Abstract
Background & objectives:
Imatinib mesylate (IM) is a reliable first line treatment for chronic myeloid leukaemia (CML). Nevertheless, despite promising results, a considerable proportion of patients develop resistance to the drug. Cytochrome P450 (CYP) enzymes play a crucial role in IM metabolism. Thus, point mutations in CYP genes may modify IM enzyme activity resulting in insufficient treatment response. This investigation was aimed to identify the functional impact of CYP3A5*3, CYP3A4*18 and CYP2B6*6 polymorphisms on the IM response in patients with CML in Azerbaijan.
Methods:
Genotyping of CYP3A5*3, CYP3A4*18 and CYP2B6*6 was performed in 153 patients (102 IM non-responders and 51 IM responders) with CML by the PCR-restriction fragment length polymorphism (RFLP) assays. The odds ratios (ORs) with 95 per cent confidence intervals (CIs) were applied to assess the association between allelic variants and IM therapy outcome. The results were validated by sequencing.
Results:
The frequency of the CYP3A4*18 allele was considerably lower in the responder’s group (97.1 vs. 100%; P=0.036). For CYP3A5*3, the allelic frequency was slightly higher among the IM responders (100 vs. 99.02%) with no significant difference. Although patients heterozygous (TC) for CYP2B6*6 demonstrated a higher risk of acquiring resistance (OR 1.04; 95% CI: 0.492-2.218), differences were not significant (P=0.909). In addition, the homozygous genotype (TT) demonstrated a lower risk of unresponsiveness (OR 0.72; 95% CI: 0.283-1.836), but associations were not significant (P=0.491).
Interpretation & conclusions:
Our results demonstrated that CYP3A4*18 was significantly associated with IM treatment response in patients with CML in Azerbaijan, whereas rather common CYP3A5*3 was identified to have no such association.
Keywords: Chronic myeloid leukaemia, CYP2B6, CYP3A4, CYP3A5, drug response, imatinib, PCR, RFLP
Imatinib mesylate (IM) is a selective tyrosine kinase inhibitor (TKI) that has become a prototype in target treatment against haematological malignancies. The management of chronic myelogenous leukaemia (CML) altered its natural course with IM introduction. It is specifically designed to block the expansion of cells expressing not only the BCR-ABL fusion gene but also the receptor of stem cell factor, c-kit tyrosine kinases and platelet-derived growth factor. In addition, IM was the first TKI approved by the Food and Drug Administration1.
Being a frontline therapy, IM has demonstrated a promising cytogenetic, haematologic and molecular response and ensured a decrease in the time of response, toxicity and the patients’ morbidity and mortality2.
Nevertheless, even though imatinib is the gold standard for treatment of CML, some patients develop resistance, which poses a major therapeutic challenge. Resistance may be orchestrated by various mechanisms such as mutation or amplification of the BCR-ABL domain, upregulation of multidrug-resistance gene 1 or genetic alterations of enzymes engaged in IM metabolism, not to mention the famous family of cytochrome P450 (CYP). Recent studies show that more than 25 per cent of patients with CML may demonstrate resistance to IM at least once through their lifetime3.
CYP complex plays a crucial part in the metabolism of various drugs and IM is not an exception. This sophisticated complex comprises a family of haemoproteins that catalyze drug metabolism reactions and the synthesis of cholesterol, steroids and other lipids. Three major CYP gene families (CYP1, CYP2 and CYP3) perform xenobiotic metabolism. CYP3A enzyme is highly inducible and plays a major role in the metabolism of drugs in the liver and gastrointestinal tract. The subfamily includes several isoforms: CYP3A4, CYP3A5, CYP3A7 and CYP3A43. In vitro studies using recombinant enzymes indicate that N-demethylation of IM is performed by CYP3A4, CYP2C8 and CYP3A5, while other enzymes play little or no role4,5.
It is important to note that IM is mainly metabolized by the CYP3A4 isoform, and to a lesser extent, by several other enzymes such as CYP3A5, CYP2C9, CYP2B6 and CYP1A2. Thus, genetic variability among the patients could explain the differences in IM bioavailability, affecting intracellular and plasma levels and eventually influencing the therapeutic response6.
The CYP3A5 gene is located on chromosome 7q21.1 position and indicates interindividual variations at expression levels. The most common isoform variant is CYP3A5*3 (A6986G, rs776746), which involves splicing defects. This polymorphism that is situated in intron 3 may decrease the expression of CYP3A5 to less than 1/1000 of what is found in carriers of the wild-type allele (CYP3A5*1)7. Moreover, several recent studies indicate its potential association with IM metabolism8-11.
Another alteration of a crucial drug metabolism contributor, the CYP3A4 gene, is CYP3A4*18 (rs28371759). One of the most frequent coding variants is located on exon 10 and involves a substitution from thymine (T) to cytosine (C) at position 878, which is known to have an association with high levels of testosterone and chlorpyrifos. Moreover, it is also associated with low bone mineral density and elevated prostate cancer risk12.
Being one of the most polymorphic CYP genes in humans, the CYP2B6 gene is situated on chromosome 19q13.2 and metabolizes different types of drugs, including cyclophosphamide, isophamide, tamoxifen, ketamine, propofol and various environmental carcinogens. Its alterations affect transcriptional regulation, splicing, mRNA and protein expression and catalytic activity. CYP2B6*6 (Q172H) is the most common variant leading to complex interactions between substrate-dependent and independent mechanisms. The frequencies of G15631T (rs3745274) vary from 15 to 60 per cent in different populations13. Recent investigations have shown this polymorphism to be associated with a genetic predisposition to acute leukaemia14-16.
Despite the fact that a significant number of investigations of CYP single-nucleotide polymorphisms (SNPs) are involved in the pharmacogenetics and pharmacodynamics of imatinib, the contribution of these variants to patients’ drug resistance remains unclear and requires further research.
This study was designed to estimate the frequency of CYP3A5*3 (G6986A), CYP3A4*18 (C878T) and CYP2B6*6 (G15631T) in patients with CML in the Azerbaijani population undergoing IM therapy along with the assessment of their impact on imatinib treatment response.
Material & Methods
Study participants: This study was carried out by the department of Haematology, Institute of Haematology and Blood Transfusion (IHB; named after B. Eyvazov), Baku, Azerbaijan, from April 2017 to December 2019. The study was approved by the Institute’s Ethical Board and carried out according to the Declaration of Helsinki of 1964, as revised in 2020. All the patients included in the investigation signed the informed consent. The study involved individuals who belonged to the Azerbaijani ethnic group and resided in Azerbaijan for three consecutive generations. Individuals who along with CML, had a history or evidence of chronic or acute disorders, other types of cancer, hepatic or haematological abnormalities and hepatitis B or C or HIV infection were excluded from the study.
A total of 748 CML patients were admitted to the IHB during the study period, of whom 404 (54%) were female and 344 (46%) male (median age, 47.4 yr; standard deviation, 15 yr and range, 4-84 yr). The resistance to IM occurred in 155 participants; based on inclusion criteria, our study involved 153 unrelated patients (102 IM resistant and 51 IM good responders) who were treated in IHB between February 2017 and December 2019. The study cohort comprised 79 females (51.6%) and 74 males (48.4%) (mean age, 46.1 yr; standard deviation, 14.4 yr and range, 19-80 yr). Diagnosis of CML was based on the standard clinical and haematological criteria and the presence of the Philadelphia chromosome and/or BCR-ABL fusion gene. Participants with CML were in chronic (n=129), accelerated (n=13) and blast (n=11) phases, treated for at least 12 months with IM on frontline treatment. Each patient had started with a standard dose of 400 mg/day; subsequently, the dose was increased to 600 or 800 mg in the absence of major molecular response (MMR) at 12 months. Among the patients included in this investigation, 123 were treated with 400 mg, 27 participants with 600 mg and three patients with 800 mg.
In addition, as risk group types define treatment strategy, patients were grouped into high, medium and low risk categories according to the Sokal scoring system. The distribution to risk groups depended on various criteria such as white blood cells and platelets counts, age, baseline spleen size, basophils, eosinophils and myeloblasts in peripheral blood (Table I).
Table I.
Patient’s characteristics
| Parameters | All patients (n=153), n (%) | IM resistant (n=102), n (%) | IM good response (n=51), n (%) | P (χ2) |
|---|---|---|---|---|
| Gender | ||||
| Male | 74 (100) | 52 (70.2) | 22 (29.8) | 0.36 (0.837) |
| Female | 79 (100) | 50 (63.2) | 29 (36.8) | |
| Age (yr) | ||||
| <50 | 102 (100) | 67 (65.7) | 35 (34.3) | 0.716 (0.132) |
| ≥50 | 51 (100) | 35 (68.6) | 16 (31.4) | |
| CML stages | ||||
| Chronic | 129 (100) | 78 (60.5) | 51 (39.5) | 0.04δ |
| Accelerated | 13 (100) | 13 (100) | 0 | |
| Blast crisis | 11 (100) | 11 (100) | 0 | |
| Risk groups | ||||
| High | 19 (100) | 14 (73.7) | 5 (26.3) | 0.785 (0.483) |
| Medium | 87 (100) | 57 (65.5) | 30 (34.5) | |
| Low | 47 (100) | 31 (66) | 16 (34) | |
| Spleen | ||||
| Normal | 21 (100) | 16 (76.1) | 5 (23.9) | 0.009 (9.243) |
| Medium (growth up to 8 cm) | 88 (100) | 50 (56.8) | 38 (43.2) | |
| Massive (>8 cm) | 44 (100) | 36 (81.8) | 8 (18.2) | |
| HB (g/l) | ||||
| Normal (120-160 g/l) | 13 (100) | 8 (61.5) | 5 (38.5) | 0.288 (3.758) |
| Mild anaemia (100-119 g/l) | 44 (100) | 30 (68.1) | 14 (31.9) | |
| Mean anaemia (80-99 g/l) | 53 (100) | 31 (58.4) | 22 (41.6) | |
| Severe anaemia (65-79 g/l) | 43 (100) | 33 (75) | 10 (25) | |
| WBC (×109/l) | ||||
| Normal (3.9-9) | 1 (100) | 1 (100) | 0 | 0.781δ |
| Leucopenia (<3.9) | 0 | 0 | 0 | |
| Leucocytosis (>9) | 152 (100) | 101 (66.4) | 51 (33.6) | |
| PLT (×109/l) | ||||
| Normal (180-400) | 92 (100) | 64 (69.5) | 28 (30.5) | 0.354 (2.074) |
| Thrombocytopenia (<180) | 21 (100) | 15 (71.4) | 6 (28.6) | |
| Thrombocytosis (>400) | 40 (100) | 23 (57.5) | 17 (42.5) | |
| Blast cell (0%) | ||||
| 0 | 68 (100) | 41 (60.2) | 27 (39.8) | 0.323δ |
| 1-10 | 81 (100) | 58 (71.6) | 23 (28.4) | |
| >10 | 4 (100) | 3 (75) | 1 (25) |
P<0.05 was considered as significant. δTest statistic not reported for Fisher’s exact test. PLT, platelet; WBC, white blood cell; HB, haemoglobin; CML, chronic myeloid leukaemia; IM, imatinib mesylate
Imatinib mesylate (IM) response assessment: Patient inclusion was performed according to the criteria defined by European Leukaemia Net recommendations for the management of chronic myeloid leukaemia17. The criteria for complete haematological response were defined on the basis of the total leucocyte, platelet and white blood cell counts, absence of immature forms and disappearance of splenomegaly18. In the given cohort, 146 (96%) patients with CML reached complete haematological response after seven weeks of therapy.
The molecular response assessment was defined, referring to the international scale (IS) as the ratio of BCR-ABL1 transcripts to ABL1 transcripts17. A BCR-ABL1 transcript level of ≤0.1 per cent corresponds with the MMR.
A complete molecular remission was attained by 107 (70%) patients after 12 months of treatment.
The BCR-ABL1 (p210) expression was evaluated using the reverse transcription-quantitative polymerase chain reaction (RT-qPCR) technique for initial diagnosis and subsequent monitoring of the efficacy of TKI therapy. The analysis of BCR-ABL1 (p210) e13a2 and e14a2 fusion transcripts in peripheral blood and bone marrow samples was performed by using Mbcr IS-MMR kit (Qiagen, Hilden, Germany) on Rotor-Gene Q (Qiagen).
Thus, IM good responders group included patients who fulfilled optimal response criteria. IM resistant group consisted of the patient who failed to achieve MMR at 12, 18 or 24 months. Treatment options for patients with inadequate IM response included a second generation TKI (nilotinib and dasatinib) or allogeneic haematopoietic stem cell transplantation (HSCT).
DNA extraction: Two millilitres of peripheral blood samples in EDTA tubes were collected from the study participants. The isolation of genomic DNA was performed with QIAamp DNA Blood Mini kit (Qiagen), according to the manufacturer’s protocol. DNA quality and quantity were analyzed using NanoDrop 2000c Spectrophotometers (Thermo Scientific, Waltham, MA, USA).
Genotyping: Genotyping was carried out at the Institute of Genetic Resources of Azerbaijan National Academy of Sciences (ANAS) by the PCR-restriction fragment length polymorphism (RFLP). Gene specific primers and PCR reaction conditions were previously published by Maddin et al11 and Berköz and Yalin14 for CYP3A5*3, CYP3A4*18 and CYP2B6*6, respectively.
The PCR reaction mixture (25 μl) included 2 μl DNA, 2.0 μl magnesium chloride (MgCl2), 0.25 μl dNTPs, 0.25 FIREPOL Taq Polymerase (Solis Biodyne, Estonia), 0.5 μl of each primer, 17 μl deionized water and 2.5 μl of ×10 PCR buffer. The electrophoresis of the products was performed on a 1.5 per cent agarose gel at 120 V for 40 min.
PCR amplification products (2.5 μl) were subjected to digestion with 0.5 units of restriction enzyme (New England BioLabs, BioLabs). The digested PCR products were run on a three per cent agarose gel for visualisation.
Sequencing: We randomly selected 10 per cent of all samples from each genotype for confirmation sequencing. The amplified DNA was purified using a QIAquick PCR (Qiagen) and sent to the Intergen Laboratory (Ankara, Türkiye) for confirmation via next generation sequencing. In addition, 10 per cent of total CYP3A5*3 allele samples were sequenced at the Genetic Resources Institute of Azerbaijan National Academy of Sciences (ANAS) via 3130XL Sanger Sequencing (Applied Biosystems, MA, USA) to validate previously obtained results.
Statistical analysis: Chi-square test (χ2) and Fisher’s exact test were applied to compare the frequencies of polymorphic genotypes between good and poor responders. Fisher’s exact test for contingency tables more than 2 × 2 was performed with Social Science Statistics (http://www.socscistatistics.com/tests/chisquare2/Default2.aspx). Binary logistic regression was carried out to calculate the odds ratios (ORs) with 95 per cent confidence intervals (CIs). All the statistical tests were two-sided; the significance level was P<0.05. Statistical analysis was performed using the SPSS package ver. 22 (Statistical Package for the Social Sciences, Chicago, IL, USA).
Results
Table I summarizes patients’ demographic profiles and clinical variables. In this study, we did not find any association between IM response and demographic parameters such as gender and age. Among clinical parameters, Sokal risk groups, white blood cells and platelets counts, haemoglobin levels and blast cells range were not associated with IM response. However, the disease phase and spleen size were considerably associated with the IM response (P=0.04 and P=0.009, respectively).
Our data revealed a significant association between CYP3A4*18 polymorphism and IM treatment response (P=0.013).
In this investigation, the frequencies of CYP3A5*3, CYP3A4*18 and CYP2B6*6 alleles were 99.3, one and 34 per cent, respectively. Between the two groups of CML participants, the frequency of the CYP3A5*3 allele was not significantly elevated (100%) among the IM responders compared to the non-responders (99.02%). Although the allelic frequency of CYP2B6*6 polymorphism in IM responders was higher (36.3%) than in the IM resistant group (32.9%), this difference was not significant. In contrast, the frequency of the CYP3A4*18 allele was significantly elevated in the poor responders (100%) compared to the good responders (97.1%; P=0.036; Table II).
Table II.
Genotype and allele frequencies of CYP3A5*3, CYP3A4*18 and CYP2B6*6
| SNP | Frequency (%) | P (χ2) | ||
|---|---|---|---|---|
|
| ||||
| IM good response (n=51), n (%) | IM resistant (n=102), n (%) | |||
| CYP3A5*3 (rs776746) | ||||
| Genotype | ||||
| Homozygous wild type (AA) | 0 | 1 (0.98) | 0.478δ | |
| Heterozygous (AG) | 0 | 0 | ||
| Homozygous mutant (GG) | 51 (100) | 101 (99.02) | ||
| Allele | ||||
| A | 0 | 2 (0.98) | 0.316δ | |
| G | 102 (100) | 202 (99.02) | ||
| CYP3A4*18 (rs28371759) | ||||
| Genotype | ||||
| Homozygous wild type (TT) | 48 (94.12) | 102 (100) | 0.013δ | |
| Heterozygous (TC) | 3 (5.88) | 0 | ||
| Homozygous mutant (CC) | 0 | 0 | ||
| Allele | ||||
| T | 99 (97.1) | 204 (100) | 0.036δ | |
| C | 3 (2.9) | 0 | ||
| CYP2B6*6 (rs3745274) | ||||
| Genotype | ||||
| Homozygous wild type (GG) | 24 (47.06) | 50 (49.02) | 0.737 (0.61) | |
| Heterozygous (GT) | 17 (33.33) | 37 (36.27) | ||
| Homozygous mutant (TT) | 10 (19.61) | 15 (14.71) | ||
| Allele | ||||
| G | 65 (63.7) | 137 (67.1) | 0.609 (0.357) | |
| T | 37 (36.3) | 67 (32.9) | ||
δTest statistic not reported for Fisher’s exact test. IM, imatinib mesylate
Interestingly, for the CYP3A5*3 polymorphism, most of the participants (99.93%) in both groups were found to possess homozygous mutant (GG) and only one subject was identified with wild type (AA). The frequency of *3/*3 (GG) genotype in good responders (100%) was not significantly different from the non-responders (99.02%). Our analysis did not reveal homozygous mutant genotype (CC) for CYP3A4*18. The frequencies of the homozygous wild type genotype (TT) were considerably lower (P=0.013) among responsive patients (94.12%) compared to resistant patients (100%). However, the frequency of heterozygous genotype (TC) was notably higher in the IM-responders (5.88 vs. 0%).
For CYP2B6, the genotypic frequencies of all three types of inheritance (GG, GT and TT) were elevated in the poor responders compared to the good responders; nevertheless, the difference was not statistically significant (Table II).
We performed a binary logistic regression to determine the risk association of polymorphisms with imatinib outcome. Heterozygous (GT) genotype carriers of CYP2B6*6 demonstrated a higher risk of acquiring resistance (OR 1.04; 95% CI: 0.492-2.218), whereas homozygous genotype (TT) demonstrated a lower risk of unresponsiveness towards the drug (OR 0.72; 95% CI: 0.283-1.836), but the associations were not significant (P=0.909 and P=0.491, respectively).
In addition, the relationship of SNPs with the epidemiological and clinical parameters was assessed of patients with CML. No significant differences were observed between CYP3A5*3 and CYP3A4*18 polymorphisms and age, gender or CML phases in the present study. However, criteria such as risk groups demonstrated an association with the given polymorphisms (P=0.035 and P=0.032, respectively) (Table III). As for CYP2B6*6, no significant differences were detected between the polymorphisms and mentioned parameters (Table III).
Table III.
Distribution of CYP3A5*3, CYP3A4*18 and CYP2B6*6 polymorphisms in chronic myeloid leukaemia patients with respect to epidemiological and clinical parameters
| Genotype | Age (yr) | Gender | Sokal risk score | CML stages | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|||||||
| <50 (n-103), n (%) | ≥50 (n=51), n (%) | Male (n=74), n (%) | Female (n=79), n (%) | High (n=19), n (%) | Medium (n=87), n (%) | Low (n=47) | Chronic (n=129), n (%) | Accelerated (n=13), n (%) | Blast crisis (n=11), n (%) | |
|
| ||||||||||
| CYP3A5 * 3 | ||||||||||
| Homozygous wild type (AA) | 1 (0.98) | 0 | 1 (1.4) | 1 (1.2) | 1 (5.2)# | 0 | 0 | 1 (0.8) | 0 | 0 |
| Heterozygous (AG) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Homozygous mutant (GG) | 101 (99.02) | 51 (100) | 73 (98.6) | 78 (98.8) | 18 (94.8)# | 87 (100)# | 47 (100)# | 128 (99.2) | 13 (100) | 11 (100) |
| CYP3A4 * 18 | ||||||||||
|
| ||||||||||
| Homozygous wild type (TT) | 100 (98.04) | 50 (98.04) | 72 (97.2) | 78 (98.8) | 19 (100)δ | 87 (100)δ | 44 (93.6)δ | 126 (97.7) | 13 (100) | 11 (100) |
| Heterozygous (TC) | 2 (1.96) | 1 (1.96) | 2 (2.8) | 1 (1.2) | 0 | 0 | 3 (6.4)δ | 3 (2.3) | 0 | 0 |
| Homozygous mutant (CC) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| CYP2B6 * 6 | ||||||||||
|
| ||||||||||
| Homozygous wild type (GG) | 49 (48) | 25 (49) | 41 (55.4) | 33 (41.7) | 7 (36.8) | 43 (49.4) | 24 (51.1) | 63 (49) | 5 (35.7) | 6 (54.6) |
| Heterozygous (GT) | 37 (36.4) | 17 (33.3) | 22 (29.7) | 32 (40.5) | 9 (47.4) | 29 (33.3) | 16 (34) | 45 (340) | 7 (50) | 2 (18.2) |
| Homozygous mutant (TT) | 16 (15.6) | 9 (17.7) | 11 (14.9) | 14 (17.8) | 3 (15.8) | 15 (17.3) | 7 (14.9) | 21 (17) | 1 (14) | 3 (27.3) |
#P<0.05 for CYP3A5*3; δP<0.05 for CYP3A4*18 polymorphism, respectively. CML, chronic myeloid leukemia
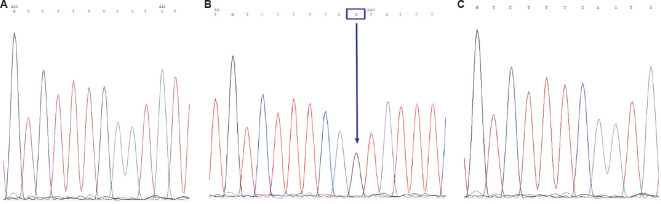
Sequencing results confirmed the successful performance of RFLP analysis of CYP3A5*3, CYP3A4*18 and CYP2B6*6 (Figure).
Figure.
Confirmation of the CYP3A5*3 allele by the Sanger sequencing method. Each peak in electropherogram represents a single nucleotide in the DNA sequence. (A) sequencing of the sample with wildtype genotype (CYP3A5*1/*1), (B) sequencing of the mutant genotype (CYP3A5*3/*3) revealed a single base substitution of adenine with guanine (A>G) in intron 3, and (C) matched reference sequence. PCR, polymerase chain reaction.
Discussion
IM possesses numerous advantageous characteristics, including a balanced dose-response relationship and very high bioavailability. However, despite being the recommended type of therapy, quite often, cancer cells develop a survival strategy causing resistance after a few months or years of therapy19. Among the identified mechanisms to develop resistance, alterations or amplification of the target protein play a significant role. Thus, mutations in major imatinib metabolizers CYP3A5 and CYP3A4 may correlate with IM treatment response.
CYP3A5*3 is the most frequent and well studied variant allele of CYP3A5. CYP3A5*1 is known to be associated with higher expression of CYP3A5 protein. Interestingly, a recent study of CYP3A5*3 polymorphism in healthy Azerbaijani participants revealed homozygous mutant genotype (*3/*3) frequency as 95 per cent, which is significantly lower than in this investigation (99%), indicating that elevated homozygous mutant genotype (*3/*3) may contribute to the condition. No significant difference was found between CYP3A5*3 and molecular response to IM in patients with CML. Our findings correlate with the results of different studies on various ethnic populations investigating this problem. For instance, recent investigations of CYP3A5*3 and dose-adjusted imatinib trough concentrations20,21 in different Asian populations did not reveal a significant correlation between the genotypes and the imatinib plasma trough concentrations and clinical response frequencies. Similar studies of the association between the polymorphism and the imatinib trough plasma levels also demonstrated an absence of the correlation22,23.
Nevertheless, our results are not in agreement with the results of recent investigations in the Egyptian and Indian populations, which concluded affirmatively about an association of CYP3A*1/*1 genotype with IM efficacy and *3/*3 genotype with inferior outcome9,24. Other related experiments with opposite outcomes were performed in the Canadian population and Indian population, where AA genotype in rs776746 was strongly associated with major or complete cytogenetic response to Imatinib8,10. Although we identified no significant relationship between CYP3A5*3 and risks of acquiring resistance to IM, studies suggest that the drug’s low clearance and high oral bioavailability may be caused by the homozygous mutant genotype. Thus, carriers of *3/*3 may have a decreased enzyme activity, leading to a better response to imatinib11.
Being one of the most abundant SNPs of CYP3A4, CYP3A4*18 may affect the overall protein structure and cause rearrangement of substrate recognition sites, which are essential for substrate recognition and substrate access to the active site. Alterations of that type may lead to disrupted metabolism of many drugs12. Since this SNP contributes to the slower metabolism of xenobiotics, it raises the suggestion of its involvement in imatinib resistance. For instance, the CYP3A4*18 variant in this study considerably influences IM therapy (P=0.013). In contrast, a study of the association between the genotypes of CYP3A4*18 and the imatinib plasma trough concentrations in the Korean population did not reveal statistically significant differences21.
Similarly, the investigation of the CYP3A4*18 and IM levels in Indian population also did not show any association. Interestingly, although high interindividual variability in the trough concentration of IM was observed, none of the patients were found to be carriers of the CYP3A4*18 allele25. Moreover, Maddin et al11 identified that patients with CML carrying the heterozygous genotype CYP3A4*18 showed a lower risk of acquiring IM resistance, but the association was not significant. An inconsistency with previously reported results in various populations highlights the demand for further studies in larger cohorts. However, similar to the studies from different ethnic populations11,26,27 and in healthy Azerbaijani volunteers28, no homozygous mutant variant was detected (CC) among our patients.
Increasing interest in G15631T has been stimulated by the revelations of a specific CYP2B6 genotype, which greatly affects the metabolism of various drugs, with the purpose of increasing drug efficacy and avoiding adverse drug reactions29. Furthermore, as this SNP causes the transition of glutamine (Glu) amino acid to histidine (His), it may block carcinogen substrates’ transformation into harmless metabolites14. This study found no statistically significant association between CYP2B6*6 polymorphism and IM therapy response in patients with CML. In accordance with our results, a study of enzyme functionality of human P450 stated that CYP2B6 had not been implicated in IM metabolism30. Differently from us, according to the investigation of the impact of CYP2B6 15631G>T on IM response and CML susceptibility, SNP has been revealed as a good therapeutic predictor due to its strong association with clinical and cytogenetical outcomes31.
To best of our knowledge, the present investigation is the first to report allelic and genotypic frequencies of CYP SNPs and their impact on IM response in the population of Azerbaijan. These findings are an attempt to provide preliminary evidence that pre-treatment genotyping of CYP3A4*18 may help predict the therapeutic response of IM in patients with CML. Nevertheless, these alterations may be population specific and should be profiled in a larger cohort to evaluate clinical relevance.
In conclusion, despite the absence of correlation with IM response, polymorphisms of CYP3A5*3 are common among the Azerbaijani population. Contrastingly, SNPs of CYP3A4*18 are significantly associated with IM response suggesting a risk of being resistant to IM therapy.
Financial support and sponsorship
None.
Conflicts of interest
None.
References
- 1.Jabbour E, Kantarjian H. Chronic myeloid leukemia:2020 update on diagnosis, therapy and monitoring. Am J Hematol. 2020;95:691–709. doi: 10.1002/ajh.25792. [DOI] [PubMed] [Google Scholar]
- 2.Minciacchi VR, Kumar R, Krause DS. Chronic myeloid leukemia: A model disease of the past, present and future. Cells. 2021;10:117. doi: 10.3390/cells10010117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yaghmaie M, Yeung CC. Molecular mechanisms of resistance to tyrosine kinase inhibitors. Curr Hematol Malig Rep. 2019;14:395–404. doi: 10.1007/s11899-019-00543-7. [DOI] [PubMed] [Google Scholar]
- 4.Nebot N, Crettol S, d’Esposito F, Tattam B, Hibbs DE, Murray M. Participation of CYP2C8 and CYP3A4 in the N-demethylation of imatinib in human hepatic microsomes. Br J Pharmacol. 2010;161:1059–69. doi: 10.1111/j.1476-5381.2010.00946.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pena MÁ, Muriel J, Saiz-Rodríguez M, Borobia AM, Abad-Santos F, Frías J, et al. Effect of cytochrome P450 and ABCB1 polymorphisms on imatinib pharmacokinetics after single-dose administration to healthy subjects. Clin Drug Investig. 2020;40:617–28. doi: 10.1007/s40261-020-00921-7. [DOI] [PubMed] [Google Scholar]
- 6.Nath A, Wang J, Stephanie Huang R. Pharmacogenetics and pharmacogenomics of targeted therapeutics in chronic myeloid leukemia. Mol Diagn Ther. 2017;21:621–31. doi: 10.1007/s40291-017-0292-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gréen H, Skoglund K, Rommel F, Mirghani RA, Lotfi K. CYP3A activity influences imatinib response in patients with chronic myeloid leukemia: A pilot study on in vivo CYP3A activity. Eur J Clin Pharmacol. 2010;66:383–6. doi: 10.1007/s00228-009-0772-y. [DOI] [PubMed] [Google Scholar]
- 8.Kim DH, Sriharsha L, Xu W, Kamel-Reid S, Liu X, Siminovitch K, et al. Clinical relevance of a pharmacogenetic approach using multiple candidate genes to predict response and resistance to imatinib therapy in chronic myeloid leukemia. Clin Cancer Res. 2009;15:4750–8. doi: 10.1158/1078-0432.CCR-09-0145. [DOI] [PubMed] [Google Scholar]
- 9.Sailaja K, Rao DN, Rao DR, Vishnupriya S. Analysis of CYP3A5*3 and CYP3A5*6 gene polymorphisms in Indian chronic myeloid leukemia patients. Asian Pac J Cancer Prev. 2010;11:781–4. [PubMed] [Google Scholar]
- 10.Vaidya S, Ghosh K, Shanmukhaiah C, Vundinti BR. Genetic variations of hOCT1 gene and CYP3A4/A5 genes and their association with imatinib response in chronic myeloid leukemia. Eur J Pharmacol. 2015;765:124–30. doi: 10.1016/j.ejphar.2015.08.034. [DOI] [PubMed] [Google Scholar]
- 11.Maddin N, Husin A, Gan SH, Aziz BA, Ankathil R. Impact of CYP3A4*18 and CYP3A5*3 polymorphisms on imatinib mesylate response among chronic myeloid leukemia patients in Malaysia. Oncol Ther. 2016;4:303–14. doi: 10.1007/s40487-016-0035-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou XY, Hu XX, Wang CC, Lu XR, Chen Z, Liu Q, et al. Enzymatic activities of CYP3A4 allelic variants on quinine 3-hydroxylation in vitro. Front Pharmacol. 2019;10:591. doi: 10.3389/fphar.2019.00591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zanger UM, Klein K. Pharmacogenetics of cytochrome P450 2B6 (CYP2B6): Advances on polymorphisms, mechanisms, and clinical relevance. Front Genet. 2013;4:24. doi: 10.3389/fgene.2013.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berköz M, Yalin S. Association of CYP2B6 G15631T polymorphism with acute leukemia susceptibility. Leuk Res. 2009;33:919–23. doi: 10.1016/j.leukres.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 15.Daraki A, Zachaki S, Koromila T, Diamantopoulou P, Pantelias GE, Sambani C, et al. The G(5)(1)(6)T CYP2B6 germline polymorphism affects the risk of acute myeloid leukemia and is associated with specific chromosomal abnormalities. PLoS One. 2014;9:e88879. doi: 10.1371/journal.pone.0088879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu LL, Zhang W, Li J, Zhao L. Association between CYP2B6 polymorphism and acute leukemia in a Han population of Northwest China. Mol Genet Genomic Med. 2020;8:e1162. doi: 10.1002/mgg3.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hochhaus A, Baccarani M, Silver RT, Schiffer C, Apperley JF, Cervantes F, et al. European LeukemiaNet 2020 recommendations for treating chronic myeloid leukemia. Leukemia. 2020;34:966–84. doi: 10.1038/s41375-020-0776-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hochhaus A, Saussele S, Rosti G, Mahon FX, Janssen JJWM, Hjorth-Hansen H, et al. Chronic myeloid leukaemia: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2018;29:iv261. doi: 10.1093/annonc/mdy159. [DOI] [PubMed] [Google Scholar]
- 19.Rosenzweig SA. Acquired resistance to drugs targeting tyrosine kinases. Adv Cancer Res. 2018;138:71–98. doi: 10.1016/bs.acr.2018.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takahashi N, Miura M, Scott SA, Kagaya H, Kameoka Y, Tagawa H, et al. Influence of CYP3A5 and drug transporter polymorphisms on imatinib trough concentration and clinical response among patients with chronic phase chronic myeloid leukemia. J Hum Genet. 2010;55:731–7. doi: 10.1038/jhg.2010.98. [DOI] [PubMed] [Google Scholar]
- 21.Seong SJ, Lim M, Sohn SK, Moon JH, Oh SJ, Kim BS, et al. Influence of enzyme and transporter polymorphisms on trough imatinib concentration and clinical response in chronic myeloid leukemia patients. Ann Oncol. 2013;24:756–60. doi: 10.1093/annonc/mds532. [DOI] [PubMed] [Google Scholar]
- 22.Adeagbo BA, Olugbade TA, Durosinmi MA, Bolarinwa RA, Ogungbenro K, Bolaji OO. Population pharmacokinetics of imatinib in Nigerians with chronic myeloid leukemia: Clinical implications for dosing and resistance. J Clin Pharmacol. 2017;57:1554–63. doi: 10.1002/jcph.953. [DOI] [PubMed] [Google Scholar]
- 23.Belohlavkova P, Vrbacky F, Voglova J, Racil Z, Zackova D, Hrochova K, et al. The significance of enzyme and transporter polymorphisms for imatinib plasma levels and achieving an optimal response in chronic myeloid leukemia patients. Arch Med Sci. 2018;14:1416–23. doi: 10.5114/aoms.2018.73538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bedewy AM, El-Maghraby SM. Do SLCO1B3 (T334G) and CYP3A5*3 polymorphisms affect response in Egyptian chronic myeloid leukemia patients receiving imatinib therapy? Hematology. 2013;18:211–6. doi: 10.1179/1607845412Y.0000000067. [DOI] [PubMed] [Google Scholar]
- 25.Gota V, Sharma A, Paradkar A, Patil A, Khattry N, Menon H, et al. Association between CYP3A4 polymorphisms, trough imatinib plasma concentration and cytogenetic response in chronic phase chronic myeloid leukemia (CML-CP) Blood. 2012;120:3788. [Google Scholar]
- 26.Hu YF, He J, Chen GL, Wang D, Liu ZQ, Zhang C, et al. CYP3A5*3 and CYP3A4*18 single nucleotide polymorphisms in a Chinese population. Clin Chim Acta. 2005;353:187–92. doi: 10.1016/j.cccn.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 27.Zaied C, Abid S, Mtiraoui N, Zellema D, Achour A, Bacha H. Cytochrome P450 (CYP3A4*18) and glutathione-S-transferase (GSTP1) polymorphisms in a healthy Tunisian population. Genet Test Mol Biomarkers. 2012;16:1184–7. doi: 10.1089/gtmb.2012.0095. [DOI] [PubMed] [Google Scholar]
- 28.Karimova N, Hasanova A, Bayramov B. CYP3A4*18 and CYP3A5*3 single nucleotide polymorphisms in an Azerbaijani population. Polymorphism. 2022;8:1–10. [Google Scholar]
- 29.Yuce-Artun N, Kose G, Suzen HS. Allele and genotype frequencies of CYP2B6 in a Turkish population. Mol Biol Rep. 2014;41:3891–6. doi: 10.1007/s11033-014-3256-9. [DOI] [PubMed] [Google Scholar]
- 30.Lewis DF. 57 varieties: The human cytochromes P450. Pharmacogenomics. 2004;5:305–18. doi: 10.1517/phgs.5.3.305.29827. [DOI] [PubMed] [Google Scholar]
- 31.Kassogue Y, Quachouh M, Dehbi H, Quessar A, Benchekroun S, Nadifi S. Functional polymorphism of CYP2B6 G15631T is associated with hematologic and cytogenetic response in chronic myeloid leukemia patients treated with imatinib. Med Oncol. 2014;31:782. doi: 10.1007/s12032-013-0782-6. [DOI] [PubMed] [Google Scholar]