Abstract
Gene transfer of the conjugative plasmid pBF1 from Pseudomonas putida to indigenous bacteria in seawater was investigated with a detection system for gene transfer based on the green fluorescent protein (GFP) (C. Dahlberg et al., Mol. Biol. Evol. 15:385–390, 1998). pBF1 was tagged with the gfp gene controlled by a lac promoter which is down regulated in the donor cell by a chromosomal repressor (lacIq). The plasmid donor cells (Pseudomonas putida KT2442) subsequently do not express gfp. Transfer to recipient strains lacking the repressor results in expression of gfp. The transconjugant can subsequently be detected by epifluorescence microscopy on a single-cell level. By using this method, transfer of pBF1::gfp and expression of the gfp gene were first shown to occur during nutrient-limiting conditions to several defined recipient bacteria in artificial seawater. Second, we measured transfer of pBF1 from P. putida to the marine bacterial community directly in seawater samples, on a single-cell level, without limiting the detection of gene transfer to the culturable fraction of bacteria. Plasmid transfer was detected on surfaces and in bulk seawater. Seawater bacteria with different morphologies were shown to receive the plasmid. Gene transfer frequencies of 2.3 × 10−6 to 2.2 × 10−4 transconjugants per recipient were recorded after 3 days of incubation.
Horizontal gene transfer is an important adaptive mechanism for bacteria that may result in increased genetic variation by bringing together genes from different genetic backgrounds. Genes can be horizontally transferred between bacteria by conjugation, transduction, and transformation. Studies on plasmid conjugation have been conducted in various environments (reviewed in references 10, 17, and 27) including some studies of marine environments or model systems mimicking marine conditions (2, 8, 11, 22–24, 26). Studies on conjugation have primarily been conducted using disruptive techniques limited to detecting transfer to the culturable fraction of the community by growth of recipients on selective media. The culturable bacteria are typically less than 1% of the total number in marine environments (3, 13, 15). By using a nondisruptive system based on the green fluorescent protein (GFP) (4) to detect plasmid transfer, problems with culturability can be circumvented (1, 7). The plasmid to be studied here was previously tagged with the gfp gene from Aequorea victoria (1, 7). The gfp gene is expressed from a lac promoter which is down regulated in the donor bacterium (Pseudomonas putida KT2442) by a chromosomal insertion of a repressor (lacIq). Consequently, the plasmid donor cells do not express gfp. When the plasmid is transferred to a recipient cell without a repressor, gfp is expressed and the transconjugants can be detected on a single-cell level by their fluorescence. The method differs from the previously published GFP-based system (5) in that no special features are needed for gfp expression in the recipient. Using this novel, nondisruptive technique we were able to study the dissemination of plasmids in different systems in situ, without limiting the detection of gene transfer to the culturable fraction of the bacteria.
In a previous study we isolated conjugative plasmids conferring mercury resistance from marine bulk water, biofilm, and air-water interface bacterial communities (8). The plasmids were transferred from bacteria in the three different marine bacterial communities to P. putida and subsequently characterized. These plasmid isolation experiments were conducted in artificial seawater without added nutrients to mimic the nutrient-limiting conditions prevailing in marine waters (19). Characterization of the plasmids further showed that they have replication and/or incompatibility determinants which are different from those of well-characterized plasmids (8). The host range for three of the plasmids (pBF1, pB7, and pB9) was further investigated by the GFP method described above (7). Transfer was determined from P. putida to several other members of the γ subclass of the class Proteobacteria and in some cases to more distantly related Proteobacteria such as Caulobacter maris and Hyphomonas neptunicum in the α subclass. pBF1 also transferred to Planctomyces maris, which is phylogenetically very distant from P. putida (7). Given the transfer potential of pBF1 during nutrient-limited conditions (8) and its broad host range (7), it was clear that further more-detailed investigations of transfer of pBF1 in marine bacterial communities could give valuable information.
Seawater in general is nutrient limited. By using the GFP transconjugant detection method, conjugation and the ability to express a plasmid gene directly in the nutrient-limited transconjugant were tested. We further studied the transfer of pBF1::gfp from P. putida to indigenous bacteria directly in situ in seawater samples to establish transfer frequencies based on single-cell transfer events. Conjugation experiments were performed on surfaces and in bulk seawater, as well as with different additions of mercury.
MATERIALS AND METHODS
Bacterial strains and seawater samples.
The bacterial strains and plasmids used in this study are listed in Table 1. Seawater was collected outside Göteborg, Sweden, on the day on which the experiment started. The water was filtered through a sterile polycarbonate filter (3-μm-pore size; Poretics Corp.) to exclude higher organisms.
TABLE 1.
Bacterial strains and plasmid used in the study
| Strain or plasmid | Relevant characteristic(s)a | Source or reference |
|---|---|---|
| P. putida KT2442 | lacIq Rifr | S. Molin |
| P. putida KT2440 | Nxr | S. Molin |
| V. fischeri | Marine isolate | L. Holmquist |
| Vibrio sp. strain S14 | Marine isolate, Smr | S. Kjelleberg |
| D. marina | Marine isolate, Smr | ATCC 25374 |
| pBF1::gfp | Hgr Kmr Plac::gfp | 7 |
Rifr, rifampin resistance; Nxr, nalidixic acid resistance; Smr, streptomycin resistance; Hgr, mercury resistance; Kmr, kanamycin resistance; gfp, gene encoding green fluorescent protein.
Culture medium, growth conditions, and cell enumeration.
P. putida, Vibrio fischeri, Vibrio sp. strain S14, and Deleya marina were grown overnight at room temperature in Luria broth supplemented with NaCl to a final concentration of 1.5% (LB15; contains the following per liter of MilliQ water: 5 g of yeast extract, 10 g of tryptone, and 15 g of NaCl). LB15 was supplemented with 25 μg of kanamycin/ml, 100 μg of streptomycin/ml, and/or 7 μg of HgCl2/ml when necessary. Solidified medium contained 1.5% (wt/vol) agar. The total number of bacteria in the seawater was determined by standard acridine orange direct counts. The number of P. putida was estimated as CFU.
Starvation conditions.
Overnight cultures of P. putida, V. fischeri, Vibrio sp. strain S14, and D. marina were centrifuged at 2,500 × g for 5 min at 20°C and washed twice with Nine Salt Solution (NSS) (18). The bacteria were then diluted 10-fold in either NSS or sterile seawater and incubated for 24 h at room temperature with shaking.
Detection of transconjugants.
The conjugative gfp-marked plasmid used in this study was pBF1::gfp, hosted in P. putida KT2442 lacIq, which does not express the gfp gene (7). When the plasmid is transferred to a recipient bacterium without the repressor the gfp gene is expressed, and the transconjugant cell can be detected by epifluorescence microscopy.
In all of the following microscopical detections, filters with bacteria were transferred to slides and covered with coverslips. The number of green fluorescent cells was determined by means of an Olympus epifluorescence microscope, at a magnification of ×1,250, using blue excitation light (excitation filter B, dichroic mirror B, and barrier filter O-530). Photos were taken with 400 ASA Fuji film.
Conjugation experiments in NSS and sterile filtered seawater.
For conjugation of pBF1::gfp from P. putida KT2442 to P. putida KT2440, V. fischeri, Vibrio sp. strain S14, and D. marina, donor and recipient cells from overnight starvation suspensions in either NSS or sterile seawater were mixed at a ratio of 1:100. One milliliter of each conjugation mixture was filtered onto sterile polycarbonate filters (pore size, 0.2 μm; diameter, 25 mm; Poretics Corp.). The mean value of donor cells for all of the experiments was 5.5 × 105 cells cm−2. The filters were left floating on 20 ml of either NSS or sterile seawater for 1 to 3 days, after which they were examined with a microscope for fluorescent cells. A total of 20 microscopic fields were examined on each filter. Vibrio sp. strain S14 and D. marina transconjugants were detected as CFU on selective medium, as well as by GFP fluorescence, to compare the two methods for transconjugant detection. Filters were in this case resuspended in NSS and the mixtures were diluted and incubated on plates selective for transconjugants. Colonies were counted after 2 days.
As controls, donor and recipient cells were deposited on separate filters and incubated for 1 to 3 days, after which the samples were examined for green fluorescence. These filters had the same amount of bacteria as those in the conjugation experiments and were examined in the same way.
Conjugation experiments in seawater.
Conjugative transfer of pBF1::gfp from P. putida KT2442 to members of the total marine bacterial community was tested. Donors that were preadapted to seawater for 24 h were mixed with seawater at different ratios (see Table 4). Samples (100 ml) of the conjugation mixtures were filtered onto sterile polycarbonate filters and left floating on 500 ml of autoclaved ultrafiltered seawater in different beakers with 0, 0.1, or 1.0 μg of HgCl2 added per ml. After 3 days of incubation at 14°C with light (30 to 60 microeinsteins s−1 m−2) the filters were examined for cells exhibiting green fluorescence. A similar experiment was made in bulk water, where conjugation mixtures were kept in flasks with 0, 0.1, or 1.0 μg of HgCl2 added per ml. After 3 days, a 100-ml sample of the conjugation mixture was filtered onto sterile polycarbonate filters and examined in the same way as that for the filter matings. A total of 100 to 200 microscopic fields were examined on each filter.
TABLE 4.
Transfer of plasmid pBF1::gfp from P. putida KT2442 to indigenous marine bacteria detected by GFP fluorescence of transconjugants after 3 days of incubationa
| Expt | Incubationb | No. of transconjugants per recipient cell with addition of HgCl2 (μg/ml)c
|
No. of replicates | No. of recipientsd | Donor/recipient ratio | ||
|---|---|---|---|---|---|---|---|
| 0 | 0.1 | 1.0 | |||||
| 1 | Filter | 1.15 × 10−5 (5.12 × 10−6) | 5.56 × 10−5 (4.09 × 10−5) | 2.56 × 10−5 (1.28 × 10−5) | 2 | 5.8 × 107 | 8:1 |
| Bulk | 3.95 × 10−6 | 3.00 × 10−6 | 4.00 × 10−6 | 1 | 2.2 × 106 | 8:1 | |
| 2 | Filter | 3.08 × 10−5 (1.98 × 10−5) | 6.27 × 10−5 (2.44 × 10−5) | 2.32 × 10−5 (1.44 × 10−5) | 3 | 4.1 × 107 | 1:1 |
| Bulk | NDe | 8.97 × 10−6 | 2.30 × 10−6 | 1 | 1.6 × 106 | 1:1 | |
| 3 | Filter | 1.42 × 10−4 | 1.01 × 10−4 | 2.24 × 10−4 | 5 | 5.5 × 107 | 1:5 |
| Bulk | 5.57 × 10−5 | 1.16 × 10−5 | 4.87 × 10−6 | 3 | 2.1 × 106 | 1:5 | |
Results of experiments with seawater from three different samplings are shown.
Donor cells were added to seawater samples and incubated on filters floating on sterile seawater (filter) or incubated in flasks (bulk).
Incubations were made in the presence of different additions of HgCl2 as indicated. Values in parentheses are numbers of aggregates with two or more fluorescent cells on the filters per recipient cell.
Numbers of marine recipients added to the mating mixtures were determined by acridine orange direct counting. Counts are numbers of cells/milliliter (bulk) or numbers of cells/cm2 (filter).
ND, not determined.
As controls, donors and recipient bacteria from the seawater samples were deposited on separate filters in separate flasks and were incubated for 3 days, after which the samples were examined for green fluorescence. These filters had the same amount of bacteria as those in the conjugation experiments and were examined in the same way.
RESULTS
Conjugation in NSS and sterile filtered seawater.
Plasmid pBF1, marked with the gfp gene, was tested for its ability to transfer from P. putida KT2442 to Vibrio sp. strain S14, V. fischeri, D. marina, and P. putida KT2440 after the cells had first been starved in artificial seawater (NSS) without added nutrients for 24 h and then mixed together on filters which were incubated and floated on NSS. Conjugation and subsequent expression of the inserted gfp gene were detected in all strains during nutrient-limiting conditions (Table 2). Frequencies of gene transfer were between 2.5 × 10−2 and 3.4 × 10−1 (transconjugants per added donor) after 3 days of mating. The intensity of the GFP fluorescence increased with incubation time in all recipient species. After 3 days of incubation the fluorescence was stable, and this incubation time was used in the in situ experiments.
TABLE 2.
Conjugation of plasmid pBF1::gfp from P. putida KT2442 to different recipient cells on filters floating on artificial seawater without added nutrients (NSS)a
| Recipient strain | No. of transconjugants per donor |
|---|---|
| Vibrio sp. strain S14 | 3.4 × 10−1 |
| V. fischeri | 2.5 × 10−2 |
| D. marina | 9.0 × 10−2 |
| P. putida KT2440 | 2.5 × 10−1 |
Transconjugants were detected microscopically by GFP fluorescence after 3 days of mating. Data are the means of two experiments.
The detection of transconjugants by fluorescence was compared with the traditional detection on selective medium for D. marina and Vibrio sp. strain S14 (Table 3). Conjugation in both NSS and sterile seawater was tested. The numbers of transconjugants that were obtained were in the same range for the two methods. None of the methods gave significantly higher transconjugant numbers when all data were compared. The variation in transfer frequencies between different experiments was high, and the mean coefficient of variation was 100% for all experiments.
TABLE 3.
Transfer of plasmid pBF1::gfp from P. putida KT2442 to Vibrio sp. strain S14 or D. marina on filters incubated on artificial seawater (NSS) or sterile seawater after different times of incubation
| Recipient strain | Detection of transconjugants | No. of transconjugants at indicated times on filters ina:
|
|||||
|---|---|---|---|---|---|---|---|
| NSS
|
Sterile seawater
|
||||||
| Day 1 | Day 2 | Day 3 | Day 1 | Day 2 | Day 3 | ||
| Vibrio sp. strain S14 | GFPb | 5.3 × 10−5 | 0.03 | 0.34e | 0.001 | NDd | ND |
| CFUc | 0.03 | 0.16 | 0.89 | 0.02 | ND | ND | |
| D. marina | GFP | 0.08 | 0.19 | 0.09e | 0.02 | 0.11 | 0.07 |
| CFU | 0.04 | 0.05 | 0.17 | 0.03 | 0.05 | 0.02 | |
Numbers of transconjugants per added donor cells are shown. Data are means of three or four experiments.
Transconjugants were detected as single cells by epifluorescence microscopy.
Transconjugants were detected as CFU on selective media.
ND, not determined.
From Table 1.
In situ detection of conjugation in seawater.
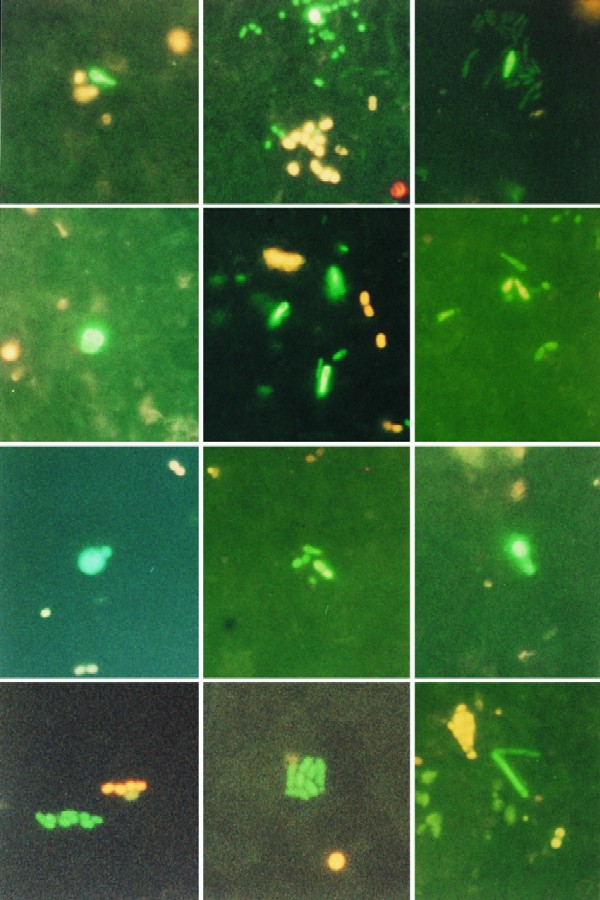
Conjugative transfer of pBF1::gfp from P. putida KT2442 to indigenous bacteria in seawater was detected by green fluorescence (Table 4). Green fluorescent cells were easy to distinguish (Fig. 1). Other indigenous fluorescent cells were present, but the emitted wavelengths did not interfere with the clear green fluorescence from GFP. Cells fluorescent at wavelengths other than that of GFP were similarly seen in the controls with only seawater bacteria, but in these controls no fluorescence similar to that of GFP could be detected. Transfer occurred to several different cellular morphotypes in all samples, and both single-transconjugant cells and aggregates of fluorescent cells were detected (Fig. 1). Gene transfer frequencies after 3 days of incubation ranged from 2.30 × 10−6 to 5.57 × 10−5 transconjugants per recipient for conjugation in bulk water and from 1.15 × 10−5 to 2.24 × 10−4 for conjugation on filters (Table 4). The variation between subsamples is shown by the coefficient of variation, which was 53% for parallel subsamples (calculated for experiment 3 where n = 5 for filters and n = 3 for bulk samples).
FIG. 1.
The different panels show GFP fluorescent transconjugant cells, formed after transfer of plasmid pBF1::gfp from P. putida KT2442 to indigenous marine bacteria in seawater. GFP transconjugants of different cell shapes are seen in the panels as follows (top, left to right): club-shaped transconjugant; aggregate of short rod-shaped transconjugants; and single dividing transconjugant; (second row, left to right) transconjugants in a tetrad form; club-shaped transconjugants; and weakly fluorescent club-shaped and rod-shaped transconjugants; (third row, left to right) budding type of transconjugant; aggregate of different transconjugant cell shapes; and large budding transconjugant cell; (bottom row, left to right) aggregate of rod-shaped transconjugants; aggregate of rod-shaped transconjugants; and long filamentous transconjugants. In all panels (except top right), transconjugants are found together with yellow or red autofluorescent cells.
The concentration of total mercury in the untreated sea water was about 0.2 μg per ml as determined by atomic fluorescence spectroscopy after reduction with SnCl2. No consistent differences in the frequencies of transconjugants were recorded after 3 days of incubation between different additions of mercury (Table 4). The number of aggregates of fluorescent cells and the number of cells in aggregates on the filters were also determined in two of the experiments. No increase in the number of fluorescent cells in aggregates could be seen for any of the mercury concentrations tested, compared with the sample without mercury addition (data not shown), nor was the number of aggregates affected by the addition of mercury (Table 4).
Growth of donors and marine recipient bacteria in seawater.
Both the number of donors and the number of recipients increased in all incubations. Growth of donors was determined by selective plating after incubation together with the indigenous flora. Growth of recipients was determined from samples containing only marine recipient bacteria. Total recipient CFU increased 3 to 10 times (mean of 6 for 11 different incubations) during the 3 days. Total donor CFU increased three to seven times. The data show that there is no excessive growth of the culturable fraction of the samples but only a slow increase. The resistance to mercury (Hgr) within the culturable fraction of the indigenous bacteria increased 6 times (mean value of the filter and bulk incubations). This was determined from incubations containing only marine recipient bacteria in experiment 3. Addition of mercury had no effect on the increase in Hgr colonies (data not shown). A similar increase in both total CFU and Hgr colonies was shown also in an independent experiment performed with only marine bacteria (data not shown).
Controls.
Controls with only donor or recipient cells were run in parallel to all experiments with the same amount of bacteria as that in the conjugation mixtures. Donor cells (P. putida KT2442 [pBF1::gfp]) in no case showed fluorescence when incubated without potential recipient bacteria, neither during starvation in NSS or seawater nor during growth in any of the experiments. Bacteria in the natural seawater community did show fluorescence, but the emitted wavelengths were clearly different from the GFP fluorescence. No plate mating was detected when donors and recipients were mixed directly on selective plates.
The repressor-less strain P. putida KT2440 (pBF1::gfp) showed no decrease in the number of fluorescent cells during 35 days in NSS. The insertion of the gfp gene in pBF1 had no effect on survival of P. putida KT2440 in NSS; the same number of CFU were scored on LB15 for P. putida (pBF1::gfp) as for P. putida (pBF1) during 21 days in NSS, and no decrease in CFU was noted during this time.
DISCUSSION
The detection of transconjugant cells by GFP fluorescence enabled us to follow transfer of the gfp-marked plasmid pBF1. Transfer from P. putida to defined recipient bacteria during nutrient limitation and to indigenous bacteria directly in seawater samples was detected on a single-cell level. Cells exhibiting GFP fluorescence were found in all mixtures of donor and recipient bacteria. No green fluorescent cells were seen when donors or recipients had been incubated separately. We therefore conclude that the fluorescent cells had received the pBF1::gfp plasmid and expressed the gfp gene. The main gene transfer mechanism is obviously conjugation in these experiments. We cannot exclude the possibility that in rare cases the gfp gene may have been transferred to new cells by transduction or transformation.
The problem of detecting gene transfer by growth of transconjugants on selective media has limited the study of transfer of plasmids in natural communities to the culturable fraction of the bacterial community. The main problem with this approach is the generally low (often less than 1%) and unpredictable culturability of indigenous bacteria. Various methods have been used to increase the sensitivity of transconjugant detection such as counterselection of the donor cells by phages (25) or by allowing the donors to enter a nonculturable state before transconjugants are grown (20, 23) or by using immunomagnetic beads to remove donors (9). The GFP detection method is likely to give a better estimate of the transfer frequencies in the total community since it does not rely on growth of transconjugants for their detection. The GFP is synthesized in several marine strains (7), but the possibility that expression of the marker gene does not occur in some potential recipient species cannot be excluded. This problem is, however, also present when resistance markers are used.
The protein takes about 4 to 6 h to develop its fluorescence (6), and we often noticed an increase in fluorescence over time for up to 3 days during nutrient limitation even if transconjugants could be detected earlier. It is possible that it takes longer to synthesize the number of GFP molecules necessary for detection under these conditions than during nutrient conditions that support higher bacterial activity. Once GFP is produced, folded, and oxidized to gain its fluorescent activity, it is stable in a starved cell for several weeks. The GFP method and the traditional detection of transconjugants on selective plates gave comparable transfer frequencies when we tested cultured donors and recipients in artificial seawater (NSS) and in filtered seawater (Table 3), which shows that under conditions with slow or no growth the GFP method estimates the transfer frequencies well. Higher transfer rates were detected by using selective media compared to that of GFP in growing donor-recipient cultures (7), which may be due to rapid gene transfer and a delay in the development of the fluorescence of the GFP.
Nutrient limitation is common in most environments, including marine waters (19). Plasmid transfer has been shown to be affected by starvation conditions where some plasmid-donor-recipient combinations showed transfer even after recipients or donors had been incubated in NSS without organic nutrients for 100 days before mating (11). The results reported in this study show that the pBF1 plasmid is transferred and developed into a functional form in the different recipient cells without addition of nutrients (Table 2). This was impossible to determine earlier with selective media to detect transconjugants.
The transfer frequencies in the experiments with indigenous marine bacteria did not vary consistently with the number of added donor cells in the concentration interval used (Table 4). The donor/recipient ratio varied between 8:1 and 1:5 and the total number of recipients was relatively stable. The highest transfer frequencies for filter conjugation were recorded in experiment 3, which had the lowest number of donor cells. This indicates that the transfer rate was probably not dependent on the cell concentration in the tested interval but may depend on factors such as a higher temporal abundance of potential recipients in certain samples. The transfer frequencies obtained in bulk seawater conjugations were higher than we expected, considering the very dilute bacterial suspension. We have speculated earlier that the bacterial concentrations in bulk seawater are too low to support significant plasmid transfer (12). However, the fact that aggregates and various surfaces are always present in a bulk water sample makes it hard to conclude if the conjugation took place on these surfaces or in the bulk.
A study of the transfer frequency of plasmid pRAS1 from Aeromonas salmonicida to indigenous recipient bacteria in marine sediment microcosms showed transfer rates of up to 3.3 × 10−4 transconjugants per recipient CFU after 8 days, detected by selective plating and colony hybridization with the plasmid as a probe (22). The high conjugation rates in sediment were explained as an effect of particle surfaces stabilizing conjugation, high numbers of sediment bacteria, and the use of an indigenous plasmid able to transfer to many marine strains (22). It is likely that the sediment system had more available nutrients than the bulk water used in this study. Plasmid transfer frequencies are difficult to compare between different systems, but in several different soils (both rhizosphere and bulk soil) frequencies were on average about 10−4 indigenous transconjugants per recipient CFU (21). When selective media are used for transconjugant detection, as in the earlier studies described above, it is not possible to distinguish between growth of transconjugants and gene transfer. With the GFP method it is possible to get a conservative measure of the transfer events. When transfer on filters was assessed, the number of aggregates with GFP fluorescent cells could be taken as a minimum number of initial gene transfer events (Table 4). But since aggregates were sometimes composed of several cell morphotypes (Fig. 1), it is likely that this may underestimate the transfer frequency.
A large morphological variation of GFP fluorescent cells, such as club-shaped cells, budding cells, and other particular cell types, was seen in all conjugation experiments with indigenous marine bacteria (Fig. 1). There was in no case one dominating cell type among the transconjugants. If these morphologically different cells are considered different species, one conclusion is that broad-host-range plasmid transfer can be common in marine bacterial communities. These results also support the broad-host-range character of pBF1 determined earlier (7). Transfer of plasmids, carrying for example resistance genes, between different species may be significant in bacterial adaptation in natural environments and may not be rare events.
Since pBF1 carries mercury resistance it was interesting to test if mercury had an effect on the frequency of transconjugant formation, that is, on the possible growth advantage of transconjugants. The number of transconjugants in aggregates, which may be taken as a measure of the growth of transconjugants, did not differ among various mercury additions (data not shown). This indicates that during these incubation times we could not detect that the plasmid-encoded mercury resistance was an immediate advantage for the recipients. Similar results were also found for additions of the antibiotic norseothricin (20) and mercury (16) to soil.
It should be noted that only transfer of the plasmid and expression of the gfp is detected. The subsequent stability of the plasmid in the fluorescent transconjugants is not known since GFP will remain stable in a cell even if pBF1::gfp is lost. The possibility of detecting such transfer events that result in a transiently maintained plasmid is important because the plasmid can still affect the genome of the recipient cell by, for example, acting as a vector for transposons.
Other microscopic methods for detection of transconjugants have been designed based on the expression of lacZ (14). The GFP, however, gives a stronger signal and allows detection on a single-cell level. An earlier method for GFP detection of transconjugants was designed by Christensen et al. (5) which used a specific positive induction of gfp in the recipient rather than a down regulation in the donor as in the system used here. The advantage with the latter is that no specific host functions are required in the recipient. The detection of transconjugants may be difficult in environments where E. coli is abundant because expression of gfp from the lac promoter may be naturally repressed in E. coli. It is unlikely that this would be the case in marine waters. The GFP method is limited to incubations in aerobic environments because the maturation of GFP requires oxygen.
The GFP method is a complement to the traditional detection of transconjugants with selective plates. The advantages of the GFP method are that the total bacterial community is assessed and the detection of gene transfer directly, in situ, will make it possible to determine accurately the actual transfer rates of plasmids in natural environments. The nondisruptive technique lends itself well to studies of surface communities where the spatial distribution of transconjugants can be recorded. In this case the frequency of initial transfer events may also be determined, which gives a conservative, minimum gene transfer frequency that is not dependent on the growth of the transconjugants.
ACKNOWLEDGMENTS
Financial support for this investigation provided by the Swedish Environmental Protection Board and The Foundation for Strategic Research through the Marine Science and Technology (MASTEC) program is gratefully acknowledged.
We thank Jonas Sommar, Departments of Chemistry, Inorganic Chemistry, Chalmers University of Technology and Göteborg University, for the mercury analysis.
REFERENCES
- 1.Andreasen M. GFP—maerking af marine plasmider. M. Sc. thesis. Denmark: University of Copenhagen; 1997. [Google Scholar]
- 2.Angles M L, Marshall K C, Goodman A E. Plasmid transfer between marine bacteria in the aqueous phase and biofilms in reactor microcosms. Appl Environ Microbiol. 1993;59:843–850. doi: 10.1128/aem.59.3.843-850.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bianchi A, Giuliano L. Enumeration of viable bacteria in the marine pelagic environment. Appl Environ Microbiol. 1996;62:174–177. doi: 10.1128/aem.62.1.174-177.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chalfie M, Tu Y, Euskirchen G, Ward W W, Prasher D C. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–804. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 5.Christensen B B, Sternberg C, Molin S. Bacterial plasmid conjugation on semi-solid surfaces monitored with the green fluorescent protein (GFP) from Aequorea victoria as a marker. Gene. 1996;173:59–65. doi: 10.1016/0378-1119(95)00707-5. [DOI] [PubMed] [Google Scholar]
- 6.Cubitt A B, Heim R, Adams S R, Boyd A E, Gross L A, Tsien R Y. Understanding, improving and using green fluorescent proteins. Trends Biochem Sci. 1995;20:448–455. doi: 10.1016/s0968-0004(00)89099-4. [DOI] [PubMed] [Google Scholar]
- 7.Dahlberg C, Bergström M, Andreason M, Christensen B B, Molin S, Hermansson M. Interspecies bacterial conjugation by plasmids from marine environments visualized by gfp expression. Mol Biol Evol. 1998;15:385–390. [Google Scholar]
- 8.Dahlberg C, Linberg C, Torsvik V L, Hermansson M. Mercury resistance plasmids isolated from bacteria in marine environments are highly variable and show no homology to probes used for replicon typing. Appl Environ Microbiol. 1997;63:4692–4697. doi: 10.1128/aem.63.12.4692-4697.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Enger Ø, Sandaa R-A. Immunomagnetic beads as a tool in conjugation experiments. J Microbiol Methods. 1994;19:111–115. [Google Scholar]
- 10.Fry J C, Day M J. Bacterial genetics in natural environments. London, United Kingdom: Chapman and Hall; 1990. [Google Scholar]
- 11.Goodman A E, Hild E, Marshall K C, Hermansson M. Conjugative plasmid transfer between bacteria under stimulated marine oligotrophic conditions. Appl Environ Microbiol. 1993;59:1035–1040. doi: 10.1128/aem.59.4.1035-1040.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hermansson M, Linberg C. Gene transfer in the marine environment. FEMS Microbiol Ecol. 1994;15:47–54. [Google Scholar]
- 13.Hoppe H G. Determination and properties of actively metabolizing heterotrophic bacteria in the sea, investigated by the means of micro-autoradiography. Mar Biol. 1976;36:291–302. [Google Scholar]
- 14.Jaenecke S, de Lorenzo V, Timmis K N, Dias E. A stringently controlled expression system for analysing lateral gene transfer between bacteria. Mol Microbiol. 1996;21:293–300. doi: 10.1046/j.1365-2958.1996.6411358.x. [DOI] [PubMed] [Google Scholar]
- 15.Jannasch H W, Jones G E. Bacterial populations in seawater as determined by different methods of enumeration. Limnol Oceanogr. 1959;4:128–139. [Google Scholar]
- 16.Kinkle B K, Sadowsky M J, Schmidt E L, Koskinen W C. Plasmids pJP4 and r68.45 can be transferred between populations of bradyrhizobia in nonsterile soil. Appl Environ Microbiol. 1993;59:1762–1766. doi: 10.1128/aem.59.6.1762-1766.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levy S B, Miller R V. Gene transfer in the environment. New York, N.Y: McGraw-Hill; 1989. [Google Scholar]
- 18.Mårdén P, Tunlid A, Malmcrona-Friberg K, Odham G, Kjelleberg S. Physiological and morphological changes during short term starvation of marine bacterial isolates. Arch Microbiol. 1985;142:326–332. [Google Scholar]
- 19.Morita R. Bioavailability of energy and the starvation state. In: Kjelleberg S, editor. Starvation in bacteria. New York, N.Y: Plenum Press; 1993. pp. 1–23. [Google Scholar]
- 20.Pukall R, Tschäpe H, Smalla K. Monitoring the spread of broad host range plasmids in soil microcosms. FEMS Microbiol Ecol. 1996;20:53–66. [Google Scholar]
- 21.Richaume A, Smit E, Faurie G, van Elsas J D. Influence of soil type on the transfer of plasmid RP4p from Pseudomonas fluorescens to introduced recipient and to indigenous bacteria. FEMS Microb Ecol. 1992;101:281–292. [Google Scholar]
- 22.Sandaa R-A. Transfer and maintenance of the plasmid RP4 in marine sediments. Microb Releases. 1993;2:115–119. [Google Scholar]
- 23.Sandaa R-A, Enger Ø. Transfer in marine sediments of the naturally occurring plasmid pRAS1 encoding multiple antibiotic resistance. Appl Environ Microbiol. 1994;60:4234–4238. doi: 10.1128/aem.60.12.4234-4238.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sandaa R-A, Torsvik V L, Goksøyr J. Transferable drug resistance in bacteria from fish-farm sediments. Can J Microbiol. 1992;38:1061–1065. [Google Scholar]
- 25.Smit E, van Elsas J D, van Veen J, de Vos W. Detection of plasmid transfer from Pseudomonas fluorescens to indigenous bacteria in soil by using bacteriophage φP2f for donor counterselection. Appl Environ Microbiol. 1991;57:3482–3488. doi: 10.1128/aem.57.12.3482-3488.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stewart K R, Koditschek L. Drug-resistance transfer in Escherichia coli in New York Bight sediment. Mar Pollut Bull. 1980;11:130–133. [Google Scholar]
- 27.Wellington E M H, van Elsas J D. Genetic interactions among microorganisms in the natural environment. London, United Kingdom: Pergamon Press; 1992. [Google Scholar]