Abstract
A tod-luxCDABE fusion was constructed and introduced into the chromosome of Pseudomonas putida F1, yielding the strain TVA8. This strain was used to examine the induction of the tod operon when exposed to benzene, toluene, ethylbenzene, and xylene (BTEX) compounds and aqueous solutions of JP-4 jet fuel constituents. Since this system contained the complete lux cassette (luxCDABE), bacterial bioluminescence in response to putative chemical inducers of the tod operon was measured on-line in whole cells without added aldehyde substrate. There was an increasing response to toluene concentrations from 30 μg/liter to 50 mg/liter, which began to saturate at higher concentrations. The detection limit was 30 μg/liter. There was a significant light response to benzene, m- and p-xylenes, phenol, and water-soluble JP-4 jet fuel components, but there was no bioluminescence response upon exposure to o-xylene. The transposon insertion was stable and had no negative effect on cell growth.
Due to the widespread use of petroleum products and the current regulations requiring underground storage tanks to be upgraded, replaced, or closed by December 1998 (4), the number of petroleum-contaminated sites has abounded. Of particular concern for drinking water quality are the more water-soluble components, benzene, toluene, ethylbenzene, and xylenes (BTEX). Natural attenuation, which relies on in situ biodegradation of pollutants, has received a large amount of attention, especially for petroleum contaminants (15). While microorganisms capable of biodegradation of BTEX compounds are usually present at these sites, there is a need to know whether or not conditions are favorable for biodegradation to occur. A recent approach to determine whether compounds are bioavailable and what conditions are favorable for degradation is the use of whole-cell bioluminescent reporters (9).
Bioluminescent reporters have been widely used for the real time nondestructive monitoring of gene expression. Heitzer et al. (8) developed a quantitative assay for naphthalene bioavailability and biodegradation by using a nah-lux reporter strain constructed by King et al. (13) and expanded its use as an on-line optical biosensor for application in groundwater monitoring (10). Other lux fusions have been constructed for monitoring the expression of catabolic genes, including those for degradation of isopropylbenzene (21) and toluene (1, 5). lux fusions have also been constructed for monitoring heat shock gene expression (24, 25), oxidative stress, (3), the presence of Hg(II) (20), and alginate production (26). In all of these cases, the lux fusions were plasmid based and were constructed by placing the promoter of interest in front of the promoterless lux genes from Vibrio fischeri contained in pUCD615 (18).
In this study, a strategy was pursued to introduce a single copy of the lux fusion into the bacterial chromosome via a transposon delivery system. A mini-Tn5 delivery vector constructed by Herrero et al. (11) provided the basic model for this work. By this approach, a tod-lux fusion was constructed and introduced into Pseudomonas putida F1 to examine the induction of the tod operon when exposed to BTEX compounds and aqueous solutions of JP-4 jet fuel constituents. Since this system contains the complete lux cassette (luxCDABE), bacterial bioluminescence was measured on-line in whole cells without addition of an aldehyde substrate. The resultant strain was also evaluated for its stability and fitness compared to those of the wild-type strain, F1.
Organisms and culture conditions.
The strains used in these experiments are shown in Table 1. All cultures were grown at 28°C with appropriate antibiotic selection, except for Escherichia coli strains, which were grown at 37°C.
TABLE 1.
Strains and plasmids used in this study
| Plasmid or strain | Relevant genotype or characteristics | Source or reference |
|---|---|---|
| Plasmids | ||
| pDTG514 | pGem3Z with a 2.75-kb EcoRI-SmaI fragment from pDTG350 containing the tod promoter, Ptod; Apr | 14 |
| pUCD615 | Promoterless luxCDABE cassette containing ori pSa and ori pBR322; Apr Kmr | 18 |
| pKK223-3 | Expression vector containing the 5S ribosomal terminator rrnB T1T2 | Pharmacia |
| pBSKS | pBluescript II KS+ with MCS KpnI-SacI; Apr | Stratagene |
| pBSMCS(−) | pBluescript without MCS (BssHII-BssHII fragment removed); Apr | This study |
| pLJS | pBSMCS(−) with added XbaI, NheI, AvrII, and SpeI sites; Apr | This study |
| pLJS-tod | pLJS containing the 1.8-kb SmaI-XhoI tod promoter fragment from pDTG514; Apr | This study |
| pLJS-lux | pLJS containing the 8.35-kb KpnI-PstI luxCDABE cassette from pUCD615; Apr | This study |
| pLJST2 | pLJS containing the 0.77-kb HindIII-HincII fragment from pKK223-3 cloned into HindIII-SmaI site; Apr | This study |
| pUC18Not | Cloning vector containing MCS flanked by NotI sites; Apr | 11 |
| pUC18Not-lux | Contains the 8.35-kb XbaI-PstI fragment from pLJS-lux; Apr | This study |
| pUC18Not-todlux | Contains the 1.8-kb SpeI-XhoI fragment from pLJS-tod; Apr | This study |
| pUT | 5.2-kb cloning vector containing mob RP4, ori R6K and Tn5 tnp lacking NotI sites; Apr | 11 |
| pCR II | 3.9-kb cloning vector for PCR products with 3′ A overhangs; Apr Kmr | Invitrogen |
| pUTK209 | pCR II containing mini-Tn5KmNX with unique NotI and XbaI sites; Apr Kmr | This study |
| pUTK210 | pUT containing mini-Tn5KmNX; Apr Kmr | This study |
| pUTK211 | pUT/mini-Tn5KmT2 containing the 0.8-kb NotI-AvrII rrnB T1T2 fragment; Apr Kmr | This study |
| pUTK214 | pUT/mini-Tn5Kmtod-lux containing the 10.2-kb NotI-XbaI fragment from pUC18Not-todlux; Apr Kmr | This study |
| Strains | ||
| E. coli | ||
| DH5α | F−ϕ80lacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rK− mK+) phoA supE44 λ− thi-1 gyrA96 relA1 | Gibco BRL |
| S17-1(λpir) | λpir recA thi pro hsdR M+ RP4:2-Tc:Mu:Km Tn7TprSmr; mobilizing strain for pUT/mini-Tn5 derivatives | 6 |
| INVαF′ | Strain used with TA cloning vector, pCR II; F′ ϕ80lacZαΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rK− mK+) phoA supE44 λ− thi-1 gyrA96 relA1 | Invitrogen |
| P. putida | ||
| F1 | Contains a chromosomally encoded tod operon for toluene degradation | 28 |
| TVA8 | F1 containing a mini-Tn5Kmtod-lux insertion in the chromosome; Kmr | This study |
DNA isolation and manipulation.
Large-scale plasmid DNA isolation was done by a modified alkaline lysis protocol (16). Chromosomal DNA was prepared by the protocol outlined by Ausubel et al. (2). All DNA preparations were further purified by CsCl-ethidium bromide ultracentrifugation (19). DNA modifications and restriction endonuclease digestions were performed as outlined by Sambrook et al. (19).
Transposon and plasmid construction.
The transposon mini-Tn5KmNX was constructed with two 58-base oligonucleotides 5′ and 3′ with respect to the kanamycin resistance gene (Kmr) in pCR II (Invitrogen, San Diego, Calif.) (I end, 5′GGGCGCTAGCGAAATGTTGACTGTCTCTTGATCAGATC TTTCAATTCAGAAGAACTCG3′; O end, 5′CGAATTCTGACTCTTATACACAAGTTCTAGATTGCGGCCGCTTGG TTAAAAAATGAGC3′). Oligonucleotides were synthesized with a Beckman Oligo 1000 DNA synthesizer (Palo Alto, Calif.). Base substitutions were made to generate both I and O insertion sequences as well as unique NotI and XbaI sites inside the transposon for cloning. An extra adenine was mistakenly added between the NotI and XbaI sites in the O primer, but it did not affect the construction. Primers were used to amplify the kanamycin resistance gene from pCR II by using touchdown PCR (7). The manufacturer’s protocol was used with the following thermocycler conditions. Initial denaturation at 94°C for 5 min, followed by five cycles of denaturation at 94°C for 1 min, 72°C annealing for 1 min, and 72°C extension for 2 min. The annealing temperature was then lowered 5°C every five cycles until 42°C, at which point, eight cycles were run, followed by a final extension of 15 min at 72°C. The resultant PCR fragment was cloned into the transposon delivery vector pUT, generating pUTK210. The cloning vector pLJS was constructed from pBluescript II (KS) (Stratagene, LaJolla, Calif.) by cleavage with BssHII removing the multicloning site (MCS). The resultant plasmid was named “pBSMCS(−).” Two oligonucleotides (a 47-mer and a 44-mer) (KpnI end, 5′CCAAGCGCGCAACTAGTCTAGACTAAAGCTAGCCTAGGCTGGGATCC3′; SacI end, 5′GTGAGCGCGCGTAATACGAGCTAGCCTAGGGCGAATTGGAGCA C3′) were synthesized to regenerate the MCS and add the restriction sites XbaI, NheI, SpeI, and AvrII. The orientation of the added sites can be seen in Fig. 1. The new MCS was amplified from pBluescript II (KS) by using the manufacturer’s protocol with the following thermocycler conditions. Initial denaturation was at 94°C for 5 min, followed by 38 cycles of denaturation at 94°C for 30 s, annealing at 42°C for 1 min, extension at 72°C for 30 s, and final extension at 72°C for 15 min. The amplified fragment was cleaved with BssHII, ligated into pBSMCS(−), and transformed into DH5α. A portion of pLJS was sequenced, confirming the base substitutions and integrity of the MCS, with an Applied Biosystems model 373A sequencer (Foster City, Calif.). Plasmid pLJST2 was generated by directional cloning of the 0.8-kb HindIII-HincII fragment containing the 5S ribosomal rrnB T1T2 transcription terminator from pKK223-3 (Pharmacia, Piscataway, N.J.) into pLJS cleaved with HindIII and SmaI. The NotI-AvrII terminator fragment from pLJST2 was subsequently cloned into the NotI-XbaI site of pUTK210, yielding pUTK211 containing mini-Tn5KmT2. This allowed for the subsequent destruction of the XbaI site by heterologous ligation and the regeneration of the NotI and XbaI unique sites downstream of the terminator. Mini-Tn5Kmtod-lux (pUTK214) was generated by directional cloning of the 10.2-kb NotI-XbaI tod-lux fragment from pUC18Not-todlux (Table 1) into the NotI-XbaI site of pUTK211. Both the insert and vector DNA were purified by agarose gel electrophoresis and electroeluted prior to cloning. Electrocompetent E. coli S17-1(λpir) cells were prepared and ligations were electroporated as outlined by the manufacturer (BTX, San Diego, Calif.). All other plasmids and relevant constructs are described in Table 1.
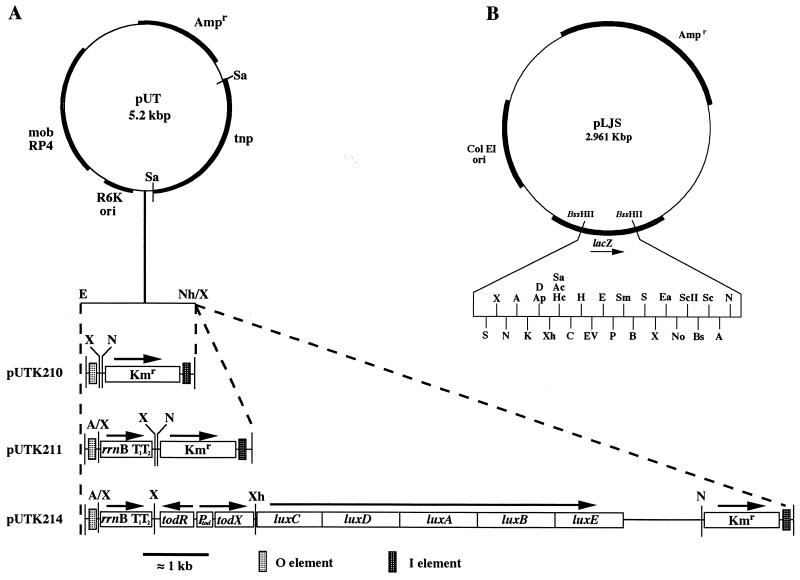
FIG. 1.
(A) Construction of mini-Tn5Kmtod-lux. A/X and Nh/X represent the AvrII-XbaI and NheI-XbaI heterologous cloning sites, respectively. N, NotI; Sa, SalI; X, XbaI. (B) Cloning plasmid pLJS with unique restriction sites. A, AvrII; Ac, AccI; Ap, ApaI; B, BamHI; Bs, BstXI; C, ClaI; D, DraII; E, EcoRI; Ea, EagI; EV, EcoRV; H, HindIII; Hc, HincII; K, KpnI; Nh, NheI; P, PstI; S, SpeI; Sc, SacI; ScII, SacII; Sm, SmaI; Xh, XhoI.
Strain construction.
Plasmid pUTK214 was conjugated into P. putida F1 from E. coli S17-1(λpir) as previously described (6). Strains carrying transposon insertions were selected on Pseudomonas isolation agar (Difco, Detroit, Mich.) supplemented with 50 μg of kanamycin/ml. Colonies which produced light upon exposure to toluene were grown in mineral salts media (MSM) (23) with toluene vapor to ascertain that the transposon had not inserted into a gene required for cell growth and also to evaluate their performance as bioreporters in liquid, growing-cell assays (8). A strain designated TVA8 was selected for further study and subjected to DNA-DNA hybridization to verify transposition, as opposed to recombination, by using a 32P-labeled probe specific for the Tn5 transposase (tnp) contained on pUT. Equal target amounts of luxA, todC, and tnp DNA were loaded onto a Biotrans nylon membrane (ICN, Irvine, Calif.) by using a Bioslot blot apparatus (Bio-Rad, Hercules, Calif.) according to the manufacturer’s protocol. The blot consisted of chromosomal DNA from F1, TVA8, and the aforementioned controls. The DNA was loaded in triplicate, the blot was subdivided, and each separate blot was hybridized with either luxA, todC, or tnp PCR-generated 32P-labeled DNA probes. Blots, hybridized and washed as previously described (1), verified that TVA8 contained luxA and todC but not tnp (data not shown). The negative transposase result confirmed that transposition had occurred.
Stability assays.
Batch stability assays were performed by transfer of 1 ml of a 100-ml overnight culture grown in Luria-Bertani (LB) broth with 50 μg of kanamycin/ml (LBKm50) to a 250-ml Erlenmeyer flask with toluene used as the sole carbon source (supplied as vapor). One milliliter of culture was transferred every day for 5 days to flasks containing 100 ml of MSM supplied with toluene vapor (without Km50). Assays were performed in triplicate. Before each transfer, cells were plated on selective (LBKm50) and nonselective (LB) media to ascertain loss of kanamycin resistance resulting from deletion or excision of the transposon. Colonies were subjected to colony hybridization with a 295-bp luxA DNA probe (12). Stability was also assayed in continuous culture with a New Brunswick Bio Flow fermentor (Edison, N.J.) with a 370-ml vessel operated at 28°C at 180 rpm. The feed consisted of MSM supplemented with toluene at approximately 100 mg/liter at a flow rate of 1.0 ml/min. Toluene was fed by simultaneously adding toluene-saturated MSM at a flow rate of 0.2 ml/min and MSM at a flow rate of 0.8 ml/min by using FMI metering pumps (Oyster Bay, N.Y.). The fermentor was operated for 14 days (100 generations) with daily bioluminescence and optical density (OD) measurements. Plate counts (at 7 and 14 days) from selective (LBKm50) and nonselective (LB) media were compared to determine if the kanamycin marker was being lost, and luxA colony blot hybridization was performed to confirm that all colonies contained the lux transposon insert. In batch and chemostat stability studies, TVA8 did not demonstrate instability when subjected to the same evaluation. From batch assays, the selective/nonselective plate count ratio was 1.12 ± 0.13 after five daily transfers, and all colonies hybridized with the luxA probe. After a 14-day continuous cultivation, the selective/nonselective plate count ratio was 1.05 ± 0.13, and all colonies from selective and nonselective plates were lux positive.
Comparison of growth of TVA8 and F1 on toluene.
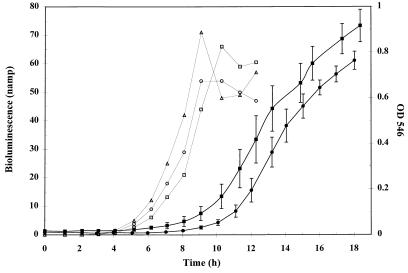
To examine the effect of bioluminescence on the fitness of TVA8, growth curves of TVA8 and F1 were obtained by growing cells in 100 ml of MSM in 250-ml Erlenmeyer flasks with toluene vapor supplied as the sole carbon source. Flasks were inoculated from a fresh overnight culture, grown to an OD at 546 nm (OD546) of 1.0 in 100 ml of LB, washed twice in 100 ml of MSM, and resuspended in 100 ml of MSM. A 1-ml aliquot of this suspension was added to the toluene flasks. The cultures were shaken at 200 rpm at 28°C and sampled approximately every hour. The OD546 was measured for each culture, and rates of increase in OD were determined from the linear portion of the curves. Growth curves for TVA8 and the parent strain F1 on toluene vapor are shown in Fig. 2. The curves show similar shapes with different lag times for TVA8 and F1 that can be attributed to slightly different inoculum concentrations. Rates were computed from the slopes of the linear portion of the growth curve for both strains. The average rates of increase in OD for F1 and TVA8 were (2.1 ± 0.3) and (2.2 ± 0.3) min−1 × 10−3, respectively and were not statistically different (α = 0.05). These results demonstrate that the bioluminescence reactions do not appear to affect cell growth.
FIG. 2.
Bioluminescence and growth of TVA8 and growth of F1 on toluene vapor under batch conditions. ○, □, and ▵ represent individual replicates of bioluminescence readings over time. The solid squares (TVA8) and circles (F1) represent the average OD546 of three replicates.
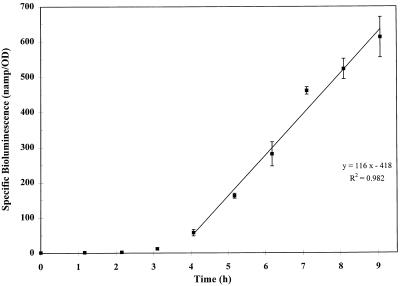
Bioluminescence of TVA8 was measured during growth on toluene and is shown along with the cell density data in Fig. 2. The graph shows that there is a relationship between an increase in biomass and an increase in light production. At higher cell densities, cells likely became limited for oxygen, resulting in decreased bioluminescence. Specific bioluminescence (nanoamperes/OD546) of TVA8 versus time shows an increase in specific bioluminescence (Fig. 3). This suggests that a steady state of luciferase in the cell is not obtained in this time frame and that luciferase is accumulating (Fig. 3).
FIG. 3.
Specific bioluminescence of TVA8 grown on toluene vapor. The regression line equation is y = 116x − 418 (r2 = 0.982). (The first four time points were not included in the linear regression because the organisms were in lag phase.)
Bioluminescence response of TVA8 to toluene, BTEX compounds, and JP-4 jet fuel.
Bioluminescence assays were conducted as described by Heitzer et al. (8). Aqueous solutions of toluene, benzene, ethylbenzene, phenol, isomers of xylene, and JP-4 jet fuel constituents were prepared by adding pure hydrocarbon or JP-4 to MSM in a 1:10 (vol/vol) ratio. The solutions were placed on a rotary shaker for 24 h. After phase separation, aqueous-phase aliquots were added to test vials. Final concentrations of dissolved toluene in growing-cell assays ranged from 0 to 50 mg/liter (based on water solubility). The final concentration of the other hydrocarbons was 50 mg/liter (based on their water solubility), and the percentage of JP-4-saturated MSM in the test samples was 2%. Vials containing test solutions and cells were shaken at 150 rpm on a rotary shaker, and bioluminescence was measured every 30 min. Sample vials were placed in a light-tight box, and light output was measured with a liquid light pipe and an Oriel photomultiplier and digital display (models 77340 and 7070; Stratford, Conn.) (8). The light detection methods for continuous culture and growth curves were similar, except that the light-tight box was modified to hold a cuvette, allowing for light measurements and OD readings to be taken consecutively.
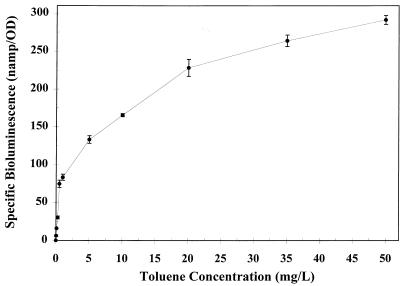
In preliminary experiments, an incubation time of 2 h was shown to provide a consistent light response and signal intensity. After 2 h, the final OD546 was measured, and values were expressed as specific bioluminescence (nanoamperes/OD546). An increase in bioluminescence was observed to correlate with increasing toluene concentrations (Fig. 4). The bioluminescence response to toluene concentrations over the range of 5 to 20 mg/liter was linear, with specific bioluminescence values of 133 to 228 nA/OD546. The fold increase in light response for concentrations above 20 mg/liter was less, showing a specific bioluminescence value of 290 nA/OD546 at 50 mg of toluene per liter. The overall bioluminescence response curve exhibited Michaelis-Menten kinetics, showing saturation at higher toluene concentrations. The toluene detection limit was determined to be 30 μg/liter (threefold increase over background bioluminescence). There was a significant light response to benzene, m- and p-xylenes, phenol, and JP-4 (Table 2). The same concentrations of toluene and benzene (50 mg/liter) resulted in a similar light response. There was no increase in bioluminescence upon exposure to o-xylene. The light response due to JP-4 was significantly greater than the additive responses for JP-4 components (i.e., BTEX compounds) present at their estimated concentrations in water saturated with JP-4 (22). The increased response may be the result of induction due to other components of JP-4 which were not tested. A significant light response was observed for ethylbenzene after 4 h. After a 2-h incubation, the cell densities for the ethylbenzene treatments were significantly less than those for the other samples, indicating that there may have been a toxicity effect. Further experiments showed that 50 mg of ethylbenzene per liter would induce the bioluminescence response without a lag period when cells were previously grown on ethylbenzene and then subjected to growing-cell assays.
FIG. 4.
Bioluminescence response of TVA8 to increasing concentrations of toluene after a 2-h exposure. Values are averages of three replicates and have been normalized to the cell density (OD546).
TABLE 2.
Effect of BTEX, phenol, and JP-4 constituents on the bioluminescence response of TVA8
| Treatmenta | Exposure time (h) | Specific bioluminescence (nA/OD)b |
|---|---|---|
| Buffer (control) | 2 | 0.2 ± 0.1 |
| Toluene | 2 | 234 ± 7 |
| Benzene | 2 | 242 ± 9 |
| Ethylbenzene | 2 | 1.0 ± 0.2 |
| 4 | 47 ± 6c | |
| o-Xylene | 2 | 0.5 ± 0.1 |
| m-Xylene | 2 | 38 ± 3 |
| p-Xylene | 2 | 24 ± 2 |
| Phenol | 2 | 70 ± 2 |
| JP-4 | 2 | 93 ± 4 |
Final concentration for BTEX and phenol treatments was 50 mg/liter (based on water solubilities), and they were added as a hydrocarbon-saturated MSM solution. The final percentage of water-soluble JP-4 constituents was approximately 2%.
Values are averages ± standard deviations of three replicate samples. Values were normalized to the final cell density (OD546).
The value for the 4-h reading was measured from a similar but separate experiment.
Conclusions.
The majority of bioluminescent reporter systems currently being used are the result of cloning of a promoter in front of either a promoterless luxAB or luxCDABE gene cassette and transfer of the plasmid construct into the strain that contained the particular promoter. Plasmid-based systems have obvious drawbacks, such as the need for constant selective pressure to ensure plasmid maintenance (17). Another important consideration is plasmid copy number. In a positively regulated system, copy number can negatively effect gene expression. Multiple copies of the promoter binding region for the regulatory protein on the plasmid compete with the binding site on the chromosome, causing less expression of the operon being studied (27). This negative effect is important when lux fusions are used for on-line monitoring of bacterial processes.
TVA8 was capable of growth on MSM with toluene as a sole carbon source, demonstrating that the transposon insertion did not disrupt a gene necessary for growth. Furthermore, TVA8 did not show loss of the transposon insertion or loss of bioluminescence after 100 generations in continuous culture or five successive transfers in batch culture without antibiotic selection. These results indicate that selective pressure is not necessary for strain integrity. This stability is important, since it eliminates the need for antibiotic selection, which, if required, would exclude the use of this bioreporter in situ. TVA8 was also compared to the wild-type strain, F1, to ascertain whether or not the bioluminescent reporter incurred a significant metabolic demand on the cell, as well as whether the site of transposition was a hindrance to the cell. No difference in growth between the two strains was seen, suggesting that neither the insertion site nor the lux fusion was a significant handicap to the cell.
The tod-lux reporter was highly sensitive, detecting 30 μg of toluene/liter. This bioreporter also showed a very low background level of bioluminescence (less than 1 nA/OD546), demonstrating its usefulness for detecting toluene present at low concentrations in aqueous solutions. Significant light levels were observed for very low ODs (Fig. 2).
TVA8 can be described as a generalized BTEX bioreporter rather than simply a toluene bioreporter, since it was responsive to benzene, ethylbenzene, and m- and p-xylene and can therefore be used as a bioreporter for hydrocarbon contamination for fuels containing BTEX compounds. TVA8 can also be used for on-line monitoring of trichloroethylene cometabolism, since the lux and tod operons are under the same regulation (toluene dioxygenase catabolizes trichloroethylene).
Bioluminescent reporters may have great potential for field use applications, since they can provide on-line and nondestructive analyses of gene expression as well as detection of chemical contaminants. The development of stable transposon insertions of lux reporter gene fusions into environmental isolates expands the utility of bioreporter strains for in situ sensing of gene expression.
Acknowledgments
We thank V. de Lorenzo for providing strain SV17-1(λpir) and plasmid pUT and for helpful comments. We are also grateful to D. T. Gibson, M. Rawlings, and C. Kado for providing strains and plasmids and S. Ripp for editing the manuscript.
This research was supported by TVA grant TV-94002V and U.S. DOE grant DE-FG05-94ER61870; Office of Health and Environmental Research and graduate fellowship support was provided by the University of Tennessee’s Waste Management Research and Education Institute (B.A.). Support was also received through Air Force grant F49620-89-C-0023 (S.K.).
REFERENCES
- 1.Applegate B, Kelly C, Lackey L, McPherson J, Kehrmeyer S, Menn F-M, Bienkowski P, Sayler G S. Pseudomonas putida B2: a tod-lux bioluminescent reporter for toluene and trichloroethylene co-metabolism. J Ind Microbiol. 1997;18:4–9. doi: 10.1038/sj.jim.2900334. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons; 1989. [Google Scholar]
- 3.Belkin S, Smulski D R, Vollmer A C, Van Dyk T K, LaRossa R A. Oxidative stress detection with Escherichia coli harboring a katG′::lux fusion. Appl Environ Microbiol. 1996;62:2252–2256. doi: 10.1128/aem.62.7.2252-2256.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brinkley D. Selecting a storage tank system. Pollut Eng. 1997;29:76–79. [Google Scholar]
- 5.Burlage R S, Palumbo A V, Heitzer A, Sayler G S. Bioluminescent reporter bacteria detect contaminants in soil samples. Appl Biochem Biotechnol. 1994;45/46:731–740. [Google Scholar]
- 6.de Lorenzo V, Fernandez S, Herrero M, Jakubik U, Timmis K. Engineering of alkyl- and haloaromatic-responsive gene expression with mini-transposons containing regulated promoters of biodegradative pathways of Pseudomonas. Gene. 1993;130:41–46. doi: 10.1016/0378-1119(93)90344-3. [DOI] [PubMed] [Google Scholar]
- 7.Don R H, Cox P T, Wainwright B J, Baker K, Mattick J S. ‘Touchdown’ PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 1991;19:4008. doi: 10.1093/nar/19.14.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heitzer A, Webb O F, Thonnard J E, Sayler G S. Specific and quantitative assessment of naphthalene and salicylate bioavailability by using a bioluminescent catabolic reporter bacterium. Appl Environ Microbiol. 1992;58:1839–1846. doi: 10.1128/aem.58.6.1839-1846.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heitzer A, Sayler G S. Monitoring the efficacy of bioremediation. Trends Biotechnol. 1993;11:334–343. doi: 10.1016/0167-7799(93)90156-4. [DOI] [PubMed] [Google Scholar]
- 10.Heitzer A, Malachowsky K, Thonnard J, Bienkowski P R, White D C, Sayler G S. Optical biosensor for environmental on-line monitoring of naphthalene and salicylate bioavailability with an immobilized bioluminescent catabolic reporter bacterium. Appl Environ Microbiol. 1994;60:1487–1494. doi: 10.1128/aem.60.5.1487-1494.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herrero M, de Lorenzo V, Timmis K N. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J Bacteriol. 1990;172:6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnston W. Fate of Pseudomonas fluorescens 5RL and its reporter plasmid for naphthalene biodegradation in soil environments. Ph.D. dissertation. Knoxville: University of Tennessee; 1996. [Google Scholar]
- 13.King J M H, DiGrazia P M, Applegate B, Burlage R, Sanseverino J, Dunbar P, Larimer F, Sayler G S. Rapid, sensitive bioluminescence reporter technology for naphthalene exposure and biodegradation. Science. 1990;249:778–781. doi: 10.1126/science.249.4970.778. [DOI] [PubMed] [Google Scholar]
- 14.Menn F-M, Zylstra G J, Gibson D T. Location and sequence of the todF gene encoding 2-hydroxy-6-oxohepta-2,4-dienote hydrolase in Pseudomonas putida F1. Gene. 1991;104:91–94. doi: 10.1016/0378-1119(91)90470-v. [DOI] [PubMed] [Google Scholar]
- 15.National Research Council. In-situ bioremediation—when does it work? Washington, D.C: National Academy Press; 1993. [Google Scholar]
- 16.Promega. Promega technical bulletin 009. Madison, Wis: Promega; 1992. [Google Scholar]
- 17.Rice J F, Fowler R F, Arrage A A, White D C, Sayler G S. Effects of external stimuli on environmental bacterial strains harboring an algD-lux bioluminescent reporter plasmid for the study of corrosive biofilms. J Ind Microbiol. 1995;15:318–328. [Google Scholar]
- 18.Rogowsky P M, Close T J, Chimera J A, Shaw J J, Kado C I. Regulation of the vir genes of Agrobacterium tumefaciens plasmid pTiC58. J Bacteriol. 1987;169:5101–5112. doi: 10.1128/jb.169.11.5101-5112.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 20.Selifonova O, Burlage R, Barkay T. Bioluminescence sensors for detection of bioavailable Hg(II) in the environment. Appl Environ Microbiol. 1993;59:3083–3090. doi: 10.1128/aem.59.9.3083-3090.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Selifonova O V, Eaton R W. Use of an ipb-lux fusion to study regulation of the isopropylbenzene catabolism operon of Pseudomonas putida RE204 and to detect hydrophobic pollutants in the environment. Appl Environ Microbiol. 1996;62:778–783. doi: 10.1128/aem.62.3.778-783.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith J H, Harper J C, Jaber H. Analysis and environmental fate of Air Force distillate and high density fuels. Tyndall Air Force Base, Fla: Air Force Engineering and Services Center; 1981. [Google Scholar]
- 23.Stanier R Y, Palleroni N J, Doudoroff M. The aerobic pseudomonads: a taxonomic study. J Gen Microbiol. 1966;41:159–271. doi: 10.1099/00221287-43-2-159. [DOI] [PubMed] [Google Scholar]
- 24.Van Dyk T K, Majarian W R, Konstantinov K B, Young R M, Dhurjati P S, LaRossa R A. Rapid and sensitive pollutant detection by induction of heat shock gene-bioluminescence gene fusions. Appl Environ Microbiol. 1994;60:1414–1420. doi: 10.1128/aem.60.5.1414-1420.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van Dyk T K, Reed T R, Vollmer A C, LaRossa R A. Synergistic induction of the heat shock response in Escherichia coli by simultaneous treatment with chemical inducers. J Bacteriol. 1995;177:6001–6004. doi: 10.1128/jb.177.20.6001-6004.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wallace W H, Fleming J T, White D C, Sayler G S. An algD-lux bioluminescent reporter plasmid to monitor alginate production in biofilms. Microb Ecol. 1994;27:225–239. doi: 10.1007/BF00182407. [DOI] [PubMed] [Google Scholar]
- 27.Wang Y, Rawlings M, Gibson D T, Labbe D, Bergeron H, Brousseau R, Lau P C K. Identification of a membrane protein and a truncated LysR-type regulator associated with the toluene degradation pathway in Pseudomonas putida F1. Mol Gen Genet. 1995;246:570–579. doi: 10.1007/BF00298963. [DOI] [PubMed] [Google Scholar]
- 28.Zylstra G J, Gibson D T. Toluene degradation by Pseudomonas putida F1. Nucleotide sequence of the todC1C2BADE genes and their expression in Escherichia coli. J Biol Chem. 1989;264:14940–14946. [PubMed] [Google Scholar]