. Then sections were denatured in degeneration buffer (70% formamide, 30% 2 × saline-sodium citrate [2×SSC]) at 78°C for 8 min and the probes were denatured in hybridization buffer (0.9 M NaCl, 0.1% SDS, 100 mM Tris, pH 7.2, 15% formamide, 10% SDS), followed by hybridization overnight at 42°C. Twenty microliters of hybridization buffer containing 15 μg probes was applied per section. This study used pmhyb449, 59-CTATTTAACAACATCCCTTC-39 (S-S-Pmul-0449-a-A-20) (Sangon Biotech, China) to detect PmCQ2 [22]. The probes were labeled with Cy3. After washed by wash buffer (50% formamide, 50% 2 × SSC) for three times with 5 min in each time, cell nuclei were determined by counterstained with 4′, 6-diamidino-2-phenylindole (DAPI, BeyotimeBiotech, China) to evaluate cellular morphology. Fluorescence was detected using a fluorescence microscope (Olympus). Six mice were included in each group and three slices were conducted in each mice lung. The strength of the fluorescent signals was quantitatively analyzed in three visions per slice.
Immunohistochemistry
The immunohistochemistry was performed as described in a previous study [7, 18]. Briefly, mouse lung tissues were collected and washed with ice-cold PBS and fixed in 4% PFA at 4°C overnight. The fixed samples were dehydrated in graded ethanol, embedded in paraffin, sliced, and blocked by 2% BSA, followed by incubation with appropriate primary (iNOS: 1:200, Abcam; Ly6G: 1:200, Abcam) and secondary antibodies. The positive areas of the sample were measured using the Image-Pro Plus 6.0 software.
Mice Primary Peritoneal Exudate Macrophages, Bronchoalveolar Lavage Macrophages, and ANA-1 Cells
Peritoneal exudate macrophages (PEMs) were isolated from mice as previously described [23]. Briefly, 3 days after the injection of 4% thioglycolate (Eiken, Japan), PEMs were collected from peritoneal cavity with cold RPMI 1640 medium (Gibco, USA). Bronchoalveolar lavage macrophages (BALs) were obtained from mice according to the method described in the literature [24]. Briefly, mice were anesthetized by intraperitoneal injection of 100 μL (1.5%) sodium pentobarbital, then the catheter was inserted into the trachea of the mouse fixed on the surgical board; the lavage fluid was collected after perfusing the lungs of mice with 1 mL of sterile balanced salt solution containing 100 μM EDTA, and the cell pellets were collected by centrifuging the lavage fluid for 7 min at 400 g and 4°C. Next, the isolated PEMs and BALs were cultured in RPMI 1640 medium with 10% FBS and 1% penicillin/streptomycin (Gibco, USA) and were incubated for 4 h at 37°C with 5% CO2. Then cells were washed with PBS to remove the nonadherent cells. ANA-1 cells were cultured in RPMI 1640 medium with 10% FBS and 1% penicillin/streptomycin (Gibco, USA). As for the treatments in macrophages, the adherent BALs, PEMs, and ANA-1 cells were cultured in RPMI 1640 medium supplemented with 10 mM glycine for 2 h, subsequently challenged with 2.2 × 105 CFUs P. multocida for 12 h. Supernatants were collected for measuring cytokines, LDH (TaKaRa), and NO (Beyotime). Cells were collected for conducting quantitative real-time-PCR, Western blot, and measuring levels of amino acids.
Quantitative Real-Time PCR
PEMs, BALs, and the lung tissues were quickly collected and stored in liquid nitrogen. Total RNA extractions were performed using an RNAprep pure Animal/Cell Kit (TIANGEN, Beijing, China) involving a gDNA elimination step. cDNAs were synthesized with an iScript cDNA synthesis kit (Bio-Rad, California, USA), and quantitative real-time PCR was performed according to previous study using a CFX96 instrument (Bio-Rad, California, USA) [7]. The primers used in this study are listed in Table 1.
Table 1.
qRT-PCR Primers used in this study
| Gene | Sequence (5′-3′) | Product, bp |
|---|---|---|
| IL-1β | F: ATGAAAGACGGCACACCCAC R: GCTTGTGCTCTGCTTGTGAG |
157 |
| IFN-γ | F: GCTTTGCAGCTCTTCCTCA R: CTTTTGCCAGTTCCTCCAG |
182 |
| TNF-α | F: AGGCACTCCCCCAAAAGAT R: TGAGGGTCTGGGCCATAGAA |
143 |
| IL-6 | F: GACAAAGCCAGAGTCCTTCAGA R: TGTGACTCCAGCTTATCTCTTGG |
76 |
| IL-12p40 | F: TGGGAGTACCCTGACTCCTG R: AGGAACGCACCTTTCTGGTT |
128 |
| Ggt1 | F: CAAAAGCCCTCCTCATCGTCT R: TTCTTCATGGCTCTGCTTCCAG |
195 |
| Gadl1 | F: GGACTCTTGTGCAGGAAGGAAC R: CTTCTGGCCACTGTCTCCTTGG |
77 |
| Hal | F: CGGCAAGCTGATATTGTGGC R: ACCTGTGGCTTTCTGCGATT |
178 |
| Glra1 | F: CGTCAACTTTGTGTCTCGGC R: TTAGCATGGGGCTCTTGTGA |
80 |
| Glra2 | F: ACATCCCTCGCAGACCCTAT R: CACTCGGTAGTCCATGGTGG |
175 |
| Glra3 | F: GGCCTCCTTACCAAAGGTGT R: TCACCTCATCATCCTTATTCTTTCT |
173 |
| Glra4 | F: GAGACCACCATGGACTACCG R: GCATGGATGGGTCTAGGTCG |
112 |
| Glrb | F: ACTTGTGCCCATCCAGTCAC R: GACCAGCAGCCTGTTCAAGA |
82 |
| Grin2b | F: CCTCCTGTGTGAGAGGAAAGA R: GGAGCGTGGTCATTCCCAAA |
119 |
| Slc6a1 | F: GGGCATTGACAGCCAGTTCT R: AAGTCTGGGGTACTCGTCCA |
70 |
| Slc6a5 | F: CCGCATCAGACATGGATTGC R: GATCCTGTTCAGGGCTGGTC |
126 |
| Slc6a9 | F: CCTTCCCCAGAACAGAATGGT R: AGGTATGGGAAACGCCAGAC |
152 |
| β-Actin | F: GTCCACCTTCCAGCAGATGT R: GAAAGGGTGTAAAACGCAGC |
117 |
qRT-PCR, quantitative real-time PCR.
Enzyme Linked Immunosorbent Assay
Cell supernatant and homogenated lung tissues were centrifugated at 12,000 rpm for 10 min at 4°C for acquiring supernatant. Cytokines (e.g., IL-1β, TFN-α, IFN-γ, IL-6, and IL-12) were detected by using ELISA kits (eBioscience, USA) in accordance with the manufacturer's protocol.
Transcriptome Analysis
To explore the anti-infection mechanism of Slc6a13−/− mice, C57BL/6 WT mice (n = 3) and Slc6a13−/− mice (n = 3) were infected by an intraperitoneal inoculation with PmCQ2 at the dose of 2.2 × 105 CFUs in 100 μL, control mice receive equal volume of PBS. Mice were euthanized at 16 h postinfection, and the lung tissues were collected and quickly frozen in liquid nitrogen. The lung tissue samples were sent to the Beijing Genomics Institute (BGI, Beijing, China) for transcriptome sequencing and analysis (HiSeq; Illumina, USA). The specific methods are as previously described [25, 26], briefly, total RNA was extracted using Trizol reagent (Invitrogen Life Technologies, USA) following the manufacturer's protocol. RNA integrity was confirmed by agarose gel electrophoresis and quantified by NanoDrop (NanoDrop 2000; Thermo Scientific). Next, rRNA was removed by Ribo-Zero rRNA Removal Kit (Illumina, San Diego, CA, USA). Then, 1 μg of each RNA sample was used to construct a strand-specific cDNA library according to the recommendations of Illumina TruSeq Stranded Kit. The cDNA library size was analyzed by using an Agilent 2100 Bioanalyzer and the effective concentration was determined using qPCR (StepOnePlus Real-Time PCR Systems; Thermo Scientific). Finally, samples were sequenced on the Illumina sequencing platform (HiSeq 4000) and 150 bp paired-end reads were generated. The reads are aligned to the rRNA reference sequence via SOAP [27], and reads were discarded with an average base quality lower than Q20, unknown base N content greater than 5% and containing linkers (linker contamination). The clean reads were mapped to the genome by HISAT software [28] and the reference gene set by using Bowtie2 software [29] (http://bowtie-bio.sourceforge.net/Bowtie2/index.shtml), and then RSEM software [30] (http://deweylab.biostat.wisc.edu/RSEM) was used to calculate gene expression levels. Differential expression analysis was performed using estimate Size Factors in DESeq (version 1.18.0) R package. p < 0.05 and |log2foldchange| ≥ 1 were set as the threshold for differentially expressed genes (DEGs) identification. Then, all DEGs were mapped to Gene ontology (GO) database and Kyoto Encyclopedia of Genes and Genomes (KEGG) ontology database to obtain respective GO terms and KEGG classification, then enrichment analysis of the DEGs was performed using R package based on hypergeometric distribution in the context of the entire reference genome. The raw data have been deposited to NCBIs Sequence Read Archive database and the accession number is PRJNA706465.
Cell-Type Enrichment
The method of lung cell-type enrichment is as previously described [25], briefly, the transcript identifiers of the DEGs for each of the two major groups were converted into their associated gene names using the BioMart software (http://uswest.ensembl.org/biomart/martview/). The lists of two DEGs were then input into the CTen database [31] to obtain the enriched cell types based on a highly expressed cell-specific gene database. One-sided Fisher's exact test was used for enrichment analysis, and the CTen enrichment score was returned from the −log10 of the Benjamini-Hochberg-adjusted p values. The CTen database is originally based on human and mouse data [32, 33].
Measurement of Amino Acids Levels
The PEMs (1 × 108 cells) and lung tissues from WT and Slc6a13−/− mice infected or uninfected by PmCQ2 were quickly collected and stored in liquid nitrogen (n = 6). The prepared samples were sent to the Beijing Amino Medical Research Co., Ltd., for free amino acid content determination. The amino acids of all samples were analyzed with isotope dilution liquid chromatography-mass spectrometry methods. Briefly, 0.5 g lung tissue was homogenized in 5 mL 0.1 M hydrochloric acid aqueous solution. The sample were centrifuged at 5,000 g for 5 min, then 0.5 mL of supernatant was added to 0.5 mL of 8% sulfosalicylic acid overnight at 4°C. The cells resuspended in PBS were centrifuged at 8,000 rpm for 10 min, and 1 mL of methanol was added to sonicate for 10 min to disrupt the cells. The PEMs (1 × 108 cells) and lung tissue supernatant were passed through 0.22 μm water phase membrane filter, and then 5 μL of the sample was injected to Hitachi L8900 High-efficiency Automatic Amino Acid Analyzer for quantitative analysis. The analysis parameters were as follows: separation column, 4.6 mm i.d × 60 mm stainless steel column; exchange resin model, No. 2622 (Special resin for amino acid analysis); column temperature, 57°C; buffer flow rate, 0.4 mL/min; ninhydrin flow rate, 0.3 mL/min; injection volume, 20 μL.
Western Blot Analysis
PEMs from WT and Slc6a13−/− mice were cultured in 12-well plates at a density of 1 × 106 cells/well in RPMI 1640 containing 10% FBS at 37°C for 2 h. After washing with PBS for three times, the culture medium was replaced with Opti-MEM (Gibco, USA), and the adherent cells were infected by P. multocida (MOI = 1:1) with or without glycine (10 mM) treatment. Supernatants were collected after 12 h postinfection, and the cells were lysed with radio-immunoprecipitation assay buffer (Beyotime, China). The culture supernatants were precipitated using 20% (w/v) trichloroacetate. The precipitates and cell lysates were subjected to SDS-PAGE and subsequently transferred to 0.2 μm polyvinylidene difluoride membranes by electroblotting. The membranes were immunoblotted with anti-HIF-1α (1:1,000)/p65 (1:500)/p-p65 (1:500)/β-actin (1:5,000) antibodies (Proteintech, UK), anti-IKK (1:200)/p-IKK (1:1,000)/AIM2 (1:1,000)/Caspase-1 (1:1,000)/NALP1 (1:1,000)/NLRP3 (1:200)/NLRC4 (1:1,000) antibodies (abcam, UK), anti-IκB (1:500)/p-IκB (1:500)/mTOR (1:500)/p-mTOR (1:1,000) antibodies (CST, USA), and anti-IL-1β (1:1,000) (Santa Cruz, CA, USA) antibody overnight at 4°C. The horseradish peroxidase-conjugated secondary antibodies (1:5,000, Proteintech, USA) were subsequently incubated for 1.5 h at 37°C. The membrane developed blots were analyzed using light imaging system (Thermo, USA). Finally, signal intensity was quantified and normalized to actin protein abundance.
Statistical Analysis
Data were shown as the means ± standard deviation. Data between two groups were analyzed using unpaired t tests (Prism 6.0) if the data were in Gaussian distribution and had equal variance, or by unpaired t test with Welch's correction (Prism 6.0) if the data are in Gaussian distribution but show unequal variance, or by nonparametric test (Mann-Whitney U test, Prism 6.0) if the data were not normally distributed. The Gaussian distribution of data was analyzed by D'Agootino-Pearson omnibus normality test (Prism 6.0) and Kolmogorov-Smirnov test (Prism 6.0). The variance of data was analyzed by Brown-Forsythe test (Prism 6.0). Differences with p < 0.05 were considered significant.
Results
Slc6a13 Deficiency Promotes Mice Resistance to P. multocida Infection
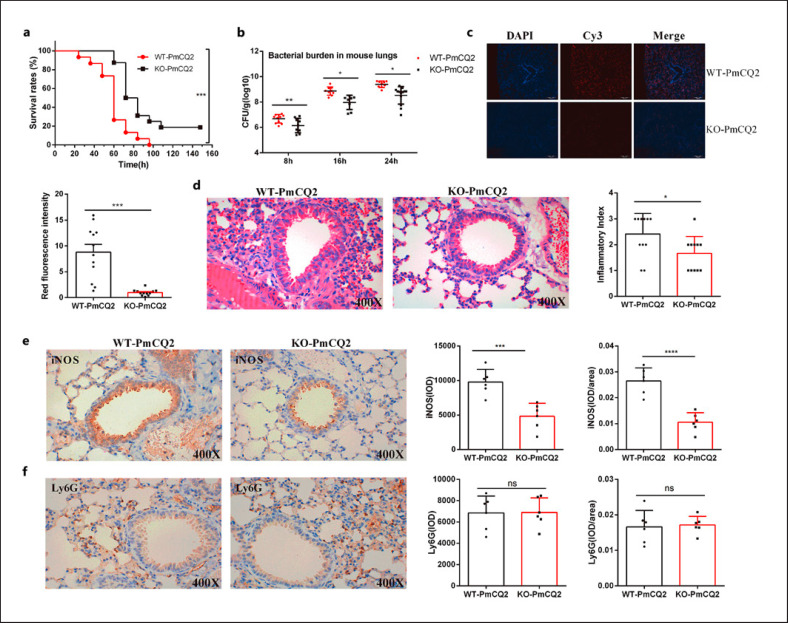
Compared to WT mice, the survival rates of Slc6a13−/− mice (hereafter KO mice) infected with PmCQ2 were significantly increased (Fig. 1a). The bacteria colonization in the lungs of KO mice was decreased at 8, 16, and 24 h postinfection (Fig. 1b); additionally, less PmCQ2 presented in the lungs of infected KO mice via FISH with specific probe of PmCQ2 (pmhyb449) (Fig. 1c). Moreover, the lower inflammation was observed in the lungs of KO mice on the basis of the hematoxylin and eosin (Fig. 1d) and IHC staining (especially for reduced iNOS abundance) (Fig. 1e, f). These aforementioned findings are closely consistent with our previous results [18]. However, the mRNA of the main virulence factors of P. multocida remains unaffected (online suppl. Fig. 1b), indicating that the anti-infective effects of Slc6a13 deficiency mice may not involve the changes of bacterial virulence, but is related to the alterations in host physiological status.
Fig. 1.
Slc6a13 deficiency reduces the hyperinflammatory responses in the lungs of mice during P. multocida infection. a The survival rates of WT and Slc6a13−/− mice infected by P. multocida (n = 10, Kaplan-Meier analysis). b The bacterial burden in mice lungs infected by P. multocida at 8, 16, and 24 h postinfection (n = 8∼13, one-way ANOVA). c The specific distribution of PmCQ2 in the lung tissues of mice at 16 h postinfection via FISH (n = 12, unpaired, two-tailed Student's t test). The probes were labeled with Cy3, and cell nuclei were determined by counterstained with DAPI. d The severe lung tissues inflammatory lesions caused by PmCQ2 at 16 h postinfection with H&E staining (n = 11, magnification ×400). e The expressions of iNOS in lungs of mice were determined by immunohistochemistry (×400) (n = 6). IOD: integrated optical density. f The expressions of Ly6G in lungs of mice were determined by immunohistochemistry (×400) (n = 6). Data are shown as means ± SD. WT, Wild-type mice; KO, Slc6a13−/− mice; H&E, hematoxylin and eosin; SD, standard deviation.
Slc6a13 Deficiency Shapes Amino Acid Metabolism in Mice during P. multocida Infection
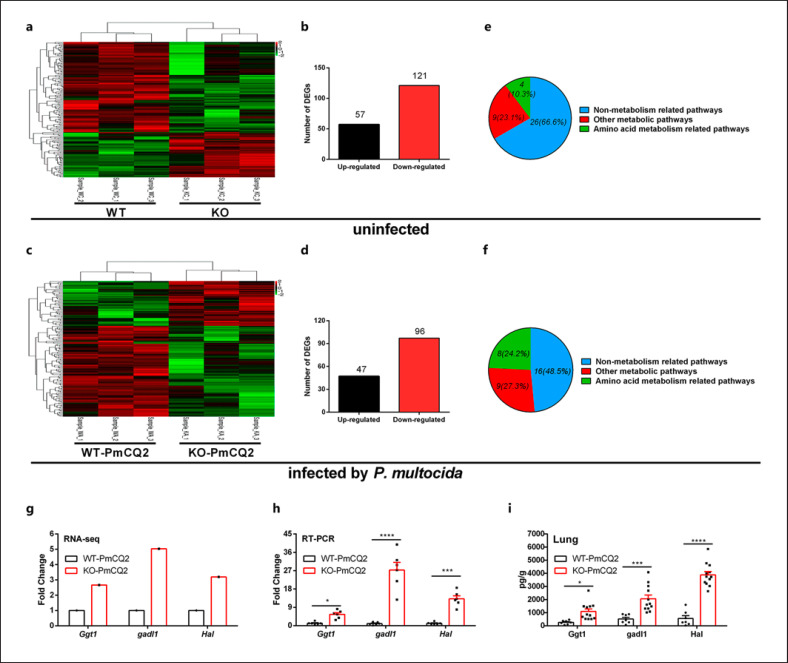
GAT2 is the transporter of GABA, however, the content of free GABA in the lung tissues of mice remain unchanged (online suppl. Fig. 1c), suggesting that the anti-infective mechanism is independent of GABA but might be related to other amino acids. In order to explore the nature of KO mice against P. multocida, we compared the DEGs of lungs from KO and WT mice by transcriptome analysis. We observed a total of 1,923 upregulated and 2,313 downregulated DEGs in WT versus WT-PmCQ2 mice [21], 2,046 upregulated and 2,047 downregulated DEGs in KO versus KO-PmCQ2 mice (online suppl. Fig. 2a, b), 57 upregulated and 121 downregulated DEGs in WT versus KO mice (Fig. 2a, b), and 47 upregulated and 96 downregulated DEGs in WT-PmCQ2 versus KO-PmCQ2 mice (Fig. 2c, d). Moreover, 1,617 upregulated and 1,654 downregulated DEGs were shared between WT versus WT-PmCQ2 and KO versus KO-PmCQ2 mice (online suppl. Fig. 2c, d), while only 5 upregulated and 10 downregulated DEGs were shared between WT versus KO and WT-PmCQ2 versus KO-PmCQ2 mice (online suppl. Fig. 2e, f). These data indicate that Slc6a13 knockdown and/or infection extensively shapes the transcriptional profilling of mouse lungs. Furthermore, KEGG pathway analysis demonstrated that there were 4 amino acid metabolism-related pathways in WT versus KO mice (Fig. 2e), and 8 amino acid metabolism-related pathways in WT-PmCQ2 versus KO-PmCQ2 mice (Fig. 2f). It should be noted that the altered amino acid metabolism-related pathways in the infected groups increased significantly from 10.3% (total 39 significant pathways) to 24.2% (total 33 significant pathways) in comparison to the uninfected groups (Fig. 2e, f). Additionally, the DEGs randomly selected from RNA-seq analysis (Fig. 2g) were validated by qRT-PCR (Fig. 2h) and ELISA (Fig. 2i) analysis. Together, these results suggest that Slc6a13 deficiency resists PmCQ2 infection which might be dependent on altering amino acid metabolism in host.
Fig. 2.
Slc6a13 deficiency shapes amino acid metabolism during P. multocida infection. a The clustering of DEGs in heat-map of WT versus Slc6a13−/− mice (n = 3). b Numbers of up/downregulated DEGs of WT versus Slc6a13−/−mice. c The clustering of DEGs in heat-map of WT versus Slc6a13−/− mice infected by P. multocida (n = 3). d Numbers of up/downregulated DEGs of WT versus Slc6a13−/− mice infected by P. multocida. e The category of KEGG enrichments of WT versus Slc6a13−/−mice. f The category of KEGG enrichments of WT versus Slc6a13−/−mice infected by P. multocida. g DEGs related to amino acids metabolism by Illumina sequencing (n = 3). h Amino acids metabolism-related DEGs by qRT-PCR (n = 6, multiple t test). i Amino acids metabolism-related DEGs using ELISA analysis (n = 12, multiple t test). All data were expressed as means ± SD. *p < 0.05, ***p < 0.001, ****p < 0.0001. GGT1, gamma-glutamyltransferase 1; GADL1, glutamate decarboxylase-like 1; HAL, histidine ammonia lyase; WT, Wild-type mice; KO, Slc6a13−/− mice; SD standard deviation.
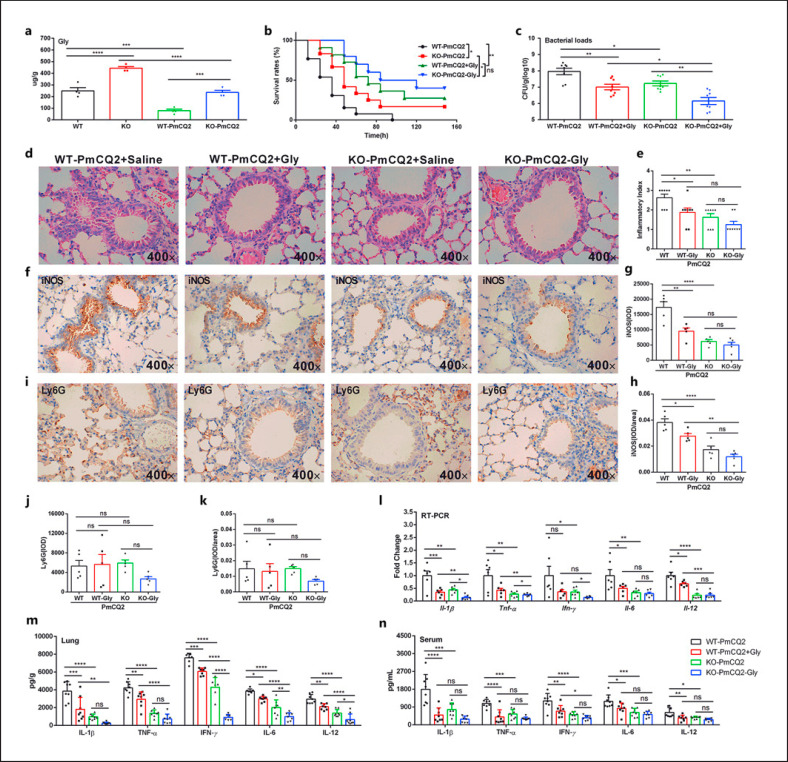
Slc6a13 Deficiency Promotes Glycine Accumulation against P. multocida Infection
To further explore the relationship between amino acid metabolism and Slc6a13 deficiency-mediated resistance to P. multocida infection, the levels of free amino acids in the lungs of WT and KO mice were determined. Vividly, Slc6a13 deficiency or PmCQ2 infection substantially shaped the levels of amino acids, and of which glycine was the most increased in KO mice (Fig. 3a; online suppl. Fig. 3a). Moreover, we found that PmCQ2 infection could lower the levels of glycine in lungs of WT and/or KO mice (Fig. 3a). Interestingly, the expression of glycine transporter (GlyT) (especially for Slc6a9) (online suppl. Fig. 3b) and receptor (GlyR) (chiefly Glra4) (online suppl. Fig. 3c) was increased in the lung tissues of KO mice with or without infection. These findings indicate that glycine accumulation might be the pivotal determinant that reducing PmCQ2 infection in our study. Subsequently, glycine (2 mg/kg) was administrated to mice by intranasal injection before P. multocida infection. It was found that exogenous glycine increased the survival rate of mice (Fig. 3b), lowered the bacteria loads (Fig. 3c), the lesions (Fig. 3d, e), and inflammation (Fig. 3f-k). Notably, exogenous glycine could mirror the suppressive effect of Slc6a13 deficiency on the mRNA expression (Fig. 3l) and secretion of inflammatory cytokines (IL-1β and TNF-α in particular) in the lungs (Fig. 3m) and serum (Fig. 3n). Collectively, these findings show that glycine accumulation might mediate the inhibitory effect of Slc6a13 deficiency on PmCQ2 infection-induced inflammation.
Fig. 3.
Slc6a13 deficiency promotes the accumulation of glycine against P. multocida infection. a The levels of glycine in lung tissues of WT and KO mice infected and uninfected with P. multocida (n = 6, one-way ANOVA) at 16 h postinfection. b The survival rates of WT and KO mice infected by P. multocida with or without 2 mg/kg glycine supplementation (n = 10, Kaplan-Meier analysis). c The bacterial burdens in WT and KO mice lungs infected by P. multocida at 16 h postinfection with or without 2 mg/kg glycine supplementation (n = 10, one-way ANOVA). d, e H&E staining of the mouse lung at 16 h postinfection by P. multocida (n = 8, magnification ×400). f–h The expressions of iNOS in lungs of mice were determined by immunohistochemistry (×400) (n = 5). IOD, integrated optical density. i–k The expressions of Ly6G in lungs of mice were determined by immunohistochemistry (×400) (n = 5). l–n Effect of 2 mg/kg glycine on the mRNA expression and protein abundance of IL-1β, TNF-α, IFN-γ, IL-6, and IL-12 in the lung and serum at 16 h postinfection by P. multocida (n = 6∼8, two-way ANOVA). All data were expressed as means ± SD. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. H&E, hematoxylin and eosin; SD, standard deviation.
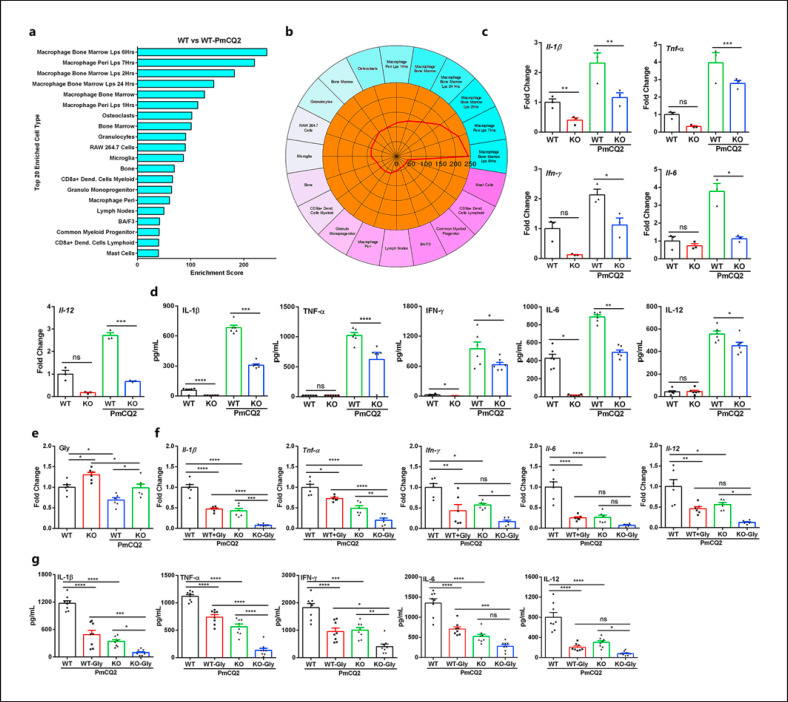
Slc6a13 Deficiency Increases the Accumulation of Glycine to Curtail Macrophage Inflammation during P. multocida Infection
Subsequently, we wanted to reveal the immune-related mechanism whereby Slc6a13 deficiency promoted mice resistance to P. multocida infection. Cell-type enrichment analysis based on the upregulated DEGs showed that macrophage was the most enriched immune cell type in both mice infected with PmCQ2 versus without PmCQ2 (Fig. 4a, b). Thus, we wanted to ask whether Slc6a13 deficiency could lessen macrophage inflammation induced by PmCQ2 infection. Interestingly, compared to WT macrophages, the mRNA expression (Fig. 4c) and secretion of IL-1β, TNF-α, IFN-γ, IL-6, and IL-12 (Fig. 4d) were substantially decreased in Slc6a13−/−macrophages. Of note, our previous study found that Slc6a13−/− mice are no longer resistant to PmCQ2 infection after clearing alveolar macrophages [18]. These findings highlight the critical roles of macrophages in mediating PmCQ2 infection-induced immune responses in vivo.
Fig. 4.
Slc6a13 deficiency enhances the accumulation of glycine to decrease the secretion of inflammatory cytokines from macrophages infected by P. multocida. a, b The 20 most enriched cell types during P. multocida infection. The enrichment scores (labeled inside circle) is displayed in the −log10 of the Benjamini-Hochberg-adjusted p values. Cell types (labeled outside circle) with an enrichment score (red line) greater than 2 were considered significant. c, d The mRNA expression and supernatant protein concentration of IL-1β, TNF-α, IFN-γ, IL-6, and IL-12 in WT and KO macrophages infected or uninfected by PmCQ2 at 12 h (n = 3 or 6, one-way ANOVA). e The levels of glycine in WT and KO macrophages uninfected or infected by PmCQ2 at 12 h (n = 6, one-way ANOVA). f, g The mRNA expression and supernatant protein concentration of IL-1β, TNF-α, IFN-γ, IL-6, and IL-12 in WT and KO BAL macrophages with or without 10 mM glycine at 12 h postinfection (n = 6 or 8, one-way ANOVA). All data were expressed as means ± SD. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. SD, standard deviation.
To explore whether glycine-mediated resistance to P. multocida infection is related to macrophages, the concentrations of free amino acids in WT and KO macrophages were determined by L8900 amino acids analyzer (online suppl. Fig. 4a). Interestingly, the levels of L-glycine were also significantly increased in KO macrophages and lowered in PmCQ2-infected macrophages (Fig. 4e), and the expression of GlyTs Slc6a9 (online suppl. Fig. 4b) and GlyR Glra4 (online suppl. Fig. 4c) was increased in KO macrophages, suggesting intracellular glycine accumulation affects inflammation. Subsequently, we investigated the direct effects of glycine on macrophages. It was found that glycine supplementation did not exhibit significant toxicity against macrophages (online suppl. Fig. 5a, b) but decreased the secretion of IL-1β, TNF-α, IFN-γ, IL-6, and IL-12 from BAL macrophages (Fig. 4f, g), PEMs (online suppl. Fig. 5c), and ANA-1 cells (online suppl. Fig. 5d). Notably, we found that glycine can even further reduce the inflammatory responses of KO macrophages. These convincing data suggest that PmCQ2 infection and Slc6a13 deficiency alter the cellular glycine, and in turn, glycine could inhibit macrophage inflammation induced by PmCQ2 infection.
Slc6a13 Deficiency Attenuates P. multocida Infection-Induced Inflammation via Glycine-Inflammasome Signaling
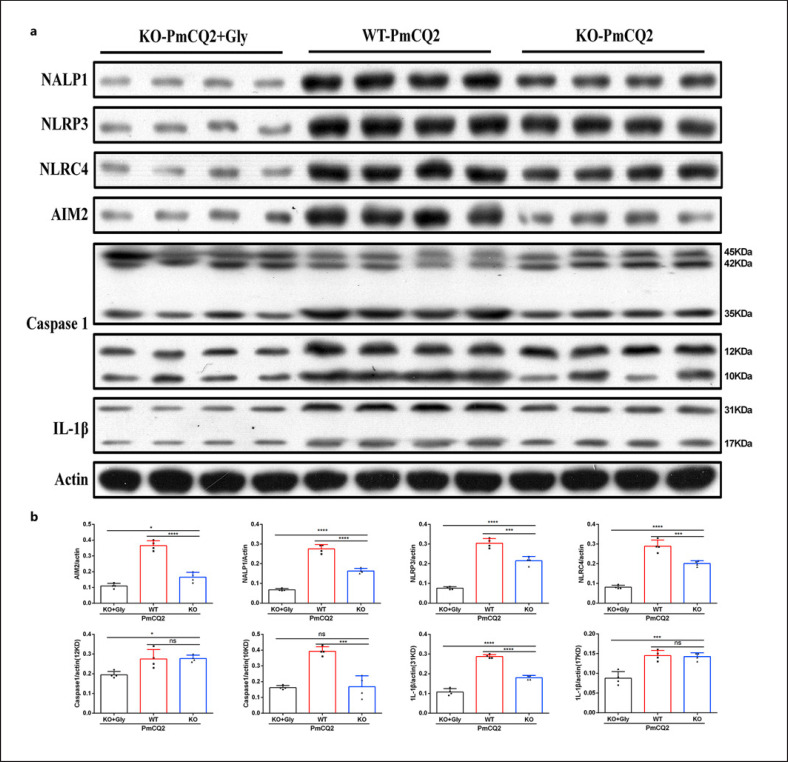
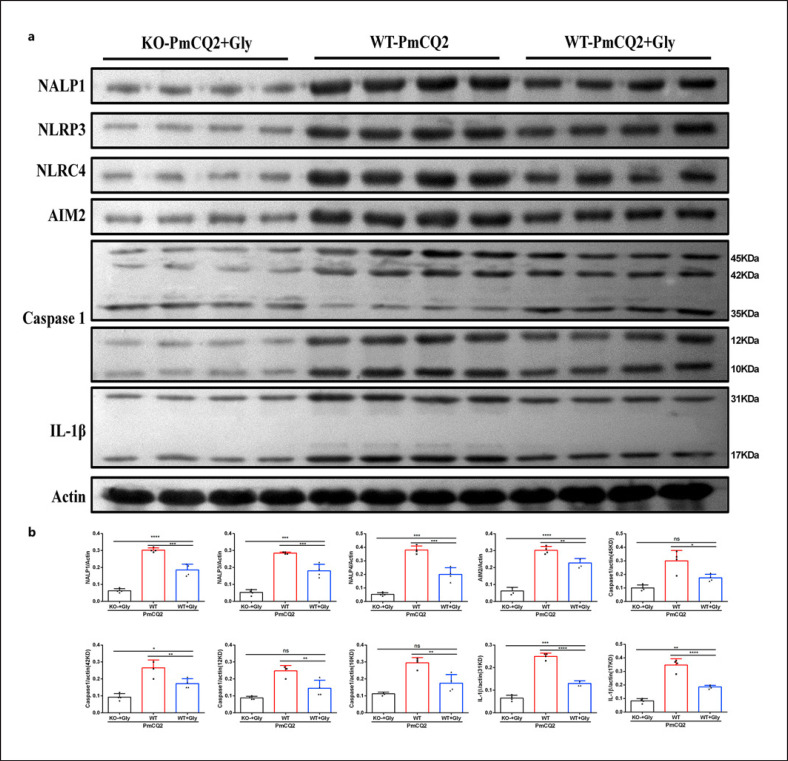
Next, we wanted to investigate how glycine mediates Slc6a13 deficiency to lessen PmCQ2 infection-triggered macrophage inflammation. Combined the results of protein abundance of inflammatory pathways from WT and KO macrophages, Slc6a13 deficiency had little effect on the activation of mTOR, HIF-1α, and NF-κB signaling during PmCQ2 infection (online suppl. Fig. 6a, b), but significantly suppressed the activation of inflammasomes (NALP1, NLRP3, NLRC4, AIM2, and Caspase-1) and the protein abundance of IL-1β in WT macrophages (Fig. 5a, b). More importantly, in the context of PmCQ2 infection, glycine supplementation even further blocked inflammasome activation (including NALP1, NLRP3, NLRC4, AIM2, Caspase-1, IL-1β) in KO macrophages (Fig. 5a, b, 6a, b). Collectively, these results implicate that Slc6a13 deficiency attenuates P. multocida infection by largely reducing the inflammatory responses of macrophages involving glycine-inflammasome signaling.
Fig. 5.
Slc6a13 deficiency reduces the secretion of inflammatory cytokines from infected macrophages through blocking inflammasomes. a, b The activation of inflammasomes (NALP1, AIM2, NLRP3, NLRC4, and Caspase-1) in WT + PmCQ2, KO + PmCQ2, and KO + PmCQ2 + glycine macrophages infected with PmCQ2 for 12 h (n = 4, one-way ANOVA). All data were expressed as means ± SD. *p < 0.05, ***p < 0.001, ****p < 0.0001. SD, standard deviation
Fig. 6.
Exogenous glycine inhibits the activation of inflammasomes in macrophages. a, b The effect of glycine on the activation of inflammasomes (NALP1, AIM2, NLRP3, NLRC4, and Caspase-1) in WT + PmCQ2, WT + PmCQ2 + glycine, and KO + PmCQ2 + glycine macrophages infected with PmCQ2 for 12 h (n = 4, one-way ANOVA). All data were expressed as means ± SD. *p < 0.05, ***p < 0.001, ****p < 0.0001. SD, standard deviation.
Discussion
We previously discovered that Slc6a13 deficiency reduces P. multocida infection [18]; however, the specific metabolic mechanism is still unclear. Therefore, this study aimed to explore the role and mechanism of Slc6a13 in P. multocida infection. It was found that Slc6a13 deficiency increases mouse survival by reducing excessive inflammation in mouse lung tissue induced by P. multocida. Furthermore, the results of transcriptome sequencing and metabolite analysis showed that Slc6a13 deficiency alters amino acid metabolism. Indeed, the interaction between host amino acid metabolism and pathogens decidedly shapes the outcomes of infection [2] and immune responses [8]. In our study, glycine, threonine, methionine, and proline in the lung tissues of KO mice are significantly higher than those in the WT mice with or without P. multocida infection. Among them, the basal levels of glycine are the most highest. Moreover, we surprisingly find that the mRNA expression of GlyTs (especially Slc6a9) is significantly increased, indicating that the accumulation and/or uptake of glycine could determine the inhibitory effect of Slc6a13 deficiency on infection in our experimental settings. However, whether Slc6a13 deficiency affects the expression of enzymes involved in amino acid metabolism that account for the different levels of amino acids in the lungs during infection still need further study. Furthermore, whether other amino acids and other metabolic pathways are responsible for reducing P. multocida infection-induced inflammation in Slc6a13−/− mice remain to be explored.
The glycine level in mouse lung tissue is greatly increased after Slc6a13 knockout, which may be related to the increased expression of GlyT and GlyR. Our previous experiments found that mouse PEMs highly express GAT2 (encoding by Slc6a13) and GAT4 (encoding by Slc6a12), and GAT4 is compensatively increased after GAT2 deletion to maintain GABA transportation [18]. Consistently, there is no difference in GABA levels between WT and KO mice in this study. In addition to transporting GABA, GAT4 also transports taurine (Tau) (IC50 = 1.6 mM), and glycine occupies an equivalent position in the related Tau transporter (TAUT) in the conserved transmembrane domain I of GAT4 [34]. Tau can be produced from cysteine catabolism in vivo, and cysteine metabolic enrichment was detected in GAT2-KO T cells consistently [35]. Therefore, upregulated GAT4 might promote Tau level. It has been demonstrated that Tau acts as a partial agonist of the GlyR in vitro and in vivo [36, 37], and a proton-dependent taurine transporter (PAT1) can transport other substrates (e.g., glycine) [38]. Therefore, Tau may act as a mediator which could link GAT2 to glycine; however, more experiments are necessary to validate the aforementioned hypothesis.
Our previous studies found that macrophages are the most abundant immune cells in the lung tissues of chickens infected by P. multocida [25], demonstrating macrophages play an important role in P. multocida infection [39, 40]. Similarly, the study found that macrophages are the most abundant immune cells in the lung tissues of mouse infected by P. multocida. Consistent with the results from in vivo, the glycine levels in Slc6a13−/− macrophages are significantly higher than those in the WT macrophages. Considering serine lowers the macrophage-and/or neutrophils-mediated inflammatory responses during P. multocida infection [7], and supports IL-1β production in macrophages through mTOR signaling and serine-glycine-glutathione axis [23, 41], we further explored whether glycine could also influence macrophage inflammation. Here, we found that exogenous glycine reduces the secretion of inflammatory cytokines from macrophages upon PmCQ2 infection. It should be mentioned that the dose of exogenous 10 mM glycine added in the experiment in vitro is far more than the physiological concentration (0.2–0.3 mM). Therefore, it is still an open question that the responses to supra-physiological concentrations of glycine have any meaning in vivo.
On the basis of the expression of specific markers, activated macrophages can be classified as classical activated (M1) or tissue-repaired (M2) macrophages [42, 43, 44, 45, 46], which are dependent of many regulators (including signaling pathways). mTOR is a central metabolic pathway that couples nutrient sensing to the regulation of metabolic processes and affects macrophage activation [47, 48]. In the study, in the context of PmCQ2 infection, mTOR pathway is not different in macrophages of WT and KO mice. HIF-1α is one of the important molecules to regulate the function of macrophages under hypoxia and can promote anaerobic glycolysis and phosphate pentose pathway metabolic reprogramming, thereby affecting macrophage polarization towards to proinflammatory phenotype [49, 50, 51]. Here, we found that there is no difference in HIF-1α expression of macrophages from WT and KO mice infected by PmCQ2. IKK/NF-κB signaling pathways involve in the regulation of the inflammatory responses [52], and NF-κB is a key transcriptional regulator of the proinflammatory macrophages [53]. However, Slc6a13 deficiency has little effect on NF-κB signaling. Notably, inflammasomes, including NALP1, NLRP3, NLRC4, and AIM2, are essential for regulating the inflammatory responses in macrophages [54]. In this study, Slc6a13 deficiency promotes accumulation of glycine to significantly inhibit the activation of inflammasomes in macrophages infected with PmCQ2. Indeed, the activation of inflammasome is known to require a two-step process of initiation and activation. The first step activates NF-κB through pattern recognition receptors such as TLR4 or cytokine receptors such as TNFRs, leading to the transcription and translation of NLRP3 and pro-IL-1β [55, 56]. The second step is the specific activation of NLRP3 triggered by the agonist, the assembly of the inflammasome complex, and, ultimately, the activation of Caspase-1 [55]. However, there was no significant difference in NF-κB pathway between WT and KO macrophages. Therefore, it is interesting to explore the underlying mechanisms, by which glycine inhibits the activation of inflammasomes during macrophage activation.
K+ efflux, Ca2+ flux, mitochondrial membrane potential (Δψm), mitochondrial ROS, ATP, and NAD production were also confirmed to be involved in NLRP3 inflammasome activation [55]. In this study, the expression of GlyTs and GlyRs was also detected, and it was found that the expression of Slc6a9 and Glra4 in KO mice and macrophages is increased. GlyR activation is known to be associated with Cl− flux [57], while GlyT is a Na+/Cl− -dependent membrane receptor [58]. Moreover, glycine acts as an co-agonist of N-methyl-D-aspartic acid receptor that can affect Na+ and Ca2+ flux [59], thus glycine may change intracellular ion flux through its transporter and/or receptor, thereby affecting inflammasome activation. Additionally, we also find that 1 mM glycine enhances mitochondrial fusion as well as mitochondrial respiration (unpublished data); therefore, glycine may also affect inflammasome activation by regulating mitochondrial activity. However, more experiments are needed to explore this specific mechanism.
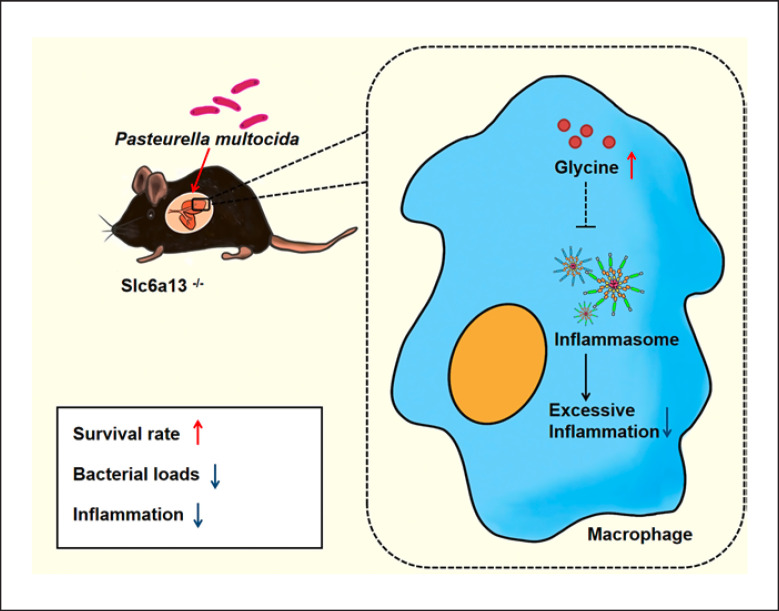
In conclusion, we find that the bacterial loads and hyperinflammatory responses of lung tissue in Slc6a13-deficient mice are significantly decreased, resulting in improved survival. Furthermore, glycine levels in the lungs of Slc6a13-deficient mice are remarkably upregulated. Notably, exogenous glycine administration lowers bacterial colonization and macrophage-mediated inflammation to enhance the survival rate of mice infected by P. multocida. Mechanically, Slc6a13 deficiency promotes glycine accumulation to curtail excessive inflammatory responses of macrophage involved in inflammasome (Fig. 7). Our findings uncover a previously unidentified amino acid metabolism-associating mechanism in guiding PmCQ2 infection.
Fig. 7.
Mechanism whereby Slc6a13 deficiency lessens PmcQ2 infection-triggered inflammation. Slc6a13 deficiency promotes glycine accumulation to attenuate P. multocida infection through reducing the excessive inflammatory responses of macrophages involving glycine-inflammasome signaling.
Statement of Ethics
The animal experiments were performed in compliance with the principles of the Basel Declaration and Recommendations of the Laboratory Animal Ethical Commission of the Southwest University (Permit No. IACUC-20190510-01), Chongqing, China.
Conflict of Interest Statement
There is no conflict of interest in this article.
Funding Sources
This work was supported by the China Agriculture Research System of MOF and MARA (Beef/Yak Cattle, CARS-37), the Chongqing Science & Technology Commission (cstc2017shms-zdyfx0036, cstc2017jcyjAX0288), and the College Student Innovation and Entrepreneurship Training Program Practical Project (S202110635157).
Author Contributions
Yuanyi Peng and Yaoyao Xia designed the experiment; Fang He, Yangyang Qiu, Xiaoyan Wu, Yaoyao Xia, Liu Yang, Chenlu Wu, Pan Li, and Rui Zhang conducted the experiment; Fang He, Yaoyao Xia, Chenlu Wu, Rendong Fang, and Nengzhang Li analyzed the data and prepared the figures; Fang He, Xiaoyan Wu, and Yaoyao Xia drafted the manuscript; Yuanyi Peng and Yaoyao Xia revised and approved the final manuscript.
Data Availability Statement
The raw data have been deposited to NCBIs Sequence Read Archive (SRA) database and the accession number is PRJNA706465.
Supplementary Material
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Acknowledgments
We thank Prof. Wenkai Ren for the kind help during the whole experimental period and the Beijing Genomics Institute (BGI, Shenzhen, China) for the sequencing service provided.
Funding Statement
This work was supported by the China Agriculture Research System of MOF and MARA (Beef/Yak Cattle, CARS-37), the Chongqing Science & Technology Commission (cstc2017shms-zdyfx0036, cstc2017jcyjAX0288), and the College Student Innovation and Entrepreneurship Training Program Practical Project (S202110635157).
References
- 1.Chauhan P, Saha B. Metabolic regulation of infection and inflammation. Cytokine. 2018 Dec;112:1–11. doi: 10.1016/j.cyto.2018.11.016. [DOI] [PubMed] [Google Scholar]
- 2.Ren W, Rajendran R, Zhao Y, Tan B, Wu G, Bazer FW, et al. Amino acids as mediators of metabolic cross talk between host and pathogen. Front Immunol. 2018;9:319. doi: 10.3389/fimmu.2018.00319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun X, Song L, Feng S, Li L, Yu H, Wang Q, et al. Fatty acid metabolism is associated with disease severity after H7N9 infection. EBioMedicine. 2018 Jul;33:218–229. doi: 10.1016/j.ebiom.2018.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang L, Wang P, Song X, Zhang H, Ma S, Wang J, et al. Salmonella typhimurium reprograms macrophage metabolism via T3SS effector SopE2 to promote intracellular replication and virulence. Nat Commun. 2021 Feb 9;12((1)):879. doi: 10.1038/s41467-021-21186-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ren W, Yin J, Gao W, Chen S, Duan J, Liu G, et al. Metabolomics study of metabolic variations in enterotoxigenic Escherichia coli-infected piglets. RSC Adv. 2015;5((73)):59550–5. [Google Scholar]
- 6.Ren W, Yin J, Xiao H, Chen S, Liu G, Tan B, et al. Intestinal microbiota-derived GABA mediates interleukin-17 expression during enterotoxigenic Escherichia coli infection. Front Immunol. 2016;7:685. doi: 10.3389/fimmu.2016.00685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He F, Yin Z, Wu C, Xia Y, Wu M, Li P, et al. L-serine lowers the inflammatory responses during Pasteurella multocida infection. Infect Immun. 2019 Dec;87((12)):e00677–19. doi: 10.1128/IAI.00677-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He F, Wu C, Li P, Li N, Zhang D, Zhu Q, et al. Functions and signaling pathways of amino acids in intestinal inflammation. Biomed Res Int. 2018;2018:9171905. doi: 10.1155/2018/9171905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wheeler MD, Thurman RG. Production of superoxide and TNF-alpha from alveolar macrophages is blunted by glycine. Am J Physiol. 1999 Nov;277((5)):L952–9. doi: 10.1152/ajplung.1999.277.5.L952. [DOI] [PubMed] [Google Scholar]
- 10.Wheeler MD, Rose ML, Yamashima S, Enomoto N, Seabra V, Madren J, et al. Dietary glycine blunts lung inflammatory cell influx following acute endotoxin. Am J Physiol Lung Cell Mol Physiol. 2000 Aug;279((2)):L390–8. doi: 10.1152/ajplung.2000.279.2.L390. [DOI] [PubMed] [Google Scholar]
- 11.Blancas-Flores G, Alarcón-Aguilar FJ, García-Macedo R, Almanza-Pérez JC, Flores-Sáenz JL, Román-Ramos R, et al. Glycine suppresses TNF-α-induced activation of NF-κB in differentiated 3T3-L1 adipocytes. Eur J Pharmacol. 2012 Aug 15;689((1–3)):270–277. doi: 10.1016/j.ejphar.2012.06.025. [DOI] [PubMed] [Google Scholar]
- 12.Xu X, Wang X, Wu H, Zhu H, Liu C, Hou Y, et al. Glycine relieves intestinal injury by maintaining mTOR signaling and suppressing AMPK, TLR4, and NOD signaling in weaned piglets after lipopolysaccharide challenge. Int J Mol Sci. 2018 Jul 6;19((7)):1980. doi: 10.3390/ijms19071980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y, Ma X, Jiang D, Chen J, Jia H, Wu Z, et al. Glycine attenuates lipopolysaccharide-induced acute lung injury by regulating NLRP3 inflammasome and NRF2 signaling. Nutrients. 2020 Feb 26;12((3)):611. doi: 10.3390/nu12030611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu C, Qin X, Du H, Li N, Ren W, Peng Y. The immunological function of GABAergic system. Front Biosci. 2017 Mar 1;22((7)):1162–1172. doi: 10.2741/4539. [DOI] [PubMed] [Google Scholar]
- 15.Prud'homme GJ, Glinka Y, Wang Q. Immunological GABAergic interactions and therapeutic applications in autoimmune diseases. Autoimmun Rev. 2015 Nov;14((11)):1048–1056. doi: 10.1016/j.autrev.2015.07.011. [DOI] [PubMed] [Google Scholar]
- 16.Crowley T, Cryan JF, Downer EJ, O'Leary OF. Inhibiting neuroinflammation: the role and therapeutic potential of GABA in neuro-immune interactions. Brain Behav Immun. 2016 May;54:260–277. doi: 10.1016/j.bbi.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 17.Ren W, Liao Y, Ding X, Jiang Y, Yan J, Xia Y, et al. Slc6a13 deficiency promotes Th17 responses during intestinal bacterial infection. Mucosal Immunol. 2019 Mar;12((2)):531–544. doi: 10.1038/s41385-018-0111-7. [DOI] [PubMed] [Google Scholar]
- 18.Xia Y, He F, Wu X, Tan B, Chen S, Liao Y, et al. GABA transporter sustains IL-1β production in macrophages. Sci Adv. 2021;7((15)):eabe9274. doi: 10.1126/sciadv.abe9274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Du H, Wu C, Li C, Fang R, Ma J, Ji J, et al. Two novel cross-protective antigens for bovine Pasteurella multocida. Mol Med Rep. 2017 Oct;16((4)):4627–4633. doi: 10.3892/mmr.2017.7153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He F, Qin X, Xu N, Li P, Wu X, Duan L, et al. Pasteurella multocida Pm0442 affects virulence gene expression and targets TLR2 to induce inflammatory responses. Front Microbiol. 2020;11((14)):1972. doi: 10.3389/fmicb.2020.01972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu C, Qin X, Li P, Pan T, Ren W, Li N, et al. Transcriptomic analysis on responses of murine lungs to Pasteurella multocida infection. Front Cell Infect Microbiol. 2017;7:251. doi: 10.3389/fcimb.2017.00251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mbuthia PG, Christensen H, Boye M, Petersen KM, Bisgaard M, Nyaga PN, et al. Specific detection of Pasteurella multocida in chickens with fowl cholera and in pig lung tissues using fluorescent rRNA in situ hybridization. J Clin Microbiol. 2001 Jul;39((7)):2627–2633. doi: 10.1128/JCM.39.7.2627-2633.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen S, Xia Y, He F, Fu J, Xin Z, Deng B, et al. Serine supports IL-1β production in macrophages through mTOR signaling. Front Immunol. 2020;11:1866. doi: 10.3389/fimmu.2020.01866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van Hoecke L, Job ER, Saelens X, Roose K. Bronchoalveolar lavage of murine lungs to analyze inflammatory cell infiltration. J Vis Exp. 2017 May;4((123)):55398. doi: 10.3791/55398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li P, He F, Wu C, Zhao G, Hardwidge PR, Li N, et al. Transcriptomic analysis of chicken lungs infected with avian and bovine Pasteurella multocida serotype A. Front Vet Sci. 2020;7:452. doi: 10.3389/fvets.2020.00452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li N, Feng T, Wang Y, Li P, Yin Y, Zhao Z, et al. A single point mutation in the hyaC gene affects Pasteurella multocida serovar A capsule production and virulence. Microb Pathog. 2021 Oct;159:105145. doi: 10.1016/j.micpath.2021.105145. [DOI] [PubMed] [Google Scholar]
- 27.Li R, Li Y, Kristiansen K, Wang J. SOAP: short oligonucleotide alignment program. Bioinformatics. 2008 Mar 1;24((5)):713–714. doi: 10.1093/bioinformatics/btn025. [DOI] [PubMed] [Google Scholar]
- 28.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015 Apr;12((4)):357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012 Mar 4;9((4)):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011 Aug 4;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shoemaker JE, Lopes TJ, Ghosh S, Matsuoka Y, Kawaoka Y, Kitano H. CTen: a web-based platform for identifying enriched cell types from heterogeneous microarray data. BMC Genomics. 2012 Sep 6;13:460. doi: 10.1186/1471-2164-13-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deist MS, Gallardo RA, Bunn DA, Dekkers JCM, Zhou H, Lamont SJ. Resistant and susceptible chicken lines show distinctive responses to Newcastle disease virus infection in the lung transcriptome. BMC Genomics. 2017 Dec 28;18((1)):989. doi: 10.1186/s12864-017-4380-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang J, Kaiser MG, Deist MS, Gallardo RA, Bunn DA, Kelly TR, et al. Transcriptome analysis in spleen reveals differential regulation of response to Newcastle disease virus in two chicken lines. Sci Rep. 2018 Jan 19;8((1)):1278. doi: 10.1038/s41598-018-19754-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Melamed N, Kanner BI. Transmembrane domains I and II of the gamma-aminobutyric acid transporter GAT-4 contain molecular determinants of substrate specificity. Mol Pharmacol. 2004 Jun;65((6)):1452–1461. doi: 10.1124/mol.65.6.1452. [DOI] [PubMed] [Google Scholar]
- 35.Ding X, Yan D, Zhang X, Liu B, Zhu G. Metabolomics analysis of the effect of GAT-2 deficiency on Th1 cells in mice. J Proteome Res. 2021 Nov 5;20((11)):5054–5063. doi: 10.1021/acs.jproteome.1c00601. [DOI] [PubMed] [Google Scholar]
- 36.Song Z, Hatton GI. Taurine and the control of basal hormone release from rat neurohypophysis. Exp Neurol. 2003 Oct;183((2)):330–337. doi: 10.1016/s0014-4886(03)00105-5. [DOI] [PubMed] [Google Scholar]
- 37.Welsh BT, Kirson D, Allen HM, Mihic SJ. Ethanol enhances taurine-activated glycine receptor function. Alcohol Clin Exp Res. 2010 Sep 1;34((9)):1634–1639. doi: 10.1111/j.1530-0277.2010.01249.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen C, Xia S, He J, Lu G, Xie Z, Han H. Roles of taurine in cognitive function of physiology, pathologies and toxication. Life Sci. 2019 Aug 15;231:116584. doi: 10.1016/j.lfs.2019.116584. [DOI] [PubMed] [Google Scholar]
- 39.Fang R, Du H, Lei G, Liu Y, Feng S, Ye C, et al. NLRP3 inflammasome plays an important role in caspase-1 activation and IL-1β secretion in macrophages infected with Pasteurella multocida. Vet Microbiol. 2019 Apr;231:207–213. doi: 10.1016/j.vetmic.2019.03.019. [DOI] [PubMed] [Google Scholar]
- 40.Fang RD, Lei GH, Jiang JL, Du HH, Liu YJ, Lei ZH, et al. High- and low-virulent bovine Pasteurella multocida induced differential NLRP3 inflammasome activation and subsequent IL-1β secretion. Vet Microbiol. 2020 Apr;243:108646. doi: 10.1016/j.vetmic.2020.108646. [DOI] [PubMed] [Google Scholar]
- 41.Rodriguez AE, Ducker GS, Billingham LK, Martinez CA, Mainolfi N, Suri V, et al. Serine metabolism supports macrophage IL-1β production. Cell Metab. 2019 Apr 2;29((4)):1003–11.e4. doi: 10.1016/j.cmet.2019.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Benoit M, Desnues B, Mege JL. Macrophage polarization in bacterial infections. J Immunol. 2008 Sep 15;181((6)):3733–3739. doi: 10.4049/jimmunol.181.6.3733. [DOI] [PubMed] [Google Scholar]
- 43.Mège JL, Mehraj V, Capo C. Macrophage polarization and bacterial infections. Curr Opin Infect Dis. 2011 Jun;24((3)):230–234. doi: 10.1097/QCO.0b013e328344b73e. [DOI] [PubMed] [Google Scholar]
- 44.Reales-Calderón JA, Aguilera-Montilla N, Corbí Á L, Molero G, Gil C. Proteomic characterization of human proinflammatory M1 and anti-inflammatory M2 macrophages and their response to Candida albicans. Proteomics. 2014 Jun;14((12)):1503–1518. doi: 10.1002/pmic.201300508. [DOI] [PubMed] [Google Scholar]
- 45.Zhou D, Huang C, Lin Z, Zhan S, Kong L, Fang C, et al. Macrophage polarization and function with emphasis on the evolving roles of coordinated regulation of cellular signaling pathways. Cell Signal. 2014 Feb;26((2)):192–197. doi: 10.1016/j.cellsig.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 46.Hesketh M, Sahin KB, West ZE, Murray RZ. Macrophage phenotypes regulate scar formation and chronic wound healing. Int J Mol Sci. 2017 Jul 17;18((7)):1545. doi: 10.3390/ijms18071545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Byles V, Covarrubias AJ, Ben-Sahra I, Lamming DW, Sabatini DM, Manning BD, et al. The TSC-mTOR pathway regulates macrophage polarization. Nat Commun. 2013;4:2834. doi: 10.1038/ncomms3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weichhart T, Hengstschläger M, Linke M. Regulation of innate immune cell function by mTOR. Nat Rev Immunol. 2015 Oct;15((10)):599–614. doi: 10.1038/nri3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Werno C, Menrad H, Weigert A, Dehne N, Goerdt S, Schledzewski K, et al. Knockout of HIF-1α in tumor-associated macrophages enhances M2 polarization and attenuates their pro-angiogenic responses. Carcinogenesis. 2010 Oct;31((10)):1863–1872. doi: 10.1093/carcin/bgq088. [DOI] [PubMed] [Google Scholar]
- 50.Tannahill GM, Curtis AM, Adamik J, Palsson-McDermott EM, McGettrick AF, Goel G, et al. Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature. 2013 Apr 11;496((7444)):238–242. doi: 10.1038/nature11986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Van den Bossche J, Baardman J, Otto NA, van der Velden S, Neele AE, van den Berg SM, et al. Mitochondrial dysfunction prevents repolarization of inflammatory macrophages. Cell Rep. 2016 Oct 11;17((3)):684–696. doi: 10.1016/j.celrep.2016.09.008. [DOI] [PubMed] [Google Scholar]
- 52.Gao LN, Feng QS, Zhang XF, Wang QS, Cui YL. Tetrandrine suppresses articular inflammatory response by inhibiting pro-inflammatory factors via NF-κB inactivation. J Orthop Res. 2016 Sep;34((9)):1557–1568. doi: 10.1002/jor.23155. [DOI] [PubMed] [Google Scholar]
- 53.Murray PJ, Wynn TA. Protective and pathogenic functions of macrophage subsets. Nat Rev Immunol. 2011 Oct 14;11((11)):723–737. doi: 10.1038/nri3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liang N, Yang YP, Li W, Wu YY, Zhang ZW, Luo Y, et al. Overexpression of NLRP3, NLRC4 and AIM2 inflammasomes and their priming-associated molecules (TLR2, TLR4, Dectin-1, Dectin-2 and NFκB) in Malassezia folliculitis. Mycoses. 2018 Feb;61((2)):111–118. doi: 10.1111/myc.12711. [DOI] [PubMed] [Google Scholar]
- 55.Sutterwala FS, Haasken S, Cassel SL. Mechanism of NLRP3 inflammasome activation. Ann N Y Acad Sci. 2014;1319((1)):82–95. doi: 10.1111/nyas.12458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sharma BR, Kanneganti TD. NLRP3 inflammasome in cancer and metabolic diseases. Nat Immunol. 2021 May;22((5)):550–559. doi: 10.1038/s41590-021-00886-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Caravagna C, Casciato A, Coq JO, Liabeuf S, Brocard C, Peyronnet J, et al. Prenatal hypoxia induces Cl- cotransporters KCC2 and NKCC1 developmental abnormality and disturbs the influence of GABAA and glycine receptors on fictive breathing in a newborn rat. Front Physiol. 2022;13:786714. doi: 10.3389/fphys.2022.786714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Marques BL, Oliveira-Lima OC, Carvalho GA, de Almeida Chiarelli R, Ribeiro RI, Parreira RC, et al. Neurobiology of glycine transporters: from molecules to behavior. Neurosci Biobehav Rev. 2020 Nov;118:97–110. doi: 10.1016/j.neubiorev.2020.07.025. [DOI] [PubMed] [Google Scholar]
- 59.Montes de Oca Balderas P. Flux-independent NMDAR signaling: molecular mediators, cellular functions, and complexities. Int J Mol Sci. 2018 Nov 29;19((12)):3800. doi: 10.3390/ijms19123800. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Data Availability Statement
The raw data have been deposited to NCBIs Sequence Read Archive (SRA) database and the accession number is PRJNA706465.