Abstract

Lipid nanoparticles (LNPs) have shown remarkable success in delivering genetic materials like COVID-19 LNP vaccines, such as mRNA-1273/SpikeVax by Moderna and BNT162b2/Comirnaty by BioNTech/Pfizer, as well as siRNA for rare inherited diseases, such as Onpattro from Alnylam Pharmaceuticals. These LNPs are advantageous since they minimize side effects, target specific cells, and regulate payload delivery. There has been a surge of interest in these particles due to their success stories; however, we still do not know much about how they work. This perspective will recapitulate the evolution of lipid-based gene delivery, starting with Felgner’s pioneering 1987 PNAS paper, which introduced the initial DNA-transfection method utilizing a synthetic cationic lipid. Our journey takes us to the early 2020s, a time when advancements in bionano interactions enabled us to create biomimetic lipoplexes characterized by a remarkable ability to evade capture by immune cells in vivo. Through this overview, we propose leveraging previous achievements to assist us in formulating improved research goals when optimizing LNPs for medical conditions such as infectious diseases, cancer, and heritable disorders.
Keywords: lipid nanoparticles, lipoplexes, gene delivery, protein corona
Introduction
Lipid nanoparticles (LNPs) have garnered attention as a valuable tool in biomedical applications, with recent studies showing their potential in fighting several medical conditions.1 LNPs have been utilized as a delivery vehicle for mRNA encoding the spike protein of SARS-CoV-2 in two COVID-19 vaccines, mRNA-1273/SpikeVax by Moderna and BNT162b2/Comirnaty by BioNTech/Pfizer, both of which have demonstrated high efficacy and obtained emergency use authorization by regulatory agencies worldwide. The lipid composition of these vaccines was similar to that of Onpattro (trade name of Patirisan by Alnylam Pharmaceuticals), an orphan drug composed of an siRNA-LNP system approved by the US FDA in August 2018 for treating hereditary transthyretin (TTR) amyloidosis.2
LNPs are versatile and capable of delivering various therapies, making them suitable for treating a range of diseases.3 LNPs are a strong candidate for targeted drug delivery in cancer therapy, offering the ability to deliver chemotherapy agents, gene therapies, or immunotherapies directly to tumor cells with precision. The application of LNPs for delivering gene-editing tools like CRISPR-Cas94 holds promise in treating genetic disorders, potentially offering breakthroughs in conditions like cystic fibrosis, sickle cell anemia, and muscular dystrophy. LNPs are being investigated for delivering RNA-based therapies to treat neurodegenerative diseases such as Alzheimer’s,5 Parkinson’s,6 and amyotrophic lateral sclerosis (ALS). Lastly, LNPs provide a valuable platform for delivering treatments for rare diseases where tailored therapies are urgently needed.
While most applications and clinical trials involving LNPs have focused on RNA delivery, there is significant promise in using LNPs for DNA delivery.7 They are a safer and more cost-effective alternative to traditional viral vectors for delivering DNA, as they are nonimmunogenic and efficiently encapsulate DNA within their lipid bilayers. However, challenges such as low transfection efficiency (TE) and immune response still need to be addressed.
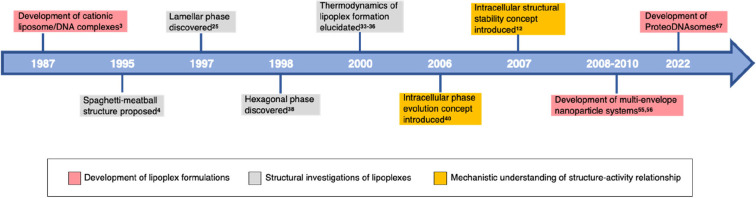
The pursuit of research and development in this domain holds pivotal significance for propelling the field of medical genetics forward and fully unleashing the capabilities of LNPs in DNA delivery. By examining both the achievements and obstacles encountered in the realm of other lipid-based DNA nanocarriers, such as lipoplexes encompassing cationic liposome (CL)/DNA complexes, researchers can steer clear of historical pitfalls and, instead, capitalize on prior accomplishments. For a visual overview of the noteworthy milestones in lipoplex research, refer to Figure 1, which outlines key achievements in the field’s evolution.
Figure 1.
Timeline of the key milestones in lipoplex research.
This perspective serves as a critical review focusing on the chemical–physical attributes of lipoplexes and their structure–function relationship. Its primary objective is to explore the potentialities of lipoplexes while also delving into the constraints that have impeded their successful translation into practical applications. Understanding the intricacies of this process is crucial to help researchers define clear and realistic research objectives for developing LNPs with enhanced therapeutic potential. The investigation will culminate in a discussion of the key areas of inquiry that merit attention in future research, including the bionano interactions that LNPs undergo in physiological milieus. By harnessing the insights garnered from prior studies, researchers can streamline the optimization procedures, expediting advancements in LNP development. Furthermore, the authors express their sincere apologies for any inadvertent omissions in acknowledging the contributions of scientists in the field. It is important to note that the nature of this article, being a perspective rather than a comprehensive review, imposes limitations on the extent to which all pertinent work can be acknowledged. Our aim is to offer a personalized perspective while acknowledging the collective efforts that have significantly influenced the field’s trajectory.
Development of Lipoplex Formulations
We recently celebrated 35 years from the pioneering PNAS paper of Philip Felgner et al. of the Stanford School of Medicine8 who reported the first use of CLs to facilitate the functional delivery of DNA in tissue culture cells. CLs were made of the cationic lipid 1,2-di-O-octadecenyl-3-trimethylammonium propane (DOTMA), encapsulated 100% DNA, and promoted gene expression in CV-1 and COS-7 cells. The protocol resulted in higher TE than available methods and rapidly became the standard of transfection experiments for the following decade. However, Felgner et al., while emphasizing the simplicity and reproducibility of the process, were the first to warn of the challenges ahead. CLs-mediated transfection could depend on numerous factors, such as the lipid composition of the CLs, the lipid concentration, the size of the nucleic acids to be delivered, and the duration of exposure. And, not least, all these parameters could be correlated with each other in a cell line-dependent manner. In the discussion section of the paper, the authors claim: “Based on our experience, it is best to optimize the various parameters described above for each cell line”. In the dissemination of future results on LNPs, it is important to remember the lesson from this influential paper: to provide a balanced perspective by not only highlighting positive results but also acknowledging potential limitations and areas that require further investigation.
It was natural that understanding the chemical–physical properties of lipoplexes became soon the subject of numerous theoretical and experimental studies.9,10 In the early 1990s, the efforts of the researchers focused on understanding the mechanism of formation of lipoplexes since this knowledge would have been useful for interpreting their biological activity and rationally designing optimized variants. Given the negatively charged nature of DNA chains juxtaposed with the positively charged character of CLs, it was hypothesized that electrostatic attraction serves as the primary driving force behind the formation of lipoplexes.
Robert C. MacDonald’s research group at Northwestern University conducted several studies that proposed models for the formation of lipoplexes.11 In accordance with the most widely accepted model, the outer coating of liposomes with DNA reduced the repulsion between the lipid vesicles, causing them to aggregate and stick together like a molecular glue. As a result of the asymmetric distribution of DNA between the exterior and interior of the vesicles, there was a pressure gradient that caused the lipid bilayer to rupture locally, facilitating the rearrangement of some vesicles into a single multilamellar lipoplex.
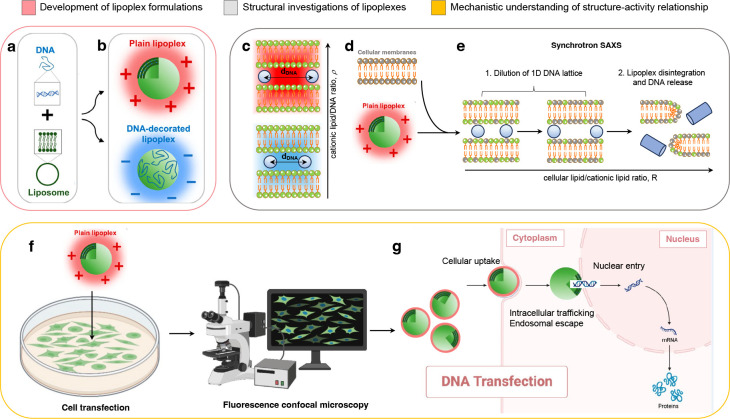
More in-depth investigations pointed out the relevance of the counterion release mechanism.12 A fraction of positive counterions, referred to as Manning counterions, are condensed to the neighborhood of the DNA. Similarly, a certain fraction of the negative counterions of CLs is confined to the lipid surface. Upon lipoplex formation, equal numbers of positive and negative Manning counterions are released to the bulk solution with a gain in translational entropy (Figure 2a). This entropic contribution to the free energy is comparable to the electrostatic contribution and is maximal at the isoelectric point that was therefore identified as the most favored thermodynamic state of lipoplexes.
Figure 2.
The optimization of lipoplex formulations involved a comprehensive understanding of their mechanism of formation, structure, and structure–activity relationship. (a) When negatively charged DNA chains interact with positively charged CLs, an equal number of positive and negative Manning counterions are released, resulting in a gain in translational entropy. The isoelectric point is identified as the most thermodynamically favored state of lipoplexes due to its maximal entropic contribution to free energy. (b) Isoelectric instability of lipoplexes allows the accommodation of additional DNA or lipid material, leading to negatively charged DNA-decorated and positively charged lipoplexes. Positively charged lipoplexes are efficient gene delivery systems, interacting effectively with oppositely charged cell membranes and yielding high transfection levels. (c) Synchrotron small-angle X-ray scattering (SAXS) revealed that lipoplex formation induces a topological transition in a lamellar phase structure composed of DNA monolayers between lipid bilayers. The 1D in-plane DNA lattice within this structure is relatively disordered, oscillating around equilibrium positions based on the cationic lipid/DNA ratio (ρ). (d) Studies propose that DNA release from lipoplexes occurs through charge neutralization by cellular lipids, which correlates with the interfacial curvature of the mesoscopic structures resulting from lipoplex–cellular lipid interaction. Lipoplexes that easily transform into nonlamellar phases when mixed with cellular lipids exhibit high transfection efficiency. (e) Cellular lipids progressively neutralize the lipid membrane of lipoplexes, leading to the increased distance between DNA molecules based on the cellular lipid/cationic lipid ratio. Structural evolution rates and susceptibility to destabilization by anionic lipids vary, influencing the dissociation of DNA from the lipid and correlating with transfection efficiency. (f) Transfection efficiency studies coupled with fluorescence confocal microscopy investigations elucidate the barriers to lipoplex-mediated transfection at the single-cell level. (g) Investigations explore cellular uptake, intracellular trafficking, endosomal escape, and nuclear entry of lipoplexes, providing mechanistic understanding for the development of multifunctional envelope-type nanodevices (MEND) and multicomponent envelope-type nanoparticle systems (MENS). A portion of the figure (panel b) was readapted from ref (16). Copyright 2022 American Chemical Society. Cartoons were created using Biorender.com.
A few years later, R. Bruinsma at the University of California Los Angeles postulated the isoelectric instability of lipoplexes.13 This means that the isoelectric point could accommodate extra DNA or lipid material due to the entropy gain obtained following the release of small ions inside the complex. This finding substantiated the concept of “overcharged lipoplexes,” denoting preparations with a surplus of either positive or negative charge, as illustrated in Figure 2b. These theoretical propositions received empirical validation from Wagner et al., who conducted direct assessments of counterion release through conductivity measurements, as documented in ref (14).
To provide a theoretical foundation for experimental findings, the group of Avinoam Ben-Shaul, Silvyo May, and Daniel Harries at The Hebrew University of Jerusalem, carried out in-depth theoretical studies regarding the phase behavior of overcharged lipoplexes and demonstrated that their equilibrium structure is regulated by a delicate interplay between the electrostatic, elastic, and mixing terms, which depend, in turn, on the lipid composition and CL/DNA ratio.15
These accomplishments were of paramount significance, as it had been apparent from the outset that the charge of lipoplexes played a central role in the process of transfection. One of the key advantages of overcharged lipoplexes is their ability to protect encapsulated nucleic acids from degradation. The cationic lipids create a protective shield around the genetic material, safeguarding it from enzymatic degradation by nucleases or exposure to harsh environmental conditions. Additionally, the positive charge of overcharged lipoplexes enhances their interaction with cellular membranes, which are typically negatively charged. This interaction promotes efficient cellular uptake, as the lipoplexes are attracted to and readily associate with the cell surface. Once inside the cell, overcharged lipoplexes face the challenge of endosomal entrapment, as they are often internalized through endocytosis. However, their positive charge comes into play again, as it allows these lipoplexes to destabilize endosomal membranes, facilitating the release of their cargo into the cytoplasm. Another crucial aspect to consider is particle size. Overcharged lipoplexes undergo re-entrant condensation17 and typically exhibit a hydrodynamic radius of approximately 200 nm or less, rendering them highly suitable for a wide array of gene delivery applications.
Conversely, neutrally charged lipoplexes do not exhibit the robust electrostatic binding affinity observed in their overcharged counterparts. Consequently, they often possess larger and more heterogeneous sizes, typically falling within the micrometric range. To efficiently encapsulate nucleic acids, additional stabilization techniques are often necessary. Furthermore, cellular uptake mechanisms for neutrally charged lipoplexes may be less efficient due to the diminished electrostatic attraction with cell surface proteoglycans.
After two decades of research, our understanding of lipoplex formation had reached a mature stage, providing us with a valuable body of knowledge and insights: experimental and theoretical evidence proved the feasibility of generating positively charged lipoplexes with small size, thus broadening their applicability as gene delivery systems.
However, a primary drawback of cationic lipoplexes arises when they are introduced into the bloodstream. They tend to become heavily coated with opsonins, including complement proteins like C3 and C4. This opsonization process triggers their swift removal from the bloodstream by immune system cells. The most effective approach to mitigate in vivo opsonization of lipoplexes was drawn from research on liposomes. It involves equipping lipoplexes with a protective shield composed of polyethylene glycol (PEG).18
Structural Investigations of Lipoplexes
Despite the multitude of physicochemical properties that can influence the TE of lipoplexes, it was proposed early on that the nanostructure could have the most significant impact on their biological activity. From the very beginning researchers tried to understand how factors such as CL/DNA molar ratio, DNA size, and lipid composition could be manipulated to influence the formation and equilibrium structure of lipoplexes.
At the University of San Francisco, Francis C. Szoka, Jr. and his team were pioneers in elucidating that, among the various factors influencing the colloidal characteristics of lipoplexes, the CL/DNA molar ratio assumed a predominant role in shaping their formation and equilibrium structure.19 From that point onward, the role of the charge ratio on the structure of lipoplexes became a cornerstone in this field of research.
The last clarification on the role of the CL/DNA ratio came about ten years later when Elena Junquera and Emilio Aicart at the Universidad Complutense de Madrid asked whether, at a fixed CL/DNA ratio, the DNA structure could affect the physical–chemical properties of lipoplexes. They showed that, when encapsulating plasmid DNA in lipoplexes, a smaller amount of cationic lipid is required compared to linear DNA.20 This phenomenon can be attributed to the way plasmids are packed within the lipoplex, which results in the retention of a substantial number of counterions and produces a lower effective negative charge.
Pitard and co-workers clarified that the morphological and structural features of lipoplexes did not depend on plasmid DNA size.21 On the other hand, gene transfer capacity of lipoplexes was affected by the size of plasmid DNA, with those containing the shortest DNA being the most efficient ones.
Cryo-transmission electron microscopy (cryo-TEM) investigations led by Leaf Huang and his team at the University of North Carolina at Chapel Hill unveiled a fascinating phenomenon. When DNA and CLs are combined in bulk, a distinctive topological shift takes place. This results in the creation of liquid-crystalline globules with optical birefringence, intriguingly termed “spaghetti-meatball” complexes. These complexes are bilayer-coated and contain supercoiled DNA.22 Further investigations using cryo-TEM showed that DNA molecules could also be found inside multilamellar structures.23 TEM findings inspired researchers to employ X-ray diffraction techniques for unveiling the nanoscale structure of lipoplexes.
In a famous Science paper, Cyrus Safinya and his team at the University of California Santa Barbara made significant progress in determining the supramolecular order of lipoplexes24 made of the cationic lipid 1,2-dioleoyl-3-trimethylammonium-propane (DOTAP) and the neutral lipid dioleoyl-phosphocholine (DOPC). The use of synchrotron small-angle X-ray scattering (SAXS) was instrumental to this end. SAXS is a nondestructive technique that uses X-rays to probe the nanoscale structure of materials. The collected X-ray scattering data produces a scattering pattern that contains information about the sample nanostructure. Researchers use mathematical analysis of the scattering pattern to extract structural parameters, such as particle size, shape, and interparticle distances.
Safinya and colleagues observed a distinctive pattern of Bragg peaks in conjunction with a broad and asymmetric diffraction peak whose position in the q-space changed with the CL/DNA ratio. These findings demonstrated that lipoplex formation induces a topological transition in a multilayer onion-like structure consisting of DNA monolayers coated between lipid bilayers. This structure was termed the lamellar phase of lipoplexes (Figure 2c and Figure S1). The lamellar spacing is determined by the thickness of the lipid bilayer and the interbilayer water layer. Meanwhile, neighboring DNA rods result in a relatively disordered 1D in-plane DNA lattice composed of DNA strands oscillating around equilibrium positions. To obtain precise measurements of the interaction between DNA and lipids, Manuel Prieto and his team at Universit Técnica de Lisboa, Portugal, utilized the technique of Fluorescence Resonance Energy Transfer (FRET).25 This approach helped to estimate the proportion of DNA that was bound and unbound to the lipids, as well as determine the distance between the opposing DNA helices. Subsequent investigations showed that DNA chains can also arrange, even if less frequently, into rectangular columnar superlattices between lipid bilayers.26
Just one year after their first paper, another Science paper by Safinya and co-workers reported for the first time lipoplexes organized in a reversed hexagonal phase27 (Figure S1). Compared to the first work dealing with the structure of DOTAP-DOPC/DNA lipoplexes, in the latter, the neutral lipid DOPC was replaced by DOPE, and lipoplexes were prepared at different weight fractions of DOPE in the lipid bilayer (i.e., 0.4< ΦDOPE < 0.85). For ΦDOPE > 0.7, the DNA molecules were coated with cylindrical micelles made up of DOTAP and DOPE in turn organized in a reverse hexagonal phase.
Subsequent investigations demonstrated that the neutral helper lipid is a major factor that determines the morphologies of the condensates and significantly affects the TE of lipoplexes. Lipids with curvature c = 0 promote the organization in the lamellar phase, while lipids with curvature c < 0 promote the formation of reverse hexagonal phases or cubic phases.28 Hexagonal systems exhibited a higher TE than the lamellar counterpart, and Safinya and co-workers ascribed this increase in efficiency to their peculiar ability to fuse with anionic lipid (AL) vesicles that were used as a model system of cell membranes.
Subsequent studies thoroughly explored the structure–function relationship and found evidence that this relationship did not exist.29 In a highly cited PNAS paper, Robert C. MacDonald and co-workers put a stop to the matter stating that “Some earlier studies suggested that the inverted hexagonal phase leads to more efficient transfection efficiency than does the lamellar phase. Recent experiments dispute this suggestion, however, and there is considerable evidence against a direct general correlation between lipoplex structure and transfection efficiency”.30
Following the investigation of the structure of the lipoplexes, researchers further explored other chemical and physical properties to identify a factor that could universally control the transfection efficiency of lipoplexes.
Among these factors, membrane density, σM, was invoked first.31 In a study examining the relationship between TE and membrane charge density, researchers discovered a common, bell-shaped curve. This curve suggested the existence of three different stages in the interactions between complexes and cells. In the first stage, when the membrane charge density is low, TE increases alongside it. In the second stage, at a moderate membrane charge density, TE reaches a plateau. Finally, in the third stage, with high membrane charge density, TE decreases. Researchers believed that, at low membrane charge density, lipoplexes remain stuck in the endosome. On the other hand, when the membrane charge density is high, lipoplexes become less efficient because the DNA struggles to separate from the highly charged membranes within the cytosol. The optimal, intermediate stage seemed to strike a balance between these two extremes, allowing for endosomal escape and successful dissociation in the cytosol. Also in this case, however, counterexamples were provided for which it was possible to have high TEs even in regimes of low surface membrane density where lipoplexes should have been poorly efficient.32
Subsequently, it was suggested that augmenting the diversity of lipid species could trigger the development of structures characterized by unconventional fusogenic properties. This was demonstrated by our group in collaboration with the Austrian SAXS beamline at the synchrotron facility of ELETTRA led by Heinz Amenitsch.33 We mixed DNA molecules to a dispersion containing two binary CL formulations, each consisting of a cationic lipid and a neutral lipid at a 1:1 molar ratio. The first formulation was made of the cationic lipid DOTAP and the neutral lipid DOPC, while the second formulation consisted of the cationic lipid 3′[N-(N′,N′-dimethylaminoethane)-carbamoyl]cholesterol (DC-Chol)34 and the neutral lipid dioleoyl-phosphatidylethanolamine (DOPE). For simplicity, we will use the terms CLA and CLB to refer to DOTAP-DOPC and DC-Chol-DOPE CLs, respectively, and CLA/DNA and CLB/DNA to denote DOTAP-DOPC/DNA and DC-Chol-DOPE/DNA lipoplexes. In the absence of DNA, the SAXS pattern displayed the exact overlap of the SAXS patterns of CLA and CLB, indicating that the coexisting CLs maintained their structural integrity in solution due to the repulsion between the charged lipid surfaces.33
However, upon the addition of DNA to the solution, the SAXS pattern showed unique features, indicating that the resulting multicomponent (MC) lipoplexes contained all four lipid species in an ideally mixed state. This experiment demonstrated that DNA promotes a uniform spatial distribution of lipid species in the membrane plane.
MC lipoplexes exhibited intermediate chemical–physical properties, including size, zeta-potential, bilayer thickness, size of the hydrophobic region, and DNA packing density between those of CLA/DNA and CLB/DNA lipoplexes. This rearrangement of lipid species is an entropic process induced by a reduction in free energy.35 The most interesting observation was provided a couple of years later when we showed that MC lipoplexes were 1 to 2 orders of magnitude more efficient than CLA/DNA and CLB/DNA lipoplexes36 with their TE increasing the greater the number of lipid species involved.37 These results prompted further investigation into the transfection behavior of MC lipoplexes. Studies conducted in the following ten years demonstrated that MC lipoplexes were often as efficient as lipofectamine, which is considered the gold standard of transfection reagents.38 Moreover, MC lipoplexes were highly efficient in cells that are difficult to transfect with lipofectamine.39 In most cases, the reduced number of positive charges compared to those of lipofectamine allowed MC lipoplexes to minimally impact cell viability, which has always been the major drawback in the use of cationic reagents.
In summary, researchers’ endeavors have illuminated the fact that the connection between the structure of lipoplexes and their transfection efficiency is not a straightforward one. To develop efficient delivery systems, it was necessary to understand the interaction of lipoplexes with cells, and it was also clear that intracellular events leading to transfection were related to the structural organization of lipoplexes. Therefore, knowledge of the lipoplex structure was a fundamental but not the only building block required to develop a mechanistic understanding of lipoplex-mediated DNA delivery.
Structure–Activity Relationship of Lipoplexes
Structural studies of lipoplexes, despite not directly contributing to the improvement of our understanding of lipofection, were of great significance. They revealed that lipofection is a complex process that is not regulated by a single parameter but rather influenced by a delicate balance of various interacting factors. These findings emphasized the need for a comprehensive understanding of the mechanisms underlying lipofection, which would be a crucial step toward advancing the field to a more mature stage.
A substantial paradigm shift was not long in coming. Experimental and theoretical investigations have shown that the entropic gain resulting from lipid mixing played a crucial role in the formation of lipoplexes. It was proposed that the intracellular evolution toward nonlamellar phases after interaction and mixing with cellular lipids could universally control the TE of lipoplexes, which is in line with the models proposed by Francis C. Szoka, Jr. for the interaction between lipoplexes and cellular membranes in the mid-1990s.9 This suggestion was also in agreement with some previous investigations performed by Dick Hoekstra at the University of Groningen in The Netherlands40 who had shown that certain lamellar lipoplexes could adopt a fusogenic-inverted hexagonal phase after interaction with acidic phospholipid-containing membranes.
The transition to a reversed hexagonal phase of lamellar lipoplexes represents a significant change in the structural arrangement of lipid bilayers and is of particular importance in the context of gene delivery. The structural transition allows for improved release of cargo molecules from the lipoplexes. When lipoplexes encounter target cells or cellular membranes, the lipid structure can adapt to facilitate the release of the cargo into the cell’s interior. This is crucial for effective drug or gene delivery. The ability of lipoplexes to transition to a reversed hexagonal phase can be engineered to respond to specific triggers or conditions, such as changes in pH or temperature.
We will continue this walk down memory lane with a reference to the studies that contributed to the maturity of the field. Rumiana Koynova, Robert MacDonald, and their colleagues contributed to the development of the field by investigating the relationship between TE and the structural evolution of lipoplexes mixed with various anionic membrane lipids such as dioleoylphosphatidylglycerol (DOPG), dioleoylphosphatidic acid (DOPA), 1,2-dioleoyl-sn-glycero-3-phospho-l-serine (DOPS), cardiolipin, and shyngomyelin.30,41 Their research demonstrated that DNA release occurs due to the charge neutralization of lipoplexes by ALs, and it correlates with the interfacial curvature of the mesoscopic structures emerging from lipoplex–AL interaction.
Highly efficient lipoplex formulations formed phases of high negative interfacial curvature (e.g., bilayer cubic, inverted hexagonal, micellar cubic, etc.) when mixed with ALs. Therefore, lipoplexes that easily transformed into nonlamellar phases when mixed with ALs would release DNA very easily, which was likely the reason for their high TE.
What our group contributed to clarifying was the relationship between TE of lipoplexes and their structural stability against interaction with cellular lipids. The method used to mimic lipoplex–cell interaction was to incubate lipoplexes with ALs as a function of the molar ratio R = AL/CL (Figure 2d). The information about the structural evolution obtained from SAXS was systematically coupled with the measurements of DNA release obtained by agarose gel electrophoresis experiments.42 By doing so, we demonstrated that the structural evolution of lipoplexes under interaction with ALs follows a two-step mechanism (Figure 2e).
In the initial stage, cationic lipids within lipoplexes engage with anionic lipids on the surface of cellular membranes. Due to the increased lipid membrane available, the distance between the DNA molecules progressively increases with R. In this phase, the DNA peak present on the SAXS spectrum is still visible and moves continuously to lower transferred momentum values q due to the increased spacing of the 1D DNA lattice in real space. This demonstrated that, until neutralization of the lipoplex is complete, the DNA remains largely trapped in the complex.28
When the lipoplex is neutralized, a second phase begins. The second step involves a more intimate interaction where lipids from the lipoplex membrane and the target lipid structure (e.g., cellular membrane) mix and merge. In this second stage, the structure of the lipoplexes is gradually disintegrated upon further addition of ALs. This process can result in membrane fusion, where the lipid bilayers of the lipoplex and the target structure merge, allowing the cargo (e.g., genetic material) to be released. Without efficient lipid mixing and fusion, the cargo may remain trapped within the lipoplex, hindering its delivery into the target cell. This step also allows the lipoplex to adapt to the lipid environment of the target cell membrane, facilitating fusion.
SAXS studies also clarified that relevant differences in the tendency of lipoplexes to be destroyed by ALs could be observed. In general, the higher the structural stability of complexes upon interaction with cellular membranes, the lower the extent of DNA release. Each lipoplex formulation exhibited its own AL/CL molar ratio at which the structure was completely disintegrated, and the gene payload entirely released.36
Another key observation was that the phase transition rates were regulated by shape coupling between lipoplex and membrane lipids.43 Lipid shape is a well-established concept in membrane biophysics, frequently used to describe the volume occupied by phospholipids.44 With the use of this approach, phospholipids can be classified as cylinders (e.g., PC), cones (e.g., phosphatidylethanolamine, PE), and inverted cones (e.g., lysophosphatidylcholine), depending on the relative volumes of their polar head-groups and fatty acyl chains. The shape and packing of lipoplexes and ALs play a crucial role in their mixing and fusion processes. When there is minimal competition for space between lipoplexes and anionic lipids, the incorporation efficiency of ALs into lipoplex membranes is enhanced. Minimal competition for space refers to the idea that when two lipid bilayers come together for fusion, there should be mixing of lipids without overcrowding or steric hindrance.43 Lipid molecules from each bilayer need adequate space to reorganize and interact effectively to achieve fusion. If there is excessive competition for space, it can lead to incomplete fusion, nonproductive intermediates, or even membrane disruption.
This ability of ALs to integrate into lipoplex membranes can regulate the destabilization of the lipoplex structure, leading to the release of DNA. Therefore, by adjusting the lipid composition of lipoplexes to align with the membrane profile, it could be possible to effectively coordinate their intracellular activities.
Lipoplexes must have appropriate lipid composition and stability to effectively interact with cellular membranes and deliver their genetic material to the cytoplasm while avoiding lysosomal degradation. To achieve this, the multilayered structure of the lipoplexes needed to be reduced. Cellular studies coupled with fluorescence confocal microscopy experiments (Figure 2f) allowed one to explore the structural evolution of lipoplexes in cellular environments (Figure, 2g).
One promising delivery strategy was proposed by Hideyoshi Harashima’s research group at Sapporo University, Japan. They created the multifunctional envelope-type nano device (MEND),45 which had a core of pDNA condensed with polycations and surrounded by a few lipid membranes which could also have functional devices added to them in an incremental process.46 Our group employed a similar approach and ended up with well-structured DNA/protamine particles covered by a lipid monolayer called multicomponent envelope-type nanoparticle systems (MENS).47
Combining the qualities of MEND and the ability of multicomponent lipid membranes to break down endosomes, MENS facilitated an outstanding release of DNA (Figure 3). Consequently, MENS overcame lipoplexes with identical lipid composition and CL/DNA ratio in their tendency for high TE in multiple types of cell lines with no observable effect on the cells’ viability. Envelope-type nanoparticle systems were a source of inspiration for developing LNPs.
Figure 3.
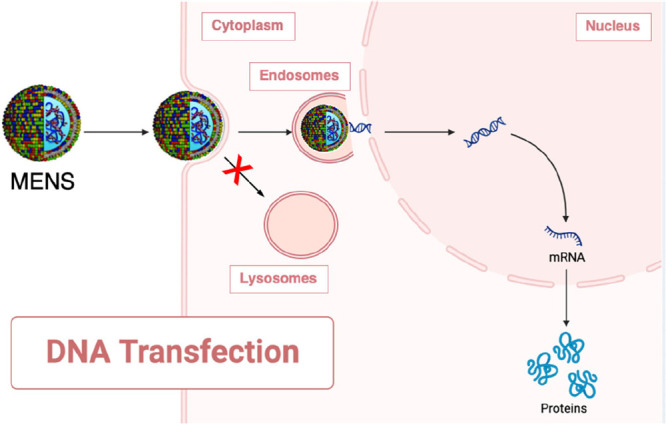
Schematic describing the performance of multicomponent envelope-type nanoparticle systems (MENS) as they navigate their way to the cell nucleus. Initially, MENS are internalized by the process of macropinocytosis, where they are taken up by the cell and enclosed within vesicles. Once inside the cell, MENS exhibit a unique characteristic: they diffuse freely within the cytosol. This diffusive behavior sets them apart from other nanoparticles and plays a crucial role in their overall performance. They possess the capability to maintain a low level of active transport, effectively avoiding pathways that would lead them to degrading lysosomes. This distinct feature allows MENS to preserve their integrity and functionality, ensuring that they remain intact throughout their journey within the cell. However, the true power of MENS lies in their exceptional DNA release from the endosomes. This process, likely due to gain in lipid mixing entropy,35 enables the DNA payload to escape the confines of the endosomes and gain access to the nucleus. The extensive nuclear entry achieved through effective DNA release is of paramount importance. It allows the DNA to reach its target destination within the cell, the nucleus, where it can exert its biological effects. The enhanced transfection efficiency resulting from successful nuclear entry is a key factor in the success of MENS as gene delivery systems. Cartoons were created using Biorender.com.
The mechanism of interaction between lipoplexes and ALs is significant for understanding how lipoplexes can undergo structural changes, and experimental approaches are extended to the understanding of LNPs as well.
Lipid Nanoparticles: Building Upon Lipoplex Milestones
In this section, we will closely examine critical facets of LNPs, encompassing their formation mechanism, lipid composition, structural characteristics, and interactions with cellular entities. Through these examinations, we aim to shed light on the progress made in applying lipoplex-derived knowledge to LNPs and emphasize the key areas requiring further attention and development.
Mechanism of Formation
Understanding how LNPs are formed is foundational to their successful application. Despite the progress made in understanding the mechanism of lipoplex formation and the factors shaping the equilibrium structure of lipoplexes, several challenges persisted in the 2010s, particularly in terms of their suitability for in vivo applications.
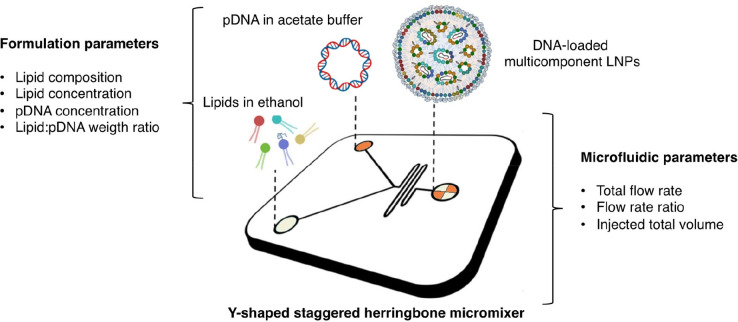
The process of bulk mixing, followed by pipetting and vortexing, yielded lipoplexes characterized by substantial heterogeneity in size, with a notable portion proving unsuitable for gene delivery purposes. Researchers aimed to tackle this problem by developing technologies that could create complexes with a uniform size and a positive charge, which would enable reliable large-scale production.48,49 LNPs were produced utilizing microfluidics, which ensures millisecond mixing at the nanoliter scale with consistency, reliability, and excellent encapsulation efficacy.50 The microfluidic system combines a stream containing nucleic acids (e.g., siRNA, mRNA, DNA, etc.) that are usually dissolved in a low pH acetate buffer (pH 4.0) with a stream containing a lipid mixture in an organic phase. Several studies explored the impact of varying operational and influential factors on the synthetic properties of LNPs in microfluidic devices.51 Microfluidic systems are adaptable, efficient, and highly customizable, accommodating various lipid compositions and optimizing LNPs for specific applications. These techniques involve lower shear stresses, preserving the integrity of sensitive molecules like mRNA. Continuous production reduced solvent usage, controlled encapsulation, and enhanced targeting capabilities further highlighting the benefits of microfluidics in producing LNPs.
Lipid Composition
The insights gleaned from the investigation of overcharged lipoplexes have been effectively harnessed in the development of FDA-approved LNPs, including mRNA-1273/SpikeVax, BNT162b2/Comirnaty, and Onpattro. While lipoplexes typically consist of a 50% cationic lipid and 50% neutral lipid composition, LNPs have evolved by substituting 50% of the cationic lipids with ionizable lipids. The introduction of ionizable lipids is of paramount importance for LNPs, ensuring the efficient encapsulation of negatively charged DNA during the mixing process. These ionizable lipids are meticulously designed with a pKa value below 7, allowing them to maintain a neutral charge at physiological pH levels. This adjustment serves a dual purpose: it reduces the potential toxicity associated with cationic charges and prolongs the circulation lifetime of LNPs.
Furthermore, researchers drew inspiration from the success of multicomponent lipoplexes and nanoparticle systems with multiple envelopes. This inspiration led them to incorporate four distinct lipid species within LNPs, with each lipid species serving a specific function.49 They include a neutral lipid such as distearoylphosphatidylcholine (DSPC), cholesterol, and a PEG-lipid.
In accordance with previous studies on lipoplexes,52 the neutral lipid and cholesterol are crucial, as they provide LNPs with bilayer stability and fusogenic properties as seen in liposomal drugs like Doxil.53 PEG lipids are responsible for particle stability, preventing aggregation, fusion, and opsonization in vivo.54
Despite the advantages of using PEG lipids, there are potential drawbacks that need to be taken into consideration. One such concern is the formation of anti-PEG antibodies,55 which can reduce the bioavailability of the drug and cause side effects such as the accelerated blood clearance (ABC) phenomenon.56 Recent studies have demonstrated that this issue can also arise when using SARS-CoV-2 LNP mRNA vaccines.57 The debate surrounding PEG lipids’ capability to prevent protein adsorption and stimulate opsonization remains unresolved among researchers. For a more in-depth understanding, we recommend readers consult Verhoef and Anchordoquy’s work.58
Structure
Conclusions about lipoplex structure inspired researchers to investigate the nanostructure of LNPs since their initial development. The structure of LNPs has been mainly studied in the context of the delivery of smaller siRNA (<30 nucleotides). The first formulations of LNPs were made in the early 2000s by macroscopic mixing of a solution containing siRNA with a solution containing ionizable aminolipids. A PEG-lipid was also required during the formulation process to prevent aggregation. These structures differed from liposomes in that they did not have an internal aqueous compartment but had a solid structure consisting of oligolamellar and multilamellar domains resembling that of lipoplexes.59
A decisive advance was achieved when Pieter Cullis and co-workers at the University of British Columbia showed that microfluidic mixing generated homogeneous and small-size LNPs with 100% encapsulation of siRNA.48 Since that turning point, many works have been published. Now we know that siRNA-loaded LNPs organize into a core–shell structure,60 wherein the core contains siRNA molecules electrostatically complexed with the ionizable lipid, with DSPC and the PEG-lipid residing primarily on the surface along with a portion of the ionizable lipid and cholesterol forming the shell of the LNPs. On the basis of the specific siRNA-to-lipid molar ratio and the lipid type, siRNA-LNPs can display various structural characteristics. These include a multilayered composition, a nanostructured core, or a uniform core–shell arrangement.61
However, the role of factors shaping the morphology of LNPs and their state within the particle interior remains unclear.62 As an instance, it has been suggested that the encapsulated size of the genetic payload affects the nanoscale arrangement of LNPs. Therefore, the question remains whether the lessons learned from siRNA-LNPs apply to mRNA-LNPs and plasmid DNA-LNPs due to their larger size (∼ 103–104 nucleotides). Using cryo-TEM, SAXS, and small-angle neutron scattering (SANS), Yanez Arteta et al. suggested that mRNA is located inside water cylinders, which are coated by cationic lipids, and proposed that this arrangement can determine instability under nonfrozen storage conditions.63 While mRNA is positioned within the particle interior, the nanoscale organization of lipids and mRNA remains ambiguous, with more research needed to confirm the morphology of mRNA-LNPs.
In a seminal investigation, Whitehead and co-workers were able to coformulate siRNA and mRNA in a single LNP formulation.64 While excellent transfection properties were obtained both in vitro and in vivo, no information about structural arrangement was provided.
We have even less information on the structure of LNPs loaded with plasmid DNA. Some recent studies demonstrated that DNA-LNPs are constituted by oligolamellar domains whose spatial extension is dependent on the lipid composition.65 Despite extensive research, a clear correlation between the structure of DNA-LNPs and their lipid composition has yet to be established.
Additionally, the effects of variables such as the CL/DNA ratio and DNA size on LNP structure remain poorly understood. In conclusion, understanding the nanoscale structure of LNPs is still in its infancy. It can also help in the determination of the stability and shelf life of these nanoparticles, which is essential for their storage and transport. Furthermore, a knowledge of the nanostructure can provide insights into the interactions of LNPs with biological membranes, which is critical for assessing their potential toxicity and safety profiles.
LNP–Cell Interactions
A pivotal aspect of this exploration entails investigating whether the principles elucidated in research concerning lipoplex–cell interactions can be effectively applied to LNPs. This inquiry is of paramount importance due to the parallels observed in their interactions with cellular membranes and the critical role that efficient lipid mixing and fusion play in ensuring successful cargo delivery. An essential facet of LNP efficacy lies in their interaction with cellular structures. In the case of lipoplexes, numerous investigations have sought to uncover the mechanisms behind their action. The cellular uptake of lipoplexes depends on their synthetic composition. Cells can internalize them through endocytosis, wherein the cell engulfs surrounding fluid, potentially carrying lipoplexes along. Different subtypes of endocytosis include clathrin-mediated endocytosis, caveolae-mediated endocytosis, and micropinocytosis.52,66 Another entry mechanism involves fusing directly with the cell membrane, releasing their cargo into the cell cytoplasm.67 This fusion is facilitated by the lipid components of the delivery systems merging with the cell membrane. The efficiency and selectivity of these mechanisms can vary, depending on the characteristics of the lipid systems (e.g., number and the types of the lipid species) and the type of target cell. Understanding the intricacies of these cellular entry mechanisms in LNP-mediated gene delivery is vital for developing effective formulations. The ability of lipoplexes to escape the autophagy-lysosomal pathway has also been investigated. It has been shown that intracellular trafficking, endosomal escape, and lysosomal degradation are interdependent processes.38 Formulations that are transported along microtubules tend to accumulate in lysosomes, while those that move through Brownian diffusion, avoiding active transport along the cytoskeleton, also evade degradation in the lysosomal compartment. Such understanding is still lacking for LNPs. Researchers have recently emphasized that the optimal proportions of various lipid species in LNP-siRNA systems can differ based on the specific ionizable cationic lipid used.60 They suggest that the ideal ratios should be fine-tuned according to the solubility of the lipid components with one another. Additionally, the lipid-to-RNA ratio has been determined through empirical methods to enhance gene silencing potency in vivo. As endosomes mature and their pH falls to 6 or lower, ionizable lipids become protonated, transitioning from a neutral to a positively charged state. This protonation is likely accompanied by a change in the lipid molecular shape, ultimately promoting the formation of inverted, highly fusogenic structures.68 Recent studies suggest that the lipid composition of LNPs is asymmetrical, with some lipid species confined to the aqueous core and others on the surface. Given that each lipid species specifically interacts with cellular lipids and may have a role as an adjuvant with cellular receptors like TLRs located on the plasma membrane or the endosomal membrane, there could be different effects even with the same lipid composition.
One effective approach for the development of LNPs with the ability to rupture endosomes involves creating a library of LNPs with systematic variations in lipid composition (Figure 4). These LNPs can then be tested by interacting them with an endosomal model membrane (EMM) at different LNP/EMM mass ratios. Lessons learned from lipoplexes suggest that EMM should closely resemble that of endosomal membranes. In accordance with the literature,69 EMM must contain a mixture of phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylinositol (PI), phosphatidylserine (PS), bis(monoacylglycero)phosphate (BMP), and sphingomyelin (SM). By performing SAXS measurements, we can analyze the structural changes in LNPs/EMM mixtures and estimate the range of R values at which LNPs start to disintegrate. Additionally, when combined with confocal microscopy experiments, this approach can help evaluate the ability of LNPs to disrupt endosomes and provide valuable insights into the optimal lipid composition for achieving endosomal rupture and facilitating large DNA release.
Figure 4.
Microfluidic manufacturing of multicomponent lipid nanoparticles (LNPs). The microfluidic manufacturing process of multicomponent LNPs loaded with plasmid DNA involves the utilization of a staggered herringbone micromixer (SHM). In this process, lipid blends dissolved in ethanol are injected through one inlet, while plasmid DNA dissolved in acetate buffer solution is injected through another inlet. At the Y-junction of the SHM, the two solutions converge and undergo chaotic mixing facilitated by the herringbone structure within the micromixer. This mixing phenomenon induces an increase in the polarity of the lipid solution, resulting in the formation of LNPs that encapsulate the plasmid DNA. Adjacent to the image, the text highlights two main categories of influential factors in the manufacturing process: synthesis and microfluidic parameters. Among these factors, lipid composition has been identified as a key parameter that governs the transfection behavior of both lipoplexes and multicomponent envelope-type nanoparticle systems (MENS). With this understanding, the hypothesis was put forth that the lipid composition of LNPs could modulate their ability to deliver the cargo when they interact and mix with cellular lipids. To explore this hypothesis, LNPs were prepared using the same lipid composition as that of MENS. Readapted with permission from ref (70). Copyright 2021 MDPI AG.
Future Research Directions
We will close this walk down memory lane with a reference to other important aspects that may contribute to bringing the field to maturity.
Like other gene delivery systems, LNPs encounter the hurdle of a brief circulation time and rapid elimination from the bloodstream. To address this issue, ionizable amino-lipids help maintain a biocompatible neutral charge, while PEG-lipids enhance stability and provide stealth properties. This shields the particle surface and limits the adsorption of serum proteins, ensuring protection against mononuclear phagocyte systemic uptake and thus contributing to increased circulation time (principles and applications are reviewed in ref (71)). However, both lipid components have their drawbacks. For instance, the ionizable cationic lipids are susceptible to temperature- and pH-dependent hydrolysis leading to detrimental effects on particle stability, while high concentrations of PEG lipids prevent the efficient delivery of RNA into cells (the “PEG dilemma”).72 Immune responses to PEG molecules have also been reported to result in accelerated blood clearance referred to as the “ABC” phenomenon.56 Future research will be aimed at understanding whether cationic lipids and alternative materials to PEG-lipids such as zwitterionic materials may provide LNPs with stealth properties.
An additional notable challenge involves the selection of suitable animal models for preclinical investigations. Mice are used for initial biodistribution and pharmacokinetics studies due to their size and ease of handling. Rats offer advantages for accessible organ studies. Rabbits are suitable for cardiovascular system research, while nonhuman primates closely mimic human responses, aiding safety assessments. Zebrafish are used for high-throughput screening due to transparency. Pigs, with human-like anatomy, are valuable for translational research, especially in relevant organ studies.
A significant aspect revolves around the capacity to accurately target selective organs and tissues. The development of targeted LNPs holds immense potential for improving the accuracy and effectiveness of drug delivery. Promising strategies for targeted LNPs encompass antibody-based targeting, peptide ligands, aptamers, and small molecules, both individually and in combination. Nevertheless, despite their potential, the clinical application of targeted LNPs has encountered hurdles. One prominent obstacle arises from the fact that, under physiological conditions, targeted LNPs become coated with biomolecules, thereby concealing their targeting ligands.
Recent studies explore the gap between laboratory findings and the clinical success of NPs. This gap is partially attributed to our limited understanding of the connection between their synthetic identity, biological identity, and physiological response.73 Upon in vivo administration (e.g., i.v., subcutaneous injection, etc.) NPs get coated by a protein corona that provides them with a new biological identity that controls physiological response.74 In previous works, we have shown that the protein corona acquired by lipid vesicles has a composition that is simultaneously regulated by the electrical charge and by the affinity of lipids for individual proteins or specific classes of plasma proteins.75 The protein corona regulates the interaction with immune system cells in vitro and ex vivo.76
These findings prompted us to create biomimetic lipoplexes with an artificial opsonin-deficient protein corona that minimizes the uptake by immune cells in vivo.16 We have also demonstrated that the protein corona of lipoplexes locks onto receptors of cancer cells, promoting internalization by a receptor-mediated mechanism. It was the case of DOTAP/DNA lipoplexes coated by a vitronectin-enriched protein corona. We showed that vitronectin promotes efficient uptake in cancer cells expressing high levels of the vitronectin ανβ3 integrin receptor.77 Accurate identification of protein corona fingerprints has allowed us to predict multiple biological interactions between lipid vesicles and cancer cells.
The development of such knowledge for LNPs will be vital for improved particle design and targeted delivery. Now that we know that the protein corona is the missing link to correlate synthetic identity and physiological response, it will be necessary to investigate the bionano interactions between LNPs and specific biological fluids.
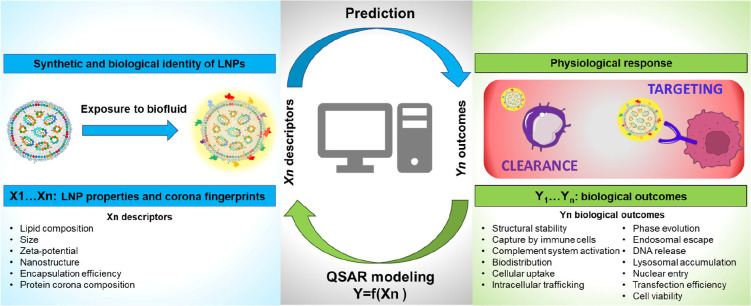
In this regard, the quantitative structure–activity relationship (QSAR) methods can be used to predict the relationships between the synthetic identity of LNPs and their mechanism of action at a cellular level. QSAR is a computational modeling technique used in nanomedicine to predict and understand the relationship between the chemical or structural properties of nanoparticles and their biological activities or effects.78 QSAR guides the design of nanoparticles by quantifying how various structural features (such as size, shape, surface charge, and composition) impact their behavior in biological environments. QSAR provides insights into the underlying mechanisms of nanoparticle interactions with biological systems. It helps researchers understand how nanoparticles are taken up by cells, influence biological pathways, and exert their therapeutic effects. QSAR approaches assume that LNPs with comparable synthetic identities will interact similarly with cells thus eliciting similar physiological responses (e.g., protein expression and production of antibodies). We envision that QSAR investigations will enable prediction of bioactivity, accelerating the development of optimized LNPs. Identifying the most appropriate set of nanodescriptors (e.g., size, shape, surface properties) representing the relevance of each factor contributing to the creation of the protein corona will require the development of novel high-throughput approaches. In this regard, QSAR is emerging as a powerful tool to describe the interactions at the nanobio interface.79 Significant new research will be therefore aimed at identifying meaningful relations between the synthetic/biological identity of NPs and their biological fate (blood circulation time and biodistribution) (Figure 5).
Figure 5.
Schematics illustrating predictive modeling workflow by quantitative structure–activity relationship (QSAR). After being administered in vivo, LNPs acquire a new biological identity through the formation of a protein corona, which can have a significant impact on their biological response, including biodistribution, toxicity, immune system activation, accumulation at the target site, and interaction with target cell receptors. To predict the biological effects of protein corona formation, QSAR models can be developed to relate LNP properties and protein corona fingerprints to biological outcomes. This can aid in identifying meaningful relationships between the synthetic and biological identities of LNPs and their effects.
A prospective avenue of research entails harnessing the potential of artificial intelligence (AI) for the refinement and enhancement of LNPs. AI plays a rapidly growing and pivotal role in the design and optimization of LNPs in several ways. AI can optimize the ratios and combinations of lipids and other components in LNPs to improve their stability, drug-loading capacity, and targeting properties. This reduces the need for time-consuming trial-and-error experiments. AI models can predict how different drug molecules interact with LNPs, helping researchers select the most suitable LNPs for specific drugs. This can accelerate the drug development process. AI can simulate the biodistribution of LNPs in the body, predicting how they interact with various tissues and cells. This aids in designing LNPs for targeted delivery and minimizing off-target effects. AI models can also assess the potential toxicity of LNPs and their components, enabling the selection of safer formulations and guiding regulatory compliance. AI can analyze data to predict the immune responses triggered by LNPs, helping researchers design LNPs that minimize immunogenicity and improve vaccine safety. AI can analyze patient-specific data to design personalized LNPs, tailoring drug delivery systems to individual genetic and physiological characteristics. AI algorithms can handle and integrate large data sets from various sources, including genomics, proteomics, and clinical trials.
In conclusion, we firmly believe that the convergence of these forthcoming advancements, when thoughtfully integrated, will yield invaluable insights and novel prospects, ultimately expediting the clinical adoption of LNPs. This transition will propel them beyond the realm of laboratory research and into practical utilization within medical settings.
Acknowledgments
The research leading to the results discussed here has received funding from the Sapienza University of Rome (Grant RM12117A87BA3B80 to G.C.) and the Italian Minister for University and Research (MUR) (Futuro in Ricerca, Grant RBFR08TLPO) (P.I. G.C.). D.P. and G.C. extend their sincere appreciation to the numerous researchers mentioned explicitly, whose work has served as a profound source of inspiration over the course of two decades. They would also like to express gratitude to all the collaborators who actively contributed to the research presented in this comprehensive article. A special acknowledgment is owed to Dr. Heinz Amenitsch from the Technical University of Graz (Austria), Dr. Augusto Amici, and Cristina Marchini from the University of Camerino (Italy), Dr. Francesco Cardarelli from the Scuola Normale Superiore di Pisa (Italy), and Dr. Enrico Gratton from the University of California, Irvine, for their invaluable contributions.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsptsci.3c00185.
Figure S1 illustrates the nanostructure of both the lamellar and inverted hexagonal phases of lipoplexes (PDF)
Author Contributions
D.P. and G.C. shared equal responsibility for conceptualization, data curation, and the initial draft writing of the manuscript. They both contributed to the critical review and editing of the content, ensuring its accuracy and coherence.
The authors declare no competing financial interest.
Supplementary Material
References
- Khurana A.; Allawadhi P.; Khurana I.; Allwadhi S.; Weiskirchen R.; Banothu A. K.; Chhabra D.; Joshi K.; Bharani K. K. Role of nanotechnology behind the success of mRNA vaccines for COVID-19. Nano Today 2021, 38, 101142 10.1016/j.nantod.2021.101142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akinc A.; Maier M. A.; Manoharan M.; Fitzgerald K.; Jayaraman M.; Barros S.; Ansell S.; Du X.; Hope M. J.; Madden T. D.; Mui B. L.; Semple S. C.; Tam Y. K.; Ciufolini M.; Witzigmann D.; Kulkarni J. A.; van der Meel R.; Cullis P. R. The Onpattro story and the clinical translation of nanomedicines containing nucleic acid-based drugs. Nature Nanotechnol. 2019, 14 (12), 1084–1087. 10.1038/s41565-019-0591-y. [DOI] [PubMed] [Google Scholar]
- Thi T. T. H.; Suys E. J.; Lee J. S.; Nguyen D. H.; Park K. D.; Truong N. P. Lipid-based nanoparticles in the clinic and clinical trials: from cancer nanomedicine to COVID-19 vaccines. Vaccines 2021, 9 (4), 359. 10.3390/vaccines9040359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazemian P.; Yu S.-Y.; Thomson S. B.; Birkenshaw A.; Leavitt B. R.; Ross C. J. Lipid-nanoparticle-based delivery of CRISPR/Cas9 genome-editing components. Mol. Pharmaceutics 2022, 19 (6), 1669–1686. 10.1021/acs.molpharmaceut.1c00916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung M.; Lee S.; Park S.; Hong J.; Kim C.; Cho I.; Sohn H. S.; Kim K.; Park I. W.; Yoon S.; Kwon S.; Shin J.; Lee D.; Kang M.; Go S.; Moon S.; Chung Y.; Kim Y.; Kim B.-S. A Therapeutic Nanovaccine that Generates Anti-Amyloid Antibodies and Amyloid-specific Regulatory T Cells for Alzheimer’s Disease. Adv. Mater. 2023, 35 (3), 2207719 10.1002/adma.202207719. [DOI] [PubMed] [Google Scholar]
- Jagaran K.; Singh M. Lipid nanoparticles: promising treatment approach for Parkinson’s disease. International Journal of Molecular Sciences 2022, 23 (16), 9361. 10.3390/ijms23169361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni J. A.; Myhre J. L.; Chen S.; Tam Y. Y. C.; Danescu A.; Richman J. M.; Cullis P. R. Design of lipid nanoparticles for in vitro and in vivo delivery of plasmid DNA. Nanomedicine: Nanotechnology, Biology and Medicine 2017, 13 (4), 1377–1387. 10.1016/j.nano.2016.12.014. [DOI] [PubMed] [Google Scholar]
- Felgner P. L.; Gadek T. R.; Holm M.; Roman R.; Chan H. W.; Wenz M.; Northrop J. P.; Ringold G. M.; Danielsen M. Lipofection: a highly efficient, lipid-mediated DNA-transfection procedure. Proc. Natl. Acad. Sci. U. S. A. 1987, 84 (21), 7413–7417. 10.1073/pnas.84.21.7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y.; Szoka F. C. Jr. Mechanism of DNA Release from Cationic Liposome/DNA Complexes Used in Cell Transfection. Biochemistry 1996, 35 (18), 5616–5623. 10.1021/bi9602019. [DOI] [PubMed] [Google Scholar]
- Zuidam N. J.; Hirsch-Lerner D.; Margulies S.; Barenholz Y. Lamellarity of cationic liposomes and mode of preparation of lipoplexes affect transfection efficiency. Biochimica et Biophysica Acta (BBA)-Biomembranes 1999, 1419 (2), 207–220. 10.1016/S0005-2736(99)00069-3. [DOI] [PubMed] [Google Scholar]
- Kennedy M. T.; Pozharski E. V.; Rakhmanova V. A.; MacDonald R. C. Factors governing the assembly of cationic phospholipid-DNA complexes. Biophysical journal 2000, 78 (3), 1620–1633. 10.1016/S0006-3495(00)76714-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning G. S.; Ray J. Counterion condensation revisited. J. Biomol. Struct. Dyn. 1998, 16 (2), 461–476. 10.1080/07391102.1998.10508261. [DOI] [PubMed] [Google Scholar]
- Bruinsma R. Electrostatics of DNA-cationic lipid complexes: isoelectric instability. European Physical Journal B-Condensed Matter and Complex Systems 1998, 4, 75–88. 10.1007/s100510050353. [DOI] [Google Scholar]
- Wagner K.; Harries D.; May S.; Kahl V.; Rädler J.; Ben-Shaul A. Direct evidence for counterion release upon cationic lipid– DNA condensation. Langmuir 2000, 16 (2), 303–306. 10.1021/la991268a. [DOI] [Google Scholar]
- Harries D.; May S.; Gelbart W. M.; Ben-Shaul A. Structure, stability, and thermodynamics of lamellar DNA-lipid complexes. Biophys. J. 1998, 75 (1), 159–173. 10.1016/S0006-3495(98)77503-4. [DOI] [PMC free article] [PubMed] [Google Scholar]; May S.; Harries D.; Ben-Shaul A. The phase behavior of cationic lipid–DNA complexes. Biophys. J. 2000, 78 (4), 1681–1697. 10.1016/S0006-3495(00)76720-8. [DOI] [PMC free article] [PubMed] [Google Scholar]; Harries D.; May S.; Ben-Shaul A. Curvature and Charge Modulations in Lamellar DNA– Lipid Complexes. J. Phys. Chem. B 2003, 107 (15), 3624–3630. 10.1021/jp026637h. [DOI] [Google Scholar]; May S.; Ben-Shaul A. Modeling of cationic lipid-DNA complexes. Curr. Med. Chem. 2004, 11 (2), 151–167. 10.2174/0929867043456142. [DOI] [PubMed] [Google Scholar]
- Giulimondi F.; Vulpis E.; Digiacomo L.; Giuli M. V.; Mancusi A.; Capriotti A. L.; Laganà A.; Cerrato A.; Zenezini Chiozzi R.; Nicoletti C.; et al. Opsonin-Deficient Nucleoproteic Corona Endows UnPEGylated Liposomes with Stealth Properties In Vivo. ACS Nano 2022, 16 (2), 2088–2100. 10.1021/acsnano.1c07687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordi F.; Cametti C.; Diociaiuti M.; Gaudino D.; Gili T.; Sennato S. Complexation of anionic polyelectrolytes with cationic liposomes: evidence of reentrant condensation and lipoplex formation. Langmuir 2004, 20 (13), 5214–5222. 10.1021/la036006u. [DOI] [PubMed] [Google Scholar]
- Papahadjopoulos D.; Allen T.; Gabizon A.; Mayhew E.; Matthay K.; Huang S.; Lee K.; Woodle M.; Lasic D.; Redemann C. Sterically stabilized liposomes: improvements in pharmacokinetics and antitumor therapeutic efficacy. Proc. Natl. Acad. Sci. U. S. A. 1991, 88 (24), 11460–11464. 10.1073/pnas.88.24.11460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y.; Hui S.-W.; Frederik P.; Szoka F. C. Jr. Physicochemical Characterization and Purification of Cationic Lipoplexes. Biophys. J. 1999, 77 (1), 341–353. 10.1016/S0006-3495(99)76894-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Úbeda M.; Misra S. K.; Barrán-Berdón A. L.; Aicart-Ramos C.; Sierra M. B.; Biswas J.; Kondaiah P.; Junquera E.; Bhattacharya S.; Aicart E. Why is less cationic lipid required to prepare lipoplexes from plasmid DNA than linear DNA in gene therapy?. J. Am. Chem. Soc. 2011, 133 (45), 18014–18017. 10.1021/ja204693f. [DOI] [PubMed] [Google Scholar]
- Kreiss P.; Mailhe P.; Scherman D.; Pitard B.; Cameron B.; Rangara R.; Aguerre-Charriol O.; Airiau M.; Crouzet J. Plasmid DNA size does not affect the physicochemical properties of lipoplexes but modulates gene transfer efficiency. Nucleic acids research 1999, 27 (19), 3792–3798. 10.1093/nar/27.19.3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg B.; Sorgi F. L.; Huang L. New structures in complex formation between DNA and cationic liposomes visualized by freeze—fracture electron microscopy. FEBS letters 1994, 356 (2–3), 361–366. 10.1016/0014-5793(94)01315-2. [DOI] [PubMed] [Google Scholar]
- Gustafsson J.; Arvidson G.; Karlsson G.; Almgren M. Complexes between cationic liposomes and DNA visualized by cryo-TEM. Biochimica et Biophysica Acta (BBA) - Biomembranes 1995, 1235 (2), 305–312. 10.1016/0005-2736(95)80018-B. [DOI] [PubMed] [Google Scholar]
- Rädler J. O.; Koltover I.; Salditt T.; Safinya C. R. Structure of DNA-cationic liposome complexes: DNA intercalation in multilamellar membranes in distinct interhelical packing regimes. Science 1997, 275 (5301), 810–814. 10.1126/science.275.5301.810. [DOI] [PubMed] [Google Scholar]
- Madeira C.; Loura L. M.; Aires-Barros M. R.; Fedorov A.; Prieto M. Characterization of DNA/lipid complexes by fluorescence resonance energy transfer. Biophysical journal 2003, 85 (5), 3106–3119. 10.1016/S0006-3495(03)74729-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artzner F.; Zantl R.; Rapp G.; Rädler J. Observation of a rectangular columnar phase in condensed lamellar cationic lipid-DNA complexes. Physical review letters 1998, 81 (22), 5015. 10.1103/PhysRevLett.81.5015. [DOI] [Google Scholar]; Koynova R.; MacDonald R. C. Columnar DNA superlattices in lamellar o-ethylphosphatidylcholine lipoplexes: mechanism of the gel-liquid crystalline lipid phase transition. Nano Lett. 2004, 4 (8), 1475–1479. 10.1021/nl049191k. [DOI] [Google Scholar]; McManus J. J.; Rädler J. O.; Dawson K. A. Observation of a rectangular columnar phase in a DNA– calcium– zwitterionic lipid complex. J. Am. Chem. Soc. 2004, 126 (49), 15966–15967. 10.1021/ja046105+. [DOI] [PubMed] [Google Scholar]; Caracciolo G.; Pozzi D.; Caminiti R.; Mancini G.; Luciani P.; Amenitsch H. Observation of a rectangular DNA superlattice in the liquid-crystalline phase of cationic lipid/DNA complexes. J. Am. Chem. Soc. 2007, 129 (33), 10092–10093. 10.1021/ja073890s. [DOI] [PubMed] [Google Scholar]
- Koltover I.; Salditt T.; Rädler J. O.; Safinya C. R. An inverted hexagonal phase of cationic liposome-DNA complexes related to DNA release and delivery. Science 1998, 281 (5373), 78–81. 10.1126/science.281.5373.78. [DOI] [PubMed] [Google Scholar]
- Koynova R.; Wang L.; MacDonald R. C. Cationic phospholipids forming cubic phases: lipoplex structure and transfection efficiency. Mol. Pharmaceutics 2008, 5 (5), 739–744. 10.1021/mp800011e. [DOI] [PubMed] [Google Scholar]
- Caracciolo G.; Pozzi D.; Caminiti R.; Congiu Castellano A. Structural characterization of a new lipid/DNA complex showing a selective transfection efficiency in ovarian cancer cells. Eur. Phys. J. E 2003, 10, 331–336. 10.1140/epje/i2002-10117-x. [DOI] [PubMed] [Google Scholar]
- Koynova R.; Wang L.; MacDonald R. C. An intracellular lamellar–nonlamellar phase transition rationalizes the superior performance of some cationic lipid transfection agents. Proc. Natl. Acad. Sci. U. S. A. 2006, 103 (39), 14373–14378. 10.1073/pnas.0603085103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin A. J.; Slack N. L.; Ahmad A.; George C. X.; Samuel C. E.; Safinya C. R. Three-dimensional imaging of lipid gene-carriers: membrane charge density controls universal transfection behavior in lamellar cationic liposome-DNA complexes. Biophysical journal 2003, 84 (5), 3307–3316. 10.1016/S0006-3495(03)70055-1. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ahmad A.; Evans H. M.; Ewert K.; George C. X.; Samuel C. E.; Safinya C. R. New multivalent cationic lipids reveal bell curve for transfection efficiency versus membrane charge density: lipid–DNA complexes for gene delivery. Journal of Gene Medicine: A cross-disciplinary journal for research on the science of gene transfer and its clinical applications 2005, 7 (6), 739–748. 10.1002/jgm.717. [DOI] [PubMed] [Google Scholar]
- Caracciolo G.; Pozzi D.; Caminiti R.; Marchini C.; Montani M.; Amici A.; Amenitsch H. Enhanced Transfection Efficiency of Multicomponent Lipoplexes in the Regime of Optimal Membrane Charge Density. J. Phys. Chem. B 2008, 112 (36), 11298–11304. 10.1021/jp803077n. [DOI] [PubMed] [Google Scholar]
- Caracciolo G.; Pozzi D.; Amenitsch H.; Caminiti R. Multicomponent Cationic Lipid–DNA Complex Formation: Role of Lipid Mixing. Langmuir 2005, 21 (25), 11582–11587. 10.1021/la052077c. [DOI] [PubMed] [Google Scholar]
- Gao X.; Huang L. A novel cationic liposome reagent for efficient transfection of mammalian cells. Biochemical and biophysical research communications 1991, 179 (1), 280–285. 10.1016/0006-291X(91)91366-K. [DOI] [PubMed] [Google Scholar]
- Caracciolo G.; Pozzi D.; Caminiti R.; Amenitsch H. Two-dimensional lipid mixing entropy regulates the formation of multicomponent lipoplexes. J. Phys. Chem. B 2006, 110 (42), 20829–20835. 10.1021/jp0620926. [DOI] [PubMed] [Google Scholar]
- Caracciolo G.; Marchini C.; Pozzi D.; Caminiti R.; Amenitsch H.; Montani M.; Amici A. Structural Stability against Disintegration by Anionic Lipids Rationalizes the Efficiency of Cationic Liposome/DNA Complexes. Langmuir 2007, 23 (8), 4498–4508. 10.1021/la063456o. [DOI] [PubMed] [Google Scholar]
- Caracciolo G.; Pozzi D.; Caminiti R.; Marchini C.; Montani M.; Amici A.; Amenitsch H. Transfection efficiency boost by designer multicomponent lipoplexes. Biochimica et Biophysica Acta (BBA)-Biomembranes 2007, 1768 (9), 2280–2292. 10.1016/j.bbamem.2007.06.027. [DOI] [PubMed] [Google Scholar]
- Cardarelli F.; Digiacomo L.; Marchini C.; Amici A.; Salomone F.; Fiume G.; Rossetta A.; Gratton E.; Pozzi D.; Caracciolo G. The intracellular trafficking mechanism of Lipofectamine-based transfection reagents and its implication for gene delivery. Sci. Rep. 2016, 6 (1), 1–8. 10.1038/srep25879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palchetti S.; Pozzi D.; Marchini C.; Amici A.; Andreani C.; Bartolacci C.; Digiacomo L.; Gambini V.; Cardarelli F.; Di Rienzo C.; et al. Manipulation of lipoplex concentration at the cell surface boosts transfection efficiency in hard-to-transfect cells. Nanomedicine: Nanotechnology, Biology and Medicine 2017, 13 (2), 681–691. 10.1016/j.nano.2016.08.019. [DOI] [PubMed] [Google Scholar]
- Zuhorn I. S.; Oberle V.; Visser W. H.; Engberts J. B. F. N.; Bakowsky U.; Polushkin E.; Hoekstra D. Phase Behavior of Cationic Amphiphiles and Their Mixtures with Helper Lipid Influences Lipoplex Shape, DNA Translocation, and Transfection Efficiency. Biophys. J. 2002, 83 (4), 2096–2108. 10.1016/S0006-3495(02)73970-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koynova R.; Tarahovsky Y. S.; Wang L.; MacDonald R. C. Lipoplex formulation of superior efficacy exhibits high surface activity and fusogenicity, and readily releases DNA. Biochimica et Biophysica Acta (BBA) - Biomembranes 2007, 1768 (2), 375–386. 10.1016/j.bbamem.2006.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caracciolo G.; Pozzi D.; Caminiti R.; Marchini C.; Montani M.; Amici A.; Amenitsch H. On the correlation between phase evolution of lipoplexes/anionic lipid mixtures and DNA release. Appl. Phys. Lett. 2007, 91 (14), 143903. 10.1063/1.2794436. [DOI] [Google Scholar]
- Pozzi D.; Caracciolo G.; Caminiti R.; De Sanctis S. C.; Amenitsch H.; Marchini C.; Montani M.; Amici A. Toward the rational design of lipid gene vectors: shape coupling between lipoplex and anionic cellular lipids controls the phase evolution of lipoplexes and the efficiency of DNA release. ACS Appl. Mater. Interfaces 2009, 1 (10), 2237–2249. 10.1021/am900406b. [DOI] [PubMed] [Google Scholar]
- Israelachvili J. N. Refinement of the fluid-mosaic model of membrane structure. Biochimica et Biophysica Acta (BBA)-Biomembranes 1977, 469 (2), 221–225. 10.1016/0005-2736(77)90185-7. [DOI] [PubMed] [Google Scholar]
- Khalil I A; Kogure K; Futaki S; Hama S; Akita H; Ueno M; Kishida H; Kudoh M; Mishina Y; Kataoka K; Yamada M; Harashima H Octaarginine-modified multifunctional envelope-type nanoparticles for gene delivery. Gene therapy 2007, 14 (8), 682–689. 10.1038/sj.gt.3302910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akita H.; Kudo A.; Minoura A.; Yamaguti M.; Khalil I. A.; Moriguchi R.; Masuda T.; Danev R.; Nagayama K.; Kogure K.; et al. Multi-layered nanoparticles for penetrating the endosome and nuclear membrane via a step-wise membrane fusion process. Biomaterials 2009, 30 (15), 2940–2949. 10.1016/j.biomaterials.2009.02.009. [DOI] [PubMed] [Google Scholar]
- Pozzi D.; Marchini C.; Cardarelli F.; Rossetta A.; Colapicchioni V.; Amici A.; Montani M.; Motta S.; Brocca P.; Cantu L.; Caracciolo G. Mechanistic understanding of gene delivery mediated by highly efficient multicomponent envelope-type nanoparticle systems. Mol. Pharmaceutics 2013, 10 (12), 4654–4665. 10.1021/mp400470p. [DOI] [PubMed] [Google Scholar]; Pozzi D.; Marchini C.; Cardarelli F.; Salomone F.; Coppola S.; Montani M.; Zabaleta M. E.; Digman M.A.; Gratton E.; Colapicchioni V.; Caracciolo G. Mechanistic evaluation of the transfection barriers involved in lipid-mediated gene delivery: interplay between nanostructure and composition. Biochimica et Biophysica Acta (BBA)-Biomembranes 2014, 1838 (3), 957–967. 10.1016/j.bbamem.2013.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belliveau N. M; Huft J.; Lin P. J.; Chen S.; Leung A. K.; Leaver T. J; Wild A. W; Lee J. B; Taylor R. J; Tam Y. K; Hansen C. L; Cullis P. R Microfluidic synthesis of highly potent limit-size lipid nanoparticles for in vivo delivery of siRNA. Molecular Therapy-Nucleic Acids 2012, 1, e37. 10.1038/mtna.2012.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kon E.; Elia U.; Peer D. Principles for designing an optimal mRNA lipid nanoparticle vaccine. Curr. Opin. Biotechnol. 2022, 73, 329–336. 10.1016/j.copbio.2021.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullis P. R.; Hope M. J. Lipid nanoparticle systems for enabling gene therapies. Molecular Therapy 2017, 25 (7), 1467–1475. 10.1016/j.ymthe.2017.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roces C. B.; Lou G.; Jain N.; Abraham S.; Thomas A.; Halbert G. W.; Perrie Y. Manufacturing considerations for the development of lipid nanoparticles using microfluidics. Pharmaceutics 2020, 12 (11), 1095. 10.3390/pharmaceutics12111095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozzi D.; Marchini C.; Cardarelli F.; Amenitsch H.; Garulli C.; Bifone A.; Caracciolo G. Transfection efficiency boost of cholesterol-containing lipoplexes. Biochimica et Biophysica Acta (BBA) - Biomembranes 2012, 1818 (9), 2335–2343. 10.1016/j.bbamem.2012.05.017. [DOI] [PubMed] [Google Scholar]
- Briuglia M.-L.; Rotella C.; McFarlane A.; Lamprou D. A. Influence of cholesterol on liposome stability and on in vitro drug release. Drug delivery and translational research 2015, 5, 231–242. 10.1007/s13346-015-0220-8. [DOI] [PubMed] [Google Scholar]
- Gabizon A.; Chemla M.; Tzemach D.; Horowitz A.; Goren D. Liposome longevity and stability in circulation: effects on the in vivo delivery to tumors and therapeutic efficacy of encapsulated anthracyclines. J. Drug Targeting 1996, 3 (5), 391–398. 10.3109/10611869608996830. [DOI] [PubMed] [Google Scholar]
- Kozma G. T.; Shimizu T.; Ishida T.; Szebeni J. Anti-PEG antibodies: Properties, formation, testing and role in adverse immune reactions to PEGylated nano-biopharmaceuticals. Advanced drug delivery reviews 2020, 154, 163–175. 10.1016/j.addr.2020.07.024. [DOI] [PubMed] [Google Scholar]
- Abu Lila A. S.; Kiwada H.; Ishida T. The accelerated blood clearance (ABC) phenomenon: clinical challenge and approaches to manage. J. Controlled Release 2013, 172 (1), 38–47. 10.1016/j.jconrel.2013.07.026. [DOI] [PubMed] [Google Scholar]
- Ju Y.; Lee W. S.; Pilkington E. H.; Kelly H. G.; Li S.; Selva K. J.; Wragg K. M.; Subbarao K.; Nguyen T. H. O.; Rowntree L. C.; Allen L. F.; Bond K.; Williamson D. A.; Truong N. P.; Plebanski M.; Kedzierska K.; Mahanty S.; Chung A. W.; Caruso F.; Wheatley A. K.; Juno J. A.; Kent S. J. Anti-PEG antibodies boosted in humans by SARS-CoV-2 lipid nanoparticle mRNA vaccine. ACS Nano 2022, 16 (8), 11769–11780. 10.1021/acsnano.2c04543. [DOI] [PubMed] [Google Scholar]
- Verhoef J. J.; Anchordoquy T. J. Questioning the use of PEGylation for drug delivery. Drug delivery and translational research 2013, 3, 499–503. 10.1007/s13346-013-0176-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semple S. C.; Klimuk S. K.; Harasym T. O.; Dos Santos N.; Ansell S. M.; Wong K. F.; Maurer N.; Stark H.; Cullis P. R.; Hope M. J.; Scherrer P. Efficient encapsulation of antisense oligonucleotides in lipid vesicles using ionizable aminolipids: formation of novel small multilamellar vesicle structures. Biochimica et Biophysica Acta (BBA)-Biomembranes 2001, 1510 (1–2), 152–166. 10.1016/S0005-2736(00)00343-6. [DOI] [PubMed] [Google Scholar]
- Kulkarni J. A.; Darjuan M. M.; Mercer J. E.; Chen S.; Van Der Meel R.; Thewalt J. L.; Tam Y. Y. C.; Cullis P. R. On the formation and morphology of lipid nanoparticles containing ionizable cationic lipids and siRNA. ACS Nano 2018, 12 (5), 4787–4795. 10.1021/acsnano.8b01516. [DOI] [PubMed] [Google Scholar]
- Viger-Gravel J.; Schantz A.; Pinon A. C.; Rossini A. J.; Schantz S.; Emsley L. Structure of lipid nanoparticles containing siRNA or mRNA by dynamic nuclear polarization-enhanced NMR spectroscopy. J. Phys. Chem. B 2018, 122 (7), 2073–2081. 10.1021/acs.jpcb.7b10795. [DOI] [PubMed] [Google Scholar]
- Pilkington E. H.; Suys E. J.A.; Trevaskis N. L.; Wheatley A. K.; Zukancic D.; Algarni A.; Al-Wassiti H.; Davis T. P.; Pouton C. W.; Kent S. J.; Truong N. P. From influenza to COVID-19: Lipid nanoparticle mRNA vaccines at the frontiers of infectious diseases. Acta biomaterialia 2021, 131, 16–40. 10.1016/j.actbio.2021.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanez Arteta M.; Kjellman T.; Bartesaghi S.; Wallin S.; Wu X.; Kvist A. J.; Dabkowska A.; Szekely N.; Radulescu A.; Bergenholtz J.; Lindfors L. Successful reprogramming of cellular protein production through mRNA delivered by functionalized lipid nanoparticles. Proc. Natl. Acad. Sci. U. S. A. 2018, 115 (15), E3351–E3360. 10.1073/pnas.1720542115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball R. L.; Hajj K. A.; Vizelman J.; Bajaj P.; Whitehead K. A. Lipid nanoparticle formulations for enhanced co-delivery of siRNA and mRNA. Nano Lett. 2018, 18 (6), 3814–3822. 10.1021/acs.nanolett.8b01101. [DOI] [PubMed] [Google Scholar]
- Quagliarini E.; Wang J.; Renzi S.; Cui L.; Digiacomo L.; Ferri G.; Pesce L.; De Lorenzi V.; Matteoli G.; Amenitsch H.; et al. Mechanistic Insights into the Superior DNA Delivery Efficiency of Multicomponent Lipid Nanoparticles: An In Vitro and In Vivo Study. ACS Appl. Mater. Interfaces 2022, 14 (51), 56666–56677. 10.1021/acsami.2c20019. [DOI] [PubMed] [Google Scholar]; Cui L.; Renzi S.; Quagliarini E.; Digiacomo L.; Amenitsch H.; Masuelli L.; Bei R.; Ferri G.; Cardarelli F.; Wang J.; et al. Efficient Delivery of DNA Using Lipid Nanoparticles. Pharmaceutics 2022, 14 (8), 1698. 10.3390/pharmaceutics14081698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Digiacomo L.; Cardarelli F.; Pozzi D.; Palchetti S.; Digman M.; Gratton E.; Capriotti A.; Mahmoudi M.; Caracciolo G. An apolipoprotein-enriched biomolecular corona switches the cellular uptake mechanism and trafficking pathway of lipid nanoparticles. Nanoscale 2017, 9 (44), 17254–17262. 10.1039/C7NR06437C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resina S.; Prevot P.; Thierry A. R. Physico-chemical characteristics of lipoplexes influence cell uptake mechanisms and transfection efficacy. PLoS One 2009, 4 (6), e6058. 10.1371/journal.pone.0006058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jadhav S. G.; Dowdy S. F. Overcoming delivery barriers with LNPs. Nat. Mater. 2021, 20 (5), 575–577. 10.1038/s41563-021-00988-3. [DOI] [PubMed] [Google Scholar]
- van Meer G.; Voelker D. R.; Feigenson G. W. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008, 9 (2), 112–124. 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quagliarini E.; Renzi S.; Digiacomo L.; Giulimondi F.; Sartori B.; Amenitsch H.; Tassinari V.; Masuelli L.; Bei R.; Cui L.; et al. Microfluidic Formulation of DNA-Loaded Multicomponent Lipid Nanoparticles for Gene Delivery. Pharmaceutics 2021, 13 (8), 1292. 10.3390/pharmaceutics13081292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suk J. S.; Xu Q.; Kim N.; Hanes J.; Ensign L. M. PEGylation as a strategy for improving nanoparticle-based drug and gene delivery. Advanced drug delivery reviews 2016, 99, 28–51. 10.1016/j.addr.2015.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakeyama H.; Akita H.; Harashima H. The polyethyleneglycol dilemma: advantage and disadvantage of PEGylation of liposomes for systemic genes and nucleic acids delivery to tumors. Biological and pharmaceutical bulletin 2013, 36 (6), 892–899. 10.1248/bpb.b13-00059. [DOI] [PubMed] [Google Scholar]
- Caracciolo G.; Farokhzad O. C.; Mahmoudi M. Biological identity of nanoparticles in vivo: clinical implications of the protein corona. Trends Biotechnol. 2017, 35 (3), 257–264. 10.1016/j.tibtech.2016.08.011. [DOI] [PubMed] [Google Scholar]
- Ke P. C.; Lin S.; Parak W. J.; Davis T. P.; Caruso F. A decade of the protein corona. ACS Nano 2017, 11 (12), 11773–11776. 10.1021/acsnano.7b08008. [DOI] [PubMed] [Google Scholar]
- Giulimondi F.; Digiacomo L.; Pozzi D.; Palchetti S.; Vulpis E.; Capriotti A. L.; Chiozzi R. Z.; Lagana A.; Amenitsch H.; Masuelli L.; Peruzzi G.; Mahmoudi M.; Screpanti I.; Zingoni A.; Caracciolo G. Interplay of protein corona and immune cells controls blood residency of liposomes. Nat. Commun. 2019, 10 (1), 1–11. 10.1038/s41467-019-11642-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giulimondi F.; Digiacomo L.; Vulpis E.; Loconte L.; Ferri G.; Cardarelli F.; Pozzi D.; Zingoni A.; Caracciolo G. In vitro and ex vivo nano-enabled immunomodulation by the protein corona. Nanoscale 2022, 14 (29), 10531–10539. 10.1039/D2NR01878K. [DOI] [PubMed] [Google Scholar]
- Caracciolo G.; Cardarelli F.; Pozzi D.; Salomone F.; Maccari G.; Bardi G.; Capriotti A. L.; Cavaliere C.; Papi M.; Laganà A. Selective Targeting Capability Acquired with a Protein Corona Adsorbed on the Surface of 1,2-Dioleoyl-3-trimethylammonium Propane/DNA Nanoparticles. ACS Appl. Mater. Interfaces 2013, 5 (24), 13171–13179. 10.1021/am404171h. [DOI] [PubMed] [Google Scholar]
- Muratov E. N.; Bajorath J.; Sheridan R. P.; Tetko I. V.; Filimonov D.; Poroikov V.; Oprea T. I.; Baskin I. I.; Varnek A.; Roitberg A.; Isayev O.; Curtalolo S.; Fourches D.; Cohen Y.; Aspuru-Guzik A.; Winkler D. A.; Agrafiotis D.; Cherkasov A.; Tropsha A. QSAR without borders. Chem. Soc. Rev. 2020, 49 (11), 3525–3564. 10.1039/D0CS00098A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigdeli A.; Palchetti S.; Pozzi D.; Hormozi-Nezhad M. R.; Baldelli Bombelli F.; Caracciolo G.; Mahmoudi M. Exploring cellular interactions of liposomes using protein corona fingerprints and physicochemical properties. ACS Nano 2016, 10 (3), 3723–3737. 10.1021/acsnano.6b00261. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.