Abstract
A method based on the use of the bacterial luciferase genes was developed in order to investigate Lactococcus lactis gene expression in the mouse digestive tract. Germfree mice were monoassociated with different strains containing transcriptional fusions of promoters with the luciferase genes. Our results demonstrate that this method is readily applicable to the study of promoter strength and physiology of bacteria in the digestive tract.
Lactic acid bacteria are widely used in food technology and may have a so-called “probiotic effect” (4, 10). However, the metabolic activities of these bacteria in the digestive tract (DT) remain uncertain, and this fact impairs our understanding of the beneficial effect of these organisms on health. The lactic acid bacteria adapt their metabolism to the environment by synthesizing proteins whose activities are required for survival and, eventually, proliferation. The level of transcription of regulated promoters reflects the need of the cells for particular functions according to the physiological state and environment stimulation. The activity of regulated promoters, therefore, reflects the physiological state of the cells.
Moreover, the use of lactic acid bacteria as vectors for new molecules with targeted activity in hosts is being investigated (5, 11, 18). Being able to measure promoter strength in the DT is a prerequisite for expression of heterologous proteins at suitable levels. This paper presents an experimental model for studying promoter activities in the DT by using the bacterial luciferase gene as a reporter gene.
Lactococcus lactis was grown in M17 (17) or chemically defined medium (CDM) (13) supplemented with erythromycin (5 μg/ml). Thermoresistant Bacillus subtilis spores were used as transit markers (2). The L. lactis strains used are derivatives of NCFB2118 (National Collection of Industrial and Marine Bacteria, Aberdeen, United Kingdom) containing integrative plasmids or transposons. These elements carry the lux genes from Vibrio harveyi fused with L. lactis promoters as described previously (15). A fragment from pJIM2366 (15) containing the fusion of lux genes and Pald, the promoter for the acetolactate decarboxylase gene from L. lactis (fragment from position 24152 to 24716 from GenBank sequence accession no. V92974), with an erythromycin marker was subcloned in the HindIII site present in tetM from Tn916 carried by pBluescript (Stratagene, CF). This construction was used to replace the tetM gene from Tn916 by transformation of a derivative of B. subtilis 168 (1). JIM5192 was obtained by introducing the transposon into NCFB2118 by conjugation. JIM5206, JIM5213, and JIM5460 are transformants of NCFB2118 containing the luxAB gene fused with the his promoter (Phis; segment fragment from position 1 to 1296 from GenBank sequence accession no. M90760), a truncated derivative of the his promoter (PhisD3; segment fragment from position 1 to 948), and the mleS promoter (Pmle; segment fragment from position 1 to 468 from GenBank sequence accession no. X75982), respectively. The plasmids used to integrate these fusions into the chromosome are derivatives of pORI28 (9), and the method used to select single crossover events has been described previously (6).
Germfree C3He/J mice (10 mice per group) were reared in a Trexler isolator fitted with a rapid transfer system (La Calhène, Vélizy, France). All materials introduced into the isolator were sterilized by irradiation or heating. Each mouse received 0.5 ml of a 24-h culture of L. lactis (about 4 × 108 CFU/ml) by the orogastric route. Malate inoculation was performed as follows: each mouse received orogastrically 0.5 ml of malate (10% in 0.15 M NaCl) at 10 and 12 a.m. Fecal samples were collected at 2 p.m., and the samples were diluted 1/3 with sterile distilled water. Luciferase activities (Lux) and L. lactis counts were estimated from these dilutions as previously described (15). Promoter strength was expressed as the logarithm of the number of relative light units per 108 CFU.
Different groups of germfree mice inoculated with a single L. lactis strain (gnotoxenic mice) were studied. One day after inoculation, the bacteria were established in the DT, and their populations remained stable (about 108 CFU/g of fecal sample) for several months. It has been shown previously that this animal model allows L. lactis to reach a stable population level in fecal samples (7). However, the metabolic activities of the bacteria were unknown.
In the present study, luciferase genes were used as biosensors to study the physiology of L. lactis established in DT. The bacterial luciferase gene has been shown to be a potential reporter gene in L. lactis (3, 15). Light emission with intact cells requires nonaldehyde as the substrate and reduced flavin mononucleotide (FMN) produced by the bacteria (12). The availability of reduced FMN depends on the physiological state of the cells, a condition that restricts the use of luxAB reporter genes to fully viable cells (8).
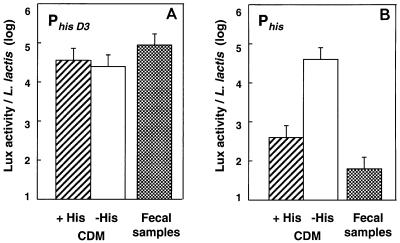
We first studied L. lactis JIM5213. In this strain the lux genes are under control of PhisD3, a truncated derivative of the histidine operon promoter which is deregulated in CDM with or without histidine (Fig. 1A). The amount of Lux activity per L. lactis cell was on the same order of magnitude in laboratory media and in feces of gnotoxenic mice colonized with L. lactis (Fig. 1A). This showed that L. lactis is metabolically active and contains nonlimiting amounts of reduced FMN when it is established in the DT of gnotoxenic mice.
FIG. 1.
Comparison of the expression of the Phis promoters in culture broth (CDM with and without histidine) and in gnotoxenic mice colonized with L. lactis. The following two forms of the Phis promoter were used: PhisD3, which is the truncated form of Phis (A), and Phis, which is the wild-type promoter (B). Lux activities and bacterial counts were determined for each mouse fecal sample. The vertical bars represent the standard errors of the means.
The lux construction JIM5206 contains the wild-type histidine promoter (Phis), which is turned off and on in CDM with and without histidine, respectively (Fig. 1B). In gnotoxenic mice, the strength of this promoter was even less than its strength in CDM with histidine (Fig. 1B). The repression of Phis was presumably mostly due to the presence of histidine in fecal samples at a concentration slightly higher than the concentration in CDM (600 and 360 nmol/g in DT and CDM, respectively).
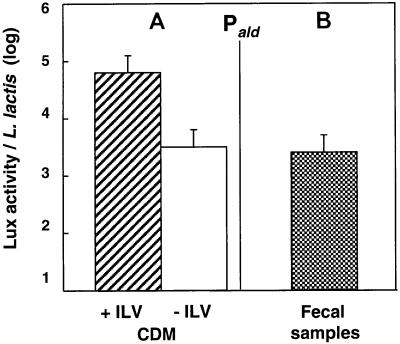
In L. lactis JIM5192, lux genes are under the control of Pald, which is induced in the presence of the three branched-chain amino acids (isoleucine, leucine, and valine [ILV]) (Fig. 2A). The Lux activity in fecal samples was similar to the Lux activity observed in CDM lacking ILV (Fig. 2B). However, the concentrations of free ILV were similar in the fecal samples and CDM (400 to 500 nmol/g in DT and CDM, respectively), suggesting that the absence of the branched-chain amino acids was not responsible for the low level of promoter expression. However, Pald has reduced activity under certain stress conditions. This response is known to be induced by amino acid starvation and low pH (14). Fusions with other promoters that reveal specifically different stress responses should be tested to determine the factor(s) involved in Pald closure in DT.
FIG. 2.
Comparison of the expression of the Pald promoter in culture broth (CDM with and without the branched-chain amino acids ILV) (A) and in gnotoxenic mice colonized by L. lactis (B). The vertical bars represent the standard errors of the means.
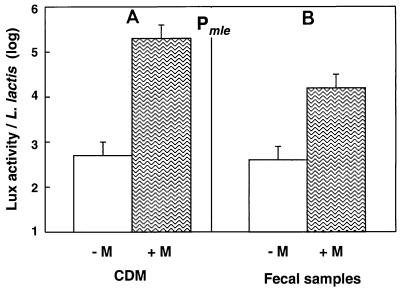
We tested our ability to stimulate L. lactis in the DT. To do this, we used JIM5460 containing a fusion with Pmle, the promoter for malolactic fermentation genes, which is induced by malate (16). Gnotoxenic mice harboring JIM5460 received malate orally, and the Lux activity of L. lactis was measured in fecal samples. The uninduced Pmle activities were low and similar in fecal samples and CDM (Fig. 3). A 40-fold increase was observed after malate was ingested. This increase was significant, although it was 10-fold lower than the increase observed in CDM with malate (Fig. 3). Activation was transient, as the Lux activity reached the uninduced level 1 day after supplementation and was again inducible another day later (data not shown). It appears that L. lactis is able to adapt its metabolism to this diet additive. The induction in the DT was not as effective as the induction in the laboratory medium, possibly because the malate given orally may have been absorbed in the small intestine, leading to reduced malate availability for L. lactis in the cecum.
FIG. 3.
Induction by malate (M) of the expression of the PmleS promoter in CDM (A) and in gnotoxenic mice colonized by L. lactis (B). The vertical bars represent the standard errors of the means.
To our knowledge, our work considering the total L. lactis population in the DT is the first work which reports an assessment of promoter strength in the DT. More information on the metabolic activity of bacterial cells in the DT could be obtained by using promoters specific for other pathways and responding to different signals. Knowledge concerning the expressed genes and metabolic activities of microorganisms ingested with food is important, as it has been proposed that some microorganisms are beneficial to human health (4, 10). This new approach for studying bacterial metabolism in vivo could help determine the role of microorganisms as probiotic organisms.
Acknowledgments
We thank G. Miranda for performing the amino acid analysis.
REFERENCES
- 1.Anagnostopoulos C, Spizizen J. Requirements for transformation in Bacillus subtilis. J Bacteriol. 1961;81:741–746. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ducluzeau R, Belier M, Raibaud P. Transit digestif de divers inoculum introduits per os chez les souris axeniques et holoxeniques (conventionnelles): effet antagoniste de la microflore du tractus gastro-intestinal. Zentbl Bakteriol Parasitenkd Infektionskr Hyg Abt 1 Orig. 1970;213:533–548. [PubMed] [Google Scholar]
- 3.Eaton T J, Shearman C A, Gasson M J. The use of bacterial luciferase genes as reporter genes in Lactococcus: regulation of the Lactococcus lactis subsp. lactis lactose genes. J Gen Microbiol. 1993;139:1495–1501. doi: 10.1099/00221287-139-7-1495. [DOI] [PubMed] [Google Scholar]
- 4.Elmer G W, Surawicz C M, McFarland L V. Biotherapeutic agents. A neglected modality for the treatment and prevention of selected intestinal and vaginal infections. JAMA. 1996;275:870–876. doi: 10.1001/jama.275.11.870. [DOI] [PubMed] [Google Scholar]
- 5.Fischetti V A, Medaglini D, Oggioni M, Pozzi G. Expression of foreign proteins on gram-positive commensal bacteria for mucosal vaccine delivery. Curr Opin Biotechnol. 1993;4:603–610. doi: 10.1016/0958-1669(93)90084-a. [DOI] [PubMed] [Google Scholar]
- 6.Godon J J, Jury K, Shearman C A, Gasson M J. The Lactococcus lactis sex-factor aggregation gene cluA. Mol Microbiol. 1994;12:655–663. doi: 10.1111/j.1365-2958.1994.tb01053.x. [DOI] [PubMed] [Google Scholar]
- 7.Gruzza M, Duval I Y, Ducluzeau R. Colonization of the digestive tract of germ free mice by genetically engineered strains of Lactococcus lactis: study of recombinant DNA stability. Microb Releases. 1992;1:165–171. [PubMed] [Google Scholar]
- 8.Lampinen J, Virta M, Karp M. Use of controlled luciferase expression to monitor chemicals affecting protein synthesis. Appl Environ Microbiol. 1995;61:2981–2989. doi: 10.1128/aem.61.8.2981-2989.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leenhouts K, Kok J, Venema G. Lactococcal plasmid pWVO1 as an integration vector for lactococci. Appl Environ Microbiol. 1991;57:2562–2567. doi: 10.1128/aem.57.9.2562-2567.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marteau P, Rambaud J C. Potential of using lactic acid bacteria for therapy and immunomodulation in man. FEMS Microbiol Rev. 1993;12:207–220. doi: 10.1111/j.1574-6976.1993.tb00019.x. [DOI] [PubMed] [Google Scholar]
- 11.Medaglini D, Pozzi G, King T P, Fischetti V A. Mucosal and systemic immune responses to a recombinant protein expressed on the surface of the oral commensal bacterium Streptococcus gordonii after oral colonization. Proc Natl Acad Sci USA. 1995;92:6868–6872. doi: 10.1073/pnas.92.15.6868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meighen E A. Molecular biology of bacterial bioluminescence. Microbiol Rev. 1991;55:123–142. doi: 10.1128/mr.55.1.123-142.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poolman B, Konings W N. Relation of growth of Streptococcus lactis and Streptococcus cremoris to amino acid transport. J Bacteriol. 1988;170:700–707. doi: 10.1128/jb.170.2.700-707.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rallu F, Gruss A, Maguin E. Lactococcus lactis and stress. Antonie Leeuwenhoek. 1996;70:243–251. doi: 10.1007/BF00395935. [DOI] [PubMed] [Google Scholar]
- 15.Renault P, Corthier G, Goupil N, Delorme C, Ehrlich S D. Plasmid vectors for Gram-positive bacteria switching from high to low copy number. Gene. 1996;183:175–182. doi: 10.1016/s0378-1119(96)00554-9. [DOI] [PubMed] [Google Scholar]
- 16.Renault P, Gaillardin C, Heslot H. Product of the Lactococcus lactis gene required for malolactic fermentation is homologous to a family of positive regulators. J Bacteriol. 1989;171:3108–3114. doi: 10.1128/jb.171.6.3108-3114.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Terzaghi B, Sandine W E. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol. 1975;29:807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wells J M, Robinson K, Chamberlain L M, Schofield K M, Le Page R W. Lactic acid bacteria as vaccine delivery vehicles. Antonie Leeuwenhoek. 1996;70:317–330. doi: 10.1007/BF00395939. [DOI] [PubMed] [Google Scholar]