Abstract
The effectiveness of SYTOX Green nucleic acid stain for measuring bacterial viability was tested on starved populations of Escherichia coli and Salmonella typhimurium. This stain underestimates the fraction of dead cells within starved populations containing cells with damaged nucleic acids or membranes. Its application to natural samples should be considered with caution.
Viability assessment of bacterial cells is a major requirement in several areas of microbiology from environmental research to industrial applications. Flow cytometry (FCM), used in conjunction with fluorescent viability probes, has great potential for rapid and accurate quantification of viable and dead cells (11, 13–15). Fluorescent dyes exist as probes for different cellular functions (9, 12). Membrane-impermeative fluorescent probes that can passively diffuse through the cell wall of a bacterium can act as an indicator of a loss in membrane integrity, which, in turn, can often act as an indicator of cell viability (3, 10). Indeed, the loss of membrane integrity results in membrane permeabilization and thus, in the degradation of nucleic acids whose maintenance is at least essential to retain viability (4, 17, 18). However, such degradation may occur with a delay after the loss of membrane integrity which may be dependent on both environmental conditions and species. Although DNA integrity is not sufficient to ensure viability of a cell, DNA degradation is sometimes used to quantify dead cells. However, fluorescent probes used to detect the fraction of dead cells with damaged DNA also have limitations. The use of DNA probes for assessment of DNA degradation should be considered with caution since apparent DNA damage can be misinterpreted due to changes in the coiling status of the DNA and/or in the permeability of membranes by these dyes (8).
Recently, SYTOX Green stain (Molecular Probes Inc., Eugene, Oreg.) has been developed for bacterial viability assessment. SYTOX Green stain is a high-affinity nucleic acid stain that does not cross the membranes of live cells and yet easily penetrates cells with compromised plasma membranes. This dead-cell indicator was recently used for rapid viability assessment after exposure of cells to disinfectant (7) or antibiotics (16). This dye was suggested as a good indicator for viability assessment (16). In this study, we have tested the efficiency of SYTOX Green dye for flow cytometric detection of dead cells of Escherichia coli and Salmonella typhimurium within starved populations.
Strains, cultures, and starvation conditions.
All experiments were conducted with E. coli ATCC 8739 and S. typhimurium ATCC 43971. Heat treatment was used as a positive control of cells with permeabilized membranes. Heat-treated cells were obtained by resuspending the cells in 1 ml of phosphate-buffered saline and placing the tube in boiling water for 10 min. To obtain starved cultures of these strains, the cells were incubated overnight in Trypticase soy broth (bioMérieux, Marcy-l’Etoile, France) at 37°C, harvested, washed twice by centrifugation at 5,000 × g for 5 min in sterile MilliQ water, and resuspended in the same medium. Erlenmeyer flasks (1-liter capacity) containing 400 ml of filtered water (Milli-Q; 0.22-μm-pore-size filter; Millipore) (pH 7.5) were autoclaved (120°C, 15 min), cooled, inoculated to a cell density of approximately 107 cells per ml, and incubated in the dark at 20 ± 0.5°C with gentle stirring. Aliquots of 20 ml were collected after 0, 8, 15, 22, 32, and 46 days of starvation. Duplicate flasks were prepared for both strains.
Enumeration of total, culturable, and dead cells.
Subsamples for total cell enumerations were fixed with 2% formaldehyde and then stained with SYBR Green I (SYBR-I) DNA stain (Molecular Probes) (11). The DNA fluorescence of stationary-phase cells which contained at least one genome was used to create a region in the FL1 histogram. This procedure was used to separate damaged-DNA (DNA−) and undamaged-DNA (DNA+) subpopulations which appear during starvation. DNA− cells were determined by cells whose fluorescence was less than that of cells containing one genome. The lower fluorescence of DNA− cells can be due to (i) damaged DNA but also to (ii) changes in the DNA coiling status which can result in a lower accessibility of the dye to the target sites and/or to a lower permeability of membranes by the dye. For enumeration of culturable cells, 100-μl volumes of serial dilutions (1/10) in phosphate-buffered saline were plated onto nutrient agar (bioMérieux). The proportion of culturable cells (CFU+) was defined as the ratio of the number of CFU+ cells to the total number of cells determined by SYBR-I staining. For enumeration of dead cells, a stock solution of SYTOX Green stain was prepared in dimethyl sulfoxide to a final concentration of 5 mM. After optimization of the staining conditions (data not shown), the cells were stained with 1 μM SYTOX Green (final concentration) for 30 min at 37°C in the dark.
FCM.
Samples were analyzed with a FACSCalibur flow cytometer (Becton Dickinson) equipped with an air-cooled laser providing 15 mW at 488 nm and with the standard filter setup. Green fluorescence from stained cells was collected in the FL1 channel (530 ± 15 nm). The sheath fluid was filtered water (Milli-Q; 0.22-μm-pore-size filter). All parameters were collected as logarithmic signals. All bacterial analyses were performed at a low flow rate setting (12 μl min−1), and acquisition was done over 1 min. The calibrated flow rate was controlled daily by measuring the volume before and after analysis. Samples were run such that the event rate was below 800 to 1,000 events s−1 to avoid coincidence. All samples were analyzed immediately after staining. Yellow-green fluorescent 0.94-μm-diameter microspheres (Polysciences Inc., Warrington, Pa.) were added to each sample as an internal fluorescence standard. Data were plotted as histograms showing the fluorescence distribution. Triplicate counts were made for each procedure.
Effect of heat treatment on the fluorescence distribution of SYTOX Green-stained cells.
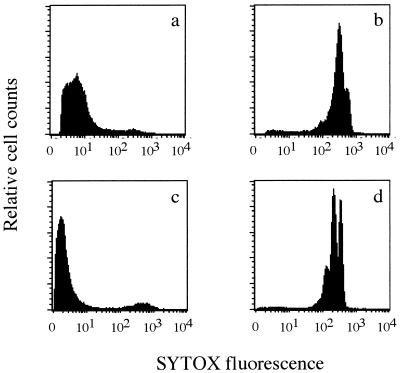
The fluorescence signal from living cells was brighter than that obtained from SYTOX Green stain alone without bacteria (data not shown). When SYTOX Green was applied to live cells, surface binding of dye resulted in a weak but detectable fluorescence emission (Fig. 1a and c). A marked enhancement of SYTOX Green fluorescence was observed for heat-treated cells (Fig. 1b and d).
FIG. 1.
FL1 histograms of live (a and c) and heat-treated (b and d) E. coli (a and b) and S. typhimurium (c and d) cells stained with SYTOX Green.
Application of SYTOX Green to starved cells.
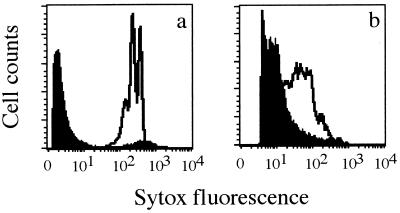
The viability of E. coli and S. typhimurium cells was monitored during prolonged starvation in sterile water (Table 1). After inoculation, the percentages of viable and culturable cells (CFU+) fell rapidly and were lower than 6% (E. coli) and 0.1% (S. typhimurium) after 46 days of starvation. As expected, this reduction in viable counts was matched by a concomitant increase in the number of dead cells as determined by SYTOX Green staining. Surprisingly, this increase was followed by a decrease in the number of dead cells. At the same time, we observed a shift in the fluorescence of a significant fraction of heat-permeabilized cells stained with SYTOX Green. At the onset of starvation, the large difference in fluorescence emission intensities between intact (live) and permeabilized (heat-treated) cells stained with SYTOX Green facilitates discrimination of both populations (Fig. 2a). This difference decreased during starvation since the fluorescence of permeabilized cells gradually decreased in brightness, resulting in an increasing overlap with the fluorescence distribution of live cells (Fig. 2b). This result suggests a decrease in the apparent DNA content of the cells. Similar trends were recorded for E. coli.
TABLE 1.
Percentages of viable cells, dead cells, and cells with apparent damaged nucleic acids of E. coli and S. typhimurium populations at different times of starvation in sterile watera
| Time (days) | % of cellsb
|
|||||
|---|---|---|---|---|---|---|
|
E. coli
|
S. typhimurium
|
|||||
| CFU+ | SYTOX+ | DNA− | CFU+ | SYTOX+ | DNA− | |
| 0 | 98 (3.2)c | 0 | 0 | 102 (5.1) | 0 | 0 |
| 8 | 59.3 (2.7) | 36.7 (4.2) | 0 | 43.7 (5.0) | 0.6 (0.1) | 6.8 (0.7) |
| 15 | 51.1 (6.1) | 41.3 (5.4) | 31.5 (1.3) | 8.4 (2.1) | 3.8 (0.1) | 30.1 (0.5) |
| 22 | NDd | ND | ND | 2.1 (0.9) | 18.8 (1.0) | 62.4 (1.1) |
| 32 | 3.25 (0.8) | 19.0 (2.4) | 64 (1.7) | 0.5 (0.3) | 5.2 (6.4) | 84.2 (0.8) |
| 46 | 5.16 (1.5) | 17.1 (3.1) | 73.5 (1.8) | 0.02 (0.01) | 3.8 (7.3) | 93.1 (1.1) |
We determined percentages of viable cells (those that were culturable [CFU+]), dead cells (those stained with SYTOX Green [SYTOX+]), and those with apparent DNA damage (DNA−).
Percentages are relative to total counts (determined by SYBR-I staining).
Values in parentheses are standard deviations determined from triplicate measurements from one experiment.
ND, not determined.
FIG. 2.
FL1 histograms of live (closed histogram) and heat-treated (open histogram) S. typhimurium cells after SYTOX Green staining at 0 (a) and 46 (b) days of starvation.
Changes in the nucleic acid content of starved cells.
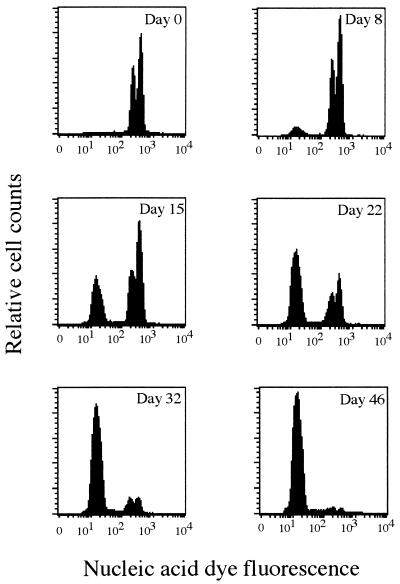
To better understand this decrease in the number of dead cells as determined by SYTOX Green staining, we analyzed the temporal evolution of the nucleic acid content of starved cells by staining the cells with SYBR-I nucleic acid dye. The degradation of nucleic acids is sometimes considered an alternative method for assessing the death of bacterial cells (5, 17). FCM analysis of the nucleic acid content of SYBR-I-stained cells revealed important changes in the course of starvation (Fig. 3). These changes may be mainly influenced by changes in the DNA content of the cells since the RNA content of starved cells may be very low (1, 2) and SYBR-I has a higher binding affinity for DNA than for RNA (11). After inoculation, stationary-phase cells exhibited a typical bimodal DNA distribution corresponding to cells containing one or two genomes (8) (Fig. 3a). After continued starvation, a subpopulation of cells emitting a lower fluorescence intensity that corresponded to cells with a lower apparent DNA content (DNA− cells) appeared. After 46 days of starvation, this subpopulation of cells with an apparent damaged DNA content represented up to 73.5 and 93.1% of total counts of E. coli and S. typhimurium populations, respectively (Table 1). At all sampling times, the sums of the viable (CFU+) and dead (DNA− and SYTOX Green-stained) cells were always less than 100%, but the results were much more coherent when dead cells were considered as the fraction of DNA− cells. The noncomplementarity at a given sampling time between both live and dead cell counts may be explained by the fact that the apparent degradation of DNA may occur after the loss of membrane integrity and, thus, with a certain time lag after the loss of culturability. This result is congruous with those of other reports (4, 17). As stated above, this apparent DNA degradation can be due to DNA degradation but can also be due to changes in the coiling status of DNA. Since an increasing fraction of heat-permeabilized cells also has a lower fluorescence intensity after SYTOX Green staining, changes in membrane permeability cannot explain this increasing fraction of DNA− cells. It is also interesting that this decrease in the fluorescence signal was not gradual but corresponded to a rapid shift from one well-characterized state to another. This DNA− state was not reversible when the cells were subjected to a 24-h resuscitation step by adding 1% Trypticase soy broth to the flask after 46 days of starvation (data not shown). Although the resuscitation conditions (time and type of nutrients) were perhaps not the most appropriate, the results suggest that DNA− cells were not viable cells. Inversely, DNA+ cells were responsive to this nutrient supply after 8 h of incubation with the nutrients.
FIG. 3.
FCM histograms demonstrating changes which occur in the DNA fluorescence distribution of S. typhimurium cells at different times of starvation in sterile water (0, 8, 15, 22, 32, and 46 days).
Limitations of SYTOX Green staining for viability assessment.
SYTOX Green and SYBR-I are nucleic acid stains with a high binding affinity for DNA. SYBR-I readily penetrates fixed cells even without membrane permeabilization, while SYTOX Green penetrates cells with permeabilized membranes. If we assume that the apparent DNA degradation occurs after the loss of membrane integrity, cells with compromised membranes may contain intact or damaged nucleic acids. The fraction of DNA− cells with a low fluorescence emission after SYBR-I staining, corresponding to cells with damaged DNA, increased with starvation time. Such cells have compromised membranes and should be stained with SYTOX Green. However, DNA degradation and/or changes in the topology of the molecule result in a shift of the fluorescence emission of SYTOX Green-stained cells which overlaps with the weak fluorescence emission of living cells (16). Thus, the weak fluorescence of these cells after SYTOX Green staining may correspond either to permeabilized cells with damaged nucleic acids or to living cells. Such dead cells with a low fluorescence, whose fraction increases during starvation, are counted as live cells since they appear in a fluorescence region corresponding to that of live cells initially defined with stationary-phase cells. This is why the number of dead cells, as determined by SYTOX Green staining, decreases during starvation. Consequently, the percentage of dead cells recorded after SYTOX Green staining corresponds only to the fraction of cells with permeabilized membranes and an as-yet-undegraded and/or topologically modified nucleic acid content.
Recently, it was suggested that SYTOX Green stain is an effective alternative to conventional methods for measuring bacterial viability and antibiotic susceptibility (16). Our results suggest that the use of SYTOX Green stain for detection and evaluation of viability in bacteria should be considered with caution and restricted to specific applications such as the analysis of antibiotic susceptibility of bacteria with undamaged nucleic acids. Although the physiology of many bacteria is unknown today, the normal state for most copiotrophic bacteria in the aquatic environment is the starvation mode (6). Recently, it was suggested that the percentage of nucleoid-containing cells within oligotrophic marine communities may be low and often lower than 50% (5, 18). Thus, the application of SYTOX Green to determine the fraction of dead cells in the natural environment may be of limited use. When SYTOX Green staining was applied to marine coastal waters from the Mediterranean Sea, it was not possible to discriminate live from dead populations (data not shown).
Concluding remarks.
The results presented here suggest that the use of viability dyes such as SYTOX Green stain which target molecules or sites which are degraded and/or modified during the starvation process may underestimate the fraction of dead cells, mainly when discrimination between labeled cells and background signals is problematic. The use of such dyes to investigate the viability of cells within natural and complex communities should be considered with caution.
Acknowledgments
This work was funded by contract ELOISE PL950439 from the European Community. The FACSCalibur flow cytometer was funded by CNRS-INSU-SDV and by contract ELOISE PL950439.
REFERENCES
- 1.Davis B D, Luger S M, Tai P C. Role of ribosome degradation in the death of starved Escherichia coli cells. J Bacteriol. 1986;166:439–445. doi: 10.1128/jb.166.2.439-445.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guindulain T, Comas J, Vives-Rego J. Use of nucleic acid dyes SYTO-13, TOTO-1, and YOYO-1 in the study of Escherichia coli and marine prokaryotic populations by flow cytometry. Appl Environ Microbiol. 1997;63:4608–4611. doi: 10.1128/aem.63.11.4608-4611.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jepras R I, Carter J, Pearson S C, Paul F E, Wilkinson M J. Development of a robust flow cytometric assay for determining numbers of viable bacteria. Appl Environ Microbiol. 1995;61:2695–2701. doi: 10.1128/aem.61.7.2696-2701.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Joux F, Lebaron P, Troussellier M. Succession of cellular states in a Salmonella typhimurium population submitted to starvation in artificial seawater microcosms. FEMS Microbiol Ecol. 1997;22:65–76. [Google Scholar]
- 5.Karner M, Fuhrman J A. Determination of active marine bacterioplankton: a comparison of universal 16S rRNA probes, autoradiography, and nucleoid staining. Appl Environ Microbiol. 1997;63:1208–1213. doi: 10.1128/aem.63.4.1208-1213.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kjelleberg S, editor. Starvation in bacteria. New York, N.Y: Plenum Press; 1993. [Google Scholar]
- 7.Langsrud S, Sundheim G. Flow cytometry for rapid assessment of viability after exposure to a quaternary ammonium compound. J Appl Bacteriol. 1996;81:411–418. doi: 10.1111/j.1365-2672.1996.tb03527.x. [DOI] [PubMed] [Google Scholar]
- 8.Lebaron P, Joux F. Flow cytometry analysis of the cellular DNA content of Salmonella typhimurium and Alteromonas haloplanktis during starvation and recovery in seawater. Appl Environ Microbiol. 1994;60:4345–4350. doi: 10.1128/aem.60.12.4345-4350.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lloyd D, Hayes A J. Vigour, vitality and viability of microorganisms. FEMS Microbiol. 1995;133:1–7. [Google Scholar]
- 10.Lopez-Amoros R, Castel S, Comas-Riu J, Vives-Rego J. Assessment of E. coli and Salmonella viability and starvation by confocal laser microscopy and flow cytometry using rhodamine 123, DiBAC4(3), propidium iodide and CTC. Cytometry. 1997;29:1–8. doi: 10.1002/(sici)1097-0320(19971201)29:4<298::aid-cyto6>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 11.Marie D, Partensky F, Jacquet S, Vaulot D. Enumeration and cell cycle analysis of natural populations of marine picoplankton by flow cytometry using the nucleic acid stain SYBR Green I. Appl Environ Microbiol. 1997;63:186–193. doi: 10.1128/aem.63.1.186-193.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McFeters G A, Yu F P, Pyle B H, Stewart P S. Physiological assessment of bacteria using fluorochromes. J Microbiol Methods. 1995;21:1–13. doi: 10.1016/0167-7012(94)00027-5. [DOI] [PubMed] [Google Scholar]
- 13.Porter J, Deere D, Pickup R, Edwards C. Fluorescent probes and flow cytometry: new insights into environmental bacteriology. Cytometry. 1996;23:91–96. doi: 10.1002/(SICI)1097-0320(19960201)23:2<91::AID-CYTO1>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 14.Porter J, Diaper J, Edwards C, Pickup R. Direct measurements of natural planktonic bacterial community viability by flow cytometry. Appl Environ Microbiol. 1995;61:2783–2786. doi: 10.1128/aem.61.7.2783-2786.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Porter J, Edwards C, Pickup R W. Rapid assessment of physiological status in Escherichia coli using fluorescent probes. J Appl Bacteriol. 1995;79:399–408. doi: 10.1111/j.1365-2672.1995.tb03154.x. [DOI] [PubMed] [Google Scholar]
- 16.Roth B L, Poot M, Yue S T, Millard P J. Bacterial viability and antibiotic susceptibility testing with SYTOX Green nucleic acid stain. Appl Environ Microbiol. 1997;63:2421–2431. doi: 10.1128/aem.63.6.2421-2431.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weichart D, McDougald D, Jacobs D, Kjelleberg S. In situ analysis of nucleic acids in cold-induced nonculturable Vibrio vulnificus. Appl Environ Microbiol. 1997;63:2754–2758. doi: 10.1128/aem.63.7.2754-2758.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zweifel U L, Hagström Ä. Total counts of marine bacteria include a large fraction of non-nucleoid-containing bacteria (ghosts) Appl Environ Microbiol. 1995;61:2180–2185. doi: 10.1128/aem.61.6.2180-2185.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]