Abstract
Recent breakthroughs in our understanding of sensorineural hearing loss etiology have encouraged the identification of novel hearing therapeutics, paving the way for precision hearing medicine. Critical to this field is the curation of health resources on hearing data. A systematic review of the literature was conducted to map existing (inter)national and regional datasets that include hearing data to inform the development of future hearing repositories. Systematic literature review was performed adhering to Preferred Reporting Items for Systematic Review and Meta-Analysis recommendations. Databases, including those from gray literature, were searched to identify publications reporting on phenotypic and/or genotypic hearing data in May 2019. The databases reviewed were Medline, PubMed, Embase databases, and Google Scholar. Publications on local datasets were excluded. All hearing datasets identified in the screening process were noted. For each dataset, geography, context, objective, period of time run, numbers and demographics of participants, genomic data, hearing measures and instruments used were extracted and cataloged. One hundred and eighty-eight datasets were identified, containing hearing data on populations ranging from 100 to 1.39 million individuals, and all extracted data have been cataloged. This searchable resource has been made accessible online. This unique catalog provides an overview of existing datasets that contain valuable information on hearing. This can be used to inform the development of national and international patient data repositories for hearing loss and guide strategic collaboration between key stakeholder groups, pivotal to the delivery and development of sensorineural hearing loss precision diagnostics and treatments.
Keywords: Hearing, hearing loss, precision medicines, sensorineural hearing loss, therapeutics
Introduction
Hearing loss is a major global health problem requiring urgent response: 466 million people worldwide have a disabling hearing loss of which 90% are adults. Owing to our aging population, over 900 million people will be affected by 2050.1 Hearing loss has considerable personal, social, and economic impact,2 the health and quality of life of people with hearing loss is poorer than that of the general population,3 and a compelling correlation has been demonstrated between adult onset hearing loss and dementia.4 The annual cost of untreated hearing loss is $750 billion globally, primarily due to unemployment.1
Hearing loss is not a diagnosis, but a symptom reflecting a range of underlying pathologies. The most common form of hearing loss is sensorineural hearing loss (SNHL), which accounts for 90% of cases and relates to the dysfunction of the inner ear and central auditory pathways.5 A multitude of genetic and molecular causes are at the root of SNHL.5,6 Recent breakthroughs in our understanding of these causes have allowed for the identification of therapeutic targets and development of innovative hearing therapeutics.6 This means that precision hearing medicine for SNHL may be on the horizon, allowing tailored treatment for individuals with hearing loss, targeted to their genotypic and phenotypic profiles.7-10
In other health fields, such as cancer and cardiovascular disease, progress has been made to this end, and collaboratives including clinicians, bioinformaticians, discovery scientists, and industry have created large-scale clinical and genomic data repositories that form the basis for precision medicine.11,12 It is important that the hearing community learns from this experience and works toward creating similar repositories for hearing loss. These will guide toward a deeper understanding of the genomic, proteomic, and metabolomic processes and environmental interactions that underlie various hearing loss phenotypes and will also accelerate the development of targeted therapeutics and patient access to innovative hearing treatments.
To inform the development of future patient data repositories for hearing loss, we set out to map what is already available in the hearing field and systematically reviewed the existing datasets including phenotypic and/or genotypic hearing data in adults and children. Unlike systematic reviews of literature, our systematic review of databases does not allow an assessment of quality nor is it amenable to classifications of levels of evidence. The aim of this review was to identify all potential valuable sources of hearing loss data. This type of review can signpost researchers toward existing hearing loss content.
Methods
We undertook a systematic review of the scientific literature to identify publications reporting on datasets that included data on hearing, and a gray literature search to identify any datasets not (yet) published in the scientific literature.
Design
Search Strategy and Selection Criteria
We performed a systematic review adhering to Preferred Reporting Items for Systematic Review and Meta-Analysis (PRISMA) recommendations13 to identify publications on international, national, and regional datasets including data on hearing in adult and pediatric populations. With expert librarian support, we designed and conducted a comprehensive search of the Medline and Embase databases using the Ovid portal in May 2019, in addition to PubMed search. The broad search string used was (database OR cohort OR dataset OR registry OR register OR repository OR catalogue) AND (hearing OR hearing loss) AND (SNHL OR sensorineural OR age related). While there is no agreed method on how to identify and report on available datasets, we have adapted the PRISMA principles of systematic literature review to systematically review the existing available datasets. Unfortunately, since this is not a systematic review of the “literature” as such, this is a limitation to our methodology, although it is a first of its kind approach to systematic reviewing.
All datasets with hearing data identified in the screening process were noted and searched in the gray literature using the Google search engine. We also approached our network of professionals from ear–nose–throat (ENT), audiology, discovery and translational hearing science, and biotechnology and pharmaceutical industries to identify additional datasets including data on hearing in adult and pediatric populations.
Three authors (PS, RM, and OY) searched the literature independently and compared the results. Abstract screening followed by full-text review was performed by 8 authors (PS, RM, OY, GH, HD, MA, ES, and MJD). The results were compared at each stage of the PRISMA flowchart, with a ninth author (AS) arbitrating any disagreements.
We included publications reporting on international, national, or regional datasets that included data on hearing in adult or pediatric populations, with no limit on the years that data were collected or on the duration of data collection. Datasets which met our inclusion criteria were those which included “hearing data,” i.e., pure tone audiometry results, speech-in-noise results, speech discrimination scores, clinical diagnoses of hearing loss, and self-reports of hearing loss. The datasets were cataloged in terms of geography, context, objective, period of time run, numbers and demographics of participants, genomic data, hearing measures, instruments used to capture hearing loss data, and open access availability of data. We discussed each publication reporting on regional and local datasets and excluded those that only had small number of individuals relative to the disease prevalence or of relevant variables. Individual publication conclusions were not analyzed in keeping with the aims and objectives of this study to identify and catalog hearing loss datasets.
Results
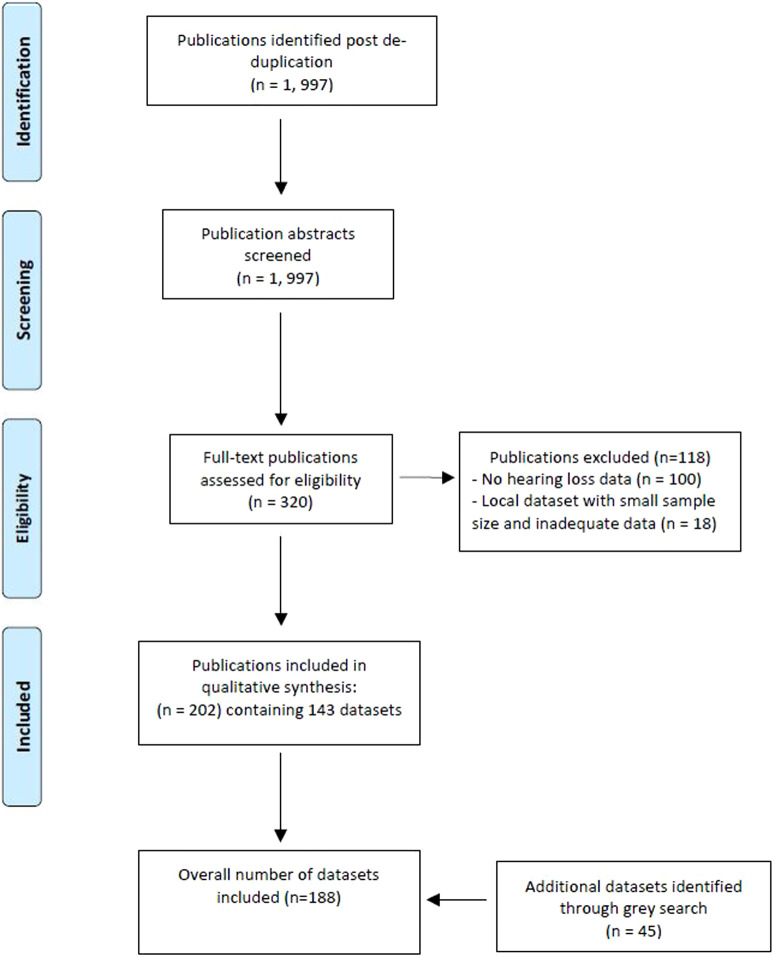
Following de-duplication, 1997 publications were identified as meeting the criteria of our systematic scientific literature search. After abstract screening, 320 publications remained that underwent full-text screening, resulting in 202 publications meeting our inclusion criteria (see PRISMA flowchart, Figure 1). These publications referred to 143 datasets, and our gray literature search revealed an additional 45 datasets not identified in the literature review. Overall, our search strategy identified 188 datasets with data on hearing in adult or pediatric populations. The electronic link in Supplementary Table 1 provides a catalog of these datasets, providing a summary of genotypic and phenotypic information held as well as geography, context, objective, period of time run, numbers and demographics of participants, hearing measures and instruments used to capture hearing data, and other investigations.
Figure 1.
Preferred reporting items for systematic review and meta-analysis diagram showing results of systematic review.
What type of data sources are hearing researchers currently using?
Of the 202 scientific publications, 33 used data from birth cohorts or population-based cohort studies (5 from the Rotterdam study,14 4 from the Shanghai Health Study,15 and 2 from the Twins UK study.16 Seven publications used data linkage between above datasets. Forty-three publications utilized data from 2 health-care databases (including 27 publications based on data from the Taiwan Longitudinal Health Insurance Database,17 5 from the Korean National Health Insurance Service18). Twenty-eight publications used data from disease registries other than hearing loss (e.g., cancer registries). The majority of the above datasets had self-report data regarding hearing only. The cohorts lacked clear audiological test data, although in some cases provided many decades’ worth of follow-up self-report data.
Six publications used data from 5 occupational datasets (routinely collected health data from occupational health visits), 4 of which were military datasets. Twenty-five publications utilized hearing-related data resources such as national hearing screening programs (n = 8), regional audiological databases (n = 3), hearing loss-specific population cohorts (6 publications from the Blue Mountains,19 3 from Nord Trondelag,20 2 from Beaver Dam,21 2 from the UK MRC National Hearing Screen, and 1 from USA’s Audiogen database).
Sixty-seven publications had used hearing loss datasets that were created to examine specific presentations of hearing loss (e.g,. age-related and sudden onset SNHL). Two publications had used the Japanese Intractable Inner Ear databank,22 3 publications had used the Swedish National Databank on Sudden SNHL, 23 and 2 the Childhood Development after Cochlear Implant dataset from Johns Hopkins, USA.24 These hearing-specific databases contained very useful hearing data, i.e. extensive audiological testing data, although they did not have either long-term or birth cohort data.
Current Hearing Data Resources
Overall, we identified 188 datasets that contained hearing-related data—ranging from 100 to 1.39 million individuals, and covering a potential hearing-related resource of 7.9 million patients; 60% were datasets that had been created for other purposes but also had hearing data (see Figure 2). Datasets specific to hearing (n = 77) were often hearing health-care datasets (n = 18), national hearing screening programs (n = 8), hearing loss-specific population datasets (n = 10) or congenital/inherited hearing loss patient datasets (n = 27) and specific hearing condition-related datasets (n = 14) (see Figure 3). All of these had objective hearing data, and many had some form of genetic testing; however, very few had repeated measures or access to the lifestyle of medical history that could allow prognosis and risk factors to be robustly identified.
Figure 2.
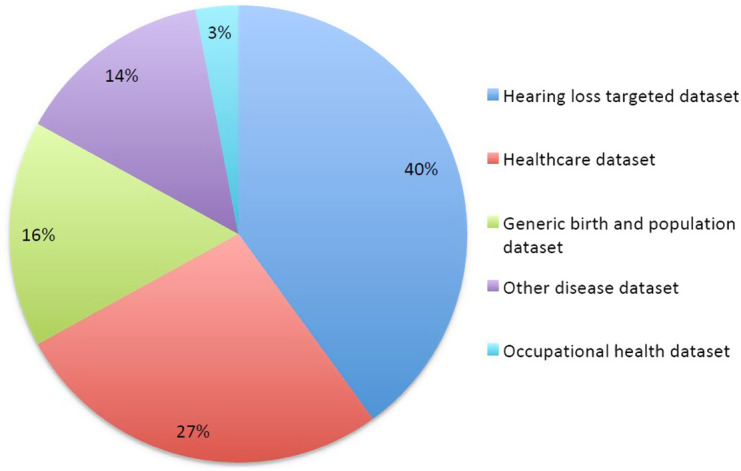
Type of dataset.
Figure 3.
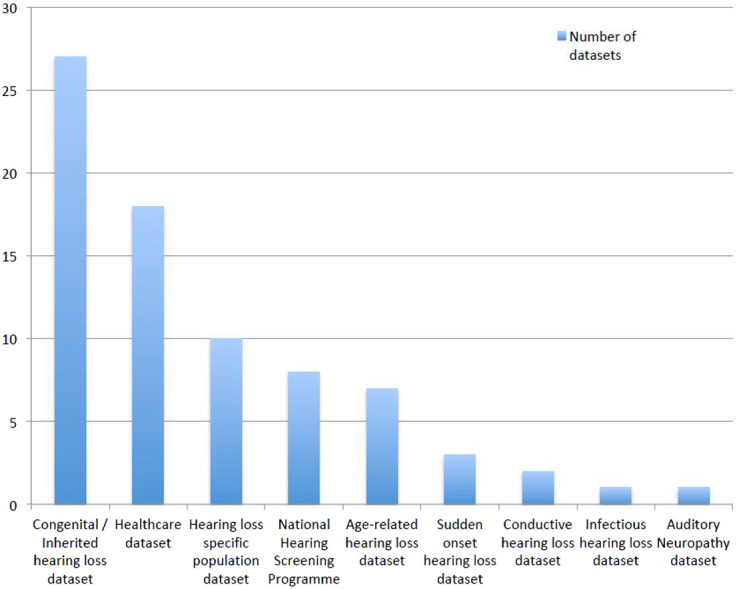
Subtypes of datasets within hearing targeted datasets.
Geographically, 96 datasets were from Europe, 51 from the Americas (only 2 from South America), 38 from Asia and Australia, and 3 from Africa. Only 6 datasets related to low-income countries.
One hundred twenty-one datasets reported pure tone audiometry, 23 had auditory-brainstem-evoked responses, 20 oto-acoustic emission, and 12 speech in noise data. Shanghai Health,15 Health ABC,25 and Blue Mountain19 cohorts used self-report to identify and stratify those with hearing loss. Diagnostic codes were used to identify hearing loss and type on the Taiwan Longitudinal Health Insurance Database.17
Eighty-one data resources had access to genetics data. Of those presenting the types of genetic data held, 22 had employed genome-wide association study (GWAS), with 5 undertaking whole exome sequencing (WES). A further 5 had undertaken GWAS and WES.26-30 29 data resources had used gene panel sequencing, with 17 using single gene sequencing. Of these studies containing genetic data, further subgroup analysis was undertaken to capture trends in hearing investigations utilized. Sixty-seven percent reported pure tone audiometry (PTA), followed by self-report hearing questionnaires (27%), auditory brainstem response (11%), and oto-acoustic emissions (10%) (see Figure 4).
Figure 4.
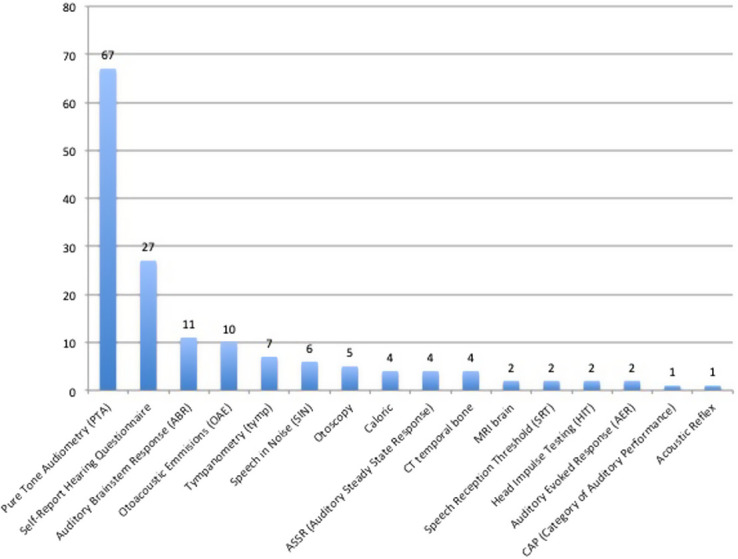
Hearing investigations utilized by studies capturing genetic data (expressed as percentages).
Discussion
Summary of Findings
This study provides an extensive catalog of existing regional, national, and international datasets containing genotypic and/or phenotypic information on populations with hearing loss. This catalog presents an overview of the epistemology of hearing data available and demonstrates: (1) Inadequate representation of hearing loss in populations from low-income countries; (2) considerable heterogeneity in the hearing-related outcomes recorded, with pure tone audiometry being the most common (but not universal) outcome reported; (3) lack of genetic and biomarker data (present in less than half of all datasets); (4) population cohorts record broader biopsychosocial data points for larger populations but have limited access to hearing specific data, with the opposite for hearing specific datasets; (5) population and birth cohorts with hearing loss data identified by gray literature searching may provide a novel source for researchers; (6) accessibility of raw data was not explicitly defined in any of the datasets. While author contact information has been provided to mitigate this, prospective dataset submission to the catalog will contain this information and is an important factor to consider in the context of registry development.
Implications
Our findings display the considerable heterogeneity among outcome measures used, making comparison difficult but more importantly, affecting the potential utility of the collected hearing loss data. We highlight the urgent need to establish consensus on key hearing-related outcome sets to develop reliable phenotypes that validly represent underlying and therapy-targetable processes.
Based on the results of this study, we have identified that PTA and self-reported hearing tools appear to be the most frequently utilized instruments capturing hearing data, followed by ABR (Auditory Brainstem Response) and OAE (Otoacoustic Emmisions) (see Figure 4) in the published literature of hearing loss cohorts containing genetic information. If this information is to be used in the formation of key hearing-related outcome sets (with a view to enable registry development), standardization of self-reported hearing tools is recommended to enable comparison of data.
Output from the available datasets has resulted in advances in understanding the epidemiology and burden of hearing loss, as well as the mechanisms of hearing loss subtypes. However, the available datasets do not have the breadth and depth of information that would be required to inform a system that aims to discover or distinguish between different targetable mechanisms of hearing loss. A potential barrier to the creation of such a dataset is cost and some investigators have avoided such costs by combining datasets (e.g., Blue Mountain study19) turning to routinely collected health-care records (e.g., Taiwan Health-care database17) or creating a centralized disease repository (e.g., Swedish Sudden Onset SNHL dataset31). Such strategies should be explored in more detail to facilitate greater data capture, especially if we hope to capture data in low-income countries.
We identified 45 datasets that contain hearing loss data but were not identified by systematic review and may prove a novel resource for researchers. To encourage researchers to utilize these new and existing datasets, we have electronically published an easily searchable file of this table on our website which is openly available to all. Furthermore, we welcome researchers to submit new datasets, following review. We hope that this resource encourages national and international collaboration that will facilitate rapid progress in the field of hearing loss and accelerate access to novel therapeutics for our patients.
Limitations
We acknowledge the potential of researcher bias in the formulation of the data extraction table. To mitigate this, key areas of data capture were derived by the immersion in the data and cross-checked by senior authors (NM, MJD, and AS) twice, together with consensus between all authors. This level of quality checking was also applied to the data extracted itself, deploying our extensive network of multi-disciplinary hearing scientists.
Conclusion
This first of its kind catalog provides an overview of existing regional, national, and international datasets that contain valuable information on hearing. This catalog can be used to inform the development of national and international repositories of hearing loss data and facilitate strategic collaboration between key stakeholder groups, pivotal to the delivery and development of precision diagnosis and treatment of hearing loss.
Funding Statement
This research was supported by the National Institute for Health Research, University College London Hospitals Biomedical Research Centre
Footnotes
Informed Consent: Peer-review: Externally peer-reviewed.
Author Contributions: Concept – P.S., R.M., A.G.M.S., N.M., M.J.D.; Design – P.S., R.M., A.G.M.S., N.M., Supervision – A.G.M.S., N.M.; Data Collection and/or Processing – All authors Analysis and/or Interpretation – P.S., N.M.; Literature Search – P.S., R.M., O.Y.; Writing – P.S., R.M., N.M.; Critical Review – A.G.M.S., N.M.
Acknowledgments: This paper follows a workshop entitled: “Translational hearing science – working towards consensus on patient selection and outcome measurement in clinical trials of innovative hearing treatments” held at the UCL Ear Institute, September 11-12, 2018. This workshop was attended by UK and French clinicians, discovery and translational scientists, investors, and biotech and pharmaceutical companies and supported by UCL Grand Challenges and the French Embassy’s Department of Science & Technology.
Declaration of Interests: The authors have no conflicts of interest to declare.
Supplementary Table 1.Summarises all datasets and publications linked to these together with the extracted data variables.
To encourage researchers to utilise these new and existing datasets, we have electronically published an easily searchable file of this table on our NIHR University College London Hospital Biomedical Research Centre website accessible via the following link: https://www.ucl.ac.uk/ear/nihr-uclh-brc-deafness-and-hearing-problems-theme. This resource is openly available to all. The file is also directly accesible via the following google doc link: https://docs.google.com/spreadsheets/d/1KHNPuWfPf53GuM1znhstvP0Mcpc_qCmb/edit?usp=sharing&ouid=112580371437801329797&rtpof=true&sd=true.
References
- 1. WHO. Glob al Costs of Unaddressed Hearing Loss and Cost-Effectiveness of Interventions A WHO Report; vol 2017; 2017. [Google Scholar]
- 2. Lancet T. Hearing loss: an important global health concern. Lancet. 2016;387(10036):2351. ( 10.1016/S0140-6736(16)30777-2) [DOI] [PubMed] [Google Scholar]
- 3. NHSEngland. Actio n Plan on Hearing Loss; 2015. [Google Scholar]
- 4. Deal JA, Betz J, Yaffe K, et al. Hearing impairment and incident dementia and cognitive decline in older adults: the health ABC study. J Gerontol A Biol Sci Med Sci. 2017;72(5):703 709. ( 10.1093/gerona/glw069) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yamasoba T, Lin FR, Someya S, Kashio A, Sakamoto T, Kondo K. Current concepts in age-related hearing loss: epidemiology and mechanistic pathways. Hear Res. 2013;303:30 38. ( 10.1016/j.heares.2013.01.021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schilder AGM, Su MP, Mandavia R, et al. Early phase trials of novel hearing therapeutics: avenues and opportunities. Hear Res. 2019;380:175 186. ( 10.1016/j.heares.2019.07.003) [DOI] [PubMed] [Google Scholar]
- 7. Macrae CA, Seidman CE. Closing the genotype-phenotype loop for precision medicine. Circulation. 2017;136(16):1492 1494. ( 10.1161/CIRCULATIONAHA.117.030831) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Robinson PN. Deep phenotyping for precision medicine. Hum Mutat. 2012;33(5):777 780. ( 10.1002/humu.22080) [DOI] [PubMed] [Google Scholar]
- 9. Rudman JR, Mei C, Bressler SE, Blanton SH, Liu XZ. Precision medicine in hearing loss. J Genet Genomics. 2018;45(2):99 109. ( 10.1016/j.jgg.2018.02.004) [DOI] [PubMed] [Google Scholar]
- 10. Schilder AGM, Su MP, Blackshaw H, et al. Hearing protection, restoration, and regeneration: an overview of emerging therapeutics for inner ear and central hearing disorders. Otol Neurotol. 2019;40(5):559 570. ( 10.1097/MAO.0000000000002194) [DOI] [PubMed] [Google Scholar]
- 11. Forrest SJ, Geoerger B, Janeway KA. Precision medicine in pediatric oncology. Curr Opin Pediatr. 2018;30(1):17 24. ( 10.1097/MOP.0000000000000570) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Giudicessi JR, Kullo IJ, Ackerman MJ. Precision cardiovascular medicine: state of genetic testing. Mayo Clin Proc. 2017;92(4):642 662. ( 10.1016/j.mayocp.2017.01.015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Moher D, Altman DG, Liberati A, Tetzlaff J. PRISMA statement. Epidemiology. 2011;22(1):128; author reply 128. ( 10.1097/EDE.0b013e3181fe7825) [DOI] [PubMed] [Google Scholar]
- 14. Hofman A, Darwish Murad SD, Van Duijn CM, et al. The rotterdam study: 2014 objectives and design update. Eur J Epidemiol. 2013;28(11):889 926. ( 10.1007/s10654-013-9866-z) [DOI] [PubMed] [Google Scholar]
- 15. Yuan J-M, Koh W-P, Gao Y-T. Shanghai cohort study | Singapore Chinese health study. Available at: https://www.schs.pitt.edu/methods/shanghai-cohort-study/ Accessed 2 May 2020. [Google Scholar]
- 16. Verdi S, Abbasian G, Bowyer RCE, et al. TwinsUK: the UK adult twin registry update. Twin Res Hum Genet. 2019;22(6):523 529. ( 10.1017/thg.2019.65) [DOI] [PubMed] [Google Scholar]
- 17. Hsieh CY, Su CC, Shao SC, et al. Taiwan’s national health insurance research database: past and future. Clin Epidemiol. 2019;11:349 358. ( 10.2147/CLEP.S196293) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ahn E. Introducing big data analysis using data from national health insurance service. Korean J Anesthesiol. 2020;73(3):205 211. ( 10.4097/kja.20129) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Joachim N, Mitchell P, Burlutsky G, Kifley A, Wang JJ. The incidence and progression of age-related macular degeneration over 15 years: the Blue Mountains Eye Study. Ophthalmology. 2015;122(12):2482 2489. ( 10.1016/j.ophtha.2015.08.002) [DOI] [PubMed] [Google Scholar]
- 20. Krokstad S, Langhammer A, Hveem K, et al. Cohort profile: the HUNT study, Norway. Int J Epidemiol. 2013;42(4):968 977. ( 10.1093/ije/dys095) [DOI] [PubMed] [Google Scholar]
- 21. Fryback DG, Dasbach EJ, Klein R, et al. The Beaver Dam Health Outcomes study: initial Catalog of Health-state Quality Factors. Med Decis Making. 1993;13(2):89 102. ( 10.1177/0272989X9301300202) [DOI] [PubMed] [Google Scholar]
- 22. Kanatani Y, Tomita N, Sato Y, Eto A, Omoe H, Mizushima H. National registry of designated intractable diseases in Japan: present status and future prospects. Neurol Med Chir (Tokyo). 2017;57(1):1 7. ( 10.2176/nmc.st.2016-0135) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nosrati-Zarenoe R, Hansson M, Hultcrantz E. Assessment of diagnostic approaches to idiopathic sudden sensorineural hearing loss and their influence on treatment and outcome. Acta Otolaryngol. 2009:1 8. [DOI] [PubMed] [Google Scholar]
- 24. Fink NE, Wang NY, Visaya J, et al. Childhood Development after Cochlear Implantation (CDaCI) study: design and baseline characteristics. Cochlear Implants Int. 2007;8(2):92 116. ( 10.1179/cim.2007.8.2.92) [DOI] [PubMed] [Google Scholar]
- 25. Helzner EP, Patel AS, Pratt S, et al. Hearing sensitivity in older adults: associations with cardiovascular risk factors in the health, aging and body composition study. J Am Geriatr Soc. 2011;59(6):972 979. ( 10.1111/j.1532-5415.2011.03444.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Requena T, Gazquez I, Moreno A, et al. Allelic variants in TLR10 gene may influence bilateral affectation and clinical course of Meniere’s disease. Immunogenetics. 2013;65(5):345 355. ( 10.1007/s00251-013-0683-z) [DOI] [PubMed] [Google Scholar]
- 27. Elhayek D, Perez de Nanclares G, Chouchane S, et al. Molecular diagnosis of distal renal tubular acidosis in Tunisian patients: proposed algorithm for northern Africa populations for the ATP6V1B1, ATP6V0A4 and SCL4A1 genes. BMC Med Genet. 2013;14:119. ( 10.1186/1471-2350-14-119) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hildebrand MS, Tack D, McMordie SJ, et al. Audioprofile-directed screening identifies novel mutations in KCNQ4 causing hearing loss at the DFNA2 locus. Genet Med. 2008;10(11):797 804. ( 10.1097/GIM.0b013e318187e106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wright J, Small N, Raynor P, et al. Cohort profile: the born in Bradford multi-ethnic family cohort study. Int J Epidemiol. 2013;42(4):978 991. ( 10.1093/ije/dys112) [DOI] [PubMed] [Google Scholar]
- 30. Lin SW, Lin YS, Weng SF, Chou CW. Risk of developing sudden sensorineural hearing loss in diabetic patients: A population-based cohort study. Otol Neurotol. 2012;33(9):1482 1488. ( 10.1097/MAO.0b013e318271397a) [DOI] [PubMed] [Google Scholar]
- 31. Nosrati-Zarenoe R, Arlinger S, Hultcrantz E. Idiopathic sudden sensorineural hearing loss: results drawn from the Swedish national database. Acta Otolaryngol. 2007;127(11):1168 1175. ( 10.1080/00016480701242477) [DOI] [PubMed] [Google Scholar]



 Content of this journal is licensed under a
Content of this journal is licensed under a