Abstract
A series of modular mini-transposon derivatives which permit the rapid cloning and mapping of the DNA flanking the minitransposon’s site of insertion has been developed. The basic plasposon, named TnMod, consists of the Tn5 inverted repeats, a conditional origin of replication, rare restriction endonuclease multiple cloning sites, and exchangeable antibiotic resistance cassettes. The broad host range and low target DNA sequence specificity of the Tn5 transposase, in combination with the flexibility afforded by the modular arrangement of TnMod, result in a versatile tool for the mapping of insertional mutations and the rapid recovery of clones from gram-negative bacteria.
Transposon mutagenesis remains one of the most extensively utilized genetic techniques available for the characterization of bacteria. Transposon mutagenesis is especially useful for bacterial species with poorly described genetic systems or when existing molecular tools are inadequate. There are many well-characterized transposons available for the mutagenesis of both gram-positive and gram-negative bacteria (for reviews, see references 3, 20, and 25). Unfortunately, there are often certain difficulties associated with their use. Typically, transposons are large (making them difficult to manipulate) and may contain antibiotic resistance determinants which are not useful for selection in some bacterial species. Many transposons are inserted nonrandomly at particular target DNA sequences (31). In addition, many transposons have a limited host range or may transpose preferentially into plasmids rather than chromosomes (15, 18). Once a transposon integrates into a target DNA, it is potentially unstable since it is still capable of undergoing additional transpositional events or promoting DNA rearrangements within the cell. Finally, once a transposon is established in the host bacterium’s genome, additional mutagenesis with a second transposon can be inhibited due to the resident transposon’s production of an inhibitory protein (3, 4).
The problems associated with the use of transposons have largely been overcome with the development of minitransposons. Minitransposons are specialized transposons which arrange the cognate transposase outside of the transposon’s inverted repeats (7, 10, 15, 18, 33). This arrangement permits the minitransposon to stably integrate into a target DNA without its transposase. Not only does this prevent further transposition and DNA rearrangements, it also allows for repeated rounds of minitransposon mutagenesis since no immunity protein is present in the cell. These synthetic minitransposons are small and stable and have been constructed to contain different antibiotic resistance determinants (7, 15). Minitransposons based on transposon Tn5 function in a wide range of gram-negative bacteria and exhibit virtually no preference for a specific target DNA sequence (4, 7).
In order to increase the cloning functionality of transposons, several investigators have included a conditional origin of replication within the basic transposon to produce “self-cloning” or “in vitro-cloning” transposons (11, 13, 17, 23, 36). The inclusion of a conditional origin of replication within the transposon allows the rapid cloning of the DNA adjacent to the transposon’s site of insertion. The in vitro cloning is performed by digesting the total genomic DNA, self-ligating it, and transforming it into a permissive Escherichia coli host. The presence of a conditional origin of replication in the transposon expedites the cloning process and decreases the chances of obtaining a noncolinear DNA fragment during the cloning process.
The unique features of minitransposons and self-cloning transposons have been combined to construct new Tn5-based minitransposons for the rapid genetic analysis of gram-negative bacterial genomes. The basic minitransposon has been modified to include a conditional origin of replication and exchangeable antibiotic resistance determinants. The modular arrangement of the new TnMod minitransposons allows for different combinations of antibiotic resistance determinants and high- or low-copy-number origins of replication. Rare restriction endonuclease sites have been incorporated near the inverted repeats in order to facilitate the localization of the minitransposon’s insertion site on a physical genome map. These rare restriction sites can also be used to construct a library of clones containing large DNA fragments surrounding the transposon’s site of insertion.
Development of TnMod plasposons.
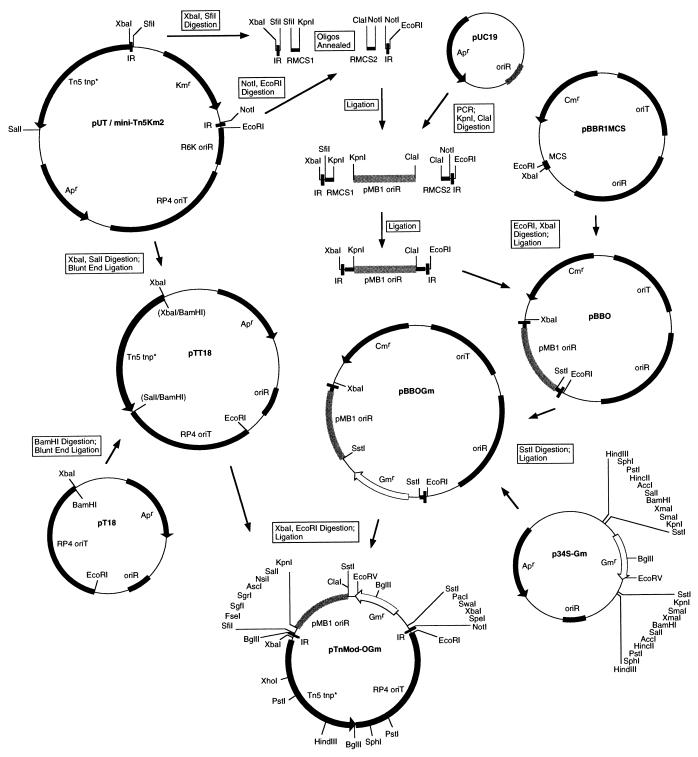
In order to construct TnMod, an XbaI-SfiI DNA fragment containing the 19-bp “inside-end” inverted repeat and a NotI-EcoRI DNA fragment containing the 19-bp “outside-end” inverted repeat were isolated from pUT/mini-Tn5Km2 (7). As shown in Fig. 1, these DNA fragments were individually ligated to two sets of complementary oligonucleotides which, when annealed, contain sites for restriction enzymes which rarely cut bacterial DNA. The Tn5 inside-end rare multiple cloning site (RMCS1) oligonucleotide contains sites for SfiI, FseI, SgfI, SgrAI, AscI, NsiI, SalI, and KpnI. The Tn5 outside-end RMCS2 oligonucleotide contains restriction sites for ClaI, SstI, PacI, SwaI, XbaI, SpeI, and NotI. A 700-bp PCR product containing the pUC (pMB1/ColE1) origin of replication was amplified from pUC19 (35) with PCR primers 5′-GGGTACCAGGAAAGAACATGTG-3′ and 5′-CCATCGATTTCGTTCCACTGAG-3′. This fragment was digested with KpnI and ClaI and in a four-way ligation was cloned into pBBR1MCS (22), creating pBBO. A gentamicin resistance cassette isolated from p34S-Gm was cloned into the SstI site of pBBO, creating pBBOGm. In order to construct the delivery DNA fragment, the origin of transfer from broad-host-range plasmid RP4 (29) was isolated from pMOB3 (27) and ligated into pUC18 (35), creating pT18. The Tn5 transposase was isolated from pUT/mini-Tn5Km2 (7) on a 1.5-kb XbaI-SalI fragment. The ends were polished with mung bean nuclease, and this fragment was blunt end ligated into pT18 digested with BamHI and mung bean nuclease, forming pTT18. The XbaI-EcoRI fragment from pTT18 containing the RP4 oriT and Tn5 transposase was isolated and ligated to the XbaI-EcoRI fragment isolated from pBBOGm. The resulting plasmid/transposon, shown in detail at the bottom of Fig. 1, was sequenced to verify its correctness and designated pTnMod-OGm. All derivatives containing different antibiotic resistance cassettes and origins of replication are modifications of this basic construct.
FIG. 1.
Construction of pTnMod-OGm. Arrows and boxed text indicate the construction manipulations. The pMB1 origin of replication is represented by a shaded box; the gentamicin resistance cassette from p34S-Gm is represented by an open arrow. Restriction sites in parentheses have been eliminated. All plasmids are drawn to scale. All commonly used restriction sites are labeled on the pTnMod-OGm plasposon illustrated at the bottom of the figure. IR, inverted repeat; oligos, oligonucleotides.
The inclusion of rare restriction sites within TnMod allows the TnMod site of insertion to be localized on a physical genome map by pulsed-field gel electrophoresis (34). Rare restriction sites for restriction endonucleases recognizing both high-GC- and low-GC-content DNA sequences were included so that TnMod could easily be mapped irrespective of the GC content of the mutagenized host’s genomic DNA. The unique common restriction sites inside of the rare restriction sites in TnMod can be utilized for the exchange of the origin-of-replication cassettes or the antibiotic resistance cassettes.
The absence of an origin of replication on the delivery DNA fragment in pTnMod forces the delivery DNA to form a nonreplicating DNA circle which is lost from the cell population once the minitransposon moves into a new replicon. Because the delivery DNA fragment cannot exist in the absence of the transposable DNA fragment and because the transposable DNA fragment is no longer functional as a mini-transposon once it has jumped away from its transposase, pTnMod cannot be described as a minitransposon situated in a delivery plasmid. As constructed, the plasmid origin of replication is an integral component of the transposable DNA fragment. Therefore, we propose the trivial name “plasposons” (plasmid minitransposons) to describe these novel genetic elements.
Construction of TnMod plasposon origin-of-replication derivatives.
In order to obtain a plasmid origin which will not replicate upon introduction into Enterobacteriaceae, a modified origin of replication from plasmid R6K was utilized (21). Since the R6K origin of replication requires the π gene product to function properly, plasmids containing an R6K origin of replication and not the π gene will not replicate unless the bacterial host can supply the π replication protein in trans (21). E. coli lambda phage lysogens which contain the π gene serve as hosts which allow the maintenance of TnMod derivatives containing the modified R6K origin of replication. TnMod variants containing the modified R6K origin of replication will not replicate in Enterobacteriaceae not expressing π, which allows the selection of cells whose plasposons have integrated into the chromosome. The R6K origin of replication was isolated from pJM703.1 (24) on a 420-bp BamHI fragment, treated with mung bean nuclease, and ligated into pGEM7Z (Promega, Madison, Wis.) digested with SmaI. The resulting plasmid, designated pJDR, contains the R6K origin of replication flanked by a KpnI site and a ClaI site. This KpnI-ClaI DNA fragment was cloned into TnMod, replacing the origin of replication from pMB1. These plasposon constructs are designated TnMod-R.
Many bacterial gene products are toxic when cloned in high copy number in a heterologous bacterial host. Decreasing the gene copy number in the cell can reduce the deleterious effects of the cloned genes. Similarly, when large DNA fragments are cloned in high copy number, illegitimate recombination can occur due to interaction between the cloned DNA sequences. Increased stability of large clones can be achieved by reducing the plasmid copy number in the host bacterium. Therefore, an origin of replication exhibiting a low copy number would facilitate the rescue of TnMod insertions in DNA containing genes for toxic proteins or of TnMod insertions in large-DNA clones. The origin of replication from narrow-host-range plasmid pSC101 (5), which has a copy number of three to five plasmids per cell, was amplified by PCR with primers designed to contain KpnI and ClaI restriction sites (5′-CCGGTACCGAAGTGGTCAGACTG-3′ and 5′-CCATCGATAAGAACCTCAGATCC-3′, respectively). This PCR fragment was cloned into pGEM7Z digested with KpnI and ClaI, resulting in plasmid pJDS, which serves as a source of the pSC101 origin of replication. TnMod variants containing the pSC101 origin of replication, designated TnMod-S, can be used to isolate large, stable DNA fragments from nonenteric gram-negative bacteria. The rare-cutting endonuclease sites in TnMod can be used in combination with the pSC101 origin of replication to generate a large DNA clone library of the target bacterium’s genome in E. coli.
Construction of exchangeable antibiotic resistance cassettes.
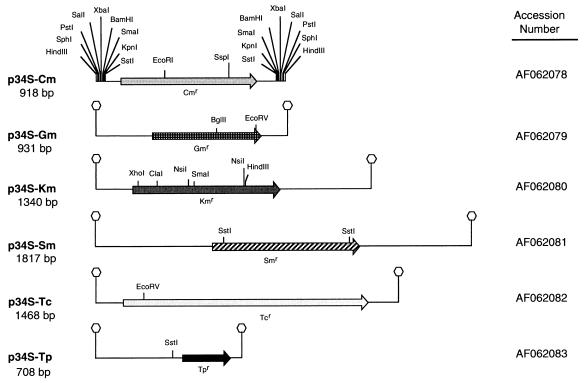
Because many gram-negative bacteria are naturally resistant to high levels of antibiotics, TnMod was constructed in such a way as to allow the addition of multiple antibiotic resistance cassettes or the exchange of different antibiotic resistance cassettes based on the antibiotic resistance characteristics of the target bacteria. Antibiotic resistance cassettes for chloramphenicol, gentamicin, kanamycin, streptomycin, tetracycline, and trimethoprim were constructed in the cassette vector p34E (30) from resistance genes originating from Tn9, Tn1619, Tn903, E. coli aac(3)-IV, pBR322, and R388, respectively. As shown in Fig. 2, the resulting antibiotic resistance cassettes are flanked by duplicate multiple cloning sites. These antibiotic resistance cassettes can be inserted into either the left or right side of the origin of replication in TnMod. Alternatively, multiple antibiotic resistance cassettes can be inserted into both ends of TnMod in order to select for TnMod insertions by using two or more antibiotics for bacteria especially resistant to antibiotics.
FIG. 2.
Restriction maps of the antibiotic resistance cassettes in the p34S plasmids. The cassette inserts in the p34S plasmids are drawn to scale. The duplicate multiple cloning sites flanking the antibiotic resistance cassettes are shown for p34S-Cm and are represented by small hexagons for the other p34S plasmids. Common restriction sites which can be used to determine the orientations of the antibiotic cassettes in pTnMod variants are shown above the DNA. The corresponding GenBank accession numbers are shown to the right of the p34S derivatives.
Specifically, the gene for chloramphenicol resistance was amplified from pUC-CML (26) by PCR with primers containing sites for SstI (5′-GGGAGCTCTTGAAATAAGATCACTAC-3′ and 5′-GGGAGCTCTTACACTTATTCAGGCG-3′). The 0.8-kb PCR fragment obtained was cloned into p34E digested with SstI. p34S-Gm was constructed by cloning the gentamicin resistance determinant directly from pUCGm (28) on a 0.9-kb SstI DNA fragment and ligating this into p34E digested with SstI. p34S-Tp was constructed by isolating a 0.6-kb EcoRI DNA fragment from p34E-Tp (9), which encodes the trimethoprim resistance determinant from R388, and blunt end ligating this fragment into p34E which had been similarly treated with EcoRI and mung bean nuclease. p34S-Km and p34S-Sm were similarly constructed from pUC4K (Pharmacia, Piscataway, N.J.) and pUT/mini-Tn5Sm (7), respectively. p34S-Tc was constructed by isolating the restriction site-modified tetracycline resistance gene from pALTER-1 (Promega) on a ClaI-StyI DNA fragment, polishing the ends with mung bean nuclease, and ligating this DNA fragment to p34E digested with EcoRI and similarly treated with mung bean nuclease. In order to restore the −35 region of the tetracycline gene lost during construction, two complementary oligonucleotides (5′-CATGTTTGACAGCTTATCAT-3′ and 5′-CGATGATAAGCTGTCAAACATGAGCT-3′) were annealed, reproducing the −35 region, and the double-stranded oligonucleotide was ligated into p34S-Tc digested with ClaI and partially digested with SstI. The ClaI site between the −10 and −35 regions was eliminated by digesting the plasmid with ClaI and filling in the ends of the DNA fragment with Klenow DNA polymerase and deoxynucleoside triphosphates. The nucleotide sequences of these plasmid constructs were verified by DNA sequence analysis.
The basic TnMod can be outfitted with multiple antibiotic resistance cassettes in a variety of ways, making a numerical designation for each derivative uninformative. Instead, TnMod derivatives are named according to the physical locations and orientations of their components. An antibiotic resistance cassette located next to the inside inverted repeat is designated with a two-letter abbreviation (Cm, Gm, Km, Sm, Tc, or Tp), which appears immediately after “TnMod-” in the name. The origin of replication, located in the center of TnMod, is abbreviated with a one-letter abbreviation (O for pMB1 oriR, R for R6K oriR, or S for pSC101 oriR). An antibiotic resistance cassette following the origin of replication, near the outside inverted repeat, is similarly designated with a two-letter abbreviation. The orientation of an antibiotic resistance cassette, which is especially important in designing primers to sequence DNA adjacent to TnMod, is denoted by a prime when antibiotic resistance cassette genes are oriented away from the origin of replication; the lack of a prime indicates the reverse orientation. By this format, a TnMod derivative containing a kanamycin resistance cassette in the KpnI site oriented toward the R6K origin and a chloramphenicol resistance cassette in the SstI site oriented away from the origin would be named TnMod-KmRCm′. If this TnMod derivative exists in plasmid form (i.e., attached to the delivery DNA fragment containing the RP4 oriT and Tn5 transposase), it is denoted pTnMod-KmRCm′.
Due to the large number of structural variations possible with the TnMod mutagenesis system, including the type of origin of replication and the number, types, and orientations of the antibiotic resistant cassettes, only a small fraction of these variants were constructed. The TnMod variants constructed, including their salient characteristics, are listed in Fig. 3. (A form for requesting the pTnMod derivatives can be downloaded from http://www.rci.rutgers.edu/~zylstra/plasposon.) In order to facilitate DNA sequencing primer design and the subcloning of the plasmids formed by the self-ligation of the excised plasposons and flanking chromosomal DNA, the complete DNA sequences of the constructed TnMod variants and the p34S antibiotic resistance cassette plasmids have been submitted to GenBank.
FIG. 3.
Schematic maps of the constructed TnMod variants. TnMod variants are listed with their origins of replication, the types and orientations of their antibiotic resistance cassettes, and their GenBank accession numbers. The flanking rare multiple cloning sites immediately inside the inverted repeats are denoted by the boxes on the left and right sides of each plasposon. Restriction sites on the left side of each plasposon are BglII, SfiI, FseI, SgfI, AscI, NsiI, SalI, and KpnI. Restriction sites on the right side of each plasposon are SstI, PacI, SwaI, XbaI, SpeI, and NotI.
TnMod mutagenesis procedure.
TnMod insertional mutagenesis and clone recovery is simple and rapid. Once the antibiotic resistance MICs have been determined for the target bacteria, pTnMod is introduced by a triparental mating. Briefly, the recipient bacterial cells, the E. coli host containing the pTnMod derivative, and E. coli HB101 containing the helper plasmid pRK2013 (12) are grown in liquid culture to mid-log phase. Equal volumes of these cells (approximately 1 ml) are combined and centrifuged at low speed at room temperature. The supernatant is discarded, and the cells are resuspended in 50 μl of Luria-Bertani (LB) broth or 10% glycerol and plated in a small pool on a nonselective LB plate. This mating plate is left for 6 to 18 h at 25 to 37°C, depending upon the optimum growth temperature of the target bacteria. The cells are then scraped off the plate and resuspended in 1 to 2 ml of buffer (e.g., 10 mM HEPES or 10% glycerol), and appropriate dilutions are plated on selective medium. This medium should contain nutrient limitations or antibiotic levels which select for exconjugant target cells and eliminate the donor E. coli strains. The plates are incubated for 1 to 2 days, and individual exconjugant colonies are picked to a second selective-medium plate. Alternatively, the recipient cells can be transformed directly with purified pTnMod by electroporation (8).
Once a desired mutation is identified, DNA flanking the insertion site can be rapidly cloned. Minichromosomal preparations are performed as described by Ausubel et al. (2). One microgram of total genomic DNA is digested with a restriction enzyme that cuts outside of or within one end of TnMod. The enzyme is removed, and the DNA is self-ligated under conditions favorable for intramolecular reactions. A portion of the ligation mixture is transformed into an appropriate E. coli host by electroporation (i.e., strain JM109 for pMB1- and pSC101-based TnMods and strain CC118 λpir (16) for R6K-based TnMods). The resulting plasmids containing TnMod are selected with the antibiotic resistance determinant(s) encoded by TnMod. In order to determine the identity of the gene into which the plasposon has been inserted, a plasmid minipreparation of the E. coli clone yields plasmid DNA ready for sequencing. This entire procedure can be performed in as little as 2 days. Once a desired clone is obtained, the entire chromosomal DNA insert can rapidly be sequenced by primer walking or by using exonuclease III to construct nested deletions in the cloned DNA fragment (6).
In order to generate a clone library by using TnMod, TnMod mutagenesis is performed as described above. The use of rare-cutting restriction enzymes will produce clones each of which contains a large segment of the genome. After selection for recipient cells by TnMod integration, genomic DNAs are isolated from pools of TnMod exconjugants, cleaved with a rare-cutting restriction enzyme, self-ligated, and transformed into a permissive E. coli host. A plasposon library generated in this fashion is a relatively rapid and simple alternative to the construction of cosmid libraries.
Applications of TnMod.
Because Tn5 has a broad host range (4), TnMod should work efficiently in gram-negative bacterial hosts such as Acinetobacter, Aeromonas, Agrobacterium, Bordetella, Caulobacter, Moraxella, Rhizobium, and Vibrio spp., as well as in the Enterobacteriaceae. We have successfully tested TnMod mutagenesis with several different genera of bacteria including Burkholderia, Escherichia, Pseudomonas, and Sphingomonas. In Pseudomonas putida F1 (14), for example, TnMod-OKm was found to be inserted in the chromosome randomly and stably, with an operational transpositional frequency of approximately 10−3 to 10−4. In all insertions examined, the transposon delivery DNA fragment did not transpose with TnMod (data not shown). We have successfully used the TnMod mutagenesis system to identify and isolate genes involved in solvent tolerance from P. putida S12 (19).
In order to demonstrate the utility of the TnMod plasposon cloning system, Burkholderia cepacia DBO1 (32) was mutagenized with pTnMod-KmO. Ten random kanamycin-resistant exconjugants were selected, and the flanking chromosomal DNAs were cloned in E. coli JM109 with restriction enzyme PstI. The individual clones were partially sequenced with primers which anneal to the two ends of TnMod-KmO and permit the sequencing of the plasposon’s point of insertion. DNA sequence analyses and comparisons by using BLAST homology searches (1) indicate that 9 of the 10 clones have TnMod-KmO inserted in genes homologous to well-characterized genes deposited in the GenBank nucleotide database. These matches are shown in Table 1. Several of these B. cepacia-derived clones are related to genes directly involved in basic metabolic functions (argF, gspA, and gpmA). Other clones appear to be related to genes involved in biodegradative pathways (tmbU) or regulation of extracellular signaling (rhlI). In addition, one isolate which was not highly similar to any well-characterized gene found in the GenBank database (f308) was obtained. Since most of the partial gene sequences listed in Table 1 have not been previously characterized for B. cepacia, the above results suggest that TnMod can be used to rapidly isolate and identify novel genes from gram-negative bacteria.
TABLE 1.
Putative identity of genes isolated by random TnMod mutagenesis of B. cepacia DBO1
| Strain | Homolog | Organism | Accession no. | Type of protein or function | % Identitya | % Similarityb | No. of gapsc |
|---|---|---|---|---|---|---|---|
| DBO1R | tmbU | P. putida | U41301 | Trimethyl benzene degradation protein | 43 (21/47) | 62 (29/47) | 2 |
| DBO2R | cspD | E. coli | D90725 | Cold shock-like protein | 57 (19/33) | 78 (26/33) | 0 |
| DBO3R | f308 | E. coli | AE000123 | Hypothetical protein | 53 (43/81) | 73 (59/81) | 1 |
| DBO4R | argF | B. subtilis | X53360 | Ornithine carbamoyltransferase | 55 (49/89) | 74 (66/89) | 0 |
| DBO5R | gspA | E. coli | M20938 | Glycerol-3-phosphate dehydrogenase | 55 (75/134) | 61 (83/134) | 0 |
| DBO6R | rhII | P. aeruginosa | U40458 | Quorum sensing autoinducer regulator | 42 (69/163) | 59 (96/163) | 4 |
| DBO7R | cobT | S. typhimurium | L12006 | Vitamin B12 biosynthesis | 56 (28/50) | 64 (32/50) | 1 |
| DBO8R | gpmA | E. coli | D90714 | Phosphoglycerate mutase | 64 (97/152) | 80 (122/152) | 1 |
| DBO9R | gltD | P. aeruginosa | U81261 | Glutamate synthase, small subunit | 45 (26/57) | 59 (34/57) | 0 |
| DBO10R | ampA | E. coli | U14003 | Leucine aminopeptidase | 73 (101/137) | 83 (115/137) | 0 |
Based on deduced amino acids matched to exact amino acids. The numbers in parentheses are the numbers of matching amino acids/the total numbers of amino acids in the alignment.
Based on deduced amino acids matched to similar groups of amino acids. Numbers in parentheses are as defined in footnote a.
Number of gaps introduced into the sequence to obtain maximal alignment between nonoverlapping segments.
The TnMod mutagenesis system described here was originally designed to provide a convenient mechanism for cloning and mapping genes in gram-negative bacteria. The placement of an origin of replication within a minitransposon combines the best features of plasmids and transposons. TnMod variants can therefore be defined as new genetic elements, which we have named plasposons. Due to the inclusion of sites for infrequently cutting restriction enzymes, the TnMod plasposons are useful not only for the rapid isolation of novel genes, but also for the mapping of these genes to physical chromosome maps and for the construction of large-DNA fragment libraries.
Nucleotide sequence accession numbers.
The sequences of p34S antibiotic resistance cassette plasmids and those of TnMod variants have been submitted to GenBank and have been assigned the accession numbers shown in Fig. 2 and 3, respectively.
Acknowledgments
This work was supported by a National Science Foundation Young Investigator Award to G.J.Z. and cooperative agreement CR822634 from the U.S. Environmental Protection Agency Gulf Breeze Environmental Research Laboratory.
We thank D. DeShazer, H. P. Schweizer, P. A. Sokol, and K. N. Timmis for sharing bacterial strains and plasmids. We thank L. Newman in this laboratory for helpful discussions and M. Murillo for excellent technical assistance.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Greene Publishing and Wiley-Interscience; 1987. [Google Scholar]
- 3.Berg C M, Berg D E, Groisman E A. Transposable elements and the genetic engineering of bacteria. In: Berg D E, Howe M M, editors. Mobile DNA. Washington, D.C: American Society for Microbiology; 1989. pp. 879–925. [Google Scholar]
- 4.Berg D E. Transposon Tn5. In: Berg D E, Howe M M, editors. Mobile DNA. Washington, D.C: American Society for Microbiology; 1989. pp. 183–208. [Google Scholar]
- 5.Cohen S N, Chang A C Y, Boyer H W, Helling R B. Construction of biologically functional bacterial plasmids in vitro. Proc Natl Acad Sci USA. 1973;70:3240–3244. doi: 10.1073/pnas.70.11.3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dale R M, McClure B A, Houchins J P. A rapid single-stranded cloning strategy for producing a sequential series of overlapping clones for use in DNA sequencing: application to sequencing the corn mitochondrial 18S rDNA. Plasmid. 1985;13:31–40. doi: 10.1016/0147-619x(85)90053-8. [DOI] [PubMed] [Google Scholar]
- 7.De Lorenzo V, Herrero M, Jakubzik U, Timmis K N. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J Bacteriol. 1990;172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dennis J J, Sokol P A. Electrotransformation of Pseudomonas. In: Nickoloff J A, editor. Methods in molecular biology. 47. Electroporation protocols for microorganisms. Totowa, N.J: Humana Press Inc.; 1995. pp. 125–133. [DOI] [PubMed] [Google Scholar]
- 9.DeShazer D, Woods D E. Broad-host-range cloning and cassette vectors based on the R388 trimethoprim resistant gene. BioTechniques. 1996;20:762–764. doi: 10.2144/96205bm05. [DOI] [PubMed] [Google Scholar]
- 10.Dodson K W, Berg D E. Factors affecting transposition activity of IS50 and Tn5 ends. Gene. 1989;76:207–213. doi: 10.1016/0378-1119(89)90161-3. [DOI] [PubMed] [Google Scholar]
- 11.Fellay R, Krisch H M, Prentki P, Frey J. Omegon-Km: a transposable element designed for in vivo insertional mutagenesis and cloning of genes in Gram-negative bacteria. Gene. 1989;76:215–226. doi: 10.1016/0378-1119(89)90162-5. [DOI] [PubMed] [Google Scholar]
- 12.Figurski D, Helinski D. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Furuichi T, Inouye M, Inouye S. Novel one-step cloning vector with a transposable element: application to the Myxococcus xanthus genome. J Bacteriol. 1985;164:270–275. doi: 10.1128/jb.164.1.270-275.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gibson D T, Koch J R, Kallio R E. Oxidative degradation of aromatic hydrocarbons by microorganisms. I. Enzymatic formation of catechol from benzene. Biochemistry. 1968;7:2653–2662. doi: 10.1021/bi00847a031. [DOI] [PubMed] [Google Scholar]
- 15.Haas R, Kahrs A F, Facius D, Allmeier H, Schmitt R, Meyer T F. TnMax—a versatile mini-transposon for the analysis of cloned genes and shuttle mutagenesis. Gene. 1993;130:23–31. doi: 10.1016/0378-1119(93)90342-z. [DOI] [PubMed] [Google Scholar]
- 16.Herrero M, de Lorenzo V, Timmis K N. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J Bacteriol. 1990;172:6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joseph-Liauzun E, Fellay R, Chandler M. Transposable elements for efficient manipulation of a wide range of gram-negative bacteria: promoter probes and vectors for foreign genes. Gene. 1989;85:83–89. doi: 10.1016/0378-1119(89)90467-8. [DOI] [PubMed] [Google Scholar]
- 18.Kahrs A F, Odenbreit S, Schmitt W, Heuermann D, Meyer T F, Haas R. An improved TnMax mini-transposon system suitable for sequencing, shuttle mutagenesis, and gene fusions. Gene. 1995;167:53–57. doi: 10.1016/0378-1119(95)00671-0. [DOI] [PubMed] [Google Scholar]
- 19.Kieboom J, Dennis J J, de Bont J A, Zylstra G J. Identification and molecular characterization of an efflux pump involved in Pseudomonas putida S12 solvent tolerance. J Biol Chem. 1998;273:85–91. doi: 10.1074/jbc.273.1.85. [DOI] [PubMed] [Google Scholar]
- 20.Kleckner N. Transposon Tn10. In: Berg D E, Howe M M, editors. Mobile DNA. Washington, D.C: American Society for Microbiology; 1989. pp. 227–268. [Google Scholar]
- 21.Kolter R, Inuzuka M, Helinski D R. Trans-complementation-dependent replication of a low molecular weight origin fragment from plasmid R6K. Cell. 1978;15:1199–1208. doi: 10.1016/0092-8674(78)90046-6. [DOI] [PubMed] [Google Scholar]
- 22.Kovach M E, Phillips P H, Elzer P H, Roop II R M, Peterson K M. pBBR1MCS: a broad-host-range cloning vector. BioTechniques. 1994;16:801–802. [PubMed] [Google Scholar]
- 23.Merriman T R, Lamont I L. Construction and use of a self-cloning promoter probe vector for Gram-negative bacteria. Gene. 1993;126:17–23. doi: 10.1016/0378-1119(93)90585-q. [DOI] [PubMed] [Google Scholar]
- 24.Miller V L, Mekalanos J J. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murphy E. Transposable elements in gram-positive bacteria. In: Berg D E, Howe M M, editors. Mobile DNA. Washington, D.C: American Society for Microbiology; 1989. pp. 269–288. [Google Scholar]
- 26.Schweizer H P. The PUC18CM plasmids: a chloramphenicol resistance gene cassette for site-directed insertion and deletion mutagenesis in E. coli. BioTechniques. 1990;8:614–616. [PubMed] [Google Scholar]
- 27.Schweizer H P. Allelic exchange in Pseudomonas aeruginosa using novel ColE1-type vectors and a family of cassettes containing a portable oriT and the counter-selectable Bacillus subtilis sacB marker. Mol Microbiol. 1992;6:1195–1204. doi: 10.1111/j.1365-2958.1992.tb01558.x. [DOI] [PubMed] [Google Scholar]
- 28.Schweizer H P. Small broad-host-range gentamicin resistance gene cassettes for site-specific insertion and deletion mutagenesis. BioTechniques. 1993;15:831–833. [PubMed] [Google Scholar]
- 29.Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 30.Tsang T, Copeland V, Bowden G T. A set of cassette cloning vectors for rapid and versatile adaptation of restriction fragments. BioTechniques. 1991;10:330. [PubMed] [Google Scholar]
- 31.Tsuda M, Nakazawa T. A mutagenesis system utilizing a Tn1722 derivative containing an Escherichia coli specific vector plasmid: application to Pseudomonas species. Gene. 1993;136:257–262. doi: 10.1016/0378-1119(93)90475-i. [DOI] [PubMed] [Google Scholar]
- 32.Walsh T A, Ballou D P. Halogenated protocatechuates as substrates for protocatechuate dioxygenase from Pseudomonas cepacia. J Biol Chem. 1983;258:14413–14421. [PubMed] [Google Scholar]
- 33.Way J C, Davis M A, Morisato D, Roberts D E, Kleckner N. New Tn10 derivatives for transposon mutagenesis and for construction of lacZ operon fusions by transposition. Gene. 1984;32:369–379. doi: 10.1016/0378-1119(84)90012-x. [DOI] [PubMed] [Google Scholar]
- 34.Wong K K, McClelland M. Dissection of the Salmonella typhimurium genome by use of a Tn5 derivative carrying rare restriction sites. J Bacteriol. 1992;174:3807–3811. doi: 10.1128/jb.174.11.3807-3811.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 36.Youngman P, Perkins J B, Losick R. A novel method for the rapid cloning in Escherichia coli of Bacillus subtilis chromosomal DNA adjacent to Tn917 insertions. Mol Gen Genet. 1984;195:424–433. doi: 10.1007/BF00341443. [DOI] [PubMed] [Google Scholar]