Abstract
The gut of the soil microarthropod Folsomia candida provides a habitat for a high density of bacterial cells (T. Thimm, A. Hoffmann, H. Borkott, J. C. Munch, and C. C. Tebbe, Appl. Environ. Microbiol. 64:2660–2669, 1998). We investigated whether these gut bacteria act as recipients for plasmids from Escherichia coli. Filter mating with E. coli donor cells and collected feces of F. candida revealed that the broad-host-range conjugative plasmid pRP4-luc (pRP4 with a luciferase marker gene) transferred to fecal bacteria at estimated frequencies of 5.4 × 10−1 transconjugants per donor. The mobilizable plasmid pSUP104-luc was transferred from the IncQ mobilizing strain E. coli S17-1 and less efficiently from the IncF1 mobilizing strain NM522 but not from the nonmobilizing strain HB101. When S17-1 donor strains were fed to F. candida, transconjugants of pRP4-luc and pSUP104-luc were isolated from feces. Additionally, the narrow-host-range plasmid pSUP202-luc was transferred to indigenous bacteria, which, however, could not maintain this plasmid. Inhibition experiments with nalidixic acid indicated that pRP4-luc plasmid transfer took place in the gut rather than in the feces. A remarkable diversity of transconjugants was isolated in this study: from a total of 264 transconjugants, 15 strains belonging to the alpha, beta, or gamma subclass of the class Proteobacteria were identified by DNA sequencing of the PCR-amplified 16S rRNA genes and substrate utilization assays (Biolog). Except for Alcaligenes faecalis, which was identified by the Biolog assay, none of the isolates was identical to reference strains from data banks. This study indicates the importance of the microarthropod gut for enhanced conjugative gene transfer in soil microbial communities.
Gene transfer is a process by which bacterial populations substantially increase their rates of evolution and adaptation (12, 59). Particularly, plasmid-located genes, which are transferred by conjugation from donor to recipient cells, can disseminate rapidly between even phylogenetically different bacterial groups (17, 36, 41) and microbial communities in different spatial habitats (34, 71). Such microbial genetic networks should be considered in risk assessments of releases of genetically engineered microorganisms into the environment (22, 37, 43). The probability and rate of plasmid transfer from a donor to indigenous microorganisms in a given habitat are influenced by plasmid-borne genes which determine the type of transfer mechanism (self-transmissible or mobilizable) and the host range of autonomous plasmid replication. Additionally, specific physicochemical conditions, such as temperature, water potential, and the availability of energy (substrates) for donor and recipient cells, are important factors influencing gene transfer rates in terrestrial and aquatic environments (23, 53, 64).
The spread of plasmid-borne genes is still extremely difficult to predict for terrestrial habitats, since a large variety of microhabitat conditions which are not well characterized exists. In bulk soil under laboratory conditions, conjugative gene transfer from recombinant bacterial donor strains to indigenous soil bacteria has been found only under specific selective conditions or on rare occasions (11, 20, 24, 27, 50, 61). Several studies failed to detect such transfer events, and it was concluded that heterogeneity and low densities of recipient cells, as well as a lack of substrates for microbial metabolism, prevented efficient plasmid transfer in bulk soil (19, 49, 54, 75). Plant exudates increased rates of gene transfer in soil (33, 48), and higher rates of gene transfer were found in rhizospheres than in bulk soil (50, 61). It was assumed that other microsites which favor gene transfer in terrestrial habitats are associated with soil invertebrates (74). However, to date little experimental evidence to prove this assumption is available.
Intraspecies transconjugants of added Enterobacter cloacae donor and recipient cells could be isolated from microcosm experiments with the variegated cutworm, Peridroma saucia, and plant material (2). The investigators in that study concluded that gene transfer events happened, most likely, in the digestive tracts or in the feces of the insects. Another recent report demonstrated that a conjugative plasmid was transferred between fed Escherichia coli strains in the guts of Rhabditis nematodes (1). Earthworms mediated transport and enhanced plasmid transfer from added donor cells to added recipients and to indigenous bacteria in soil (14, 15). High rates of intraspecies plasmid transfer, comparable to those obtained in pure broth cultures, were detected with Bacillus thuringiensis in infected lepidopterous larvae (31).
Microarthropods (collembolans and mites) are the most abundant invertebrate group in the majority of soils (5) but have not been recognized, so far, for their impact on microbial gene transfer. There are some indications that microarthropods harbor a large variety of microorganisms in their guts and thereby contribute to microbial biodiversity in terrestrial environments (7, 9, 57). In the accompanying paper, we have described the gut of Folsomia candida (Collembola) as a habitat and species-specific vector for microorganisms (67). The gut of this soil-dwelling insect, which has a volume of only several nanoliters, was found to be densely colonized, predominantly by rod-shaped bacterial cells. We were interested to know whether such bacterial cells act as recipients for plasmids and thereby promote gene transfer in microbial communities. F. candida feeds, under natural conditions, on bacteria (3), fungal mycelia (6, 66), and nematodes (35). Here, we report on the results of experiments in which plasmid-bearing E. coli strains were fed to F. candida in microcosms. Self-transferable plasmids, as well as mobilizable plasmids with different host ranges, and a nonmobilizable plasmid were included in this study in order to determine the specific capacities of these different classes of plasmids to spread into indigenous bacterial populations. For detection purposes, all plasmids were engineered by the insertion of the luciferase-encoding marker gene luc or lux (30, 47).
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth media.
E. coli donor strains and plasmids used in this study are shown in Table 1. Cloning of the luciferase genes did not affect maintenance or transfer functions of the individual plasmids. All strains were cultivated on Luria-Bertani (LB) medium (51). In order to maintain the plasmids, filter-sterilized antibiotic stock solutions were added to the autoclaved media to the following final concentrations for the listed plasmids: 10 μg of tetracycline ml−1 for pRP4-luc and pUTluxAB, 100 μg of ampicillin ml−1 for pUC18-luc, 100 μg of ampicillin and 50 μg of chloramphenicol ml−1 for pSUP202-luc, and 50 μg of chloramphenicol ml−1 for pSUP104-luc.
TABLE 1.
E. coli strains and plasmids used in this study
| Strain or plasmid | Description | Reference and/or source |
|---|---|---|
| E. coli strains | ||
| S17-1 | recA derivative of E. coli 294 (F−thi pro hsdR) carrying a modified derivative of IncPα plasmid pRP4 (Aps Tcs Kms) integrated in the chromosome, Tpr | 58 |
| S17-1 (λpir) | λ lysogenic S17-1 derivative producing π protein for replication of plasmids carrying oriR6K | K. N. Timmis |
| CC118 (λpir) | Routine strain for maintenance of π-dependent plasmids carrying oriR6K | 28 |
| NM522 | hsdR strain with F′ plasmid | Promega Corporation |
| HB101 | F−hsdS (rB− mB−) thi pro leu lacY ara xyl supE recA13 Smr | 10; obtained from Promega Corporation |
| Plasmids | ||
| pUC18 | Cloning vector, Apr | 77 |
| pUC18-luc | pUC18 carrying a 2.3-kb npt-2p–luc cassette inserted into a multicloning site as an HindIII fragment, confers a bioluminescence phenotype | J. Schiemann |
| pSUP202 | Tra− Mob+ derivative of plasmid pBR325, narrow host range of replication, Apr Cmr Tcr | 58 |
| pSUP202-luc | pSUP202 carrying a 2.3-kb npt-2p–luc cassette inserted into an HindIII site, confers a bioluminescence phenotype, Apr Cmr | J. Schiemann |
| pSUP104 | Tra− Mob+ derivative of plasmid pACYC184, broad host range of replication, Cmr Tcr | 46 |
| pSUP104-luc | pSUP104 carrying a 2.3-kb npt-2p–luc cassette inserted into an HindIII site, Cmr | This study |
| pUT-luxAB | Tra− Mob+ derivative of pUT (oriR6K) with a miniTn5 transposon and carrying promoterless luxAB and luxB genes, Tcr | 18 |
| pRP4 | Tra+, IncPα broad-host-range plasmid, Apr Kmr Tcr | 68 |
| pRP4-luc | pRP4 carrying a 2.3-kb npt-2p–luc cassette cloned into an HindIII site, confers a bioluminescence phenotype, Apr Tcr | This study |
All media inoculated with feces from F. candida were amended with cycloheximide (100 μg ml−1) in order to inhibit growth of eukaryotic microorganisms. The total number of bacteria (heterotrophic, aerobic, and culturable) from feces and the gut of F. candida was determined, in this study, on nonamended LB agar. Agar medium cultures were incubated at 28°C for 2 days. Donor strains were enumerated after growth on LB agar with the appropriate antibiotic additions at 37°C. Recipients were obtained on M9 minimal growth medium (51) with purified agar (Merck, Darmstadt, Germany) and benzoic acid (2.5 mM) as the sole source of organic carbon. Transconjugants were grown on M9 benzoic acid growth agar with plasmid-selective antibiotics. Antibiotic concentrations were identical to those indicated above for LB medium. Transconjugants were cultivated routinely onto M9 agar with benzoic acid and plasmid-selective antibiotics.
Construction of pRP4-luc.
A 2.3-kb nptII promoter–luc cassette (56) was ligated as an HindIII fragment to HindIII-digested pRP4, and the ligation mixture was transformed into E. coli HB101. Transformants were selected on LB agar with tetracycline. Of 100 tetracycline-resistant clones tested, 20 conferred a bioluminescence phenotype. Restriction enzyme analysis of a selected plasmid designated pRP4-luc confirmed the presence of a single 2.3-kb HindIII fragment in pRP4-luc (data not shown).
Filter mating experiments with E. coli donor strains and feces of F. candida.
Fecal pellets were collected from petri dish microcosms with water-agar (1.5%, pH 7.0), in which a total of 100 specimens of F. candida were incubated with one YTP (2.0 g each of yeast extract, tryptone, and peptone liter−1) agar cube (1.7-cm2 surface, 0.5-cm thick) as a sole nutrient source. Microcosms were incubated for 10 days at 18°C in the dark. After the insects and the remaining YTP agar cube were removed, the feces were suspended from the water-agar twice with 1 ml of 0.85% NaCl solution each time. The suspensions were collected in Eppendorf tubes, with one tube per microcosm, and centrifuged for 5 min at 2,700 × g in a microcentrifuge. The supernatant was removed, and the pellet was suspended with 200 μl of a donor cell suspension. The donor cell suspension was obtained by the following procedure. Overnight cultures were grown in 75 ml of LB broth with the appropriate antibiotic(s) (dependent upon the plasmid) at 37°C and shaken at 200 rpm on a rotary shaker (TM-3; Infors, Basel, Switzerland). Cells were harvested by centrifugation (5 min at 2,700 × g and 4°C), resuspended in 75 ml of 0.85% NaCl solution, centrifuged again, and, finally, resuspended in 2 ml of 0.85% NaCl solution. Suspensions of feces and donor strains (200 μl) were transferred directly onto LB agar (1.2% agar), which had been covered previously with a presterilized nylon filter (8.2 cm in diameter, Hybond-N; Amersham, Braunschweig, Germany). After an incubation at 28°C for 24 h, filters were removed carefully with a sterile forceps and transferred into 50-ml Falcon tubes, and grown cells were suspended in 2 ml of NaCl solution by vortexing at the highest setting. Dilutions were inoculated onto the appropriate media for determination of total numbers of bacteria and donor, recipient, and transconjugant cells. Donor and recipient cells were also inoculated onto separate filters, incubated overnight, diluted, and cultivated on the appropriate media in order to correlate the effect of the transconjugant detection technique with the occurrence of transconjugants (plate mating). This control is required for plasmids, such as pRP4, which have high transfer potentials (63).
Feeding experiments.
Microcosms for feeding experiments of F. candida with E. coli strains consisted of petri dishes with water-agar (1.5%, pH 7.0) and a YTP agar cube (see above) in the center. A total of 200 μl of donor cells (approximately 1010 cells), obtained as described above for the filter mating experiments, were loaded carefully onto the YTP agar cube and air dried for 30 min under sterile conditions. Each microcosm was inoculated with a total of 100 specimens of F. candida taken from a breeding stock (67) and preincubated (starved) for 24 h in petri dishes with sterile water-agar. Microcosms were kept at 18°C in the dark. All treatments were tested with three replicate microcosms. Before analysis of the microbial populations in the feces, all animals were anesthetized by a CO2 fumigation and were counted, and specimens were stored at −20°C for further analyses. The YTP agar cube with remaining donor cells was then removed carefully from the water-agar, and the feces which lay on the surface of the water-agar were extracted twice with 1 ml of 0.85% NaCl solution, as described above for the filter mating.
Detection of reporter genes.
Two different types of reporter genes were used in this study: (i) a firefly-derived luciferase (luc) gene (30, 44) and (ii) a bacterial luciferase-encoding luxAB gene (47). Bioluminescence of grown colonies was detected after transfer onto nylon membranes (Hybond-N), as described by Selbitschka et al. (55). The luxAB-encoded luciferase was detected by a procedure similar to that used for detection of the luc luciferase, with the exception that instead of the substrate luciferin being loaded onto the surfaces of the membranes, 4 drops (20 μl each) of the undiluted substrate n-decyl aldehyde (Sigma, St. Louis, Mo.) were placed next to the filter in opposite positions. To detect light emission of luc or luxAB reporter gene-encoded enzymes, a film (Kodak T-Mat plus DG; Kodak-Pathé, Paris, France) was placed onto the membranes and incubated in a light-tight film cassette at room temperature overnight in a dark room. After development of the film, the number of bioluminescent colonies could be determined.
The luc luciferase gene was also detected in some experiments by PCR by amplifying an internal 302-bp fragment of the gene with primers P2 and P3 under the conditions described by Dammann-Kalinowski et al. (16).
Characterization of transconjugants.
Transconjugants isolated from the gut or feces of F. candida were diluted for purification and subcultured on selective growth agar. Grown colonies were transferred, in two replicates for each strain, onto selective agar in order to exclude plasmid segregants from further analyses. Colonies grown on one replicate plate were analyzed for reporter gene expression, as described above, and the corresponding colonies on replicate plates were subcultured further and subjected to amplified ribosomal DNA restriction analysis (ARDRA) (73).
For ARDRA, single colonies with confirmed reporter gene activity were suspended in 50 μl of lysis buffer (0.05 M NaOH, 0.25% sodium dodecyl sulfate) in reaction tubes and incubated at 95°C for 15 min. Subsequently, 450 μl of water was added and the suspension was microcentrifuged, at the highest setting, for 5 min at room temperature. A total of 450 μl was then transferred to a new reaction tube and served as a template for PCR amplifications. PCR was conducted by targeting a 1.2-kb region of the 16S rRNA gene with universal eubacterial primers. Primer sequences were obtained from R. Simon and H.-V. Tichy, Freiburg, Germany. The forward primer (41f) sequence was (5′ to 3′) GCT CAG ATT GAA CGC TGG CG, and the reverse primer (1066r) sequence (5′ to 3′) was ACA TTT CAC AAC ACG AGC TG. PCR was carried out in 50-μl volumes consisting of 5 μl of 10× PCR buffer (100 mM Tris-HCl, 25 mM MgCl2, 500 mM KCl), 1 μl of a deoxynucleoside triphosphate (dATP, dCTP, dGTP, and dTTP [2.5 mM each]) mixture, 1 μl of each primer (10 mM each), of 0.2 μl of Taq polymerase (1 U; Pharmacia, Freiburg, Germany), 2 μl of template DNA, and water to reach the final volume. Solutions were covered with 10 μl of Chill-out 14 liquid wax (MJ Research, obtained from Biozym, Hessisch, Oldendorf, Germany). Tubes were incubated in a thermocycler (Omni Gene; Hybaid Limited, Teddington, United Kingdom) for 2 min at 95°C, followed by 30 cycles, each consisting of 1 min at 95°C, 1 min at 50°C, and 1 min at 72°C. Final primer extension was 5 min at 72°C. PCR products were taken directly for restriction enzyme digestions.
A total of 5 μl of PCR product was incubated with 10 U of a restriction endonuclease (CfoI and AluI, separately; both were obtained from Boehringer Mannheim, Mannheim, Germany) and the manufacturer-recommended incubation buffer in a final volume of 20 μl overnight at 37°C. Restriction fragment length polymorphisms were analyzed after agarose gel electrophoreses (2% low-melting-point, ultra-pure agarose; Gibco BRL, Life Technologies Inc., Gaithersburg, Md.) and staining with ethidium bromide (51) on a UV (312 nm)-illuminated table. Representative isolates of each ARDRA group, consisting of isolates with identical fragment length profiles, were then characterized by Gram staining (42).
Phenotypic characterization of the isolates was performed by a microtiter plate-bound substrate utilization assay (Biolog; Biolog Inc., Hayward, Calif.) (8). Substrate utilization patterns were recorded with a microtiter plate spectrophotometer (Vmax; Molecular Devices, Menlo Park, Calif.) and compared to patterns in a database (MicroLog GN 3.50) with Microlog2 software (Biolog Inc.).
Determination and analysis of 16S rRNA gene sequences of transconjugants.
Genomic DNAs were prepared from individual colonies picked from agar medium, resuspended in 100 μl of TE buffer (50 mM Tris-HCl, 1 mM EDTA [pH 8.0]), and incubated at 95°C for 5 min. After cooling on ice, the cell debris was pelleted in a microcentrifuge for 30 s. The genomic DNA in the supernatant was concentrated with Microcon-100 spin concentrators (Amicon GmbH, Witten, Germany) and resuspended in 10 μl of TE buffer.
The 16S rRNA genes were targeted for amplification by PCR with aliquots (between 2 and 5 μl) of the DNA suspension, a forward primer hybridizing at the complement of positions 8 to 27, a reverse primer hybridizing at positions 1525 to 1541 (E. coli 16S rRNA gene sequence numbering), and reaction conditions described in detail previously (32). The 16S rRNA gene PCR products were purified with Microcon-100 spin concentrators and sequenced directly with a Perkin-Elmer/Applied Biosystems, Inc. (Weiterstadt, Germany) model 373A DNA sequencer according to the protocols of the manufacturer for Taq cycle sequencing with fluorescent-dye-labelled dideoxynucleotides.
Sequence data were aligned with reference rRNA and rRNA gene sequences (40, 72) with evolutionarily conserved primary sequence and secondary structure as references (26). Cluster analyses were carried out with programs contained in the Phylogeny Inference Package (PHYLIP), version 3.5c (J. Felsenstein, University of Washington).
RESULTS
Plasmid transfer from E. coli to fecal bacteria.
Filter matings with E. coli donor strains and bacteria extracted directly from collected feces of F. candida were carried out in order to detect plasmid transfer potentials. Donor counterselection was achieved on minimal medium with benzoic acid as the sole source of carbon, a substrate which could not be used by the E. coli strains in this investigation. Table 2 indicates that a significant proportion of fecal bacteria (37.1% ± 33.7% of CFU obtained on nonselective yeast tryptone medium) was capable of using benzoic acid as the sole carbon source.
TABLE 2.
Results of filter mating experiments with different plasmid-bearing E. coli donor strains and feces collected from F. candida after an incubation period of 24 h
| E. coli donor strain | Plasmid | log CFU per filter
|
Estimated plasmid transfer frequency, no. of trans- conjugant cells per:
|
||||
|---|---|---|---|---|---|---|---|
| Total fecal cells | Recipient cells | Donor cells | Transconjugants | Recipient cell | Donor cell | ||
| S17-1 | pUC18-luc | 9.20 | 8.74 | 8.80 | NDa | <1.0 × 10−9 | <1.0 × 10−9 |
| pSUP202-luc | 10.27 | 10.20 | 8.38 | ND | <6.3 × 10−11 | <4.0 × 10−9 | |
| pSUP104-luc | 10.44 | 9.70 | 6.45 | 6.58 | 7.7 × 10−4 | 1.35 × 100 | |
| pRP4-luc | 9.43 | 9.40 | 8.15 | 7.60 | 1.6 × 10−2 | 5.4 × 10−1 | |
| HB101 | pSUP104-luc | 9.37 | 8.42 | 8.60 | ND | <3.0 × 10−9 | <2.0 × 10−9 |
| NM522 | pSUP104-luc | 9.74 | 9.14 | 9.18 | 6.93 | 6.2 × 10−3 | 5.6 × 10−3 |
| S17-1 λPir | pUTluxAB | 8.57 | 8.43 | 4.74 | ND | <3.0 × 10−9 | <1.8 × 10−5 |
| CC118 λPir | pUTluxAB | 10.39 | 9.35 | 9.15 | ND | <4.5 × 10−10 | <7.1 × 10−10 |
ND, not detected.
A total of five different plasmids, two of them in various E. coli host strains, were selected. Transconjugants were detected in matings with the self-transferable, broad-host-range IncP1 plasmid pRP4-luc and with the IncQ plasmid pSUP104-luc (Table 2). The latter plasmid was mobilized from E. coli S17-1, a strain with chromosomally inserted tra (transfer) genes of the IncP plasmid pRP4 (see also Table 1). The IncF′ E. coli strain NM522 was also capable of promoting IncQ plasmid transfer into indigenous fecal bacteria. Transfer rates, calculated as transconjugants per donor cell and also as transconjugants per recipient cell, indicated that pSUP104-luc mobilization from E. coli NM522 was less efficient than that from E. coli S17-1. Plasmid pSUP104-luc was not transferred from the nonmobilizing strain HB101. Narrow-host-range plasmid pSUP202-luc was also not transferred to fecal bacteria. Filter matings with the broad-host-range mobilizing strain S17-1 (λPir) and miniTn5 plasmid pUTluxAB, which aimed at detecting transconjugants with chromosomally inserted luxAB genes among the fecal bacteria, failed. Additionally, no transconjugants were detected after pUTluxAB transfer experiments with CC118λPir as the donor and S17-1 as the mobilizing strain.
Transfer frequencies could only be estimated, because clonal expansion of early occurring transconjugants during the 24-h incubation period could not be excluded. Transfer rates above 1, as determined for E. coli S17-1 with pSUP104-luc (Table 2), are indicative of growth of transconjugant populations. Generally, however, the frequencies shown in Table 2 indicate that transfer efficiencies for both pRP4-luc and pSUP104-luc from E. coli S17-1 were comparable to those which are observed for intraspecies transfer with added E. coli recipients under optimum conditions.
Plasmid transfer to F. candida-associated microorganisms in feeding experiments.
F. candida specimens were fed with the same selection of donor strains as that used in the previously described filter matings. Bacterial populations were analyzed after 7 and 14 days of incubation. After 7 days, 91 specimens (±7.7; n = 12) and, after 14 days, 72 specimens (±21.1) of 100 initial specimens were still alive in the microcosms. The specimens which were alive could be distinguished by their behavior: one group was actively feeding and another group, which was occupied with molting, was not feeding. The analysis of feces after 7 and 14 days indicated a high proportion of recipient cells among all culturable (LB) medium cells (Table 3). In contrast to the results of the filter mating experiments, donor cells were recovered only at very low concentrations, indicating that the E. coli cells were digested in the gut of F. candida. Transconjugants were recovered from microcosms with strain S17-1 harboring pRP4-luc and pSUP104-luc. In contrast to the results obtained from the filter mating experiments, mobilization of pSUP202-luc from E. coli S17-1 to gut bacteria was detected. Except for pSUP202-luc, the incidence of transconjugants in the feces was higher after 14 days than after 7 days. Strain NM522 did not mobilize pSUP104-luc in these experiments. No transconjugants of the miniTn5 delivery plasmid, pUTluxAB, were observed.
TABLE 3.
Results of plasmid transfer from E. coli donor strains to gut bacteria in feeding experiments with F. candida
| E. coli donor strain | Plasmid | Fecal bacteria (log CFU per specimen) extracted from microcosms
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| After 7 days of feeding
|
After 14 days of feeding
|
||||||||
| Total cells | Recipient cells | Donor cells | Transconjugant cells | Total cells | Recipient cells | Donor cells | Transconjugant cells | ||
| S17-1 | pUC18-luc | 7.07 | 6.75 | NDa | ND | 6.46 | 5.97 | 1.85 | ND |
| pSUP202-luc | 6.85 | 7.36 | ND | −0.19 | 6.29 | 5.45 | 1.57 | −0.98 | |
| pSUP104-luc | 6.27 | 6.00 | 0.87 | 0.77 | 6.74 | 6.00 | 2.69 | 2.82 | |
| pRP4-luc | 6.27 | 5.88 | 3.75 | 0.36 | 6.39 | 5.48 | 4.73 | 1.61 | |
| HB101 | pSUP104-luc | 6.02 | 5.89 | 2.08 | ND | 7.02 | 4.15 | ND | ND |
| NM522 | pSUP104-luc | 6.16 | 6.16 | 3.21 | ND | 6.93 | 6.21 | 3.76 | ND |
| S17-1 λPir | pUTluxAB | 6.20 | 6.08 | ND | ND | 6.63 | 4.61 | ND | ND |
| CC118 λPir | pUTluxAB | 6.00 | 4.28 | 1.41 | ND | 6.22 | 6.13 | 2.5 | ND |
ND, not detectable.
Kinetics of pRP4-luc transconjugant occurrence in feeding experiments.
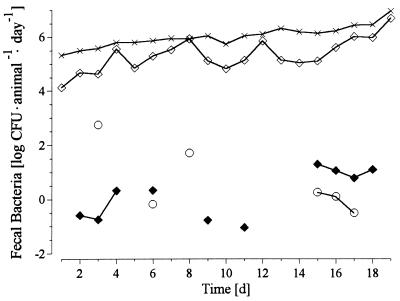
A more detailed analysis of the time-dependent occurrence of transconjugants in feces was performed with E. coli S17-1 pRP4-luc. F. candida specimens were transferred from one microcosm to another every day. Thus, data shown in Fig. 1 indicate the number of fecal bacteria released from the insects over a period of 24 h. Due to the mobility of the insects in the microcosms, it cannot be excluded that some of the donor cells detected on the water-agar were not of fecal origin but were contaminants from the food supply. This may explain the high numbers of donor cells detected after 3 and 8 days. Fluctuations in the numbers of recipients recovered from feces were repetitive every 4 days, which, most likely, was influenced by the molting cycles of the insects. The numbers of total bacteria in the feces, however, did not follow this pattern. Transconjugants occurred for the first time 48 h after the beginning of the experiment and were detected infrequently over the entire period of incubation. The cumulative number of transconjugants detected between days 7 and 14 was higher than between days 1 and 7, which was in accordance with the data shown in Table 3. At the end of the experiment, more transconjugants than donor cells were recovered from the microcosms.
FIG. 1.
Feeding of E. coli pRP4-luc to F. candida in petri dish microcosms. Levels of occurrence, as measured daily, of total bacteria (×), recipients (◊), donor cells (○), and transconjugants (⧫) in feces are represented. d, day.
Higher numbers of transconjugant cells than donor cells were also detected when the total gut contents of F. candida specimens, fed for 10 days with E. coli S17-1 pRP4-luc, were analyzed. An average of 2.32 × 104 CFU animal−1 was detected on LB medium. Benzoic acid-degrading bacteria (recipients) occurred at 1.30 × 104 CFU animal−1 (56%). Only 1.9 CFU of donor cells animal−1 was detected. Transconjugants occurred at 4.5 CFU animal−1 (not shown).
Localization of plasmid transfer events.
The selected method for detecting transconjugants involved, inevitably, the cultivation of a donor-recipient-containing suspension on nutrient agar. Under such conditions, plasmid transfer (plate mating) may occur (60). In order to determine whether transconjugants arose as a result of plate mating or before mating in the gut or feces of F. candida, controls in which E. coli pRP4-luc and recipient cells were separately preincubated under filter mating conditions were combined and inoculated onto the transconjugant-selective growth agar. No transconjugants could be detected under these conditions. Thus, plate mating did not influence the detection of transconjugants in our investigation (data not shown).
An experiment was conducted in which F. candida was fed with E. coli pRP4-luc for 7 days in order to determine whether the formation of transconjugants occurred mainly in the gut of F. candida or subsequently in the feces. Under these conditions, feces were incubated on water-agar for 1 to 7 days, depending on the experiment. In one set of microcosms, water-agar contained nalidixic acid, an antibiotic which inhibits DNA replication, conjugative gene transfer, and plate mating (39, 60, 65). The selected nalidixic acid concentration (10 μg ml−1) inhibited the growth of E. coli S17-1 pRP4-luc. The MICs for eight randomly selected transconjugant strains, determined in separate experiments, were in the range 1 to 50 μg ml−1, with four strains being more resistant than E. coli (data not shown). Feces extracted from both types of microcosms, i.e., with and without nalidixic acid, were analyzed for the occurrence of total cells, donor cells, recipients, and transconjugants. The numbers of total cells and recipients were reduced by an order of magnitude of approximately 0.5 (Table 4). This result was in accordance with the previously described tolerance (MIC) of fecal bacteria to this antibiotic. No donor cells were detected on water-agar supplemented with nalidixic acid, indicating efficient diffusion of the antibiotic into the fecal depositions. Transconjugants, however, could be isolated from both types of microcosms, i.e., with and without nalidixic acid. Thus, since donor cells were completely inhibited, it could be concluded that pRP4-luc was transferred from E. coli to recipient cells in the gut of F. candida and not in its feces.
TABLE 4.
Effects of nalidixic acid on the recovery of fecal bacteria of F. candidaa
| Cell type | No. of cells (log CFU animal−1) extracted from water-agar
|
|
|---|---|---|
| Without nalidixic acid | With nalidixic acid | |
| Total fecal | 6.39 | 6.06 |
| Recipient | 6.03 | 5.54 |
| Donorb | 1.17 | NDc |
| Transconjugantb | 0.15 | 0.25 |
Nalidixic acid (10 μg ml of water-agar−1) was added or not added to petri dish microcosms of fecal bacteria from F. candida in feeding experiments with E. coli S17-1 pRP4-luc as a donor strain.
Growth occurred on the appropriate selective medium and was confirmed by luciferase colony detection.
ND, not detected.
Phylogenetic diversity of transconjugants.
Segregation was observed frequently with all plasmid-bearing transconjugant strains isolated in this study under nonselective conditions. Nevertheless, subcultures which stably maintained the expression of their respective marker genes could be obtained with all transconjugants except those with the narrow-host-range plasmid pSUP202-luc. In the case of transconjugants with pSUP202-luc, bioluminescence faded away during the first three subcultures and detection of the luc gene by PCR was positive only during the first five subcultures (data not shown).
The diversity of transconjugants was assessed with a total of 264 pure-culture isolates from both feeding and filter mating experiments. Purified colonies, grown on agar plates, were differentiated first by restriction fragment length polymorphism analysis of their 16S rRNA genes (ARDRA). Fifteen pattern types could be differentiated with two DNA restriction enzyme endonucleases, CfoI and AluI. These ARDRA types were analyzed at a physiological level by microtiter plate-bound substrate utilization assays (Biolog) and at the phylogenetic level by sequencing and comparison of the nearly complete, PCR-amplified 16S rRNA gene. All isolates were gram negative and belonged to the alpha, beta, or gamma subclass of the class Proteobacteria (Table 5). All three plasmids transferred from E. coli to a variety of species. There was accordance between the results obtained from data bank comparisons with Biolog and 16S rRNA gene sequencing. However, at the DNA level no isolate was identical and by Biolog only one isolate (HR4; Alcaligenes faecalis) was completely identical to described strains in the data banks.
TABLE 5.
Comparison of transconjugants isolated from F. candida with data bank strains
| Subclass of Proteobacteria | Strain | Acquired plasmid | Biolog identification
|
16S rRNA gene sequencing
|
|||
|---|---|---|---|---|---|---|---|
| Closest relative | Similarity coefficienta | Closest relative | % Similarity | EMBL accession no. | |||
| Alpha | HS1 | pSUP202-luc | Ochrobactrum anthropi | 0.93 | Ochrobactrum anthropi | 99.8 | AJ002812 |
| Beta | H151 | pSUP104-luc | Alcaligenes xylosoxidans | 0.86 | Alcaligenes xylosoxidans | 98.6 | AJ002802 |
| H158 | Comamonas acidovorans | 0.90 | Comamonas acidovorans | 99.4 | AJ002803 | ||
| H159 | Alcaligenes xylosoxidans | 0.98 | Alcaligenes xylosoxidans | 97.9 | AJ002804 | ||
| HR4 | pRP4-luc | Alcaligenes faecalis | 1.00 | Alcaligenes faecalis | 98.7 | AJ002815 | |
| HR5 | Alcaligenes xylosoxidans | 0.74 | Alcaligenes xylosoxidans | 98.5 | AJ002808 | ||
| HR6 | Alcaligenes xylosoxidans | 0.97 | Alcaligenes xylosoxidans | 97.8 | AJ002809 | ||
| HR7 | Comamonas testosteroni | 0.82 | Comamonas acidovorans | 96.1 | AJ002810 | ||
| Gamma | HS2 | pSUP202-luc | Pseudomonas cichorii | 0.51 | Pseudomonas agarici | 98.0 | AJ002813 |
| HS3 | Stenotrophomonas maltophilia | 0.88 | Stenotrophomonas maltophilia | 99.5 | AJ002814 | ||
| H150 | pSUP104-luc | Pseudomonas fluorescens | 0.91 | Pseudomonas fluorescens | 99.1 | AJ002801 | |
| HR1 | pRP4-luc | “Pseudomonas maculicola” | 0.80 | Pseudomonas putida | 99.1 | AJ002805 | |
| HR2 | Brevundimonas diminuta | 0.65 | Stenotrophomonas maltophilia | 97.6 | AJ002806 | ||
| HR3 | Aquaspirillum dispar | 0.71 | Steonotrophomonas maltophilia | 97.6 | AJ002807 | ||
| HR8 | Enterobacter cloacae A | 0.82 | Pantoea agglomerans | 99.4 | AJ002811 | ||
Readings were taken after 24, 48, and 72 h. A coefficient of 1.0 indicates complete homology to a data bank species.
DISCUSSION
The sensitive detection of gene transfer from donor strains to indigenous microorganisms in soil requires both efficient marker genes and donor counterselection (13, 63). Antibiotic resistance genes, as carried on the plasmids used in our study, may not be sufficient for the detection of gene transfer because natural resistances of indigenous recipients can mask the detection of transconjugants (64). The insertion of constitutively expressing luciferase genes into the plasmids used in our study provided the crucial technique to distinguish transconjugants from a background of antibiotic-resistant or -tolerant indigenous bacteria. By this means we were able to detect even transconjugants which could not stably maintain their acquired plasmid, such as pSUP202 in strain HS2 (Pseudomonas agarici). In environmental settings, unstable transconjugants might be able to act as transient hosts and propagate plasmids to other recipients (52). Pseudomonas spp. were found to be more persistent in the gut of F. candida than E. coli (67), and therefore the survival of a plasmid in this particular habitat might be extended, even in transient hosts, compared to its survival in the fed donor strain.
Most of the E. coli cells fed to F. candida were digested, but counterselection was still necessary to exclude contamination by E. coli donor cells. Counterselection was achieved by selecting growth media with benzoic acid as a sole source of carbon. In contrast to results with E. coli, a rather high proportion of the culturable gut bacteria were able to use benzoic acid, but this method of donor counterselection inevitably excluded the proportion of indigenous bacteria incapable of growing with benzoic acid. Types of counterselection used in other studies, however, suffered the same disadvantages (61, 69). While recipient exclusion might lead to an underestimation of gene transfer frequencies, as described above, growth and clonal expansion can tend to produce overestimations of transconjugants (19). Since, in some experiments of our study, feces were incubated in microcosms several days before analyses, we could not calculate exact transfer frequencies of the feeding experiments. However, the diversity of indigenous transconjugants, as well as the number of transconjugants detected in experiments by daily analyses, clearly indicated that plasmid transfers in the gut of F. candida were not rare events.
Feeding experiments with E. coli donor strains were incubated for a period of 7 and 14 days in order to allow for intensive contact between the animals and the donor strains. Also, we anticipated that plasmid transfer events might be restricted to certain periods, correlating with the molting cycles of F. candida. Due to the incubation period of 7 days, some of the feces were relatively old before analysis, and it might be speculated that conjugation events may have occurred mostly on the agar surface and not during passage through the gut. One might speculate that the insects contributed to the generation of transconjugants only by providing the ecosystem to mix recipient with donor cells. However, in our study we were able to inhibit plasmid transfer (pRP4-luc) in feces by supplementing water-agar with nalidixic acid but transconjugants could still be isolated. Additionally, data from the feeding experiments indicated that only a few donor cells survived the gut passage. Smit and van Elsas (60) found that plasmid transfer on agar plates (plate mating) did not occur when donor cell numbers were low. Thimm et al. (67) observed with F. candida that the gut passage reduced ingested E. coli cells over 60,000-fold. Thus, the donor/recipient ratio in the feces was relatively low. Donor/recipient ratios were probably much higher immediately after donor cells were ingested, and thus conditions for conjugative gene transfer were much more favorable than in the feces. In the accompanying paper we have determined that the gut of F. candida had an average volume of 10 nl (67). The numbers of recipients in this investigation were approximately 106 to 107 CFU animal−1. Thus, recipient cell concentrations (1011 to 1012 CFU ml−1) were comparable to those of stationary-phase batch broth cultures.
Digestion of the ingested E. coli cells should result in the release of DNA into the gut, which would thus be a potential substrate for transformation. However, in our investigation, transformation as a process of gene transfer was unlikely because nonmobilizing non-self-transferable pUC18-luc, a cloning vector which can be transferred efficiently by artificial transformation in the laboratory (51), was not transferred from E. coli to indigenous gut bacteria of F. candida. Our data do not show whether these results were a consequence of the lack of DNA uptake, DNA protection, or expression by the gut bacteria. The narrow host range of replication of pUC (ori of ColE1) (4) might also have prevented the detection of transformation.
IncQ plasmids are able to be mobilized by plasmids of different incompatibility groups, e.g., IncP (21). In our investigation, the IncQ mobilizable plasmid pSUP104-luc was mobilized to indigenous bacteria in filter mating and feeding experiments from E. coli S17-1, a strain with chromosomally integrated IncP transfer genes of pRP4 (58). In filter matings, the E. coli strain NM522, with an F′ plasmid (IncF1), was also capable of mobilizing pSUP104-luc. The efficiency of IncQ mobilization by IncF1 was orders of magnitude below that of IncP mobilization. This result is in accordance with results of a study by Willetts and Crowther (76). However, other pure-culture studies did not detect IncQ mobilization by F plasmids (29). In feeding experiments, NM522 could not mobilize pSUP104-luc, probably because survival rates of the donor strain and therefore the period of contact between donor and recipient cells was, due to digestion, drastically reduced compared to that under filter mating conditions. Mobilization of plasmids in our investigation occurred only when the mobilizing functions were provided by the donor cells themselves. In other studies, IncQ plasmid mobilization in soil was observed from a nonmobilizing donor strain when E. coli cells harboring pRP4 were added (48, 62). Recently, IncQ mobilization by indigenous bacteria could be detected in a field experiment with manure-amended soil (25). In this investigation, IncQ mobilization could not be detected in the gut or with fecal bacteria of F. candida. However, other studies have shown that self-transferable plasmids and plasmids with mobilization potentials are present in the environment and able to interact with released bacterial cells (38, 45, 70). We assume, therefore, that further analysis will also detect such mobilizing functions in habitats associated with soil microarthropods.
This work demonstrates that the gut of the selected soil microarthropod provided appropriate-to-ideal conditions for conjugation and, thus, should be regarded as a hot spot for gene transfer in soil. If the host range of a plasmid is broad and the biodiversity in a hot spot habitat is uncharacterized, as for most soil microarthropods, the spread of recombinant genes may occur at much higher rates than anticipated by previous risk assessment studies performed with bulk soil microcosms.
ACKNOWLEDGMENTS
We thank Claudia Wahrenburg for excellent technical assistance and Wilfried Vahjen for contributing suggestions during the initiation of this project. Werner Selbitschka, Bielefeld, Germany, has supported this study with his helpful comments, which we gratefully acknowledge.
This work was supported by the German Ministry of Education and Research (grant 0310664).
REFERENCES
- 1.Adamo J A, Gealt M A. A demonstration of bacterial conjugation within the alimentary canal of Rhabditis nematodes. FEMS Microbiol Ecol. 1996;20:15–22. [Google Scholar]
- 2.Armstrong J L, Wood N D, Porteous L A. Transconjugation between bacteria in the digestive tract of the cutworm Peridroma saucia. Appl Environ Microbiol. 1990;56:1492–1493. doi: 10.1128/aem.56.5.1492-1493.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bakonyi G. Effect of Folsomia candida (Collembola) on the microbial biomass in a grassland soil. Biol Fertil Soils. 1989;7:138–141. [Google Scholar]
- 4.Balbás P, Soberón X, Merino E, Zurita M, Lomeli H, Valle F, Flores N, Bolivar F. Plasmid vector pBR322 and its special purpose derivatives—a review. Gene. 1986;50:3–40. doi: 10.1016/0378-1119(86)90307-0. [DOI] [PubMed] [Google Scholar]
- 5.Bardgett R D, Griffiths B S. Ecology and biology of soil protozoa, nematodes and microarthropods. In: van Elsas J D, Trevors J T, Wellington E M H, editors. Modern soil microbiology. New York, N.Y: Marcel Dekker Inc.; 1997. pp. 129–163. [Google Scholar]
- 6.Bengtsson G, Erlandsson A, Rundgren S. Fungal odour attracts soil collembola. Soil Biol Biochem. 1988;20:25–30. [Google Scholar]
- 7.Bignell D E. The arthropod gut as an environment for microorganisms. In: Anderson J M, Rayner A D M, Walton D W H, editors. Invertebrate-microbial interactions. Cambridge, United Kingdom: Cambridge University Press; 1984. pp. 205–227. [Google Scholar]
- 8.Bochner B. Breathprints at the microbial level. ASM News. 1989;55:536–539. [Google Scholar]
- 9.Borkott H, Insam H. Symbiosis with bacteria enhances the use of chitin by the springtail, Folsomia candida (Collembola) Biol Fertil Soils. 1990;9:126–129. [Google Scholar]
- 10.Boyer H W, Rouland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969;41:459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- 11.Brokamp A, Schmidt F R J. Survival of Alcaligenes xylosoxidans degrading 2,2-dichloropropionate and horizontal transfer of its halidohydrolase gene in a soil microcosm. Curr Microbiol. 1991;22:299–306. [Google Scholar]
- 12.Cohan F M. The role of genetic exchange in bacterial evolution. ASM News. 1996;62:631–636. [Google Scholar]
- 13.Cresswell N, Wellington E M H. Detection of genetic exchange in the terrestrial environment. In: Wellington E M H, van Elsas J D, editors. Genetic interactions among microorganisms in the natural environment. Oxford, United Kingdom: Pergamon Press; 1992. pp. 59–82. [Google Scholar]
- 14.Daane L L, Molina J A E, Berry E C, Sadowsky M J. Influence of earthworm activity on gene transfer from Pseudomonas fluorescens to indigenous soil bacteria. Appl Environ Microbiol. 1996;62:515–521. doi: 10.1128/aem.62.2.515-521.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Daane L L, Molina J A E, Sadowsky M J. Plasmid transfer between spatially separated donor and recipient bacteria in earthworm-containing soil microcosms. Appl Environ Microbiol. 1997;63:679–686. doi: 10.1128/aem.63.2.679-686.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dammann-Kalinowski T, Niemann S, Keller M, Selbitschka W, Tebbe C C, Pühler A. Characterization of two bioluminescent Rhizobium meliloti strains constructed for field releases. Appl Microbiol Biotechnol. 1996;45:509–512. doi: 10.1007/BF00578463. [DOI] [PubMed] [Google Scholar]
- 17.De Flaun M F, Levy S B. Genes and their varied hosts. In: Levy S B, Miller R V, editors. Gene transfer in the environment. New York, N.Y: McGraw-Hill; 1989. pp. 1–32. [Google Scholar]
- 18.De Lorenzo V, Herrero M, Jakubzik U, Timmis K N. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J Bacteriol. 1990;172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Rore H, Top E, Houwen F, Mergeay M, Verstraete W. Evolution of heavy metal resistant transconjugants in a soil environment with a concomitant selective pressure. FEMS Microbiol Ecol. 1994;14:263–274. [Google Scholar]
- 20.De Rore H, Demolder K, De Wilde K, Top E, Houwen F, Verstrate W. Transfer of the catabolic plasmid RP4::Tn4371 to indigenous soil bacteria and its effect on respiration and biphenyl breakdown. FEMS Microbiol Ecol. 1994;15:71–78. [Google Scholar]
- 21.Fry J, Bagdasarian M. The molecular biology of IncQ plasmids. In: Thomas C M, editor. Promiscuous plasmids of gram-negative bacteria. London, United Kingdom: Academic Press; 1989. pp. 79–94. [Google Scholar]
- 22.Fry J C, Day M J. Plasmid transfer and the release of genetically engineered bacteria in nature: a discussion and summary. In: Fry J C, Day M J, editors. Bacterial genetics in natural environments. London, United Kingdom: Chapman and Hall; 1990. pp. 243–250. [Google Scholar]
- 23.Gealt M A, Chai M D, Alpert K B, Boyer J C. Transfer of plasmids pBR322 and pBR325 in wastewater from laboratory strains of Escherichia coli to bacteria indigenous to the waste disposal system. Appl Environ Microbiol. 1985;49:836–841. doi: 10.1128/aem.49.4.836-841.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glew J G, Angle J S, Sadowsky M J. In vivo transfer of pR68.45 from Pseudomonas aeruginosa into indigenous soil bacteria. Microb Releases. 1993;1:237–241. [PubMed] [Google Scholar]
- 25.Götz A, Smalla K. Manure enhances plasmid mobilization and survival of Pseudomonas putida introduced into field soil. Appl Environ Microbiol. 1997;63:1980–1986. doi: 10.1128/aem.63.5.1980-1986.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gutell R R, Weiser B, Woese C R, Noller H F. Comparative anatomy of 16S-like ribosomal RNA. Prog Nucleic Acid Res Mol Biol. 1985;32:155–216. doi: 10.1016/s0079-6603(08)60348-7. [DOI] [PubMed] [Google Scholar]
- 27.Henschke R B, Schmidt F R J. Plasmid mobilization from genetically engineered bacteria to members of the indigenous soil microflora in situ. Curr Microbiol. 1990;20:105–110. [Google Scholar]
- 28.Herrero M, de Lorenzo V, Timmis K N. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J Bacteriol. 1990;172:6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hughes V M, Datta N. Conjugative plasmids in bacteria of the ‘preantibiotic’ era. Nature. 1983;302:725–726. doi: 10.1038/302725a0. [DOI] [PubMed] [Google Scholar]
- 30.Jansson J K. Tracking engineered microorganisms in nature. Curr Opin Biotechnol. 1995;6:275–283. doi: 10.1016/0958-1669(95)80048-4. [DOI] [PubMed] [Google Scholar]
- 31.Jarrett P, Stephenson M. Plasmid transfer between strains of Bacillus thuringiensis infecting Galleria mellonella and Spodoptera littoralis. Appl Environ Microbiol. 1990;56:1608–1614. doi: 10.1128/aem.56.6.1608-1614.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karlson U, Dwyer D F, Hooper S W, Moore E R B, Timmis K N, Eltis L D. Two independently regulated cytochromes P-450 in a Rhodococcus rhodochrous strain that degrades 2-ethoxyphenol and 4-methoxybenzoate. J Bacteriol. 1993;175:1467–1474. doi: 10.1128/jb.175.5.1467-1474.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klingmüller W. Plasmid transfer in natural soil, as stimulated by sucrose, wheat rhizosphere, and ground sugar beets: a study with nitrogen-fixing Enterobacter. Microb Releases. 1993;1:229–235. [Google Scholar]
- 34.Kruse H, Sørum H. Transfer of multiple drug resistance plasmids between bacteria of diverse origin in natural microenvironments. Appl Environ Microbiol. 1994;60:4015–4021. doi: 10.1128/aem.60.11.4015-4021.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee Q, Widden P. Folsomia candida a “fungivorous” collembolan feeds preferentially on nematodes rather than soil fungi. Soil Biol Biochem. 1996;28:689–690. [Google Scholar]
- 36.Levy S B, Marshall B M. Genetic transfer in the natural environment. In: Sussman M, Collins C H, Skinner F A, Stewart-Tull D E, editors. The release of genetically-engineered micro-organisms. London, United Kingdom: Academic Press; 1988. pp. 61–76. [Google Scholar]
- 37.Levy S B, Miller R V. Gene transfer in the environment. New York, N.Y: McGraw-Hill; 1989. [Google Scholar]
- 38.Lilley A K, Bailey M J. The acquisition of indigenous plasmids by a genetically marked pseudomonad population colonizing the sugar beet phytosphere is related to local environmental conditions. Appl Environ Microbiol. 1997;63:1577–1583. doi: 10.1128/aem.63.4.1577-1583.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lloyd R G, Hart J, Johnson S. Loss of Hfr DNA from Escherichia coli merozygotes during inhibition of conjugation by nalidixic acid. Genet Res. 1980;36:69–79. doi: 10.1017/s0016672300019674. [DOI] [PubMed] [Google Scholar]
- 40.Maidac B L, Olsen G J, Larsen N, Overbeek R, McCaughey M J, Woese C R. The RDP (Ribosomal Database Project) Nucleic Acids Res. 1997;25:109–110. doi: 10.1093/nar/25.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mazodier P, Davis J. Gene transfer between distantly related bacteria. Annu Rev Genet. 1991;25:147–171. doi: 10.1146/annurev.ge.25.120191.001051. [DOI] [PubMed] [Google Scholar]
- 42.Murray R G E, Doetsch R N, Robinow C F. Determinative and cytological light microscopy. In: Gerhardt P, Murray R G E, Wood W A, Krieg N R, editors. Methods in general and molecular bacteriology. Washington, D.C: ASM Press; 1994. pp. 21–41. [Google Scholar]
- 43.Olson B H, Ogunseitan O A, Rochelle P A, Tebbe C C, Tsai Y L. The implications of horizontal gene transfer for the environmental impact of genetically engineered microorganisms. In: Levin M, Strauss H, editors. Risk assessment in genetic engineering. New York, N.Y: McGraw-Hill; 1991. pp. 163–188. [Google Scholar]
- 44.Ow D W, Wood K V, De Luca M, de Wet J R, Helinski D R, Howell S H. Transient and stable expression of the firefly luciferase genes in plant cells and transgenic plants. Science. 1986;234:856–859. doi: 10.1126/science.234.4778.856. [DOI] [PubMed] [Google Scholar]
- 45.Powell B J, Purdy K J, Thompson I P, Bailey M J. Demonstration of tra+ plasmid activity in bacteria indigenous to the phyllosphere of sugar beet; gene transfer to a recombinant pseudomonad. FEMS Microbiol Ecol. 1993;12:195–206. [Google Scholar]
- 46.Priefer U, Simon R, Pühler A. Extension of the host range of Escherichia coli vectors by incorporation of RSF1010 replication and mobilization functions. J Bacteriol. 1985;163:324–330. doi: 10.1128/jb.163.1.324-330.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prosser J I, Rattray E A S, Killham K, Glover L A. lux as a marker gene to track microbes, section 6.1.1. In: Akkermans A D L, van Elsas J D, de Brujn F J, editors. Molecular microbial ecology manual. Dordrecht, The Netherlands: Kluwer Academic Press; 1996. pp. 1–17. [Google Scholar]
- 48.Pukall R, Tschäpe H, Smalla K. Monitoring the spread of broad host and narrow host range plasmids in soil microcosms. FEMS Microbiol Ecol. 1996;20:53–66. [Google Scholar]
- 49.Ramos-Gonzales M-I, Duque E, Ramos J L. Conjugational transfer of recombinant DNA in cultures and in soils: host range of Pseudomonas putida TOL plasmids. Appl Environ Microbiol. 1991;57:3020–3027. doi: 10.1128/aem.57.10.3020-3027.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Richaume A, Smit E, Faurie G, van Elsas J D. Influence of soil type on the transfer of plasmid pRP4p from Pseudomonas fluorescens to introduced recipient and to indigenous bacteria. FEMS Microbiol Ecol. 1992;101:281–292. [Google Scholar]
- 51.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 52.Saunders J R, Saunders V A. The estimation of gene transfer in natural environments. In: Wellington E M H, van Elsas J D, editors. Genetic interactions among microorganisms in the natural environment. Oxford, United Kingdom: Pergamon Press; 1992. pp. 258–263. [Google Scholar]
- 53.Saye D J, Miller R V. The aquatic environment: consideration of horizontal gene transmission in a diversified habitat. In: Levy S B, Miller R V, editors. Gene transfer in the environment. New York, N.Y: McGraw-Hill; 1989. pp. 223–259. [Google Scholar]
- 54.Schilf W, Klingmüller W. Experiments with Escherichia coli on the dispersal of plasmids in environmental samples. Recomb DNA Tech Bull. 1983;6:101–102. [PubMed] [Google Scholar]
- 55.Selbitschka W, Pühler A, Simon R. The construction of recA deficient Rhizobium meliloti and R. leguminosarum strains marked with gusA or luc cassettes for use in risk assessment studies. Mol Ecol. 1992;1:9–19. [Google Scholar]
- 56.Selbitschka W, Dresing U, Hagen M, Niemann S, Pühler A. A biological containment system for Rhizobium meliloti based on the use of recombination-deficient (recA−) strains. FEMS Microbiol Ecol. 1995;16:223–232. [Google Scholar]
- 57.Seniczak S, Stefaniak O. The microflora of the alimentary canal of Oppia nitens (Acarina, Orbatei) Pedobiologia. 1978;18:110–119. [Google Scholar]
- 58.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 59.Slater J H. Gene transfer in microbial communities. In: Halvorson H O, Pramer D, Rogul M, editors. Engineered organisms in the environment: scientific issues. Washington, D.C: ASM Press; 1985. pp. 89–98. [Google Scholar]
- 60.Smit E, van Elsas J D. Determination of plasmid transfer frequency in soil: consequences of bacterial mating on selective agar media. Curr Microbiol. 1990;21:151–157. [Google Scholar]
- 61.Smit E, van Elsas J D, van Veen J A, de Vos W M. Detection of plasmid transfer from Pseudomonas fluorescens to indigenous bacteria in soil by using bacteriophage φR2f for donor counterselection. Appl Environ Microbiol. 1991;57:3482–3488. doi: 10.1128/aem.57.12.3482-3488.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smit E, Venne D, van Elsas J D. Mobilization of a recombinant IncQ plasmid between bacteria on agar and in soil via cotransfer and retrotransfer. Appl Environ Microbiol. 1993;59:2257–2263. doi: 10.1128/aem.59.7.2257-2263.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Smit E, van Elsas J D. Detection of gene transfer in the environment: conjugation in soil, section 5.2.3. In: Akkermans A D L, van Elsas J D, de Bruijn F J, editors. Molecular microbial ecology manual. Dodrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 1–10. [Google Scholar]
- 64.Stotzky G. Gene transfer among bacteria in soil. In: Levy S B, Miller R V, editors. Gene transfer in the environment. New York, N.Y: McGraw-Hill; 1989. pp. 165–222. [Google Scholar]
- 65.Sugino A, Pebbles C L, Kreuzer K N, Cozzavelli N R. Mechanism of action of nalidixic acid: purification of Escherichia coli nalA gene product and its relationship to DNA gyrase and a novel nicking-closing enzyme. Proc Natl Acad Sci USA. 1977;47:4767–4771. doi: 10.1073/pnas.74.11.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thimm T, Larink O. Grazing preferences of some collembola for endomycorrhizal fungi. Biol Fertil Soils. 1995;19:266–268. [Google Scholar]
- 67.Thimm T, Hoffmann A, Borkott H, Munch J C, Tebbe C C. The gut of the soil microarthropod Folsomia candida (Collembola) is a frequently changeable but selective habitat and a vector for microorganisms. Appl Environ Microbiol. 1998;64:2660–2669. doi: 10.1128/aem.64.7.2660-2669.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thomas C M, Smith C A. Incompatibility group P plasmids: genetics, evolution, and use in genetic manipulation. Annu Rev Microbiol. 1987;41:77–101. doi: 10.1146/annurev.mi.41.100187.000453. [DOI] [PubMed] [Google Scholar]
- 69.Top E, Mergeay M, Springael D, Verstraete W. Gene escape model: transfer of heavy metal resistance genes from Escherichia coli to Alcaligenes eutrophus on agar plates and in soil samples. Appl Environ Microbiol. 1990;56:2471–2479. doi: 10.1128/aem.56.8.2471-2479.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Top E, de Smet I, Verstraete W, Dijkmans R, Mergeay M. Exogenous isolation of mobilizing plasmids from polluted soils and sludges. Appl Environ Microbiol. 1994;60:831–839. doi: 10.1128/aem.60.3.831-839.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tschäpe H. The spread of plasmids as a function of bacterial adaptability. FEMS Microbiol Ecol. 1994;15:23–32. [Google Scholar]
- 72.Van de Peer Y, Jansen J, De Rijk P, De Wachter R. Database on the structure of small ribosomal subunit RNA. Nucleic Acids Res. 1997;25:111–116. doi: 10.1093/nar/25.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vaneechoutte M, Roussau R, De Vos P, Gillis M, Janssens D, Paepe N, De Rouck A, Fiers T, Claeys G, Kersters K. Rapid identification of bacteria of the Comamonadaceae with amplified ribosomal DNA-restriction analysis (ARDRA) FEMS Microbiol Lett. 1992;93:227–234. doi: 10.1111/j.1574-6968.1992.tb05102.x. [DOI] [PubMed] [Google Scholar]
- 74.Van Elsas J D. Antibiotic resistance gene transfer in the environment: an overview. In: Wellington E M H, van Elsas J D, editors. Genetic interactions among microorganisms in the natural environment. Oxford, United Kingdom: Pergamon Press; 1992. pp. 17–39. [Google Scholar]
- 75.Van Elsas J D, Trevors J T. Plasmid transfer to indigenous bacteria in soil and rhizosphere: problems and perspectives. In: Fry J C, Day M J, editors. Bacterial genetics in natural environments. London, United Kingdom: Chapman and Hall; 1990. pp. 188–199. [Google Scholar]
- 76.Willetts N, Crowther C. Mobilization of the non-conjugative IncQ plasmid RSF1010. Genet Res. 1981;37:311–316. doi: 10.1017/s0016672300020310. [DOI] [PubMed] [Google Scholar]
- 77.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]