Abstract
Plants are often exposed not only to short-term (S-) but also to long-term (L-)heat stress over several consecutive days. A few Arabidopsis mutants defective in L-heat tolerance have been identified, but the molecular mechanisms are less understood for this tolerance than for S-heat stress tolerance. To elucidate the mechanisms of the former, we used a forward genetic screen for sensitive to long-term heat (sloh) mutants and isolated sloh3 and sloh63. The mutants were hypersensitive to L- but not to S-heat stress, and sloh63 was also hypersensitive to salt stress. We identified the causal genes, SLOH3 and SLOH63, both of which encoded splicing-related components of the MOS4-associated complex (MAC). This complex is widely conserved in eukaryotes and has been suggested to interact with spliceosomes. Both genes were induced by L-heat stress in a time-dependent manner, and some abnormal splicing events were observed in both mutants under L-heat stress. In addition, endoplasmic reticulum (ER) stress and subsequent unfolded protein response occurred in both mutants under L-heat stress and were especially prominent in sloh63, suggesting that enhanced ER stress is due to the salt hypersensitivity of sloh63. Splicing inhibitor pladienolide B led to concentration-dependent disturbance of splicing, decreased L-heat tolerance, and enhanced ER stress. These findings suggest that maintenance of precise mRNA splicing under L-heat stress by the MAC is important for L-heat tolerance and suppressing ER stress in Arabidopsis.
Keywords: heat tolerance, long-term heat stress, selective splicing, ER stress, Arabidopsis
Significance Statement.
Plants are exposed not only to short-term (S-) but also to long-term (L-)heat stress, however, the L-heat stress response is not yet well understood. Using forward genetic screening, we isolated sloh3 and sloh63 mutants, which are hypersensitive to L-heat but not to S-heat stress. SLOH3 and SLOH63 were identical to the splicing-related factor genes MAC9 and MAC17, respectively. Splicing inhibitor pladienolide B led to concentration-dependent disturbance of splicing, decreased L-heat tolerance, and enhanced ER stress. These findings suggest that maintenance of appropriate mRNA splicing and suppression of ER stress under L-heat stress are important for L-heat tolerance in Arabidopsis.
Introduction
High temperature is a serious stress that affects crop growth and production. The yield of major cereal crops has reportedly decreased due to recent temperature increases [1], so it is important to elucidate the mechanisms of plant responses to heat stress and to generate heat-tolerant crops. In Arabidopsis thaliana, a dynamic transcriptional network is induced within 1 h through proteins of the heat shock factor A1 (HsfA1) group. Under short-term (S-) heat stress (42°C for 30–50 min), this group regulates most heat stress-responsive genes including those for heat shock proteins (HSPs). In Arabidopsis, the HsfA1 group consists of HsfA1a, -b, -d, and -e. The hsfa1a/b/d triple mutation impairs the global induction of heat-responsive genes and reduces heat tolerance [2–4]. Conversely, HsfA1 overexpression induces the expression of various heat-responsive genes and improves S-heat tolerance [5]. Plants exhibit S-heat tolerance after heat acclimation, in which exposure to moderately high temperatures enables plants to acquire tolerance to subsequent otherwise lethal high temperatures [3]. In Arabidopsis, HsfA2 is necessary for the maintenance of acquired thermotolerance. The transient binding of HsfA2 to HSP genes results in a sustained increase in histone marks (such as histone H3 lysine 4 trimethylation; H3K4me3), which promotes HSP gene expression [6, 7]. After heat exposure, HsfA2 levels gradually decrease, while H3K4me3 levels and HSP expression remain high. JMJ protein-mediated sustained H3K27me3 demethylation on small chaperone-encoding HSP genes that function as memory genes controls heat memory [8].
Plants are also exposed to long-term (L-) heat stress, which can involve temperatures up to 37°C over a period of several days. While the molecular mechanisms for responding to S-heat stress are well-characterized as described above, the regulation of L-heat stress tolerance is not fully understood. To investigate the contribution of HSPs and Hsfs to L-heat tolerance, we previously evaluated the L-heat tolerance of the hsp90, hsp101, hsfa1d, hsfa1e, and hsfa2 mutants (Isono et al. [9]). Only hsfa1d was slightly more sensitive to L-heat stress than the WT, whereas the other mutants were comparable to WT, implying that different genes and mechanisms contribute to S-heat and L-heat tolerances.
A forward genetic approach identified a few Arabidopsis mutants including heat intolerant (hit) mutants hypersensitive to L-heat stress [10–13]. To better understand the mechanism for L-heat tolerance, we have also isolated a sensitive to long-term heat (sloh) mutant from EMS-mutagenized seeds of Col-0, an L-heat-tolerant A. thaliana accession [9, 14]. The causal gene of sloh4 is identical to MIP3, which encodes a member of the MAIGO2 (MAG2) tethering complex localized at the endoplasmic reticulum (ER) membrane; that of hit1-1 encodes a protein identical to the homolog of yeast Vsp53p, a member of the GARP tethering complex localized at the Golgi [9, 10, 12]. Both mutants are hypersensitive to L-heat and salt stresses but not to S-heat stress [9, 12]. Under L-heat stress, ER stress and the following unfolded protein response (UPR) are more pronounced in sloh4 than in wild type (WT) [9].
Alternative splicing occurs in all eukaryotes and enhances transcriptional plasticity and proteome diversity in response to various growth and stress cues including heat stress [15]. Alternative splicing is classified into five types: exon skipping, mutually exclusive exons, alternative 5′ splice site, alternative 3′ splice site, and intron retention [15]. Exon skipping is the major type in mammals, whereas intron retention is the major type in plants [16]. Intron retention is thought to enrich proteome diversity in mammals, whereas it produces nonfunctional isoforms in plants [17–19]. The spliceosome is composed of five core subcomplexes: the U1, U2, U4, U5, and U6 small ribonucleoproteins (snRNPs) [20]. Each U snRNP forms a ring structure composed of a corresponding snRNA and an SM-like (LSM) family protein [21]. The activation and catalysis of splicing are largely mediated by the NineTeen complex in humans and yeast or MOS4-associated complex (MAC) in Arabidopsis [22, 23]. The MAC is necessary to define the specificity of the interaction of U5 and U6 with the 5′ splice site [24]. The knockout mutant of LSM5 is hypersensitive to abscisic acid (ABA), NaCl, and S-heat stress (42°C for 90 min) [25, 26]. The mutant of a U5-snRNP-interacting protein homolog, STABILIZED1 (STA1), is hypersensitive to heat (38°C for 1 day) [27]. In both mutants, premature RNA levels of heat shock factor A3 (HsfA3) are increased, while its mature RNA levels are decreased. However, the overexpression of HsfA3 in the sta1-1 background is unable to complement the heat-sensitive phenotype of this mutant, suggesting that STA1 affects broad transcriptional regulation other than that of the HsfA3 regulon [27]. Thus, knowledge of the regulatory mechanism underlying premature mRNA splicing under environmental stress conditions in plants is still fragmentary.
In this study, to elucidate the mechanisms of L-heat stress responses, we used a forward genetic screen for sloh mutants and isolated sloh3 and sloh63. The causal genes both encoded splicing-related factors identified as components of the MOS4-associated complex (MAC), which is widely conserved in eukaryotes and has been suggested to interact with spliceosomes. Under L-heat stress, sloh3 and sloh63 exhibited some abnormal splicing events and enhanced ER stress responses. Our findings suggest that maintenance of precise mRNA splicing under L-heat stress is important for L-heat tolerance and suppressing ER stress in Arabidopsis.
Results
Isolation and characterization of sloh3 and sloh63 mutants
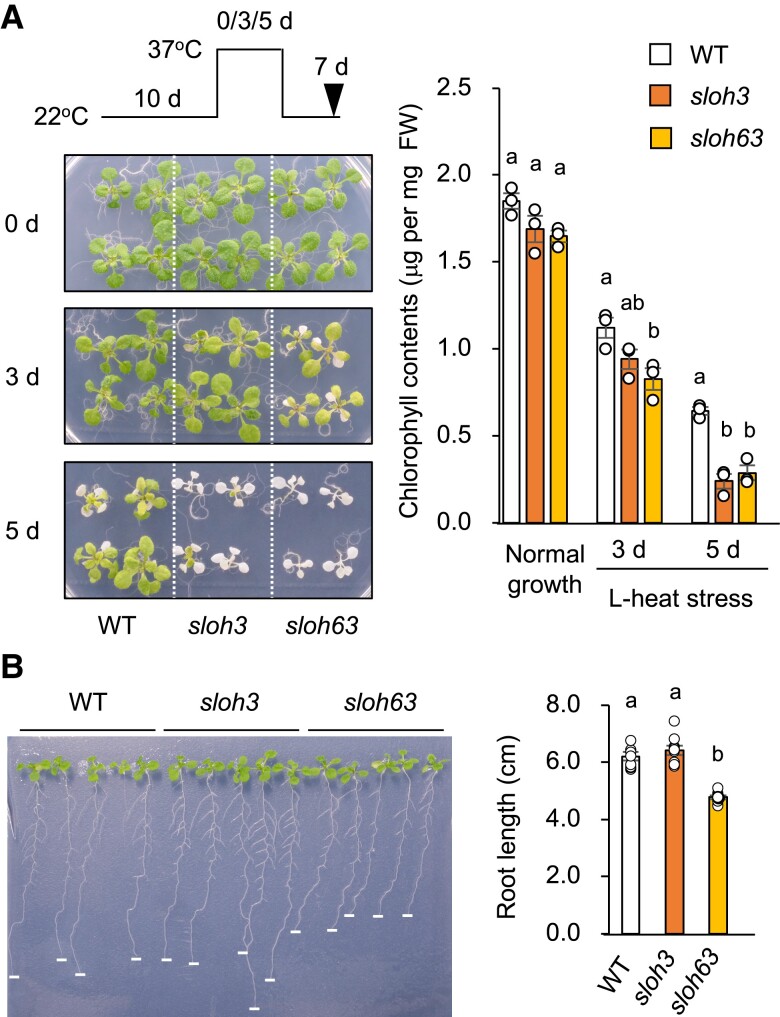
When 10-day-old seedlings were exposed to 37°C for 3 days, A. thaliana Col-0 WT did not show significant growth inhibition (Fig. 1A). We generated an EMS-mutagenized Col-0 population and screened 3,000 M2 seedlings for sloh mutants, which exhibits growth inhibition for 3 days at 37°C. We isolated sloh3 and sloh63 as L-heat-sensitive mutants by the forward genetic screening (Fig. 1A). The sensitivity to L-heat stress was slightly and not significantly higher in sloh63 than in sloh3 (Fig. 1A). Roots were shorter in sloh63 than in WT or sloh3 (Fig. 1B).
Fig. 1.
Identification of the sloh3 and sloh63 mutants. A) L-heat tolerance of the mutants. Ten-day-old wild-type (WT) and mutant seedlings grown at 22°C were placed at 37°C for 0, 3, or 5 days, as indicated, and then returned to 22°C for another 7 days. Left: plant images at the time point indicated with an arrowhead. Right: chlorophyll contents. B) Root length. Seedlings were grown vertically at 22°C for 14 days. Data are mean ± SE, n = 3. The same letters above columns indicate no significant difference within a time point in one-way ANOVA with post hoc Tukey HSD test, P < 0.05.
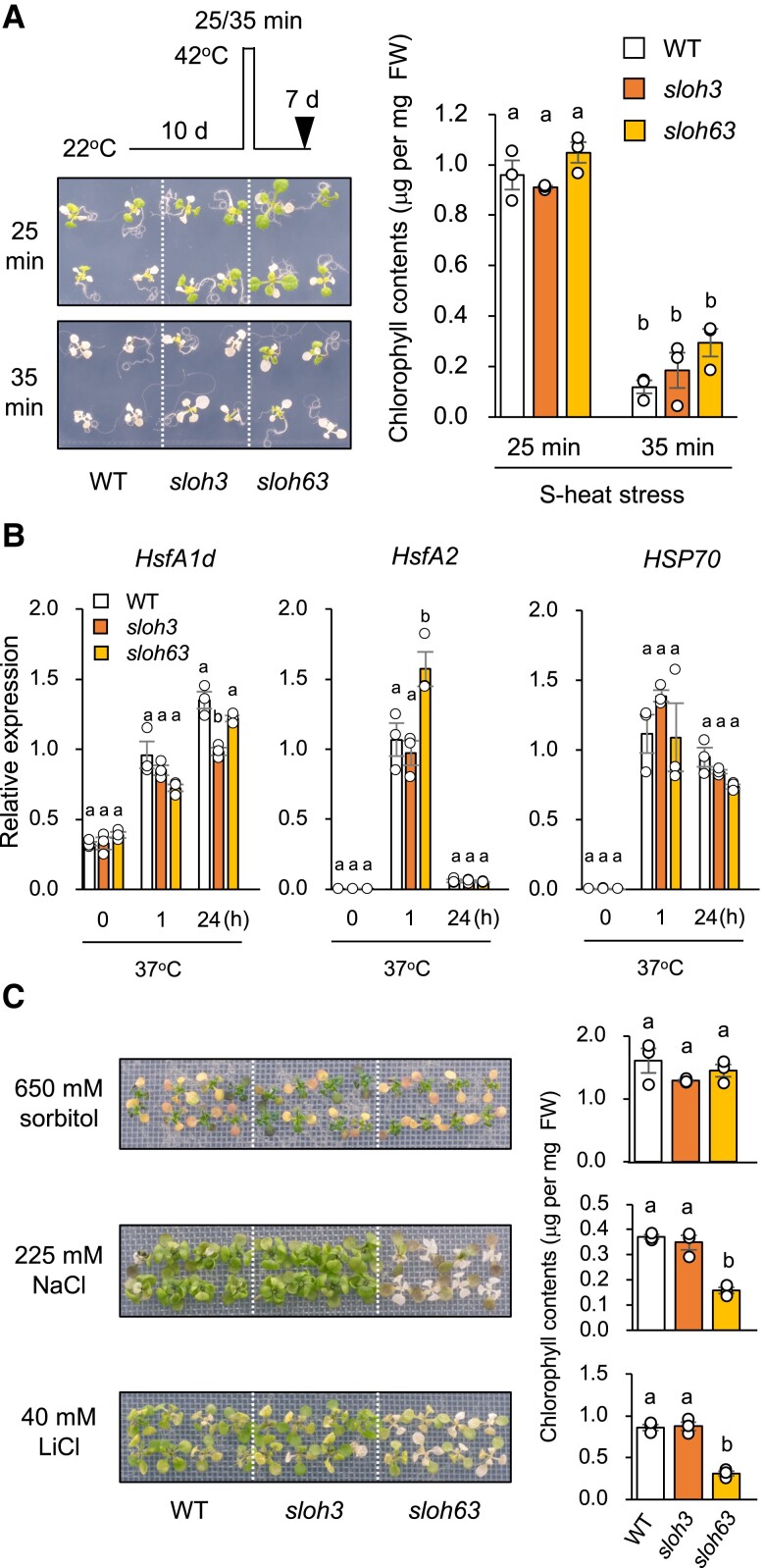
Both mutants did not differ significantly from WT in their tolerance to S-heat stress or to S-heat stress after heat acclimation (Figs. 2A, S1) or in the expression profiles of HsfA1d (except in sloh3 at 24 h), HsfA2 (except in sloh63 at 1 h), and HSP70 under heat stress (Fig. 2B). Sensitivity to NaCl and LiCl stress (Li+ is more toxic than Na+) was higher in sloh63 than in WT and sloh3, but osmo-tolerance did not differ significantly among them (Fig. 2C). These results suggest that sloh3 is specifically defective in L-heat stress response, and that sloh63 is defective in L-heat stress response, salt stress response, and root elongation.
Fig. 2.
Characterization of the sloh3 and sloh63 mutants. A) S-heat tolerance. Ten-day-old wild-type (WT) and mutant seedlings grown at 22°C (normal conditions) were placed at 42°C for 25 or 35 min and returned to 22°C for 7 days. Left: plant images. Right: chlorophyll contents (mean ± SE, n = 3). B) Expression of HsfA1d, HsfA2, and HSP70 under normal and heat-stress conditions; expression relative to ACTIN2 was determined by qRT-PCR. Ten-day-old seedlings grown at 22°C (0 h) were placed at 37°C for 1 or 24 h. C) Osmotic, NaCl, and LiCl stress tolerance. Ten-day-old seedlings grown at 22°C were mesh-transferred onto MS agar plates containing 650 mM sorbitol for 21 days (osmotic tolerance), 225 mM NaCl for 11 days (NaCl tolerance), or 40 mM LiCl for 8 days (LiCl tolerance). Left: plant images. Right: chlorophyll content. Data are mean ± SE, n = 3. The same letters above columns indicate no significant difference within a time point in one-way ANOVA with post hoc Tukey HSD test, P < 0.05.
Identification of the causal genes of sloh3 and sloh63
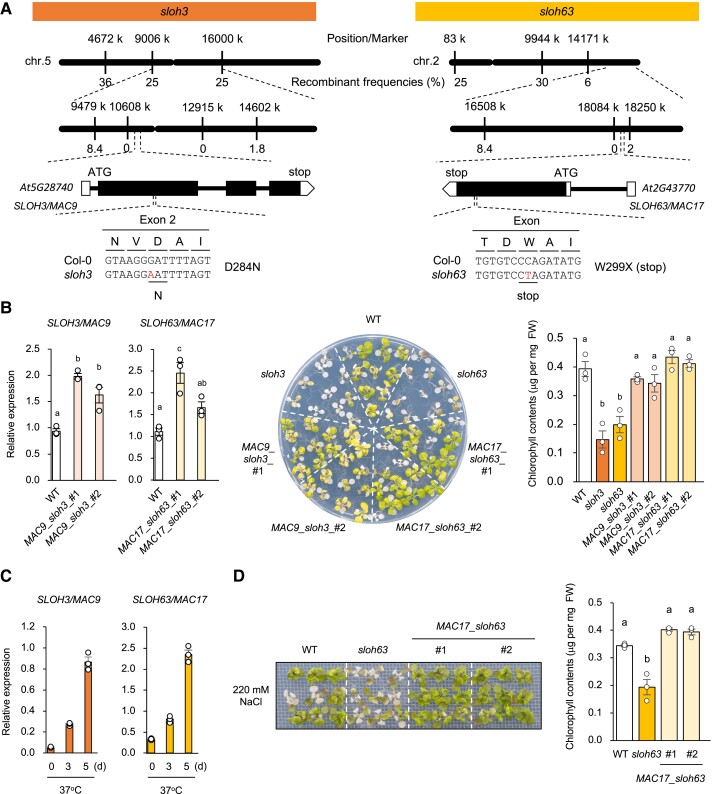
To identify the causal genes of sloh3 and sloh63, each mutant was crossed with Da (1)-12, an Arabidopsis accession moderately tolerant to L-heat. Using F2 progeny, we mapped the causal loci near the simple-sequence-length polymorphism (SSLP) markers at 10,608 kb in the middle of chromosome 5 for sloh3 and at 18,084 kb close to the distal end of the long arm of chromosome 2 for sloh63 (Fig. 3A). To detect all mutations within the mapped regions, we sequenced the sloh3 and sloh63 genomes and found only one nonsynonymous mutation in the coding region of each mutant: a mutation in the second exon of At5g28740 led to a D284N substitution in sloh3, and a nonsense mutation in the exon of At2g43770 led to W299X (X, stop codon) and a truncated protein in sloh63 (Fig. 3A). To perform a complementation test, we transformed sloh3 and sloh63 with At5g22350 and At2g43770, respectively, including its native promoter region. In each complementation line, the expression level was about 2-fold higher than that of the WT, which is reasonable as the sum of the expression of the native gene-derived expression and that of the transgene (Fig. 3B). The introduction of At5g28740 into sloh3 and At2g43770 into sloh63 restored the L-heat tolerance to that of WT, indicating that these genes were the causal genes of interest (Fig. 3B). To understand whether SLOH3 and SLOH63 are involved in the L-heat stress response, we examined their expression profiles under L-heat stress. The levels of both transcripts were increased by L-heat stress in a time-dependent manner (Fig. 3C). In our previous studies, defects in ER-Golgi transport and very-long-chain fatty acid synthesis impaired not only L-heat tolerance but also other abiotic stress tolerances such as salt and osmotic stress [9, 28]. This suggests that the mechanism for L-heat tolerance in Arabidopsis is related to other stress tolerance mechanisms. To determine whether the sloh3 and sloh63 phenotypes are specific to L-heat tolerance, we evaluated tolerance to osmotic (sorbitol) and salinity (NaCl and LiCl) stresses. SLOH63 also complemented the NaCl and LiCl hypersensitivity and shorter roots of sloh63, suggesting that the mutation in this gene is responsible for these phenotypes (Figs. 3D, S2, S3).
Fig. 3.
Identification of the causal genes of sloh3 and sloh63. A) High-resolution mapping of the causal loci. Mutations are shown in red. B) L-heat tolerance of wild-type (WT), sloh3, sloh63, sloh3 lines complemented by transformation with MAC9 (MAC9_sloh3_#1, 2), and sloh63 lines complemented by transformation with MAC17 (MAC17_sloh63_#1, 2). Left: Relative expression of SLOH3/MAC9/At5g22350 or SLOH63/MAC17/At2g43770 in the complementation lines under normal conditions; expression levels were determined by quantitative real-time PCR relative to those of Actin2 (mean ± SE, n = 3). Middle: plant images. Right: Chlorophyll contents of WT, mutants, and complemented lines. C) Expression of SLOH3/MAC9 and SLOH63/MAC17 in WT under L-heat stress. ACTIN2 was used as an internal standard. D) NaCl tolerance of WT, sloh63, and sloh63 lines complemented by transformation with MAC17 (MAC17_ sloh63_#1, 2). Left: plant images. Right: chlorophyll contents. B–D) Data are mean ± SE, n = 3. The same letters above columns indicate no significant difference within a time point in one-way ANOVA with post hoc Tukey HSD test, P < 0.05.
Effects of SLOH3 and SLOH63 on transcriptional regulation
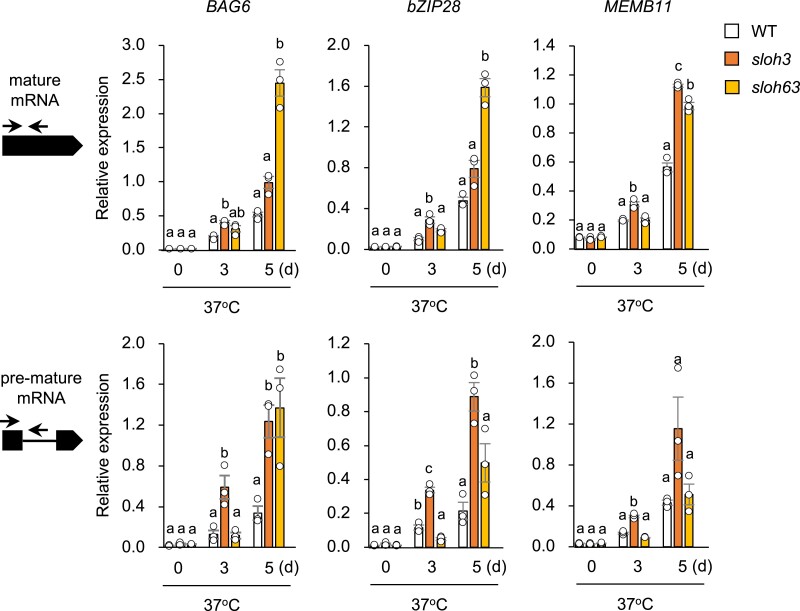
SLOH3 and SLOH63 were identical to MOS4-ASSOCIATED COMPLEX 9 and 17 (MAC9 and MAC17), respectively, which encode an RNA-binding proteins with an RNA helicase domain and an intron-binding domain. MAC proteins are widely conserved in eukaryotes and interact with spliceosomes [22, 29, 30]. MAC9 and MAC17 regulate the expression levels of plant disease response genes and are involved in miRNA biosynthesis and mRNA splicing, but their involvement in responses to heat and other abiotic stresses is unknown. As both genes encode RNA-binding proteins, the loss of their function may affect alternative splicing under L-heat stress. To test this hypothesis, we performed qRT-PCR of the BAG6, bZIP28, and MEMB11 genes, which retain their introns under heat stress [28]. All three genes had higher premature mRNA levels in sloh3 and sloh63 than in WT under L-heat stress (Fig. 4); sloh3 showed the earliest increase in premature mRNA level under L-heat stress.
Fig. 4.
Detection of retained introns in heat-responsive genes under L-heat stress in sloh3 and sloh63. Expression of mature mRNAs (upper panels) and premature mRNAs (lower panels) of BAG6, bZIP28, and MEMB11 was measured in wild type (WT), sloh3, and sloh63 Ten-day-old seedlings grown at 22°C (0 day) were placed at 37°C for 3 or 5 days. Data are mean ± SE, n = 3. The same letters above columns indicate no significant difference within a time point in one-way ANOVA with Tukey test, P < 0.05.
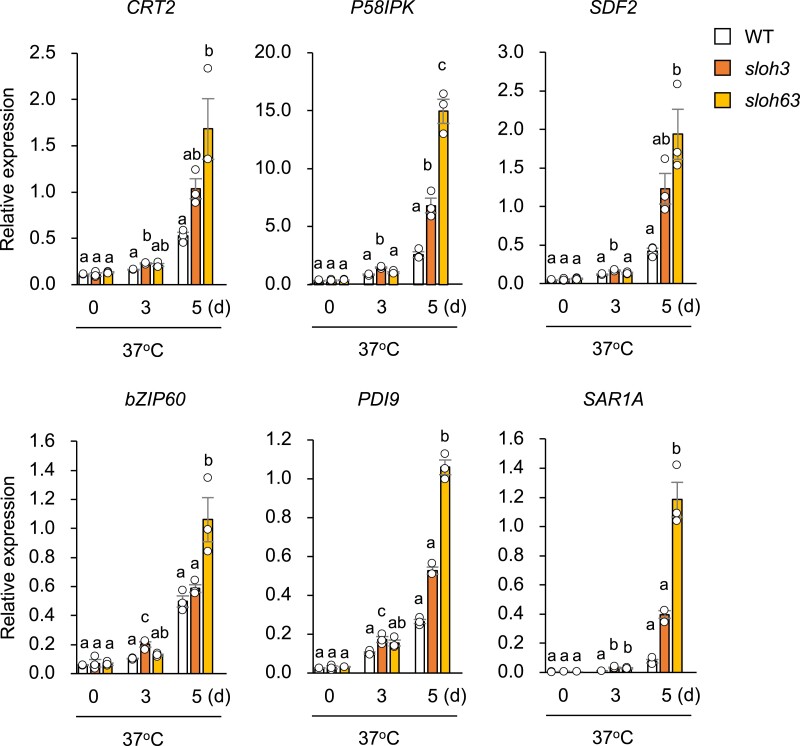
The mature mRNA level of bZIP28, a key activator of the ER stress response [31], was higher in sloh63 than in WT and sloh3 under L-heat stress (Fig. 4). Defects in ER-to-Golgi transport and biosynthesis of very-long-chain fatty acids, which enhance ER stress, impair not only L-heat tolerance but also salt tolerance in Arabidopsis [9, 28]. To examine differences in the ER stress response among WT and mutants under normal and L-heat stress conditions, we examined the transcript levels of bZIP28-regulated genes (CRT2, P58IPK, and SDF2) and other key activators of the ER stress response (bZIP60 and bZIP60-regulated genes PDI9 and SAR1A) [32]. The transcript levels of these genes were highest at day 3 of heat stress in sloh3 and at day 5 in sloh63 (Fig. 5). bZIP60 induces the UPR triggered by splicing of bZIP60 [33, 34]. The transcript level of the spliced (active) bZIP60 form detected by RT-PCR was only slightly higher in sloh3 and sloh63 than in WT under L-heat stress (Fig. S2).
Fig. 5.
ER stress responses in sloh3 or sloh63 under L-heat stress. Expression levels of genes regulated by bZIP28 (CRT2, P58IPK, and SDF2), genes regulated by bZIP60 (PDI9 and SAR1A) and bZIP60 were measured in 10-day-old wild-type (WT) and mutant seedlings grown at 22°C (0 day) and then placed at 37°C for 3 or 5 days. Data are mean ± SE, n = 3. The same letters above columns indicate no significant difference within a time point in one-way ANOVA with Tukey test, P < 0.05.
Effects of a splicing inhibitor on L-heat stress tolerance and ER stress response
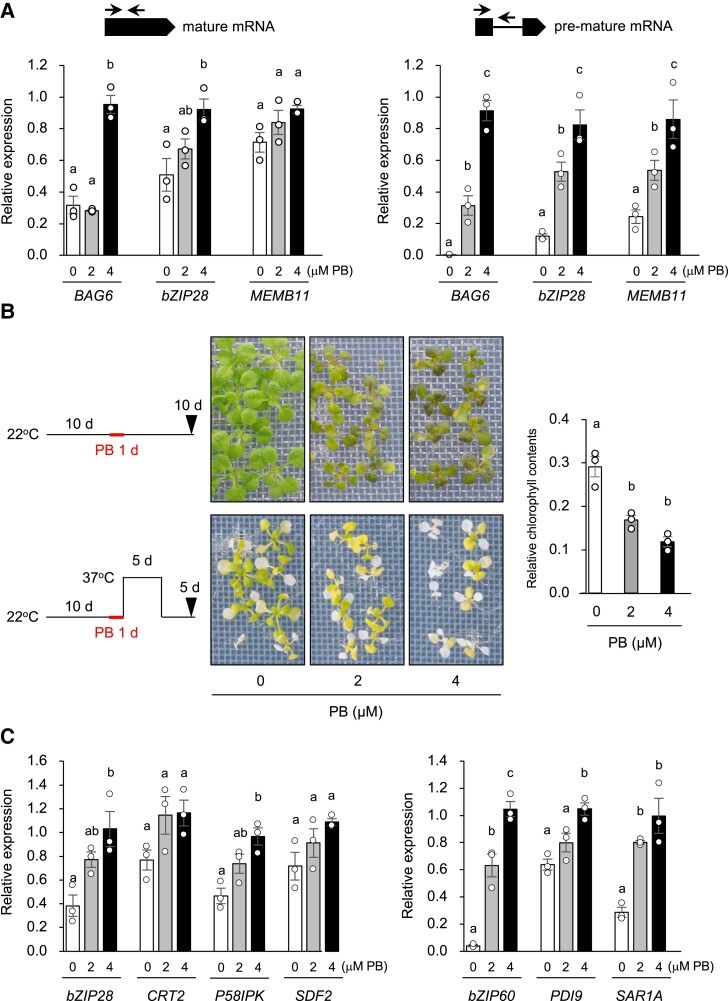
To determine whether disturbing splicing machinery impairs L-heat tolerance and whether an increase in premature mRNAs induces ER stress, we evaluated the effects of a splicing inhibitor on L-heat tolerance and ER stress. Pladienolide B (PB) targets the splicing factor SF3b in humans, an ortholog of MAC18 in Arabidopsis, and inhibits constitutive and alternative splicing [35, 36]. The levels of premature mRNAs of BAG6, bZIP28, and MEMB11 and mature mRNA of BAG6 and bZIP28 were increased by PB in a concentration-dependent manner (Fig. 6A). We treated WT plants with PB for 1 day and subjected them to L-heat stress for 5 days and found that L-heat tolerance was impaired by PB (Fig. 6B). To determine whether ER stress is induced by splicing inhibition, we analyzed the expression levels of ER stress response genes in PB-treated WT plants. The transcript levels of all six genes tested were upregulated by PB; bZIP60 and PDI9 were upregulated in a PB concentration-dependent manner (Fig. 6C). These findings suggest that inhibition of splicing induces ER stress.
Fig. 6.
Effects of splicing inhibitor on alternative splicing, L-heat tolerance, and ER stress response in wild-type (WT) seedlings. A) Effects of pladienolide B (PB) treatment on alternative splicing. Expression of mature and premature mRNAs of BAG6, bZIP28, and MEMB11 was measured in 10-day-old seedlings grown at 22°C and then mesh-transferred onto water plates containing 2 or 4 μM PB for 1 day at 22°C. B) Effects of PB treatment on L-heat tolerance. Seedlings were grown and treated with PB as in (A), subjected to L-heat stress for 5 days, and then grown at 22°C for 5 days; control plants were grown for 10 days at 22°C after PB treatment. Left: plant images. Right: chlorophyll contents relative to those in control seedlings. Data are mean ± SE, n = 3. C) Effects of PB treatment on ER stress and the following unfolded protein response. Expression levels of genes regulated by bZIP28 (CRT2, P58IPK, and SDF2) or bZIP60 (PDI9 and SAR1A) in seedlings grown and treated with PB as in (A). All data are mean ± SE, n = 3. The same letters above columns indicate no significant difference within a time point in one-way ANOVA with Tukey test, P < 0.05.
Effects of L-heat stress on gene expression in sloh3 and sloh63
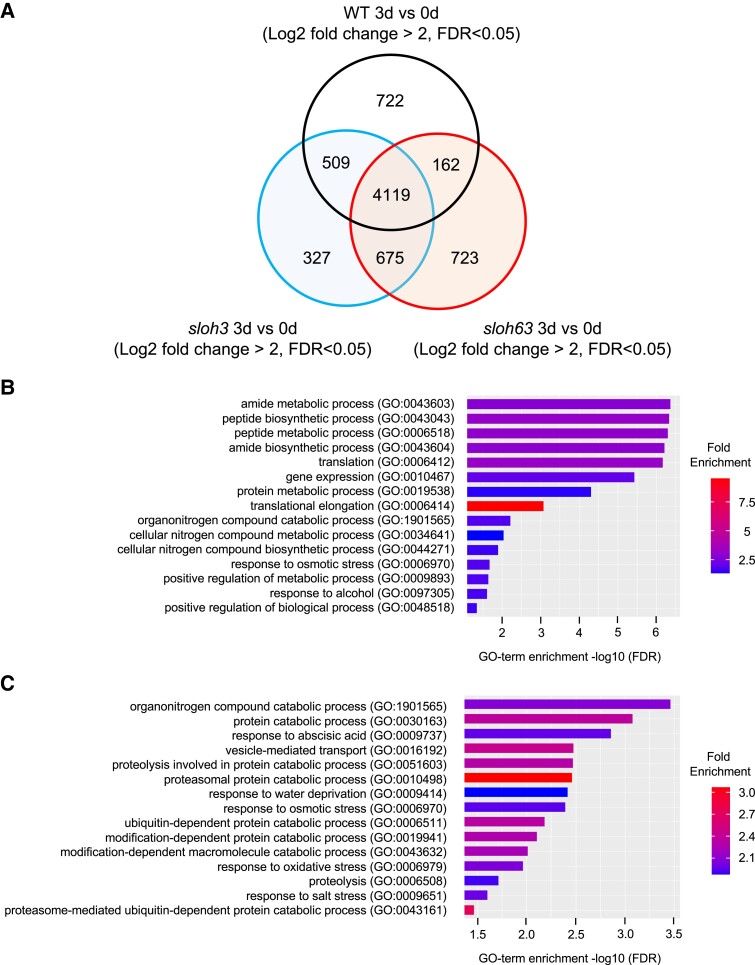
To investigate the mechanisms underlying the heat sensitive phenotype of sloh3 and sloh63, the RNAseq analysis was performed on WT, sloh3 and sloh63 seedlings under normal or heat stress (37°C 3 day) conditions. The differentially expressed genes (DEGs) between normal and heat stress conditions in WT, sloh3 and sloh63 were identified and classified them as shown in the Venn diagram in Fig. 7 (Fig. 7A, Table S1). To characterize DEGs in sloh3 and sloh63, a GENEONTOLOGY (GO) enrichment analysis was performed for sloh3-specific, sloh63-specific, or sloh3 and sloh63 common DEGs (Fig. 7B, C). As a result, no significant GO terms were detected for sloh3-specific DEGs, but significant GO terms were successfully obtained for sloh63-specific or sloh3 and sloh63 common DEGs. GO terms related to protein, peptide, and amide metabolism were enriched for the sloh3 and sloh63 common DEGs (Fig. 7B). It is known that abnormal splicing results in ER stress due to the accumulation of abnormal proteins, which in turn results in their degradation [37–40]. Both sloh3 and sloh63 mutants appear to promote a response that degrades abnormal proteins under L-heat stress. On the other hand, for sloh63 specific DEGs, vesicle-mediated transport related to ER stress response and GO terms related to osmotic and salt stress response were enriched (Fig. 7C). As shown in Fig. 5, a significantly enhanced ER stress response was observed in sloh63 compared to WT and sloh3. Furthermore, consistent with the finding that sloh63 was highly sensitive to salt stress (Figs 2C and 3D), the GO terms for osmotic and salt stress responses were enriched, suggesting that the defect of SLOH63/MAC17 affects not only L-heat stress but also salt stress responses.
Fig. 7.
Effects of L-heat stress on gene expression in sloh3 and sloh6. A) Venn diagrams show the number of differentially expressed genes (DEG) between normal and L-heat stress (37°C for 3 days) conditions (Log2 fold change > 2, FDR < 0.05) in WT, sloh3 and sloh63. B, C) Applying GO enrichment analysis to 675 common DEGs in sloh3 and sloh63 (B) and 723 specific DEGs in sloh63 (C) at 3 days after heat stress for top 15 biological processes based on positive fold enrichment. The DEGs were classified according to the GO terms in GENEONTOLOGY software (https://geneontology.org/) into categories based on biological processes. The colour represents the fold enrichment of the DEGs.
Discussion
Using mutants highly sensitive to L-heat stress, we here isolated the causal genes of sloh3 and sloh63 and identified them as MAC9 and MAC17, respectively. Both genes are involved in mRNA splicing. In sloh3, a possible nonsynonymous substitution was found in the half-a tetratricopeptide (HAT) domain. This domain is found in proteins involved in RNA processing and contributes to protein–protein interactions. Mutations in the HAT domain affect RNA processing [41, 42]. The mutation in SLOH3/MAC9 resulted in the loss of function. In sloh63, a possible nonsense mutation in the WD40 domain, which is involved in protein–protein interactions [43], resulted in a truncated protein with one WD40 domain missing. In these mutants, splicing was disturbed and ER stress under L-heat stress was increased; this may explain their L-heat stress sensitivity. These findings suggest that maintenance of appropriate mRNA splicing and suppression of ER stress under L-heat stress are important for stress tolerance.
The sloh63 mutant, but not the sloh3 mutant, was significantly more sensitive to salt stress than WT. We have reported that enhanced ER stress impairs not only L-heat tolerance but also salt tolerance [9]. The ER stress was enhanced more in sloh63 than in sloh3, which may impair the salt tolerance of sloh63. Both genes encode MAC proteins, which are involved in splicing, and sloh63 may contribute to splicing that more specifically affects ER stress responses. Alternatively, sloh63 may contribute to splicing broadly, as it also confers slightly higher sensitivity to L-heat stress than does sloh3. In the sloh63 mutant, root elongation was inhibited. The lack of MAC5A, a member of the same MAC protein complex as SLOH63/MAC17, inhibits root elongation [44]. The mac5a mutant shows a hypersensitive phenotype in response to a DNA damage inducer, methyl methanesulfonate, suggesting that MAC5 is important in maintaining the proper response to DNA damage that regulates plant growth and stress adaptation [44]. SLOH63 may also play an important role in orchestrating growth and adaptation to L-heat and salt stresses.
An increase in the levels of premature mRNAs and expression of ER stress-responsive genes was observed in WT plants under L-heat stress (Figs 4 and 5). Premature mRNA levels and expression of ER-stress marker genes were increased in a PB concentration-dependent manner. Disturbance of splicing such as intron retention caused by L-heat stress may result in the production of structurally abnormal proteins, which in turn may induce ER stress.
We found a single locus responsible for the difference in L-heat tolerance between Ms-0 and Col-0, which we named Long-term Heat Tolerance1 (LHT1) (Isono et al., submitted as a back-to-back article). It is identical to MAC7 and encodes a putative RNA helicase involved in mRNA splicing as a component of MAC. The mos4-2, cdc5-1, mac3a mac3b, and prl1 prl2 mutants, which are deficient in core components of MAC, are hypersensitive to L-heat stress in comparison with WT, suggesting that MAC proteins are important in L-heat stress responses. Interestingly, LHT1/MAC7 interacts with SLOH3/MAC9 and SLOH63/MAC17 [45]. Both quantitative trait locus (QTL) analysis using natural variation in L-heat tolerance (Isono et al., submitted as a back-to-back article) and the forward genetic screening of L-heat-sensitive mutants revealed the importance of proper alternative splicing under L-heat stress in Arabidopsis. An LHT1 ortholog, EMB4, was also found to contribute to heat tolerance of Caenorhabditis elegans (Sato et al., submitted as a back-to-back article). These findings indicate that maintenance of proper splicing under heat stress is a common high-temperature adaptation mechanism in plants and animals.
Materials and methods
Plant materials and growth conditions
Col-0 seeds were sown on plates (90 mm × 20 mm; BIO-BIK, Osaka, Japan) with 1% (w/v) agar containing full-strength Murashige and Skoog (MS) salts, vitamin mixture (10 mg L−1 myoinositol, 200 µg L−1 glycine, 50 µg L−1 nicotinic acid, 50 µg L−1 pyridoxine hydrochloride, 10 µg L−1 thiamine hydrochloride, pH 5.7), and 1% sucrose. The plates were sealed with surgical tape. The seeds were stratified at 4°C for 4–7 days and then transferred to a growth chamber (80 µmol photons m−2 s−1; 16/8-h light/dark cycle; 22°C) for germination and growth. Seeds of A. thaliana mac5a (SALK_132881), mac11 (SALK_122548), mac13 (SALK_032761), and mac14 (SAIL_873_E04) mutants and of the L-heat-stress-tolerant accession Da (1)-12 (CS917) were obtained from the Arabidopsis Biological Resource Center (Ohio State University).
Mutagenesis and mutant screening
Seedlings were mutagenized according to the web site http://cocoabiotech.koko.gov.my/Mutagenesis5.html. For mutant screening, 10-day-old seedlings grown at 22°C were transferred to 37°C for 5 days and then to 22°C. We isolated candidate mutants on the basis of their sensitivity to L-heat stress (37°C for 5 days) and selected sloh3 and sloh63 for further characterization.
Genetic mapping of the causative genes of sloh3 and sloh63
The sloh3 and sloh63 mutants were crossed with Da (1)-12, and the F1 progeny were self-fertilized to generate F2 populations. Genomic DNA was prepared from individual F2 plants with the recessive L-heat-sensitive phenotype for use as PCR templates. The SSLP markers listed in Table S2 were used for mapping. PCR conditions were as follows: 94°C for 2 min; 35 cycles of 94°C for 15 s, 58–60°C (depending on primers) for 15 s, and 72°C for 10 s; and a final 72°C for 2 min. Microsatellites were fractionated in 5–7% agarose gels, and the recombination value was calculated from the band pattern.
DNA library construction and sequencing
DNA library construction was performed as described in Isono et al. [9].
All libraries were diluted to 10 nM, mixed in equal amounts, and subjected to 1 × 100-bp single-read sequencing on an Illumina HiSeq 2500 platform (Illumina, San Diego, CA, USA). Reads in FASTQ format were generated in bcl2fastq2 Conversion v. 2.18 software (Illumina). The read data were submitted to the DDBJ Read Archive (accession number DRA015832).
Detection of mutations
Mutations were detected as described [14].
Plasmid construction and plant transformation
For complementation tests, the genomic region of At5g28740 or At2g43770 (1 kb upstream of the ATG initiation codon to 1 kb downstream of the terminator) was amplified by PCR with the SLOH3/MAC9 or SLOH63/MAC17 primers (Table S2) and cloned into the pGH35S-mGFP vector. The plant transformation and subsequent selection of transgenic plants were performed as described in Isono et al. [9].
Abiotic stress assays
Seeds were sown on agar plates containing 20 mL MS medium at 22°C. For the L-heat-tolerance assay, plates with 10-day-old seedlings were moved to 37°C for 3 or 5 days and then back to 22°C for 7 days [9]. For the S-heat-tolerance assay, plates with 10-day-old seedlings were placed into a water bath at 42°C for 25 or 35 min and then moved back to 22°C for 7 days. For the S-heat stress after heat acclimation, plates with 10-day-old seedlings were placed at 37°C for 1 h, moved back to 22°C for 2 h, placed into a water bath at 42°C for 150 or 180 min, and moved back to 22°C for 7 days. For the osmotic, NaCl, or LiCl stress assay, 10-day-old seedlings grown on nylon mesh on MS agar plates at 22°C were mesh-transferred to a plate supplemented with 650 mM sorbitol for 21 days; 220 mM NaCl for 7 days or 225 mM NaCl for 11 days; or 30 mM LiCl for 10 days or 40 mM LiCl for 8 days. Chlorophyll content was determined as in [46].
RNA extraction and RT-qPCR or RT-PCR
Total RNA was isolated with a RNeasy Plant Mini Kit (Qiagen, Hilden, Germany) and used as a template to synthesize first-strand cDNA with a ReverTra ACE kit (Toyobo, Osaka, Japan). qRT-PCR was performed in a Thermal Cycler Dice Real Time System III (Takara Bio, Shiga, Japan) with Thunderbird SYBR qPCR Mix (Toyobo) in a total volume of 20 μL as follows: 95°C for 30 s, 60°C for 10 s, and 72°C for 10 s; 50 cycles of 95°C for 10 s, 60°C for 15 s, and 72°C for 30 s; and a final 95°C for 10 s, 65°C for 1 min, and 97°C for 1 s. ACTIN2 was used as an internal standard for qRT-PCR. Primers are listed in Table S2.
PB assay
Seeds were sown on nylon mesh on MS agar plates and placed at 22°C; 10-day-old seedlings grown on nylon mesh on MS agar plates were mesh-transferred to water containing 2 μM or 4 μM pladienolide B dissolved in 0.01% DMSO (Funakoshi, Japan) or 0.01% DMSO (vehicle) for 1 day. Plants were transferred onto MS agar plates, held at 37°C for 5 days, and moved back to 22°C for 5 days.
cDNA library construction and RNA sequencing
Total RNA was isolated with RNAiso reagent (Takara) from 10-day-old seedlings under normal growth (22°C) and heat stress (37°C for 3 days) conditions. All other procedures were performed according to Enoki et al. [47]. The read data were submitted to the DDBJ Read Archive.
GO enrichment analysis
The DEGs between normal and L-heat stress (37°C for 3 days) conditions (Log2 fold change > 2, FDR < 0.05) in WT, sloh3 and sloh63 were applied to GENEONTOLOGY software (https://geneontology.org/) into categories based on biological processes to obtain the GO terms. The obtained GO terms were ordered in descending order for the top 15 biological processes based on positive fold enrichment. These top 15 GO terms are then arranged in descending order of the FDR.
Supplementary Material
Acknowledgments
The authors thank Mr. Seitaro Ito of Tokyo University of Agriculture for assisting in physiological analyses.
Contributor Information
Naoya Endo, Department of Bioscience, Tokyo University of Agriculture, 1-1-1 Sakuragaoka, Setagaya, Tokyo 156-8502, Japan.
Ryo Tsukimoto, Department of Bioscience, Tokyo University of Agriculture, 1-1-1 Sakuragaoka, Setagaya, Tokyo 156-8502, Japan.
Kazuho Isono, Department of Bioscience, Tokyo University of Agriculture, 1-1-1 Sakuragaoka, Setagaya, Tokyo 156-8502, Japan.
Akito Hosoi, NODAI Genome Research Center, Tokyo University of Agriculture, 1-1-1 Sakuragaoka, Setagaya, Tokyo 156-8502, Japan.
Ryo Yamaguchi, Department of Bioscience, Tokyo University of Agriculture, 1-1-1 Sakuragaoka, Setagaya, Tokyo 156-8502, Japan.
Keisuke Tanaka, NODAI Genome Research Center, Tokyo University of Agriculture, 1-1-1 Sakuragaoka, Setagaya, Tokyo 156-8502, Japan.
Satoshi Iuchi, RIKEN BioResource Research Center, 3-1-1 Koyadai, Tsukuba, Ibaraki 305-0074, Japan.
Izumi Yotsui, Department of Bioscience, Tokyo University of Agriculture, 1-1-1 Sakuragaoka, Setagaya, Tokyo 156-8502, Japan.
Yoichi Sakata, Department of Bioscience, Tokyo University of Agriculture, 1-1-1 Sakuragaoka, Setagaya, Tokyo 156-8502, Japan.
Teruaki Taji, Department of Bioscience, Tokyo University of Agriculture, 1-1-1 Sakuragaoka, Setagaya, Tokyo 156-8502, Japan.
Supplementary Material
Supplementary material is available at PNAS Nexus online.
Funding
Research funded by Japan Society for the Promotion of Science (21H05668, 23H04206, and 23H00334 to T.T.).
Author contributions
N.E. and T.T. conceived, initiated, and coordinated the project; R.T. and K.I. isolated the sloh3 and sloh63 mutants; N.E. identified the causal genes of these mutants; N.E. performed physiological analyses; A.H. and R.Y. performed the RNAseq analysis; K.T. performed genome sequencing; N.E. detected alternative splicing and ER stress responses under PB treatment; T.T. wrote the manuscript with assistance from I.Y. and Y.S.
Data availability
All data that support the findings of this study are available within its Supplementary Materials.
References
- 1. Lobell DB, Schlenker W, Costa-Roberts J. 2011. Climate trends and global crop production since 1980. Science. 333:616–620. [DOI] [PubMed] [Google Scholar]
- 2. Yoshida T, et al. 2011. Arabidopsis HsfA1 transcription factors function as the main positive regulators in heat shock-responsive gene expression. Mol Genet Genomics. 256:321–332. [DOI] [PubMed] [Google Scholar]
- 3. Ohama N, Sato H, Shinozaki K, Yamaguchi-Shinozaki K. 2017. Transcriptional regulatory network of plant heat stress response. Trends Plant Sci. 22:53–65. [DOI] [PubMed] [Google Scholar]
- 4. Liu H-C, Liao H-T, Charng Y-Y. 2011. The role of class A1 heat shock factors (HSFA1s) in response to heat and other stresses in Arabidopsis. Plant Cell Environ. 34:738–751. [DOI] [PubMed] [Google Scholar]
- 5. Higashi Y, et al. 2013. Hsfa1d, a protein identified via FOX hunting using thellungiella salsuginea cDNAs improves heat tolerance by regulating heat-stress-responsive gene expression. Mol Plant. 6:411–422. [DOI] [PubMed] [Google Scholar]
- 6. Charng Y, et al. 2006. A heat-inducible transcription factor, HsfA2, is required for extension of acquired thermotolerance in Arabidopsis. Plant Physiol. 143:251–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Friedrich T, et al. 2021. Heteromeric HSFA2/HSFA3 complexes drive transcriptional memory after heat stress in Arabidopsis. Nat Commun. 12:3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yamaguchi N, et al. 2021. H3k27me3 demethylases alter HSP22 and HSP17.6C expression in response to recurring heat in Arabidopsis. Nat Commun. 12:3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Isono K, et al. 2020. An ER–Golgi tethering factor SLOH4/MIP3 is involved in long-term heat tolerance of Arabidopsis. Plant Cell Physiol. 62:272–279. [DOI] [PubMed] [Google Scholar]
- 10. Lee C-F, et al. 2006. Mutation in a homolog of yeast Vps53p accounts for the heat and osmotic hypersensitive phenotypes in Arabidopsis hit1-1 mutant. Planta. 224:330–338. [DOI] [PubMed] [Google Scholar]
- 11. Wu S-J, Wang L-C, Yeh C-H, Lu C-A, Wu S-J. 2010. Isolation and characterization of the Arabidopsis heat-intolerant 2 (hit2) mutant reveal the essential role of the nuclear export receptor EXPORTIN1A (XPO1A) in plant heat tolerance. New Phytol. 186:833–842. [DOI] [PubMed] [Google Scholar]
- 12. Wang L-C, et al. 2011. Involvement of the Arabidopsis HIT1/AtVPS53 tethering protein homologue in the acclimation of the plasma membrane to heat stress. J Exp Bot. 62:3609–3620. [DOI] [PubMed] [Google Scholar]
- 13. Wang L-C, et al. 2013. Arabidopsis HIT4 encodes a novel chromocentre-localized protein involved in the heat reactivation of transcriptionally silent loci and is essential for heat tolerance in plants. J Exp Bot. 64:1689–1701. [DOI] [PubMed] [Google Scholar]
- 14. Tsukimoto R, et al. 2021. Mitochondrial fission complex is required for long-term heat tolerance of Arabidopsis. Plant Cell Physiol. 63:296–304. [DOI] [PubMed] [Google Scholar]
- 15. Dikaya V, et al. 2021. Insights into the role of alternative splicing in plant temperature response. J Exp Bot. 72:7384–7403. 10.1093/jxb/erab234. [DOI] [PubMed] [Google Scholar]
- 16. Chaudhary S, et al. 2019. Alternative splicing and protein diversity: plants versus animals. Front Plant Sci. 10:708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Reddy ASN, Ali GS. 2011. Plant serine/arginine-rich proteins: roles in precursor messenger RNA splicing, plant development, and stress responses. Wiley Interdiscip Rev RNA. 2:875–889. [DOI] [PubMed] [Google Scholar]
- 18. Staiger D, Brown JWS. 2013. Alternative splicing at the intersection of biological timing, development, and stress responses. Plant Cell. 25:3640–3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Filichkin SA, et al. 2015. Environmental stresses modulate abundance and timing of alternatively spliced circadian transcripts in Arabidopsis. Mol Plant. 8:207–227. [DOI] [PubMed] [Google Scholar]
- 20. Wahl MC, Will CL, Lührmann R. 2009. The spliceosome: design principles of a dynamic RNP machine. Cell. 136:701–718. [DOI] [PubMed] [Google Scholar]
- 21. Veretnik S, Wills C, Youkharibache P, Valas RE, Bourne PE. 2009. Sm/Lsm genes provide a glimpse into the early evolution of the spliceosome. PLoS Comput Biol. 5:e1000315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Johnson K, Dong O, Li X. 2011. The evolutionarily conserved MOS4-associated complex. Open Life Sci. 6:776–784. [Google Scholar]
- 23. Chanarat S, Sträßer K. 2013. Splicing and beyond: the many faces of the Prp19 complex. Biochim Biophys Acta. 1833:2126–2134. [DOI] [PubMed] [Google Scholar]
- 24. Chan S-P, Cheng S-C. 2005. The Prp19-associated complex is required for specifying interactions of U5 and U6 with pre-mRNA during spliceosome activation. J Biol Chem. 280:31190–31199. [DOI] [PubMed] [Google Scholar]
- 25. Xiong L, et al. 2001. Modulation of abscisic acid signal transduction and biosynthesis by an Sm-like protein in Arabidopsis. Dev Cell. 1:771–781. [DOI] [PubMed] [Google Scholar]
- 26. Okamoto M, et al. 2016. Sm-like protein-mediated RNA metabolism is required for heat stress tolerance in Arabidopsis. Front Plant Sci. 7:1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kim G-D, Cho Y-H, Lee B-H, Yoo S-D. 2017. STABILIZED1 Modulates Pre-mRNA splicing for thermotolerance. Plant Physiol. 173:2370–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fukuda N, et al. 2022. ECERIFERUM 10 encoding an enoyl-CoA reductase plays a crucial role in osmotolerance and cuticular wax loading in Arabidopsis. Front Plant Sci. 13:898317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Monaghan J, et al. 2009. Two Prp19-like U-box proteins in the MOS4-associated complex play redundant roles in plant innate immunity. PLoS Pathog. 5:e1000526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Koncz C, deJong F, Villacorta N, Szakonyi D, Koncz Z. 2012. The spliceosome-activating complex: molecular mechanisms underlying the function of a pleiotropic regulator. Front Plant Sci. 3:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pastor-Cantizano N, Ko DK, Angelos E, Pu Y, Brandizzi F. 2020. Functional diversification of ER stress responses in Arabidopsis. Trends Biochem Sci. 45:123–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kim J-S, Yamaguchi-Shinozaki K, Shinozaki K. 2018. ER-anchored transcription factors bZIP17 and bZIP28 regulate root elongation. Plant Physiol. 176:2221–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Deng Y, et al. 2011. Heat induces the splicing by IRE1 of a mRNA encoding a transcription factor involved in the unfolded protein response in Arabidopsis. Proc Natl Acad Sci USA. 108:7247–7252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nagashima Y, et al. 2011. Arabidopsis IRE1 catalyses unconventional splicing of bZIP60 mRNA to produce the active transcription factor. Sci Rep. 1:519–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kotake Y, et al. 2007. Splicing factor SF3b as a target of the antitumor natural product pladienolide. Nat Chem Biol. 3:570–575. [DOI] [PubMed] [Google Scholar]
- 36. Ling Y, et al. 2017. Pre-mRNA splicing repression triggers abiotic stress signaling in plants. Plant J. 89:291–309. [DOI] [PubMed] [Google Scholar]
- 37. Smith MH, Ploegh HL, Weissman JS. 2011. Road to ruin: targeting proteins for degradation in the endoplasmic reticulum. Science. 334:1086–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu Y, et al. 2015. EBS7 is a plant-specific component of a highly conserved endoplasmic reticulum-associated degradation system in Arabidopsis. Proc Natl Acad Sci USA. 112:12205–12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Su K-H, et al. 2016. HSF1 Critically attunes proteotoxic stress sensing by mTORC1 to combat stress and promote growth. Nat Cell Biol. 18:527–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chhipi-Shrestha JK, et al. 2022. Splicing modulators elicit global translational repression by condensate-prone proteins translated from introns. Cell Chem Biol. 29:259–275.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Champion EA, Lane BH, Jackrel ME, Regan L, Baserga SJ. 2008. A direct interaction between the Utp6 half-a-tetratricopeptide repeat domain and a specific peptide in Utp21 is essential for efficient Pre-rRNA processing. Mol Cell Biol. 28:6547–6556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Champion EA, Kundrat L, Regan L, Baserga SJ. 2009. A structural model for the HAT domain of Utp6 incorporating bioinformatics and genetics. Protein Eng Des Sel. 22:431–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Stirnimann CU, Petsalaki E, Russell RB, Müller CW. 2010. WD40 proteins propel cellular networks. Trends Biochem Sci. 35:565–574. [DOI] [PubMed] [Google Scholar]
- 44. Meng X, et al. 2022. RNA-binding protein MAC5A interacts with the 26S proteasome to regulate DNA damage response in Arabidopsis. Plant Physiol. 191:446–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jia T, et al. 2017. The Arabidopsis MOS4-associated complex promotes microRNA biogenesis and precursor messenger RNA splicing. Plant Cell. 29:2626–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Porra RJ, Thompson WA, Kriedemann PE. 1989. Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim Biophys Acta. 975:384–394. [Google Scholar]
- 47. Enoki S, et al. 2018. Physiological characterization of leaf and internode after bud break in Japanese indigenous Koshu grape by comparative RNA sequencing analysis. PLoS One. 13:e0194807–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data that support the findings of this study are available within its Supplementary Materials.