Abstract
The microbial community composition of Wadden Sea sediments of the German North Sea coast was investigated by in situ hybridization with group-specific fluorescently labeled, rRNA-targeted oligonucleotides. A large fraction (up to 73%) of the DAPI (4′,6-diamidino-2-phenylindole)-stained cells hybridized with the bacterial probes. Nearly 45% of the total cells could be further identified as belonging to known phyla. Members of the Cytophaga-Flavobacterium cluster were most abundant in all layers, followed by the sulfate-reducing bacteria.
Marine sediments cover 70% of the total earth; consequently, they play an important role in the global cycling of carbon and nutrients (36). Early diagenetic processes are catalyzed mainly by the microorganisms that colonize the marine sediments (41). However, despite their environmental importance, the bacterial community structures of marine sediments remain poorly studied (23). Several attempts to describe marine sediment microbial communities have already been made. Most of these have been based on cultivation (see, e.g., references 6, 20, and 30) and were therefore subject to restrictions and biases leading to a distorted representation of the true community composition (2). Molecular techniques have greatly increased our knowledge of marine microbial diversity. For example, 16S rDNA libraries of marine plankton (see, e.g., references 7, 8, 15, and 33) and sediment (see, e.g., references 16 and 25) suggested the presence of hitherto-uncultured organisms. Techniques such as reassociation analysis of DNA (45), denaturing gradient gel electrophoresis (44), and restriction fragment length polymorphism (25) have yielded insight into bacterial diversity and community composition. However, phylogenetically based oligonucleotide hybridization techniques permit not only the monitoring of individual phylogenetic groups but also a quantification of their abundance in the natural habitats (2). Marine sediment microbial diversity has been studied by using the quantitative slot blot hybridization technique (9, 24, 35). However, these results cannot be directly translated into cell numbers because of the differences in absolute rRNA content per cell among the different members of the community (2). In situ hybridization with rRNA-targeted fluorescent oligonucleotide probes, in contrast, permits the identification and quantification of individual cells (2) and has demonstrated great power in the analysis of bacterial community composition in several environments (14, 32, 40, 42, 51). To date this method has not been tested with marine sediments.
In the present work we describe, to our knowledge for the first time, the community composition and vertical distribution of a marine sediment determined by using the in situ hybridization technique. The sampling area is located within the Jadebusen Bay, which is a part of the German Wadden Sea that forms the southern boundary of the North Sea, extending from The Netherlands to Denmark. The area of study is under the influence of the fluvial input of the river Weser, although it is not exposed to the extreme seasonal changes of salinity observed in the Weser estuary (37). The sediment is silty and experiences tides which expose it to air for about 5 h and leave it inundated for about 7 h, with some variability due to the wind velocity and direction (37).
Two cores were obtained from the near shore intertidal mud and sand flats of Dangast on 9 November 1997. One core was completely composed of mud (mud core). The second core originated from an artificial beach which contained a superficial layer (1 to 2 cm) of a thick-grained sand (beach core). Samples were transported at 4°C and processed immediately upon return to the laboratory, within 1 h of sampling. Sediment cores were sliced in 0.5-cm sections and fixed directly in ethanol (96%) or in 4% formaldehyde–phosphate-buffered saline (PBS) (composed of 0.13 M NaCl, 7 mM Na2HPO4, and 3 mM NaH2PO4 [pH 7.2 in water]) for 2 to 4 h on ice. The formaldehyde-fixed samples were then washed in PBS and stored in ethanol-PBS (1:1) at −20°C. Samples were diluted and treated by mild sonication with an MS73 probe at a setting of 20 for 30 s (Sonopuls HD70; Bandelin, Berlin, Germany). Samples were then mixed with 0.05% agarose, and 10 μl was dropped onto glass slides and dried at room temperature. Glass slides were immersed in 50, 80, and 96% ethanol for 3 min each. The inclusion in agarose did not result in artifacts such as autofluorescence and did not impede the access of the probes to the sample, in contrast to what was observed in similar studies using antibodies (3).
Oligonucleotide probes were synthesized with Cy3 fluorochrome at the 5′ end (Interactiva Biotechnologie GmbH, Ulm, Germany). Hybridizations and microscopy counts of hybridized and DAPI (4′,6-diamidino-2-phenylindole)-stained cells were performed as previously described (40). The probes and formamide concentrations used are given in Table 1. The slides were examined with an Axiophot II microscope (Zeiss, Jena, Germany). For each probe and sample, between 700 and 1,000 DAPI-stained cells and the respective hybridized cells in 10 to 20 independent fields were counted. Standard deviations of counts ranged between 2 and 8%. They are relatively high for those probes that gave low cell counts, and this is mainly due to the heterogeneity of the sample. Counting results were always corrected by subtracting signals observed with the probe NON338 (40).
TABLE 1.
Total DAPI cell counts and relative percentages of hybridized cells with specific probes
| Samples and depth (cm) | Absolute DAPI counts (cells/cm3 [108]) (mean ± SD) | % of cells hybridized with probea:
|
% Affiliated bacteriab | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EUB338 (mean ± SD) | ALF1b | BET42a | GAM42a | CF319a | PLA30 | SRB385 | HGC69a | ARC94 | DNMA657 | |||
| Mud core | ||||||||||||
| 0.5 | 44 ± 8 | 73.0 ± 6 | 3.5 | 4.1 | 3.6 | 18.1 | 3.5 | 4.4 | 3.2 | 1.1 | 0.3 | 41.5 |
| 1.0 | 45 ± 4 | 65.0 ± 3 | 4.7 | 4.0 | 4.7 | 16.3 | 5.7 | 4.3 | 3.6 | 1.0 | 0.2 | 44.3 |
| 1.5 | 46 ± 4 | 54.0 ± 5 | 4.5 | 4.3 | 3.5 | 13.0 | 5.4 | 5.1 | 2.3 | 1.1 | 0.4 | 39.2 |
| 2.0 | 45 ± 3 | 43.5 ± 11 | 4.3 | 3.7 | 3.4 | 12.1 | 4.5 | 6.5 | 3.1 | 1.3 | 0.6 | 38.9 |
| 2.5 | 43 ± 2 | 54.5 ± 3 | 4.4 | 4.0 | 3.4 | 13.6 | 4.4 | 5.3 | 2.6 | 0.9 | 0.5 | 38.6 |
| 3.0 | 45 ± 6 | 41.8 ± 7 | 3.9 | 3.8 | 3.3 | 12.0 | 4.0 | 4.3 | 2.4 | 0.1 | 0.5 | 33.8 |
| 3.5 | 33 ± 5 | 41.3 ± 4 | 3.1 | 3.0 | 3.4 | 11.8 | 3.3 | 6.1 | 2.5 | 0.01 | 0.3 | 33.2 |
| 4.0 | 24 ± 5 | 40.0 ± 8 | 2.5 | 2.8 | 3.5 | 8.1 | 2.2 | 5.6 | 1.7 | 0.02 | 0.2 | 26.4 |
| 4.5 | 27 ± 5 | 37.0 ± 9 | 2.7 | 2.0 | 3.4 | 6.4 | 1.3 | 3.5 | 1.0 | 0.01 | 0.4 | 20.3 |
| 5.0 | 18 ± 4 | 28.6 ± 7 | 2.3 | 2.0 | 3.3 | 6.2 | 1.3 | 2.0 | 0.1 | 0.01 | 0.3 | 17.2 |
| Beach core | ||||||||||||
| 0.5 | 6.1 ± 9 | 40.0 ± 6 | 1.5 | 1.0 | 0.8 | 7.5 | 2.8 | 2.9 | 0.9 | 0.9 | 0.2 | 18.3 |
| 1.0 | 7.8 ± 7 | 45.0 ± 3 | 2.0 | 1.2 | 1.3 | 7.7 | 3.0 | 3.5 | 1.4 | 0.9 | 0.3 | 21.1 |
| 1.5 | 25 ± 3 | 54.0 ± 2 | 3.5 | 3.8 | 3.0 | 8.5 | 3.7 | 3.8 | 1.6 | 1.5 | 0.4 | 29.4 |
| 2.0 | 32 ± 4 | 50.0 ± 7 | 3.3 | 3.5 | 3.2 | 8.7 | 4.1 | 4.2 | 1.3 | 1.6 | 0.3 | 29.9 |
| 2.5 | 22 ± 5 | 43.5 ± 5 | 3.4 | 3.6 | 3.1 | 8.3 | 3.2 | 5.2 | 1.2 | 1.3 | 0.2 | 29.3 |
| 3.0 | 23 ± 6 | 41.2 ± 2 | 2.9 | 3.3 | 2.8 | 6.2 | 3.4 | 4.9 | 1.0 | 1.0 | 0.4 | 25.5 |
| 3.5 | 20 ± 7 | 40.0 ± 11 | 2.5 | 3.0 | 2.7 | 5.7 | 3.1 | 3.6 | 0.4 | 0.8 | 0.2 | 21.8 |
| 4.0 | 12 ± 4 | 35.0 ± 9 | 2.3 | 2.8 | 2.6 | 5.0 | 2.8 | 2.3 | 0.8 | 0.3 | 0.3 | 18.9 |
Oligonucleotide probes (formamide concentration used for experiments): domain Eukarya, EUK516 (20%); domain Archaea, ARCH915 (20%); domain Bacteria, EUB338 (35%); α subclass of Proteobacteria, ALF1b (20%); β subclass of Proteobacteria, BET42a (35%); γ subclass of Proteobacteria, GAM42a (35%); sulfate reducers of the δ subclass of Proteobacteria, SRB385 (20%); Cytophaga-Flavobacterium cluster, CF319a (35%); Planctomycetes, PLA30 (20%); gram-positive bacteria with high G+C content, HGC69a (20%); Arcobacter spp., ARC94 (20%) (40, 51); Desulfonema magnum, D. limicola, and D. ishimotoei, DNMA657 (35%) (12). Probes BET42a, GAM42a, and PLA30 were used with competitor (22, 27).
Results from the addition of counts with the following specific probes: ALF1b, BET42a, GAM42a, SRB385, CF319a, PLA30, HGC69a, and ARC94.
Regarding the specificity of DAPI staining and fluorescent probes, a few cells (<1%) showed very weak staining with DAPI after hybridization with EUB338-Cy3. However, by taking micrographs of those cells, it became clear that they were stained with DAPI, and we regarded this phenomenon to be a result of effective absorption of DAPI emission by Cy3. Furthermore, we observed an effect similar to that reported by Zarda et al. (51). After 3 months of storage of fixed sediments in PBS-ethanol at −20°C, an increased detection of members of the α subclass of Proteobacteria with probe ALF1b was observed. Values increased from about 1% to a maximum of 4.1% of the DAPI counts. For the identification of members of the δ subclass of Proteobacteria, we used the probe SRB385, which is targeted to most of the known sulfate-reducing bacteria of this subclass (1). We are aware that this probe does not target all members of the δ subclass and that it is complementary to some organisms which are not affiliated with the δ subclass, such as numerous gram-positive bacteria (32, 50). Fluorescence in situ hybridization (FISH) was done on formaldehyde-fixed cells, which renders most gram-positive bacteria unreactive with fluorescent oligonucleotide probes, and consequently it is likely that most of the organisms detected by the SRB385 probe belong to the δ subclass.
Total cell counts and domain-specific probing.
Total cell counts determined by DAPI staining in the surface sediments were in accordance with what has been reported previously for similar environments (21, 38, 48). As shown in Table 1 and Fig. 1, DAPI-stained-cell counts were relatively constant in the top 3 cm of the mud core, i.e., 4.3 × 109 to 4.5 × 109 cells/cm3, and decreased with depth to 1.8 × 109 cells/cm3. In contrast, we detected many fewer microorganisms (6 × 108 to 8 × 108 cells/cm3) in the top 1 cm of the beach core, which corresponded to the sand layer. This observation is in accordance with a lower microbial load in sand flats than in mud flats (19, 23). The total cell counts increased below the sand-mud boundary at the 1.5-cm depth to 2.5 × 109 cells/cm3, although they remained lower than in the mud core.
FIG. 1.
Vertical profiles of the mud core (left) and beach core (right). In both cases the absolute numbers of bacteria detected are given. The drafted diagram of the cores is given on the left of each DAPI-EUB profile, indicating the transition zone to the iron-sulfide precipitation layer (black).
FISH resulted in the detection of a large fraction of the microbial community living in the top 5 cm of the Wadden Sea sediment. Up to 73% of the DAPI-stained cells hybridized with our set of probes (Table 1). Our detection yields are comparable to those obtained for activated sludge (40) and freshwater (31) but are higher than those obtained for soil (51) or seawater (32). In addition, hybridized cells were visualized with strong fluorescent signals (Fig. 2), which directly demonstrates a high cellular rRNA content (2). The Wadden Sea sediments of the German North Sea coast are highly influenced by the discharges of eutrophic freshwater from the Ems, Weser, and Elbe rivers. They are among the most active areas for decomposition of organic material in the German Bight (17). These environmental conditions would explain the high abundance and activity of microorganisms observed.
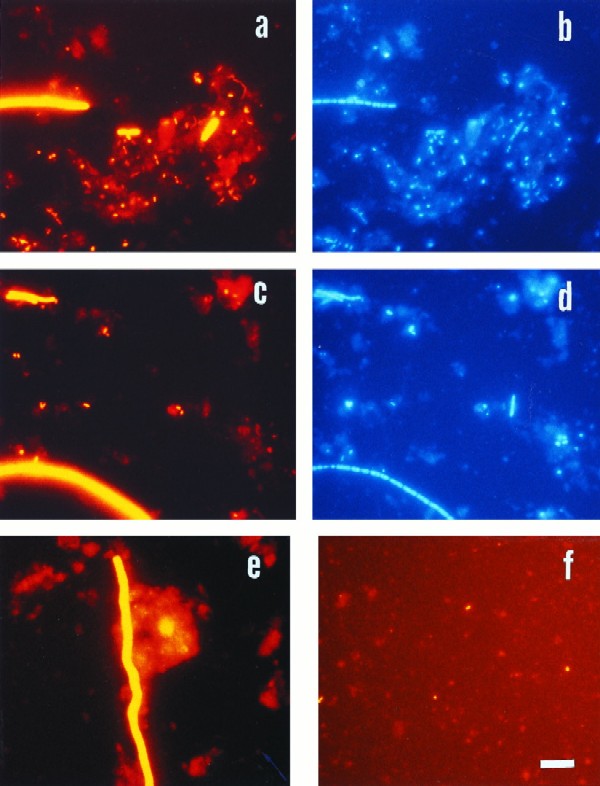
FIG. 2.
Epifluorescence micrographs of bacteria in sediment samples from the Jadebusen Bay of the German Wadden Sea. (a) Hybridization with probe EUB338, specific for Bacteria. (b) Same microscopic field as in panel a with UV excitation (DAPI staining). (c and d) Identical microscopic fields with probe SRB385 (c) and DAPI staining (d). (e) Hybridization with probe DNMA657, specific for Desulfonema. (f) Specific hybridization for Arcobacter with probe ARC94. Bar, 10 μm (applies to all panels).
The microbial communities analyzed were dominated by Bacteria (probe EUB338), whereas Archaea (probe ARCH915) and Eucarya (probe EUK516) were detected only in numbers that remained below the detection limit, set at 1% of the DAPI-stained cells. Although similar low counts of members of the domain Archaea were observed in bulk soil (51), results of other molecular studies on marine and nonmarine environments indicated higher abundance (8, 11, 29). With the still relatively large amount of cells that are not detected by FISH, we currently cannot exclude the possibility that Archaea make up a large part of those DAPI-stained cells which did not hybridize with ARCH915.
The fraction of detectable bacteria in the mud core strongly decreased over the vertical profile (Table 1; Fig. 1), from 73% in the uppermost layer to 28.6% at the 5-cm depth. Similar results were obtained for the beach core. However, we found lower detection rates in the sand layer of the core (40 to 45%) than in the mud layers. These yield differences may be related to the organic-matter content of the sediment (19, 23). Below the sand-mud interface, detection yields were similar to those for the mud core (Table 1; Fig. 1). Thus, not only the absolute cell number but also the fraction of cells detectable by FISH decreased thorough the profiles. Our results are in accordance with the correlation between total cell number and total bacterial production and activity, with the uppermost layers of mud flats showing the highest values (5, 48).
Abundances of major bacterial groups.
With a set of eight probes for major phyla within the domain Bacteria, we could affiliate between 17 and 44% of the total DAPI cell counts with known bacterial groups (Table 1; Fig. 1). This means that the majority of the detectable bacteria could be affiliated to a known group, and only between 8.5 to 35.8% of the EUB338 counts remained unaffiliated.
The most abundant phylogenetic group in Wadden Sea sediments was the Cytophaga-Flavobacterium cluster. This is remarkable, since high numbers of Cytophaga-Flavobacterium had so far not been found in marine sediments by either molecular methods (16, 36) or culture-based analysis (6, 10). Most of the cells identified within this cluster showed a homogeneous morphology of thin long rods. Their relative abundance ranged from 5 to 6.2% of the DAPI counts in the deepest layers to 18.1% in the uppermost layer of the mud core. This result means that between 15 and 25% of the total detectable bacteria could be affiliated to this group. Significant numbers of Cytophaga-Flavobacterium members in marine environments have so far been found only in the water column associated with macroscopic marine aggregates (7) or with alga blooms in sea ice (4). The members of the Cytophaga-Flavobacterium cluster are mainly aerobic, gram-negative bacteria which are specialized for the degradation of complex macromolecules (18, 34). Since the bacterial use of electron acceptors in sediments is stratified according to decreasing redox potentials (41) and since the oxygen depletion in the Wadden sediments occurs within the first 5 mm (37), we can only speculate on the energy metabolism of the Cytophaga-Flavobacterium cells found below the oxic zone. However, considering the brightness of the hybridization (2), the cells detected by probe CF319a seem to be intact and metabolically highly active.
The sulfate-reducing members of the δ subclass of Proteobacteria detected with the probe SRB385 (SRBs) made up the second-largest group, with a maximum of 6.5% of DAPI counts. We observed positive signals through the whole vertical profile, with a maximum at the 2-cm depth. The relative abundance of SRB counts through the sediment profile, together with their relatively high amounts in the upper layers of the sediment where sulfate reduction should not be the predominant process (37), was in accordance with results obtained for comparable environments (9, 20, 39). The morphology of SRBs was quite variable (Fig. 2). Among them were large filamentous bacteria whose affiliation with the genus Desulfonema (49) was confirmed with the probe DNMA657 (Fig. 2). A maximum of 2.7 × 107 cells cm−3 was found at a depth of 2 to 2.5 cm in the mud core (Table 1). Although Desulfonema organisms made up only 9% of all SRBs, these bacteria contribute significantly to the total bacterial biomass in the Wadden sediments due to their large size (Fig. 2).
In contrast to the high abundance of members of the Cytophaga-Flavobacterium cluster, we found relative low numbers of Proteobacteria (α, β, and γ subclasses), Planctomycetes, and gram-positive bacteria with a high G+C content (each accounted for 1 to 5.7%). The relatively low numbers of members of α-subclass Proteobacteria in sediments was unexpected, since they have been described as a predominant group in marine plankton (13, 15, 26).
One of our most surprising results was the presence of members of the genus Arcobacter at >107 cells cm−3 (1.3% of DAPI counts) (Table 1) in the upper layers of the sediments. All cells detected with the probe ARC94 showed the small, bow-shaped rod morphology (Fig. 2) characteristic of the members of this genus in the ɛ subclass of Proteobacteria (46). There was a clear stratification, with higher counts in the upper 3 cm (Fig. 1). Almost no Arcobacter organisms were detected at below 3.5 cm. Although members of this genus have recently been detected in different natural ecosystems (40, 43, 46, 47), they have not previously been reported to be significant in marine sediments. Arcobacters represent the most aerotolerant of the former campylobacters (46). An ability for denitrification has been reported (43). It is therefore not surprising that Arcobacter spp. could be found only in the upper layers. Even though the relative abundance is low, the total cell counts for a single genus (exceeding 107 cm−3) are high, and the number is nearly identical to that for Arcobacter organisms observed in activated sludge (40).
Overall, these results indicate that the community structure in the sediment differs significantly from that in the overlying water column. However, there should be a direct interaction between the water phase and the sediment, which has implications for the community development. In this respect, we think that one of the most interesting findings in our work is the unexpected high abundance of members of the Cytophaga-Flavobacterium cluster. This group has also been found to be a major constituent of the macroaggregate-attached bacterial communities in marine environments (7). Indirect observations (28) indicate that the sedimentation of microbial aggregates plays an essential role in the formation of the microbial communities in marine sediments, not only by the input of organic material but also by the input of established microbial communities. Our results support this hypothesis, but there is a question as to whether a high abundance of Cytophaga and Flavobacterium is common in marine sediments.
Future investigations on marine sediments will combine a larger set of specific probes with the analysis of biogeochemical processes to more fully understand the structure and function of marine sediments.
Acknowledgments
This study was supported by funds of the Max Planck Society.
Bo Barker Jørgensen and Jakob Pernthaler are acknowledged for critically reading the manuscript and for helpful comments on earlier versions.
REFERENCES
- 1.Amann R I, Binder B J, Olsen R J, Chisholm S W, Devereux R, Stahl D A. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann R I, Ludwig W, Schleifer K-H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anguish L J, Ghiorse W C. Computer-assisted laser scanning and video microscopy for analysis of Cryptosporidium parvum oocysts in soil, sediment, and feces. Appl Environ Microbiol. 1997;63:724–733. doi: 10.1128/aem.63.2.724-733.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowman J P, McCammon S A, Brown M V, Nichols D S, McMeekin T A. Diversity and association of psychrophilic bacteria in Antarctic sea ice. Appl Environ Microbiol. 1997;63:3068–3078. doi: 10.1128/aem.63.8.3068-3078.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cammen L M. Annual bacterial production in relation to benthic microalgal production and sediment oxygen uptake in an intertidal sandflat and an intertidal mudflat. Mar Ecol Prog Ser. 1991;71:13–25. [Google Scholar]
- 6.Delille D. Seasonal changes of subantarctic benthic bacterial communities. Hydrobiologia. 1995;310:45–57. [Google Scholar]
- 7.DeLong E F, Franks D G, Alldredge A L. Phylogenetic diversity of aggregate-attached vs. free-living marine bacterial assemblages. Limnol Oceanogr. 1993;38:924–934. [Google Scholar]
- 8.DeLong E F, Wu K Y, Prézelin B B, Jovine R V M. High abundance of Archaea in Antarctic marine picoplankton. Nature. 1994;371:695–697. doi: 10.1038/371695a0. [DOI] [PubMed] [Google Scholar]
- 9.Devereux R, Winfrey M R, Winfrey J, Stahl D A. Depth profile of sulfate-reducing bacterial ribosomal RNA and mercury methylation in an estuarine sediment. FEMS Microbiol Ecol. 1996;20:23–31. [Google Scholar]
- 10.Do H K, Kogure K, Simidu U. Identification of deep-sea-sediment bacteria which produce tetrodotoxin. Appl Environ Microbiol. 1990;56:1162–1163. doi: 10.1128/aem.56.4.1162-1163.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuhrman J A, Davis A A. Widespread Archaea and novel bacteria from the deep sea as shown by 16S rRNA gene sequences. Mar Ecol Prog Ser. 1997;150:275–285. [Google Scholar]
- 12.Fukui, M., G. Muyzer, A. Teske, B. Assmus, and F. Widdel (Max Planck Institut for Marine Microbiology, Bremen, Germany). 1998. Personal communication.
- 13.Giovannoni S J, Britschgi T B, Moyer C L, Field K G. Genetic diversity in Sargasso Sea bacterioplankton. Nature. 1990;345:60–63. doi: 10.1038/345060a0. [DOI] [PubMed] [Google Scholar]
- 14.Glöckner F O, Amann R, Alfreider A, Pernthaler J, Psenner R, Trebesius K, Schleifer K-H. An in situ hybridization protocol for detection and identification of planktonic bacteria. Syst Appl Microbiol. 1996;19:403–406. [Google Scholar]
- 15.González J M, Moran M A. Numerical dominance of a group of marine bacteria in the α-subclass of the class Proteobacteria in coastal seawater. Appl Environ Microbiol. 1997;63:4237–4242. doi: 10.1128/aem.63.11.4237-4242.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gray J P, Herwig R P. Phylogenetic analysis of the bacterial communities in marine sediments. Appl Environ Microbiol. 1996;62:4049–4059. doi: 10.1128/aem.62.11.4049-4059.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heyer K, Engel M, Brockmann U H, Rick H-J, Dürselen C-D, Hünnerfuss H, Kammann U, Steinhart H, Kienz W, Krause M, Karbe L, Faubel A, Regier S. Local studies in the German Bight during winter/spring 1988/1989. In: Sündermann J, editor. Circulation and contaminant fluxes in the North Sea. Berlin, Germany: Springer-Verlag; 1994. pp. 190–249. [Google Scholar]
- 18.Holmes B. The genera Flavobacterium, Sphingobacterium, and Weeksella. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes. Berlin, Germany: Springer-Verlag; 1991. pp. 3620–3630. [Google Scholar]
- 19.Hyllerberg J, Riis-Vestergaard H. Marine environments; the fate of detritus. Copenhagen, Denmark: Akademisk Forlag; 1984. [Google Scholar]
- 20.Jørgensen B B, Bak F. Pathways and microbiology of thiosulfate transformations and sulfate reduction in marine sediment (Kattegat, Denmark) Appl Environ Microbiol. 1991;57:847–856. doi: 10.1128/aem.57.3.847-856.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jørgensen B B, Revsbech N P. Oxygen uptake, bacterial distribution, and carbon-nitrogen-sulfur cycling in sediments from the Baltic Sea-North Sea transition. Ophelia. 1989;31:29–49. [Google Scholar]
- 22.Manz W, Amann R, Ludwig W, Wagner M, Schleifer K-H. Phylogenetic oligodeoxynucleotide probes for the major subclasses of Proteobacteria: problems and solutions. Syst Appl Microbiol. 1992;15:593–600. [Google Scholar]
- 23.Meyer-Reil L-A. Mikrobielle besiedlung und Produktion. In: Meyer-Reil L-A, Köster M, editors. Mikrobiologie des Meeresbodens. Jena, Germany: Gustav Fischer Verlag; 1993. pp. 38–81. [Google Scholar]
- 24.Moran M A, Rutherford L T, Hodson R E. Evidence for indigenous Streptomyces populations in a marine environment determined with a 16S rRNA probe. Appl Environ Microbiol. 1995;61:3695–3700. doi: 10.1128/aem.61.10.3695-3700.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moyer C, Dobbs F C, Karl D M. Estimation of diversity and community structure through restriction fragment length polymorphism distribution analysis of bacterial 16S rRNA genes from a microbial mat at an active, hydrothermal vent system, Loihi Seamount, Hawaii. Appl Environ Microbiol. 1994;60:871–879. doi: 10.1128/aem.60.3.871-879.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mullins T D, Britschgi T B, Krest R L, Giovannoni S J. Genetic comparisons reveal the same unknown bacterial lineages in Atlantic and Pacific bacterioplankton communities. Limnol Oceanogr. 1995;40:148–158. [Google Scholar]
- 27.Neef A. Anwendung der in situ-Einzelzell-Identifizierung von Bakterien zur Populationsanalyse in komplexen mikrobiellen Biozönosen. Ph.D. thesis. Munich, Germany: Technical University of Munich; 1997. [Google Scholar]
- 28.Novitsky J A. Evidence for sedimenting particles as the origin of the microbial community in coastal marine sediment. Mar Ecol Prog Ser. 1990;60:161–167. [Google Scholar]
- 29.Ogram A, Sun W, Brockman F J, Fredrickson J K. Isolation and characterization of RNA from low-biomass deep-subsurface sediments. Appl Environ Microbiol. 1995;61:763–768. doi: 10.1128/aem.61.2.763-768.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parkes R J, Cragg B A, Bale S J, Getllff J M, Goodman K, Rochelle P A, Fry J C, Weightman A J, Harvey S M. Deep bacterial biosphere in Pacific Ocean sediments. Nature. 1994;371:410–413. [Google Scholar]
- 31.Pernthaler J, Alfreider A, Posch T, Andreatta S, Psenner R. In situ classification and image cytometry of pelagic bacterial from a high mountain lake (Gossenköllesee, Austria) Appl Environ Microbiol. 1997;63:4778–4783. doi: 10.1128/aem.63.12.4778-4783.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramsing N B, Fossing H, Ferdelman T G, Andersen F, Thamdrup B. Distribution of bacterial populations in a stratified fjord (Mariager Fjord, Denmark) quantified by in situ hybridization and related to chemical gradients in the water column. Appl Environ Microbiol. 1996;62:1391–1404. doi: 10.1128/aem.62.4.1391-1404.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rappé M S, Kemp P F, Giovannoni S J. Phylogenetic diversity of marine coastal picoplankton 16S rRNA genes cloned from the continental shelf off Cape Hatteras, North Carolina. Limnol Oceanogr. 1997;42:811–826. [Google Scholar]
- 34.Reisenbach H. The order Cytophagales. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes. Berlin, Germany: Springer-Verlag; 1991. pp. 3631–3675. [Google Scholar]
- 35.Risatti J B, Capman W C, Stahl D A. Community structure of a microbial mat: the phylogenetic dimension. Proc Natl Acad Sci USA. 1994;91:10173–10177. doi: 10.1073/pnas.91.21.10173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rochelle P A, Cragg B A, Fry J C, Parkes R J, Weightman A J. Effect of sample handling on estimation of bacterial diversity in marine sediments by 16S rRNA gene sequence analysis. FEMS Microbiol Ecol. 1994;15:215–226. [Google Scholar]
- 37.Sageman J. Saisonale variationen von porenwasserprofilen, nährstoff-flüssen und reaktionen in intertidalen sedimenten des Weser ästuares. Ph.D. thesis. Bremen, Germany: Bremen University; 1994. [Google Scholar]
- 38.Sander B C, Kalff J. Factors controlling bacterial production in marine and freshwater sediments. Microb Ecol. 1993;26:79–99. doi: 10.1007/BF00177045. [DOI] [PubMed] [Google Scholar]
- 39.Sass H, Cypionka H, Babenzien H-D. Vertical distribution of sulfate-reducing bacteria at the oxic-anoxic interface in sediments of the oligotrophic Lake Stechlin. FEMS Microbiol Ecol. 1997;22:245–255. [Google Scholar]
- 40.Snaidr J, Amann R, Huber I, Ludwig W, Schleifer K-H. Phylogenetic analysis and in situ identification of bacteria in activated sludge. Appl Environ Microbiol. 1997;63:2884–2896. doi: 10.1128/aem.63.7.2884-2896.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sørensen J, Jørgensen B B. Early diagenesis in sediments from Danish coastal waters: microbial activity and Mn-Fe-S geochemistry. Geochim Cosmochim Acta. 1987;51:1583–1590. [Google Scholar]
- 42.Spring S, Amann R, Ludwig W, Schleifer K-H, van Germerden H, Petersen N. Dominating role of an unusual magnetotactic bacterium in the microaerobic zone of a freshwater sediment. Appl Environ Microbiol. 1993;59:2397–2403. doi: 10.1128/aem.59.8.2397-2403.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Teske A, Sigalevich P, Cohen Y, Muyzer G. Molecular identification of bacteria from a coculture by denaturing gradient gel electrophoresis of 16S ribosomal DNA fragments as a tool for isolation in pure cultures. Appl Environ Microbiol. 1996;62:4210–4215. doi: 10.1128/aem.62.11.4210-4215.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Teske A, Wawer C, Muzyer G, Ramsing N B. Distribution of sulfate-reducing bacteria in a stratified fjord (Mariager Fjord, Denmark) as evaluated by most-probable-number counts and denaturing gradient gel electrophoresis of PCR-amplified ribosomal DNA fragments. Appl Environ Microbiol. 1996;62:1405–1415. doi: 10.1128/aem.62.4.1405-1415.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Torsvik V, Sørheim R, Goksøyr J. Total bacterial diversity in soil and sediment communities—a review. J Ind Microbiol. 1996;17:170–178. [Google Scholar]
- 46.Vandamme P, Vancanneyt M, Pot B, Mels L, Hoste B, Dewettinck D, Vlaes L, Van den Borre C, Higgins R, Hommez J, Kersters K, Butzler J-P, Goossens H. Polyphasic taxonomic studies of the emended genus Arcobacter with Arcobacter butzleri comb. nov. and Arcobacter skirrowii sp. nov., an aerotolerant bacterium isolated from veterinary specimens. Int J Syst Bacteriol. 1992;42:344–356. doi: 10.1099/00207713-42-3-344. [DOI] [PubMed] [Google Scholar]
- 47.Voordouw G, Armstrong S M, Reimer M F, Fouts B, Telang A J, Shen Y, Gevertz D. Characterization of 16S rRNA genes from oil field microbial communities indicates the presence of a variety of sulfate-reducing, fermentative, and sulfide-oxidizing bacteria. Appl Environ Microbiol. 1996;62:1623–1629. doi: 10.1128/aem.62.5.1623-1629.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wellsbury P, Herbert R A, Parkes R J. Bacterial activity and production in near-surface estuarine and freshwater sediments. FEMS Microbiol Ecol. 1996;19:203–214. [Google Scholar]
- 49.Widdel F, Bak F. Gram-negative mosophilic sulfate-reducing bacteria. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes. Berlin, Germany: Springer-Verlag; 1991. pp. 3352–3378. [Google Scholar]
- 50.Young-Tae P, Nishimura M, Ohwada K. Detection and enumeration of marine sulfate-reducing bacteria using in situ hybridization with 16S rRNA oligonucleotide probes. Fish Sci. 1997;63:99–104. [Google Scholar]
- 51.Zarda B, Hahn D, Chazinotas A, Schönhuber W, Neef A, Amann R I, Zeyer J. Analysis of bacterial community structure in bulk soil by in situ hybridization. Arch Microbiol. 1997;168:185–192. [Google Scholar]