Abstract
Purpose:
Nuclear Overhauser Enhancement mediated saturation transfer effect, termed NOE(−3.5 ppm), is a major source of chemical exchange saturation transfer (CEST) MRI contrasts at 3.5 ppm in the brain. Previous phantom experiments have demonstrated that both proteins and lipids, two major components in tissues, have substantial contributions to NOE(−3.5 ppm) signals. Their relative contributions in tissues are informative for the interpretation of NOE(−3.5 ppm) contrasts that could provide potential imaging biomarkers for relevant diseases, which remain incompletely understood.
Methods:
Experiments on homogenates and supernatants of brain tissues collected from healthy rats, that could isolate proteins from lipids, were performed to evaluate the relative contribution of lipids to NOE(−3.5 ppm) signals. On the other hand, experiments on ghost membranes with varied pH, and reconstituted phospholipids with different chemical compositions were conducted to study the dependence of NOE(−3.5 ppm) on physiological conditions. Besides, CEST imaging on rat brains bearing 9L tumors and healthy rat brains was performed to analyze the causes of the NOE(−3.5 ppm) contrast variations between tumors and normal tissues, and between gray matter and white matter.
Results:
Our experiments reveal that lipids have dominant contributions to the NOE (−3.5 ppm) signals. Further analysis suggests that decreased NOE(−3.5 ppm) signals in tumors and higher NOE(−3.5 ppm) signals in white matter than in gray matter are mainly explained by changes in membrane lipids, rather than proteins.
Conclusion:
NOE(−3.5 ppm) could be exploited as a highly sensitive MRI contrast for imaging membrane lipids in the brain.
Keywords: Chemical exchange saturation transfer (CEST), nuclear Overhauser enhancement (NOE), lipids, magnetization transfer (MT), tumor
INTRODUCTION
Chemical exchange saturation transfer (CEST) is an MRI contrast mechanism for indirectly detecting dilute solute molecules with enhanced sensitivity by measuring changes in water signals (1-5). In CEST imaging, a Z-spectrum is usually created by plotting water signals as a function of the frequency offset of RF saturation pulse, in which CEST effects sourced from multiple molecular pools can be investigated. For instance, in the central nervous system, amide proton transfer (APT) effect at 3.5 ppm, amine-water exchange effect at 3 ppm, guanidinium proton transfer effect at 2 ppm, Nuclear Overhauser Enhancement (NOE) medicated CEST effect at −1.6 ppm and −3.5 ppm (abbreviated as NOE(−1.6 ppm) and NOE(−3.5 ppm) in this study), along with a broad semi-solid MT effect can be observed in the Z-spectrum. Most of the CEST effects observed in the Z-spectrum have been assigned to certain molecular sources. Specifically, the APT effect originates from backbone amide protons on mobile proteins/peptides (6); the amine-water exchange effect originates from lysine protons on proteins, glutamate and free amino acids (7,8); the guanidinium proton transfer effect originates from creatine and arginine residues of mobile proteins (9-11); the NOE(−1.6 ppm) effect originates from choline head groups on phospholipids (12,13); the MT effect originates from immobile macromolecules (e.g. myelin) (14,15). In recent studies, although the origins of the NOE(−3.5 ppm) effect have been assigned to aliphatic protons associated with mobile proteins and lipids (12,13,16-19), their relative contributions to the in vivo NOE(−3.5 ppm) signals remain incompletely understood. Further studies on this issue would be highly rewarding since the NOE(−3.5 ppm) imaging has shown great potential for applications on brain tumors (12,17,20-24), Alzheimer’s disease (25) and spinal cord injury (26), etc.
NOE is a dipolar cross-relaxation between two nuclear spins at a close distance on a molecule (27-29). The cross-relaxation rate (or NOE coupling rate) depends on the speed of the molecule tumbling due to thermal motions. NOE usually are seen from macromolecules or molecules that have aliphatic protons with limited mobility. The proteins and lipids that are the major sources of the NOE signals play important roles in physiological functions of the brain. Proteins perform many functions in cells including catalyzing metabolic reactions and DNA replications, responding to stimuli, as well as providing structure to cells and organisms; lipids (i.e., phospholipids) are major components of cellular membranes, which are critical to maintaining the structure and functions of cells. Changes in the two molecules in specific diseases represent a different pathological process. Thus, further evaluation of their relative contributions to the NOE(−3.5 ppm) signal in biological tissues is informative for the interpretation of the NOE(−3.5 ppm) contrast.
Previously, the NOE(−3.5 ppm) signal from bovine serum albumin (BSA) phantoms (22,30) and reconstituted phospholipid phantoms (12,13) have been reported. Also, it was reported that NOE(−3.5 ppm) signal from proteins is sensitive to the structural integrity of proteins (22,30), and the NOE(−3.5 ppm) signal from phospholipids is sensitive to the spatial conformations and chemical compositions (12,13). However, the relative contributions of proteins and lipids to NOE(−3.5 ppm) signals in tissues cannot be assessed by these simple phantom experiments for the following reasons. First, BSA cannot represent the proteins in biological tissues that have different molecular structures and weights which could alter the NOE signals in human cells. Second, reconstituted phospholipids in the phantom experiments have a limited biophysical reality with respect to the cell membranes in tissues.
In this study, tissue homogenates are used to investigate the NOE(−3.5 ppm) effect to ensure a biophysical reality. Furthermore, the supernatant of the homogenates with the removal of most lipids is used to estimate the net contribution of the NOE(−3.5 ppm) effect from proteins. Although absolute concentrations of proteins in the supernatant may be lower than that in the tissue homogenates due to incomplete wall-broken of cells, the relative amplitudes of protein-sourced NOE(−3.5 ppm) signal and protein-sourced APT signal could keep unchanged since the cells with incomplete wall-broken have the similar protein components as that with complete wall-broken. Thus, a comparison of the ratio of the NOE(−3.5 ppm) signal to the APT signal between the tissue homogenates and the supernatant is performed to assess the relative contributions of proteins and lipids to the NOE(−3.5 ppm) signals. Besides, experiments on ghost membranes with varied pH, and reconstituted phospholipids with different chemical compositions were conducted to study the dependence of NOE(−3.5 ppm) on physiological conditions.
MATERIALS AND METHODS
Animal preparation
Three rats bearing 9L tumors and three healthy rats were included in this study. For brain tumor induction, each rat was injected with 1 × 105 9L glioblastoma cells in the right brain hemisphere and was then imaged after 2 to 3 weeks. During data acquisition, all rats were immobilized and anesthetized with a 2%/98% isoflurane/oxygen mixture. Respiration was monitored to be stable, and a constant rectal temperature of 37°C was maintained throughout the MRI imaging using a warm-air feedback system (SA Instruments, Stony Brook, NY, USA). All animal procedures were approved by the Animal Care and Usage Committee of Vanderbilt University Medical Center.
Phantom preparation
The homogenates and the supernatant of brain tissues were prepared from two freshly sacrificed healthy rats. Brain tissues were weighed and washed quickly with 1 × phosphate buffered saline (PBS) to remove residual blood after being removed from the rats. Then, after adding 4-times 1 × PBS (w/w), the tissues were homogenized with a motor-driven blade-type homogenizer (Brinkmann Polytron PT3000; Kinematica, Malters, Switzerland) at a speed of 20,000 rpm for 2 × 30 second bursts. The homogenates were divided into two aliquots, one of which is used for the homogenate sample. The supernatant of brain tissues was separated from the other aliquot by removing sediments after centrifugation (20,000 × g, 30 min, 4°C). Finally, the two aliquots were titrated to 7.0 at the room temperature by using NaOH/HCl.
Ghost membranes were used to further investigate the origin of the NOE(−3.5 ppm) signal from the plasma membrane and to study the dependence of the NOE(−3.5 ppm) signal on tissue pH. The ghost membranes were prepared according to the method that was modified from previous reports (31,32). Specifically, 15 mL of bovine blood was diluted with an equal volume of 1 × PBS. Then, red cells were separated from plasma by centrifugation (1000 × g, 20 min, 4°C), and the plasma and the buffy coat were removed by aspiration. To remove plasma proteins, the cells were further washed twice with 2 volumes of 172 mM Tris buffer (pH 7.6) by centrifugation (1000 × g, 20 min, 4°C). The washed cells were suspended in isotonic 172 mM Tris buffer (pH 7.6) to an approximate hematocrit of 50% and mixed gently by inversion for approximately 1 min. 10ml aliquots of the above 50% cell suspension were transferred to 50ml centrifuge tubes. 40 ml of 11.35 mM Tris buffer (pH 7.6, 4 °C) was added to the cell suspension and mixed well by stirring for 5 min. Finally, ghost membranes were harvested by centrifugation (20,000 × g, 30 min, 4°C). The ghost membranes were washed ten times with 11.35 mM Tris buffer (pH 7.6, 4 °C) to sufficiently remove proteins and then resuspended in 1× PBS for MRI scans.
Reconstituted phospholipids were used to investigate the dependence of NOE(−3.5 ppm) signal on the chemical composition. Two phantoms containing two types of synthetic phospholipids, DSPC (1,2-Distearoyl-sn-glycero-3-phosphocholine) and DEPC (1,2-Dierucoyl-sn-glycero-3-phosphocholine), were prepared respectively. Note that DEPC (22 × 2 chain length) has a longer aliphatic chain than DSPC (18 × 2 chain length). The reconstituted phospholipid phantoms were prepared by adding the synthetic phospholipids and cholesterol with a molar ratio of 1:1 to the chloroform solution. Chloroform was then removed in the environment of air stream and vacuum. The phospholipids were resuspended in water with the ratio of 1:3 by weight. The samples were then placed in a bath sonicator, and subsequently frozen and thawed for a few cycles to produce liposomes.
MRI
A continuous wave (CW)-CEST sequence with a 5-second hard saturation pulse followed by single-shot spin-echo echo planar imaging (SE-EPI) acquisitions with a TR of 7 second was used on animal imaging. Images were acquired with matrix size 64 × 64, field of view 30 mm × 30 mm, 2 mm slice thickness, and one acquisition. A non-imaging CW-CEST sequence with free induction decay (FID) readout and 8-second hard saturation pulse and TR of 10 second was used for phantom imaging. Z-spectra were acquired with RF offsets (Δω) from −10 ppm to 10 ppm. Control images were acquired with RF offsets of 250 ppm. R1w and MT pool size ratio were obtained using a selective inversion recovery quantitative MT method (33). All in vivo and phantom imaging were performed at 37°C with saturation power of 0.5 μT. Experiments on all animals and phantoms were performed on a Varian 4.7-T magnet, and the reconstituted phospholipids were performed on a Varian 9.4-T magnet.
Multiple-pool Lorentzian fit and asymmetric analysis of CEST Z-spectra
We used multiple-pool Lorentzian fitting to process the CEST Z-spectrum from animals, tissue homogenates, and the supernatant. Eq. (1) gives the model function of the Lorentzian fit method.
| (1) |
Here, is the label signal and is the control signal; is the RF frequency offset; , which represents a Lorentzian line with a central frequency offset relative to the bulk water (), peak full width at half maximum (), peak amplitude (); N is the number of fitted pools. A five-pool model Lorentzian fit including the amide, amine, water, NOE(−3.5 ppm), and MT was performed to process the CEST Z-spectra on tissue homogenates and the supernatant. Note that variable frequency offsets of the pools were used in the multiple-pool Lorentzian fit (see Table 1). The NOE(−1.6 ppm) pool was not included in the five-pool model for processing the data from phantoms since the NOE(−1.6 ppm) signals are ignorable in postmortem tissues (34). The fitting was performed to achieve the lowest root mean square (RMS) of residuals between the measured data and the modeled signals. the starting points and boundaries of the amplitude, width, and offset of the fit were list in Table 1. The goodness of the fit was estimated with the sum of RMS of residuals averaged across the whole Z-spectrum.
Table 1.
Starting points and boundaries of the amplitude (A) and width (W) of all pools in the Lorentzian fit. The unit of peak width is ppm.
| Start | Lower | Upper | |
|---|---|---|---|
| Awater | 0.9 | 0.02 | 1 |
| Wwater | 1.4 | 0.1 | 10 |
| Aamide | 0.025 | 0 | 0.2 |
| Wamide | 0.5 | 0.4 | 3 |
| Aamine | 0.01 | 0 | 0.2 |
| Wamine | 1.5 | 0.5 | 5 |
| ANOE(−3.5 ppm) | 0.02 | 0 | 1 |
| WNOE(−3.5 ppm) | 3 | 1 | 5 |
| Asemi-solid MT | 0.1 | 0 | 1 |
| Wsemi-solid MT | 25 | 10 | 100 |
The reference signals (Sref) for the APT, NOE(−3.5 ppm), and semi-solid MT signals were obtained by the sum of all Lorentzians except the corresponding pool estimated (35). An apparent exchange-dependent relaxation (AREX) method (36-38), which inversely subtracts Slab from Sref together with T1 correction, was used to quantify the APT/NOE(−3.5 ppm) effects.
| (2) |
where R1w is the water longitudinal relaxation rate; the subscript ‘mfit’ represents the multiple-pool Lorentzian fit. () was added to Eq. (2) for all animal imaging to make the inverse subtraction method more specific (38). For all phantom imaging, was set to 0 due to the low MT pool size ratio. The AREX determined semi-solid MT effect was obtained by , where is the Lorentzian line for the MT effect (39). The AREXmfit quantified APT and NOE(−3.5 ppm) values were obtained by choosing the maximum value between 4 ppm and 3 ppm in the fitted APT spectra and between −3 ppm and −4 ppm in the fitted NOE(−3.5 ppm) spectra, respectively. The AREXmfit determined MT signal was obtained at −2.3 ppm in the fitted MT spectrum.
We used the asymmetric analysis, in which was acquired on the symmetrical side relative to the water peak, to process the CEST Z-spectrum from ghost membranes and reconstituted phospholipid samples.
| (3) |
where (+) represents the resonance frequency offset of the labeled protons; (−) is the offset on the opposite side relative to the water peak; the subscript ‘asym’ represents the asymmetric analysis.
Data analysis and statistics
Regions of interest (ROIs) of tumors, contralateral normal tissues, the corpus callosum and the caudate putamen were delineated in T2 weighted images. The corpus callosum and the caudate putamen were selected as representative regions to investigate the NOE(−3.5 ppm) signals in white matter and gray matter, respectively. The student’s t-test was employed to compare the ROI-averaged signals. It was considered to be statistically significant if P < 0.05. All data analysis and statistical analyses were performed using MATLAB (Mathworks, Natick, RMA, USA).
RESULTS
Figure 1 shows the CEST Z-spectra, the corresponding residual spectra as well as the AREXmfit spectra acquired from the homogenates and the supernatant of brain tissues prepared from healthy rats. Figure S1 shows the corresponding magnetization transfer ratio (MTR) spectra in Supplementary information, which is derived from the direct subtraction of Sref and Slab from the multiple-pool Lorentzian fit. The RMS of the residues averaged across the whole Z-spectrum are 0.37% and 0.63% for the homogenates and the supernatant, respectively, which are smaller than the MTR quantified APT signals and NOE(−3.5 ppm) signals from homogenates (APT: 2.16%, NOE(−3.5 ppm): 5.40%) and supernatant (APT: 1.38%, NOE(−3.5 ppm): 0.89%). The low RMS of the residuals indicate high goodness of the multiple-pool Lorentzian fit. Table S1 shows results of the multiple-pool Lorentzian fit from the homogenates and the supernatant in Supplementary information.
Figure 1.
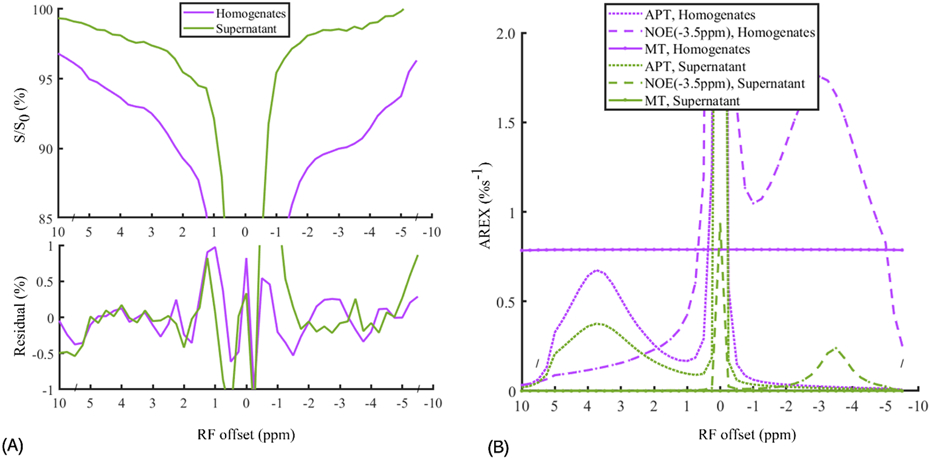
CEST Z-spectra and the corresponding residual spectra (A) and AREXmfit determined APT, NOE(−3.5 ppm), and MT spectra (B) from brain tissue homogenates (purple lines) and the supernatant (green lines). Note that the AREX determined MT in the supernatant is close to 0; the relatively small residuals (< ±0.02%) at ± 3.5 ppm compared with the AREXmift determined APT and NOE(−3.5 ppm) signals indicate high goodness of the multiple-pool Lorentzian fitting.
In this study, the ratios of the AREXmfit NOE(−3.5 ppm) signal to the APT signal are derived to assess the relative contributions of proteins and lipids in the brain tissues as below. For the in vitro homogenates, the ratio of NOE(−3.5 ppm) signal (1.76 %s−1) to APT signal (0.68 %s−1) is around 2.59. For the supernatant without lipids, the ratio of the NOE(−3.5 ppm) signal (0.24 %s−1) to the APT signal (0.38 %s−1) is around 0.63. A reasonable assumption is that the ratio of protein-sourced NOE(−3.5 ppm) signal to protein-sourced APT signal is identical for the homogenates and the supernatant. In this context, the relative contributions of the lipids and proteins to the NOE(−3.5 ppm) signals are derived as around 76% and 24%, respectively. Note that the AREXmfit spectra show no obvious MT effects in the supernatant, which suggests the successful removal of not only semi-solid components but also the membrane lipids, considering that membrane lipids could generate the MT effects (see the Z-spectra beyond ±5 ppm in Figure 2A). The CEST signals at 2 ppm in the tissue homogenates and the supernatant are greater than that measured in the in vivo experiments, which may be due to the increased creatine from dephosphorylation of phosphocreatine after the death of the rats (10,40).
Figure 2.
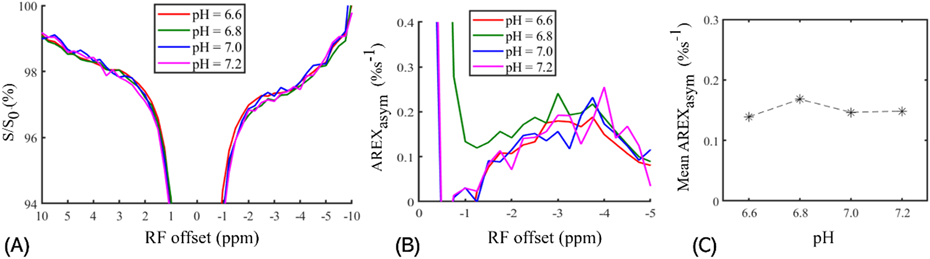
CEST Z-spectra (A), AREXasym spectra (B), and the mean values of AREXasym signals (C) measured from the ghost membrane phantoms with pH of 6.6, 6.8, 7.0, and 7.2. Note that the mean values of AREXasym signals are derived by averaging the AREXasym signals from −5 ppm to −2 ppm in B.
Figure 2 and Figure 3 demonstrate the dependence of the NOE(−3.5 ppm) on two physiological conditions (i.e., pH and the length of aliphatic chain of phospholipid). FigureFigure 2 shows CEST Z-spectra, AREXasym spectra and the mean AREXasym value derived from −5 ppm to −2 ppm measured from the phantom of ghost membranes with varied pH values of 6.6, 6.8, 7.0 and 7.2. The mean AREXasym value derived from −5 ppm to −2 ppm in FigureFigure 2C was used to improve the low SNR of signals measured in the Z-spectra, which demonstrates that the NOE(−3.5 ppm) signal has nearly no dependence on pH. Figure 3 compares CEST Z-spectra and AREXasym spectra measured from DSPC and DEPC. Note that the NOE(−3.5 ppm) signal measured in DEPC is higher than that in DSPC. The aliphatic chain of DEPC is longer than that of DSPC, which may explain its higher NOE(−3.5 ppm) signals.
Figure 3.
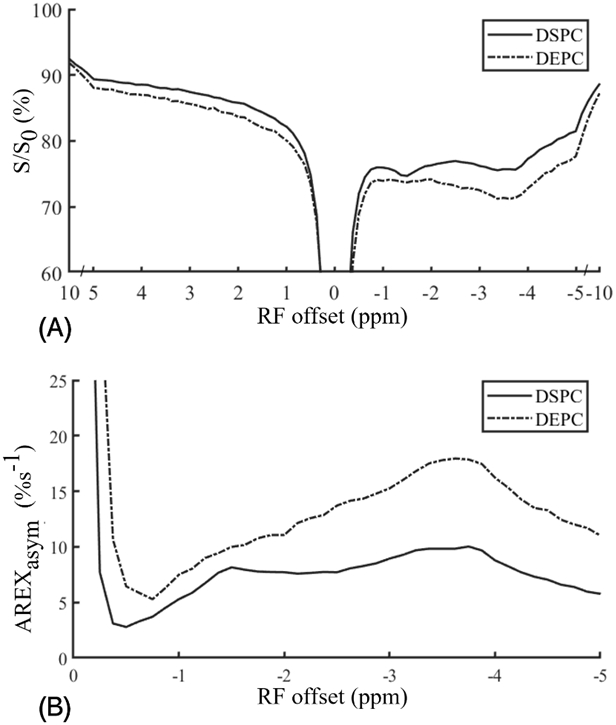
CEST Z-spectra (A) and AREXasym spectra (B) measured from the reconstituted membranes that are prepared with DSPC and DEPC, respectively.
Figure 4 displays the ROI-averaged CEST Z-spectra, the fitting residuals and the AREXmfit spectra acquired from brain tumors and the contralateral normal tissues, as well as from the corpus callosum and the caudate putamen in the brains of rats. The APT signals at 3.5 ppm, the amine signals at 2 ppm, the NOE signals −3.5 ppm and the broad semi-solid MT signals can be clearly observed in the CEST Z-spectra. Figure S2 shows the corresponding MTR spectra in Supplementary information. The RMS of the residues averaged across the whole Z-spectrum are 0.22% and 0.19% for the normal tissues and tumors, respectively, which are much smaller than the MTR quantified APT and NOE(−3.5 ppm) signals from normal tissues (APT: 3.04%, NOE(−3.5 ppm): 8.84%) and tumors (APT: 5.10%, NOE(−3.5 ppm): 8.87%). The RMS of the residues averaged across the whole frequency offset range is 0.18% and 0.21% for the CP and CC, respectively, which are much smaller than the MTR quantified APT signals and NOE(−3.5ppm) signals from CP (APT: 2.82%, NOE(−3.5 ppm): 8.32%) and CC (APT: 2.47%, NOE(−3.5 ppm): 8.47%). The low RMS of the residuals indicate high goodness of the multiple-pool Lorentzian fitting. Table S2 and S3 shows the multiple-pool Lorentzian fitted results from the tumors and contralateral normal tissues as well from the CP and CC in Supplementary information.
Figure 4.
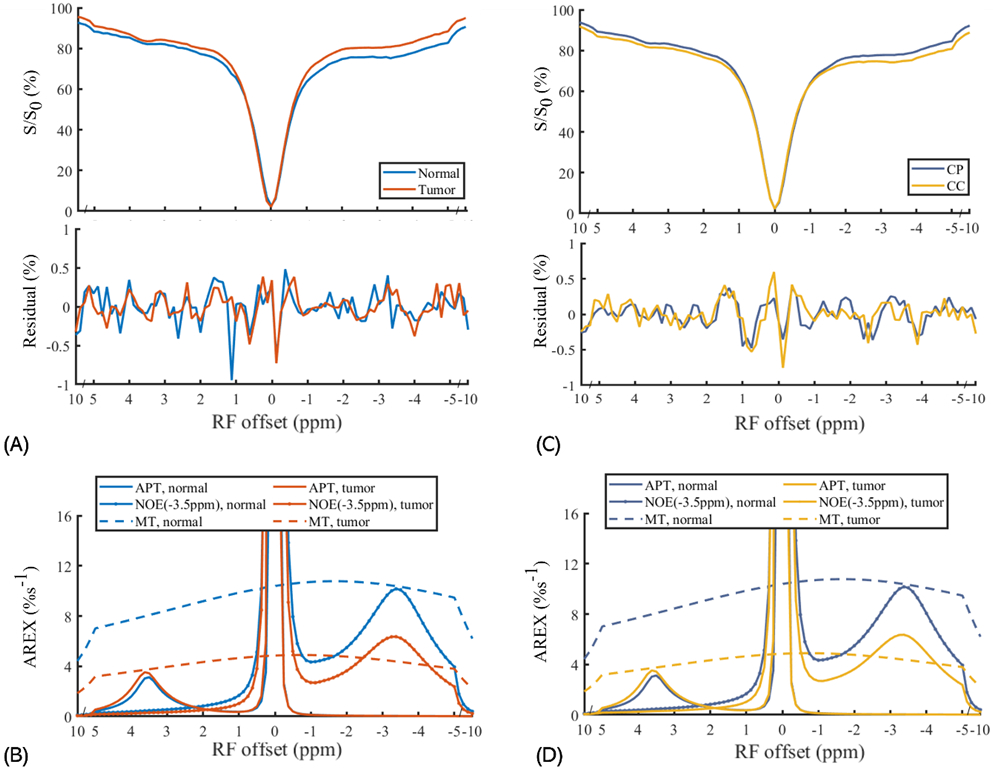
Average CEST Z-spectra and the corresponding residual spectra (A, B) and AREXmfit spectra (C, D) detected from tumors and contralateral normal tissues (left panels), and from the caudate putamen (CP) and the corpus callosum (CC) (right panels). Note that relatively small residuals (< ±0.02%) at ±3.5ppm compared with the AREXmift determined APT and NOE(−3.5 ppm) signals indicate high goodness of the multiple-pool Lorentzian fitting.
Figure 5 demonstrates maps of T2 weighted anatomy, R1w, MT pool size ratio, AREXmfit APT at 3.6 ppm, AREXmfit NOE at −3.5 ppm, and AREXmfit MT at −2.3 ppm from a representative rat brain that bears 9L tumors and a representative healthy rat brain. The normal tissues and tumor tissues, the corpus callosum and the caudate putamen in the rat brain are delineated in the T2 weighted image for the intuitive comparisons of the image contrasts. Summarily, the maps of T2 weighted anatomy and the AREXmfit APT display higher intensities in tumor tissues than in normal tissues while the maps of R1w, MT pool size ratio, AREXmfit NOE(−3.5 ppm), and AREXmfit MT show lower intensities; the maps of R1w, MT pool size ratio, AREXmfit NOE(−3.5 ppm), and AREXmfit MT show higher intensities in the corpus callosum than in the caudate putamen. Note that the previous studies have also demonstrated reduced NOE(−3.5 ppm) signals in the cancer tissues (12,17,18,39,41-44), which guarantees the reproducibility of our experiments.
Figure 5.
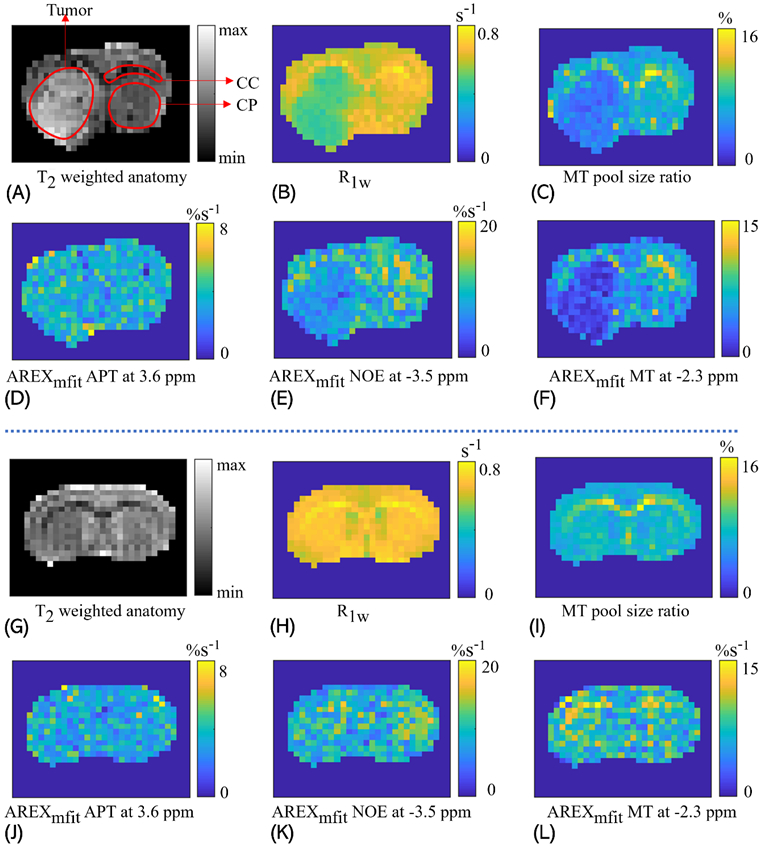
Maps of T2 weighted anatomy (A, G), R1w (B, H), MT pool size ratio (C, I), AREXmfit for APT at 3.6ppm (D, J), AREXmfit for NOE(−3.5 ppm) at −3.5ppm (E, K), and AREXmfit for MT at −2.3ppm (F, L), from the brain of a representative rat that bears 9L tumors (A-F) and a representative healthy rat (G-L). In the T2 weighted image, the tumor tissues, the caudate putamen (CP) and the corpus callosum (CC) are delineated by red lines.
Figure 6 shows the comparisons of R1w, MT pool size ratio, and the quantified CEST signals between tumors and contralateral normal tissues, as well as between the corpus callosum and the caudate putamen. The t-test shows significant differences (P < 0.05) of R1w and MT pool size ratio between tumors and normal tissues as well as between the corpus callosum and the caudate putamen; significant differences of the AREXmfit NOE(−3.5 ppm) between tumors and normal tissues, and between the corpus callosum and the caudate putamen; and significant difference of the AREXmfit MT between tumor and normal tissues. To determine the origins of differences in NOE(−3.5 ppm) signals between tumors and normal tissues and between gray matter and white matter, the ratios of the region-averaged AREXmfit signals are derived as below. The ratio of NOE(−3.5 ppm) signal in normal tissues (10.17%) to that in tumors (6.36%) is around 1.60. As inferred above, membrane lipids have a contribution of around 76% to NOE(−3.5 ppm) and proteins have a contribution of around 24%, thus the substantial change in NOE(−3.5 ppm) signal should be mainly explained by lipids. Besides, the ratio of NOE(−3.5 ppm) signal in the corpus callosum (11.85%) to that in the caudate putamen (9.56%) is around 1.24; it also suggestions that the NOE(−3.5 ppm) signals mainly originate from membrane lipids, considering that the white matter (i.e., the corpus callosum) has more membrane lipids and fewer proteins than the gray matter (i.e., the caudate putamen). Note that the AREXfit quantified APT has no significant differences between tumors and normal tissues, which agrees with previous studies (41,43). However, the differences in the MTR quantified APT signals have been observed in other previous studies (45-47), which may be due to confounding factors including R1w and ‘shine through’ effect from direct water saturation and MT that are involved in the MTR metric (48).
Figure 6.
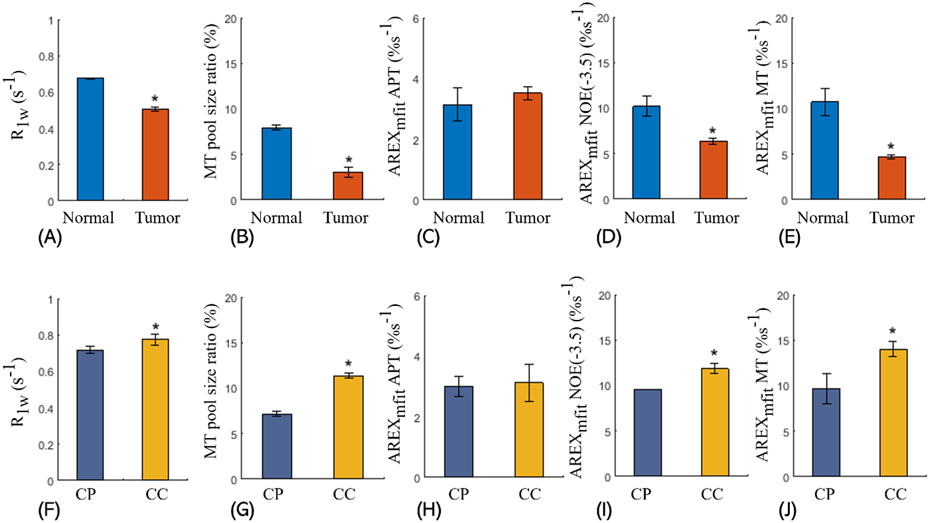
Comparisons between the tumors and the contralateral normal tissues for R1w (A), MT pool size ratio (B), AREXmfit APT at 3.6 ppm (C), AREXmfit NOE(−3.5 ppm) at −3.5 ppm (D), and AREXmfit determined MT at −2.3 ppm (e); and comparisons between and the caudate putamen (CP) and the corpus callosum (CC) for R1w (F), MT pool size ratio (G), AREXmfit determined APT at 3.6 ppm (H), AREXmfit determined NOE(−3.5 ppm) at −3.5 ppm (I), and AREXmfit determined MT at −2.3 ppm (J). The significant differences between two signals are indicated by asterisks (*P < 0.05).
DISCUSSION
In this study, the results of in vitro experiments on tissue homogenates and the supernatant of brain tissues suggest that the NOE(−3.5 ppm) signals in the brain mainly originate from membrane lipids. Further studies on ghost membranes and reconstituted phospholipids reveal that the NOE(−3.5 ppm) signals are independent on pH and dependent on the chemical composition of lipids. The independence of lipid-sourced effect on pH along with the similar property of protein-sourced NOE(−3.5 ppm) demonstrated in a recent study (49) can be explained by the mechanism of relayed NOE that NOE is relayed by chemical exchange to achieve saturation transfer from protons on aliphatic chains to the water proton pool (16). Specifically, NOE mediated magnetization transfer is much slower than chemical exchange from relay sites to the water proton pool. Thus, the efficiency of the relayed NOE is dominantly determined by NOE coupling rather than chemical exchange, leading to that the NOE effect is unsusceptible to pH that is associated with the chemical exchange rate. The biophysical mechanism of the dependence on the chemical compositions is hard to be clearly determined in this study, considering that the NOE process is modulated by the chemical composition that varies the number of aliphatic protons and the mobility of molecules in a complex way. The dependence of NOE on the chemical composition is worth a further study since the changes in the chemical composition of membrane lipids are engaged in the pathogenesis of many diseases (50).
The in vivo experiments on the rat brain show that the NOE(−3.5 ppm) signals in normal tissues are significantly higher than that in tumors, and the NOE(−3.5 ppm) signals in the corpus callosum (white matter) are significantly higher than that in the caudate putamen (gray matter), which agrees with previous reports (17,20,22,23,42). Given that our in vitro experiments demonstrate membrane lipids being the major source of the NOE(−3.5 ppm) signals, the differences in NOE(−3.5 ppm) signals could be largely accounted for by the membrane lipids rather than proteins, which is analyzed as below. The decreases of NOE(−3.5 ppm) signals in tumors should be mainly due to the change in the properties of lipids rather than proteins for the following three reasons. (1) the higher expression of proteins in the tumor tissues (51) could lead to an increase in protein-sourced NOE(−3.5 ppm) signals. (2) since the proteins account for ~24% of NOE signals as inferred in this study, the protein changes in tumors have a limited influence on NOE signals (note that the relative decrease of the NOE signal in tumor is up to 38%); note that a previous study (22,30) reported that the changes in the spatial conformation of proteins could reduce NOE(−3.5 ppm) signals; however, there is no evidence supporting that the spatial conformation of proteins account for the reduced NOE(−3.5 ppm) signals in tumors; (3) numerous reports have demonstrated that membrane lipids in tumors are significantly less rigid than that of normal tissues (51-55); the increased mobility (fluidity) of the cell membranes could reduce the NOE coupling rate, which could explain the reduced NOE(−3.5 ppm) signals in tumors. The higher NOE(−3.5 ppm) signals in white matter than that in gray matter should also mainly result from the variations of membrane lipids, since the dry weight mass fraction of proteins in white matter is 10-15% lower than that in gray matter in proteins while the dry weight mass fraction of lipids in white matter is 60% higher than that in gray matter (56).
The relative contributions of membrane phospholipids and proteins to the NOE(−3.5 ppm) signals are estimated with the reasonable assumption that the ratio of the protein-sourced NOE(−3.5ppm) signal to the protein-sourced APT signal is roughly the same for the homogenates and supernatants. In principle, for the solvent system, the solute (i.e., soluble proteins) is uniformly distributed within the solvent (i.e., water). Hence, the supernatants contain the same soluble proteins as the homogenates, considering that the preparation of supernatants only involves the removal of the insoluble solid components. Note that only soluble proteins could generate the CEST signals with narrow line widths. Also note that there may be some differences in the structure and molecular size of proteins between the supernatant and the homogenates. Considering that the preparation of the supernatant is very quick, the structure and molecular size of proteins should remain unchanged. A previous study indicates that the NOE(−3.5 ppm) signal changes only when there are substantial changes in the protein structure(30). As inferred from the in vitro experiments, membrane lipids have a contribution of around 70% to NOE(−3.5 ppm) and proteins have a contribution of around 30%. In spite of the potential experimental bias mentioned above, a reasonable inference is that the membrane phospholipids have a dominant contribution to NOE(−3.5 ppm) signals.
Besides phospholipids, ghost membranes also contain a large proportion of membrane proteins (57), which have trivial contributions to the APT and NOE(−3.5 ppm) signals. As shown in Figure 2, there are no APT peaks in the Z-spectrum from the ghost membrane phantom, which demonstrates that membrane proteins cannot generate CEST signal peaks with narrow line widths. It may be explained by that the water-insoluble membrane proteins with high molecular weights have limited mobility due to motion restrictions from cell membranes that also have high molecular weights. On the other hand, amide protons inside membrane proteins with large molecular sizes have no access to water protons and thus cannot generate CEST effects(46). Note that phospholipids in cell membranes could generate the NOE(−3.5 ppm) signals with a narrow linewidth since their low molecular weights ensure high mobility.
Protease inhibitor that could inhibit the potential protein degeneration was not used in the preparation of the homogeneities in this study since the most widely used inhibitor (e.g., ethylenediaminetetraacetic acid) has a cation (Ca++/Mg++ chelator) that could change the structures of proteins (i.e., degeneration of proteins) and thus alter the NOE signals as well as the CEST signals. To avoid the degradation of proteins due to proteases, the preparation of tissue samples was performed within a short time at a low temperature (4°C) to minimize protease activities. In addition, MRI measurements of these samples were also performed within 0.5 h to reduce the protease-induced decomposition of proteins.
The multiple-pool Lorentzian fit was used to quantify the CEST, NOE and MT signals, in which the limitations of this method could introduce the relevant bias in the study. However, the relevant bias of the fitting can be reduced to a low level under the following specific conditions. First, although the semi-solid MT can be better fitted by super-Lorentzian absorption line shape (58-60), we fit CEST signals obtained with RF offsets within ± 10 ppm, where the semi-solid MT can be fitted by the Lorentzian function with small errors (61). In biological tissues, besides the five pools used in the multiple-pool Lorentzian fit, the amines at 3 ppm and the hydroxyls from 1 ppm to 3 ppm also contribute signals in the Z-spectrum. However, by observing the CEST Z-spectrum, these pools can be roughly treated as a single pool centered at 2 ppm. Previous studies have shown that the APT can be still quantified at relatively low saturation power with the presence of fast exchanging amines at 3 ppm (41). In addition, aliphatic NOE signals include many peaks spanning from −5 ppm to −1 ppm. However, by observing the CEST Z-spectrum, these pools can be roughly treated as two peaks at around −1.6 ppm and −3.5 ppm (12). The CEST signals at 3.5 ppm may consist of the APT and the aromatic NOE (62), both of which should originate from proteins.
In the brain tissues, lipids can be cataloged into three types: myelin lipids, membrane phospholipids, and neutral lipids (56). Myelin lipids, that are the major components of the myelin sheath in nerve fibers in white matter, have the lowest mobility of the three types of lipids, which can be manifested by its very broad line width observed in the conventional MT Z-spectrum. Neutral lipids have a very low concentration in brain (50), which exist in the form of lipid droplets as energy reserves in bio tissues and cannot produce CEST effects. Note that neutral lipids in tissues could generate artifacts in Z-spectrum, which need to be carefully removed in CEST imaging (63-65). The NOE(−3.5ppm) signals arise from the aliphatic chains of phospholipids. Note that the NOE(−1.6 ppm) is another NOE effect, which is sourced from the choline head group of phospholipids (12,34,42,66,67). To date, myelin lipids can be detected by MT imaging (14,15), and neutral lipids can be detected by magnetic resonance imaging with a postprocess of fat-water separation (68), and magnetic resonance spectroscopy (MRS) (69). However, the in vivo detection of membrane lipids is challenging because of the short transverse relaxation time of the membrane lipids. In this study, we demonstrate that NOE(−3.5 ppm) imaging could serve as an MRI tool for monitoring membrane phospholipid.
In this preliminary work, although we investigated the partial properties of the membrane-lipid-sourced NOE(−3.5 ppm) signals (i.e., regarding the chemical composition and the pH), its complete biophysical properties warrant further systematic studies that are could be beneficial to interpreting its underlying contrast mechanism of membrane lipids associated with the pathogenesis of diseases at molecular levels. The length of aliphatic chains of lipid molecule could change the number of protons and thus affect the NOE signals via a concentration effect. Besides, it also alters the fluidity of membrane (70) and thus modulate the efficiency of the NOE. In addition, the unsaturation of membrane lipids, the concentration of cholesterol in the membrane and the behavior of the membrane-bound proteins could also modulate the efficiency of the NOE by altering the fluidity of membrane (71-73). Note that the changes in the chemical composition of membrane lipids in the pathogenesis of diseases are companied with multiple factors mentioned above (50,74). In future studies, univariate experiments based on reconstituted membrane phantoms can be used to investigate these factors, separately.
CONCLUSION
In this study, we demonstrate that the NOE(−3.5 ppm) signals in the brain mainly originate from membrane lipids, which suggests that membrane lipids account for the origins of the NOE(−3.5 ppm) contrast differences between tumors and normal tissues, and between gray matter and white matter. NOE(−3.5 ppm) imaging could be exploited as a new MRI method to monitor membrane lipids in vivo, which has great potential for clinical applications.
Supplementary Material
Grant Sponsor:
R21 AR074261, R03 EB029078, R01 EB029443
REFERENCES
- 1.Zhou JY, van Zijl PCM. Chemical exchange saturation transfer imaging and spectroscopy. Prog Nucl Mag Res Sp 2006;48(2-3):109–136. [Google Scholar]
- 2.van Zijl PCM, Yadav NN. Chemical Exchange Saturation Transfer (CEST): What is in a Name and What Isn't? Magnetic Resonance In Medicine 2011;65(4):927–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim J, Wu Y, Guo Y, Zheng H, Sun PZ. A review of optimization and quantification techniques for chemical exchange saturation transfer MRI toward sensitive in vivo imaging. Contrast media & molecular imaging 2015;10(3):163–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu B, Warnock G, Zaiss M, Lin C, Chen M, Zhou Z, Mu L, Nanz D, Tuura R, Delso G. An overview of CEST MRI for non-MR physicists. Ejnmmi Phys 2016;3(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Zijl PCM, Lam WW, Xu JD, Knutsson L, Stanisz GJ. Magnetization Transfer Contrast and Chemical Exchange Saturation Transfer MRI. Features and analysis of the field-dependent saturation spectrum. Neuroimage 2018;168:222–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou J, Payen JF, Wilson DA, Traystman RJ, van Zijl PC. Using the amide proton signals of intracellular proteins and peptides to detect pH effects in MRI. Nature medicine 2003;9(8):1085–1090. [DOI] [PubMed] [Google Scholar]
- 7.Cai K, Haris M, Singh A, Kogan F, Greenberg JH, Hariharan H, Detre JA, Reddy R. Magnetic resonance imaging of glutamate. Nature medicine 2012;18(2):302–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cui J, Zu ZL. Towards the molecular origin of glutamate CEST (GluCEST) imaging in rat brain. Magnetic Resonance In Medicine 2020;83(4):1405–1417. [DOI] [PubMed] [Google Scholar]
- 9.Haris M, Singh A, Cai KJ, Kogan F, McGarvey J, DeBrosse C, Zsido GA, Witschey WRT, Koomalsingh K, Pilla JJ, Chirinos JA, Ferrari VA, Gorman JH, Hariharan H, Gorman RC, Reddy R. A technique for in vivo mapping of myocardial creatine kinase metabolism. Nature medicine 2014;20(2):209–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang XY, Xie JP, Wang F, Lin EC, Xu JZ, Gochberg DF, Gore JC, Zu ZL. Assignment of the molecular origins of CEST signals at 2ppm in rat brain. Magnetic Resonance In Medicine 2017;78(3):881–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen L, Zeng HF, Xu X, Yadav NN, Cai SH, Puts NA, Barker PB, Li T, Weiss RG, van Zijl PCM, Xu JD. Investigation of the contribution of total creatine to the CEST Z-spectrum of brain using a knockout mouse model. NMR in biomedicine 2017;30(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang XY, Wang F, Jin T, Xu JZ, Xie JP, Gochberg DF, Gore JC, Zu ZL. MR imaging of a novel NOE-mediated magnetization transfer with water in rat brain at 9.4T. Magnetic Resonance In Medicine 2017;78(2):588–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zu ZL, Lin EC, Louie EA, Xu JZ, Li H, Xie JP, Lankford CL, Chekmenev EY, Swanson SD, Does MD, Gore JC, Gochberg DF. Relayed nuclear Overhauser enhancement sensitivity to membrane Cho phospholipids. Magnetic Resonance In Medicine 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gass A, Barker GJ, Kidd D, Thorpe JW, Macmanus D, Brennan A, Tofts PS, Thompson AJ, Mcdonald WI, Miller DH. Correlation Of Magnetization-Transfer Ratio with Clinical Disability In Multiple-Sclerosis. Annals of neurology 1994;36(1):62–67. [DOI] [PubMed] [Google Scholar]
- 15.Armstrong CL, Traipe E, Hunter JV, Haselgrove JC, Ledakis GE, Tallent EM, Shera D, van Buchem MA. Age-related, regional, hemispheric, and medial-lateral differences in myelin integrity in vivo in the normal adult brain. Am J Neuroradiol 2004;25(6):977–984. [PMC free article] [PubMed] [Google Scholar]
- 16.van Zijl PCM, Zhou J, Mori N, Payen JF, Wilson D, Mori S. Mechanism of magnetization transfer during on-resonance water saturation. A new approach to detect mobile proteins, peptides, and lipids. Magnetic Resonance In Medicine 2003;49(3):440–449. [DOI] [PubMed] [Google Scholar]
- 17.Jones CK, Huang A, Xu JD, Edden RAE, Schar M, Hua J, Oskolkov N, Zaca D, Zhou JY, McMahon MT, Pillai JJ, van Zijl PCM. Nuclear Overhauser enhancement (NOE) imaging in the human brain at 7 T. Neuroimage 2013;77(0):114–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou Y, Bie CX, van Zijl PCM, Yadav NN. The relayed nuclear Overhauser effect in magnetization transfer and chemical exchange saturation transfer MRI. NMR in biomedicine 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goerke S, Zaiss M, Longo D, Garello F, Gregorio ED, Breitling J, Ladd ME, Bachert P. CEST signals of lipids. ISMRM 25th Annual Meeting, Honolulu, HI, USA 2017:0201. [Google Scholar]
- 20.Shen YY, Xiao G, Shen ZW, Zhang XL, Tang XY, Hu W, Zhang ZY, Guan JT, Wu RH. Imaging of nuclear Overhauser enhancement at 7 and 3T. NMR in biomedicine 2017;30(9). [DOI] [PubMed] [Google Scholar]
- 21.Heo HY, Jones CK, Hua J, Yadav N, Agarwal S, Zhou JY, van Zijl PCM, Pillai JJ. Whole-Brain Amide Proton Transfer (APT) and Nuclear Overhauser Enhancement (NOE) Imaging in Glioma Patients Using Low-Power Steady-State Pulsed Chemical Exchange Saturation Transfer (CEST) Imaging at 7T. Journal Of Magnetic Resonance Imaging 2016;44(1):41–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zaiss M, Kunz P, Goerke S, Radbruch A, Bachert P. MR imaging of protein folding in vitro employing Nuclear-Overhauser-mediated saturation transfer. NMR in biomedicine 2013;26(12):1815–1822. [DOI] [PubMed] [Google Scholar]
- 23.Paech D, Zaiss M, Meissner JE, Windschuh J, Wiestler B, Bachert P, Neumann JO, Kickingereder P, Schlemmer HP, Wick W, Nagel AM, Heiland S, Ladd ME, Bendszus M, Radbruch A. Nuclear Overhauser Enhancement Mediated Chemical Exchange Saturation Transfer Imaging at 7 Tesla in Glioblastoma Patients. Plos One 2014;9(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mehrabian H, Desmond KL, Soliman H, Sahgal A, Stanisz GJ. Differentiation between Radiation Necrosis and Tumor Progression Using Chemical Exchange Saturation Transfer. Clin Cancer Res 2017;23(14):3667–3675. [DOI] [PubMed] [Google Scholar]
- 25.Chen L, Wei ZL, Chan KWY, Cai SH, Liu GS, Lu HZ, Wong PC, van Zijl PCM, Li T, Xu JD. Protein aggregation linked to Alzheimer's disease revealed by saturation transfer MRI. Neuroimage 2019;188:380–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang F, Zu ZL, Wu TL, Yan XQ, Lu M, Yang PF, Byun NE, Reed JL, Gore JC, Chen LM. Sensitivity and specificity of CEST and NOE MRI in injured spinal cord in monkeys. Neuroimage-Clin 2021;30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anderson WA, Freeman R. Influence Of a Second Radiofrequency Field on High-Resolution Nuclear Magnetic Resonance Spectra. J Chem Phys 1962;37(1):85-&. [Google Scholar]
- 28.Solomon I. Relaxation Processes In a System Of 2 Spins. Phys Rev 1955;99(2):559–565. [Google Scholar]
- 29.Overhauser AW. Polarization Of Nuclei In Metals. Phys Rev 1953;92(2):411–415. [Google Scholar]
- 30.Goerke S, Zaiss M, Kunz P, Klika KD, Windschuh JD, Mogk A, Bukau B, Ladd ME, Bachert P. Signature of protein unfolding in chemical exchange saturation transfer imaging. NMR in biomedicine 2015;28(7):906–913. [DOI] [PubMed] [Google Scholar]
- 31.Al-Jafari AA, Kamal MA, Greig NH, Alhomida AS, Perry ER. Kinetics of human erythrocyte acetylcholinesterase inhibition by a novel derivative of physostigmine: Phenserine. Biochem Bioph Res Co 1998;248(1):180–185. [DOI] [PubMed] [Google Scholar]
- 32.Schwoch G, Passow H. Preparation And Properties Of Human Erythrocyte-Ghosts. Mol Cell Biochem 1973;2(2):197–218. [DOI] [PubMed] [Google Scholar]
- 33.Gochberg DF, Gore JC. Quantitative magnetization transfer imaging via selective inversion recovery with short repetition times. Magnetic Resonance in Medicine 2007;57(2):437–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zu ZL. Toward more reliable measurements of NOE effects in CEST spectra at around-1.6 ppm (NOE (−1.6)) in rat brain. Magnetic Resonance In Medicine 2019;81(1):208–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Windschuh J, Zaiss M, Meissner JE, Paech D, Radbruch A, Ladd ME, Bachert P. Correction of B1-inhomogeneities for relaxation-compensated CEST imaging at 7T. Nmr in Biomedicine 2015;28(5):529–537. [DOI] [PubMed] [Google Scholar]
- 36.Zaiss M, Bachert P. Exchange-dependent relaxation in the rotating frame for slow and intermediate exchange - modeling off-resonant spin-lock and chemical exchange saturation transfer. NMR in biomedicine 2013;26(5):507–518. [DOI] [PubMed] [Google Scholar]
- 37.Zaiss M, Bachert P. Chemical exchange saturation transfer (CEST) and MR Z-spectroscopy in vivo: a review of theoretical approaches and methods. Physics in medicine and biology 2013;58(22):R221–269. [DOI] [PubMed] [Google Scholar]
- 38.Zaiss M, Zu ZL, Xu JZ, Schuenke P, Gochberg DF, Gore JC, Ladd ME, Bachert P. A combined analytical solution for chemical exchange saturation transfer and semi-solid magnetization transfer. NMR in biomedicine 2015;28(2):217–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cui J, Sun C, Zu Z. NOE-weighted imaging in tumors using low-duty-cycle 2π-CEST. Magn Reson Med 2022;(published online):DOI: 10.1002/mrm.29475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hackett MJ, Lee J, El-Assaad F, McQuillan JA, Carter EA, Grau GE, Hunt NH, Lay PA. FTIR Imaging of Brain Tissue Reveals Crystalline Creatine Deposits Are an ex Vivo Marker of Localized Ischemia during Murine Cerebral Malaria: General Implications for Disease Neurochemistry. Acs Chem Neurosci 2012;3(12):1017–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang XY, Wang F, Li H, Xu JZ, Gochberg DF, Gore JC, Zu ZL. Accuracy in the quantification of chemical exchange saturation transfer (CEST) and relayed nuclear Overhauser enhancement (rNOE) saturation transfer effects. NMR in biomedicine 2017;30(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zu ZL. Ratiometric NOE(−1.6) contrast in brain tumors. NMR in biomedicine 2018;31(12). [DOI] [PubMed] [Google Scholar]
- 43.Xu JZ, Zaiss M, Zu ZL, Li H, Xie JP, Gochberg DF, Bachert P, Gore JC. On the origins of chemical exchange saturation transfer ( CEST) contrast in tumors at 9.4 T. NMR in biomedicine 2014;27(4):406–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zaiss M, Windschuh J, Paech D, Meissner JE, Burth S, Schmitt B, Kickingereder P, Wiestler B, Wick W, Bendszus M, Schlemmer HP, Ladd ME, Bachert P, Radbruch A. Relaxation-compensated CEST-MRI of the human brain at 7 T: Unbiased insight into NOE and amide signal changes in human glioblastoma. Neuroimage 2015;112:180–188. [DOI] [PubMed] [Google Scholar]
- 45.Zhou J, Lal B, Wilson DA, Laterra J, van Zijl PC. Amide proton transfer (APT) contrast for imaging of brain tumors. Magnetic Resonance in Medicine 2003;50(6):1120–1126. [DOI] [PubMed] [Google Scholar]
- 46.Zhou JY, Tryggestad E, Wen ZB, Lal B, Zhou TT, Grossman R, Wang SL, Yan K, Fu DX, Ford E, Tyler B, Blakeley J, Laterra J, van Zijl PCM. Differentiation between glioma and radiation necrosis using molecular magnetic resonance imaging of endogenous proteins and peptides. Nature medicine 2011;17(1):130–U308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou JY, Heo HY, Knutsson L, van Zijl PCM, Jiang SS. APT-weighted MRI: Techniques, current neuro applications, and challenging issues. Journal Of Magnetic Resonance Imaging 2019;50(2):347–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zu ZL. Towards the complex dependence of MTRasym on T-1w in amide proton transfer (APT) imaging. NMR in biomedicine 2018;31(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jin T, Kim SG. Role of chemical exchange on the relayed nuclear Overhauser enhancement signal in saturation transfer MRI. Magnetic Resonance In Medicine 2022;87(1):365–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Butler LM, Perone Y, Dehairs J, Lupien LE, de Laat V, Talebi A, Loda M, Kinlaw WB, Swinnen JV. Lipids and cancer: Emerging roles in pathogenesis, diagnosis and therapeutic intervention. Adv Drug Deliv Rev 2020;159:245–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mori S, Eleff SM, Pilatus U, Mori N, van Zijl PCM. Proton NMR spectroscopy of solvent-saturable resonances: A new approach to study pH effects in situ. Magnetic Resonance In Medicine 1998;40(1):36–42. [DOI] [PubMed] [Google Scholar]
- 52.Kuriyama S, Theveneau E, Benedetto A, Parsons M, Tanaka M, Charras G, Kabla A, Mayor R. In vivo collective cell migration requires an LPAR2-dependent increase in tissue fluidity. J Cell Biol 2014;206(1):113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao WN, Prijic S, Urban BC, Tisza MJ, Zuo Y, Li L, Tan Z, Chen XL, Mani SA, Chang JT. Candidate Antimetastasis Drugs Suppress the Metastatic Capacity of Breast Cancer Cells by Reducing Membrane Fluidity. Cancer Res 2016;76(7):2037–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nakazawa I, Iwaizumi M. A Role Of the Cancer Cell-Membrane Fluidity In the Cancer Metastases - an Esr Study. Tohoku J Exp Med 1989;157(3):193–198. [DOI] [PubMed] [Google Scholar]
- 55.Zouaoui J, Trunfio-Sfarghiu AM, Brizuela L, Piednoir A, Maniti O, Munteanu B, Mebarek S, Girard-Egrot A, Landoulsi A, Granjon T. Multi-scale mechanical characterization of prostate cancer cell lines: Relevant biological markers to evaluate the cell metastatic potential. Bba-Gen Subjects 2017;1861(12):3109–3119. [DOI] [PubMed] [Google Scholar]
- 56.O'Brien JS, Sampson EL. Lipid composition of the normal human brain: gray matter, white matter, and myelin. Journal of lipid research 1965;6(4):537–544. [PubMed] [Google Scholar]
- 57.Guidotti G. The composition of biological membranes. Archives of internal medicine 1972;129(2):194–201. [PubMed] [Google Scholar]
- 58.Tozer D, Ramani A, Barker GJ, Davies GR, Miller DH, Tofts PS. Quantitative magnetization transfer mapping of bound protons in multiple sclerosis. Magnetic Resonance In Medicine 2003;50(1):83–91. [DOI] [PubMed] [Google Scholar]
- 59.Morrison C, Stanisz G, Henkelman RM. Modeling Magnetization-Transfer for Biological-Like Systems Using a Semisolid Pool with a Super-Lorentzian Lineshape And Dipolar Reservoir. J Magn Reson Ser B 1995;108(2):103–113. [DOI] [PubMed] [Google Scholar]
- 60.Morrison C, Henkelman RM. A Model for Magnetization-Transfer In Tissues. Magnetic Resonance In Medicine 1995;33(4):475–482. [DOI] [PubMed] [Google Scholar]
- 61.Zaiss M, Schmitt B, Bachert P. Quantitative separation of CEST effect from magnetization transfer and spillover effects by Lorentzian-line-fit analysis of z-spectra. J Magn Reson 2011;211(2):149–155. [DOI] [PubMed] [Google Scholar]
- 62.Jin T, Kim SG. In vivo saturation transfer imaging of nuclear overhauser effect from aromatic and aliphatic protons: implication to APT quantification. Proceedings of International Society for Magnetic Resonance in Medicine 2013;Salt Lake City, Utah, USA:p 2528. [Google Scholar]
- 63.Zhang S, Seiler S, Wang X, Madhuranthakam AJ, Keupp J, Knippa EE, Lenkinski RE, Vinogradov E. CEST-Dixon for human breast lesion characterization at 3 T: A preliminary study. Magnetic Resonance In Medicine 2018;80(3):895–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhao Y, Yan X, Zhang Z, Zhao W, Liu Z, Li J. Self-adapting multi-peak water-fat reconstruction for the removal of lipid artifacts in chemical exchange saturation transfer (CEST) imaging. Magn Reson Med 2019;82(5):1700–1712. [DOI] [PubMed] [Google Scholar]
- 65.Zhao Y, Zu Z, Wang Z, Liu Z, Guo B, Yan X, Gochberg DF, Li J. Effectiveness of fat suppression using a water-selective binomial-pulse excitation in chemical exchange saturation transfer (CEST) magnetic resonance imaging. Magma 2020;33(6):809–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang XY, Wang F, Afzal A, Xu JZ, Gore JC, Gochberg DF, Zu ZL. A new NOE-mediated MT signal at around-1.6 ppm for detecting ischemic stroke in rat brain. Magn Reson Imaging 2016;34(8):1100–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cui J, Zhao Y, Wang F, Gochberg DF, Zu ZL. Contribution of blood to nuclear Overhauser effect at-1.6 ppm. Magnetic Resonance In Medicine 2022;87(1):409–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hernando D, Kramer JH, Reeder SB. Multipeak fat-corrected complex R2* relaxometry: theory, optimization, and clinical validation. Magn Reson Med 2013;70(5):1319–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Podo F, de Certaines JD. Magnetic resonance spectroscopy in cancer: phospholipid, neutral lipid and lipoprotein metabolism and function. Anticancer research 1996;16(3B):1305–1315. [PubMed] [Google Scholar]
- 70.Bianchetti G, Azoulay-Ginsburg S, Keshet-Levy NY, Malka A, Zilber S, Korshin EE, Sasson S, De Spirito M, Gruzman A, Maulucci G. Investigation of the Membrane Fluidity Regulation of Fatty Acid Intracellular Distribution by Fluorescence Lifetime Imaging of Novel Polarity Sensitive Fluorescent Derivatives. Int J Mol Sci 2021;22(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lenaz G. Lipid fluidity and membrane protein dynamics. Bioscience reports 1987;7(11):823–837. [DOI] [PubMed] [Google Scholar]
- 72.Winocour PD, Watala C, Perry DW, Kinloughrathbone RL. Decreased Platelet Membrane Fluidity Due To Glycation Or Acetylation Of Membrane-Proteins. Thromb Haemostasis 1992;68(5):577–582. [PubMed] [Google Scholar]
- 73.Ballweg S, Sezgin E, Doktorova M, Covino R, Reinhard J, Wunnicke D, Hanelt I, Levental I, Hummer G, Ernst R. Regulation of lipid saturation without sensing membrane fluidity. Nat Commun 2020;11(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vigh L, Escriba PV, Sonnleitner A, Sonnleitner M, Piotto S, Maresca B, Horvath I, Harwood JL. The significance of lipid composition for membrane activity: New concepts and ways of assessing function. Prog Lipid Res 2005;44(5):303–344. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


