Summary
Misregulation of histone lysine methylation is associated with several human cancers and with human developmental disorders. DOT1L is an evolutionarily conserved gene encoding a lysine methyltransferase (KMT) that methylates histone 3 lysine-79 (H3K79) and was not previously associated with a Mendelian disease in OMIM. We have identified nine unrelated individuals with seven different de novo heterozygous missense variants in DOT1L through the Undiagnosed Disease Network (UDN), the SickKids Complex Care genomics project, and GeneMatcher. All probands had some degree of global developmental delay/intellectual disability, and most had one or more major congenital anomalies. To assess the pathogenicity of the DOT1L variants, functional studies were performed in Drosophila and human cells. The fruit fly DOT1L ortholog, grappa, is expressed in most cells including neurons in the central nervous system. The identified DOT1L variants behave as gain-of-function alleles in flies and lead to increased H3K79 methylation levels in flies and human cells. Our results show that human DOT1L and fly grappa are required for proper development and that de novo heterozygous variants in DOT1L are associated with a Mendelian disease.
Keywords: DOT1 Like histone lysine methyltransferase, DOT1L, histone lysine methyltransferase, H3K79 methylation, Drosophila, grappa, gpp, gain of function, developmental delay, congenital anomalies
This study identified individuals with de novo DOT1L variants associated with developmental delay and congenital anomalies. DOT1L encodes a histone methyltransferase and the variants led to increased methylation in flies and human cells. Hence, DOT1L now joins the list of histone lysine methyltransferases associated with Mendelian developmental disorders.
Introduction
Dynamic changes in chromatin state through post-translational modifications (PTMs) of histone proteins play a key role in gene expression regulation and genomic stability.1 One of the most common types of chromatin modifications is the methylation of lysine residues at histone tails, which is known to affect DNA repair, transcription, stress response, and differentiation.2 Misregulation of histone lysine methylation is associated with several human cancers and other disorders.3
Methylation of different types of histones at specific lysine residues is catalyzed by specific histone lysine methyltransferases (KMTs) and there are thirty-four KMT-encoding genes in humans.4 Of these, dysregulation (typically haploinsufficiency) of sixteen genes encoding KMTs have been previously associated with dominant human developmental disorders (Table S1). Most of these disorders of epigenetic regulation are also now known to have unique patterns of genome-wide DNA methylation alterations (“DNAm signatures”) in peripheral blood (Table S1). A review of the orthologs in available animal models of genes encoding KMTs demonstrates that both disease-associated and non-disease-associated genes encoding KMTs are important in animal development.5 Therefore, it is likely that additional KMTs will be associated with Mendelian disease over time.
DOT1 Like histone lysine methyltransferase (DOT1L [MIM: 607375]) is an evolutionarily conserved KMT found in a broad range of eukaryotic species including yeast, fruit fly, zebrafish, mice, and human.6 Compared to other KMTs, DOT1L has a number of distinct characteristics. It is the only KMT that does not contain the SET catalytic domain; instead, it contains an N-terminal Dot1 catalytic domain.7,8 Unlike SET-domain containing KMTs, which target lysine residues in the histone tails where most modifications occur, DOT1L methylates a lysine residue (K79) in the core globular domain of histone H3, and to our knowledge, it is the only enzyme known to do so.9 DOT1L, in contrast to other KMTs, seems to be a distributive enzyme that sequentially adds methyl groups to H3K79.10,11 It also differs from other KMTs as it methylates H3K79 only in the form of intact chromatin and not as free histones.12,13,14,15
DOT1L and H3K79 methylation have been implicated in several fundamental cellular processes, including gene expression,6 cell cycle regulation,16 DNA damage response,17 telomeric silencing,18 proliferation,19 and differentiation.20 Distribution of H3K79 methylation throughout the chromatin as well as DOT1L are strongly associated with actively transcribed genes.21,22,23,24 Indeed, DOT1L directly interacts with elongating RNA Polymerase II and gets recruited to actively transcribed genes.25 Possibly owing to its aforementioned cellular roles, DOT1L is a critical regulator during development, particularly in hematopoiesis of some metazoans. In mice, germline deletion of Dot1l leads to embryonic lethality due to defects in the yolk sack and heart and failure of primitive hematopoiesis.26 Similarly, dot1l-depleted zebrafish morphants show impaired growth, defective angiogenesis, and cardiac dilatation.27 Loss-of-function (LoF) mutants for the fly ortholog of DOT1L, called grappa (gpp), exhibit slow growth and die at larval stages.28 Furthermore, knockdown of DOT1L orthologs in the above listed animal models decreases expression of several Wnt signaling target genes.29,30,31 This suggests that loss of DOT1L affects Wnt signaling in multiple species but the precise mechanism as to how DOT1L regulates development of specific tissues and organs remains elusive.
Here, we present a cohort of nine individuals with a total of seven different de novo missense variants in DOT1L. Their phenotypes include central nervous system (CNS) dysfunction and a range of congenital anomalies, consistent with the variability associated with other KMT disorders (Table S1). We provide functional data in flies showing that six of the variants cause an increase in DOT1L enzymatic activity and disrupt development. We also show the remaining variant alters DOT1L activity in human cells. Our data support that de novo missense variants in DOT1L cause a KMT disorder through a gain-of-function mechanism.
Subjects and methods
Subject recruitment and sequencing
The two index families were identified through the Undiagnosed Disease Network (UDN)32,33 and the SickKids Complex Care genomics project,34,35 respectively. The five additional participants were identified through GeneMatcher.36,37 The initial GeneMatcher entry was made on October 17, 2017, and all matches made until 2020 were considered in this study (Table S2). Two additional individuals (probands #5 and #8) were identified through a pre-existing collaboration. Two families (three total individuals) with rare DOT1L missense variants suspected to be non-diagnostic also participated in aspects of this study for comparison purposes; they were identified through GeneMatcher (Table S2) and through a prior publication,5,38 respectively.
Genome- and exome-wide sequencing was performed at each participating center in either a clinical diagnostic laboratory or a research laboratory, using standard methods for sequencing, quality control, and analysis (Table S3). All DOT1L variants were high-confidence calls, and maternity and paternity were confirmed by a trio genome- or exome-wide sequencing design and/or other standard methods. This research was conducted with the voluntary, informed consent of any research participants, free of coercion or coercive circumstances, and received Institutional Review Board (IRB) or Research Ethics Committee (REC) approval consistent with the principles of research ethics and the legal requirements of the lead authors’ jurisdiction(s) (The Hospital for Sick Children, Canada; Baylor College of Medicine and NIH, USA).
DNAm profiling and data processing
Genomic DNA was extracted from peripheral blood and bisulfite converted using the EpiTect Bisulfite Kit (EpiTect PLUS Bisulfite Kit, QIAGEN). Sodium-bisulfite-converted DNA was then hybridized to the Illumina Infinium Human Methylation EPIC BeadChip to interrogate more than 850,000 CpG sites in the human genome at The Center for Applied Genomics (TCAG), Hospital for Sick Children Research Institute, Toronto, Ontario, Canada. On each microarray chip, cases and controls were randomly assigned a chip position. The minfi Bioconductor package in R was used to preprocess data including quality control, Illumina normalization, and background subtraction, followed by extraction of beta (β) values.39 Standard quality control metrics in minfi were used, including median intensity QC plots, density plots, and control probe plots. Probes with detection flaws (n = 691), probes near SNPs with minor allele frequencies above 5% (n = 91,246), cross-reactive probes (n = 39,371), probes with raw beta of 0 or 1 in >0.25% of samples (n = 236), non-CpG probes (n = 2,635), and X and Y chromosome probes (n = 18,653) were removed, resulting in a total of n = 716,401 probes remained for differential methylation analysis.
DNAm signature exploration
To assess DNAm patterns, we identified differentially methylated sites in whole-blood-derived DNA from individuals with suspected deleterious variants in DOT1L, sex- and age-matched typically developing controls, and individuals with suspected non-deleterious variants in DOT1L (Table S7). For all samples, we applied the blood cell-type proportion estimation tool in minfi based on Illumina EPIC array data from FACS-sorted blood cells.40 Two of the samples were not useable as part of a discovery cohort for technical reasons (age outliers), corresponding to the variants p.Cys45Gly (DNA sample obtained when the individual was an infant) and p.Lys1025Met (DNA sample from adult), and no sample was available for the deceased study participant. The DOT1L discovery cohort (n = 4) therefore included 2 females and 2 males with mean age at sample collection of 4.2 ± 2.2 years (range 2–8 years). The 12 sex- and age-matched control subjects included 8 females and 4 males and mean age at sample collection of 4.8 ± 2.3 years (range 1.3–9 years). We identified differentially methylated CpG sites using Limma41 regression modeling with age and PC1 as covariates. The thresholds for differentially methylated CpG sites were false discovery rate (FDR)-adjusted p value < 0.3 and a |Δβ| > 0.10. Δβ represents the difference in average DNAm (β) between groups. Principal component analysis (PCA) and hierarchical clustering were generated using Qlucore Omics Explorer (QOE, www.qlucore.com).
Fly husbandry and fly stocks
All flies were raised at room temperature, unless otherwise noted, on standard cornmeal and molasses medium in plastic vials.
All fly strains used in this study were created in house or obtained from the Bloomington Drosophila Stock Center (BDSC) or Kyoto Stock Center. The gppTG4 allele was created by insertion of an artificial exon containing a T2AGAL4 cassette into the third coding intron of gpp via CRISPR-mediated homologous recombination.42,43 gppxxv allele was created via X-ray mutagenesis and characterized previously.28 For complementation tests, immunostaining, and RNAi assays, see genotypes in Table S4. UAS-Empty is an empty pGW-HA.attB plasmid injected into the identical docking site as a control and has been published previously.44
Generation of human UAS-DOT1L transgenic lines
The DOT1L cDNA clone used in this study corresponds to GenBank: NM_032482.3, which encodes the canonical full-length DOT1L isoform and is defined as reference here. DOT1L variants were created via site directed mutagenesis (Q5 Site-Directed Mutagenesis Kit, NEB) on a pENTR223.1-DOT1L cDNA construct (Horizon Discovery, clone ID 100062384). Primers are detailed in Table S5. Reference and mutagenized cDNA products were carried into pGW-HA-attB- gateway expression vector45 using LR clonase II (ThermoFisher). Since the DOT1L cDNA clone does not contain a translational stop codon, all constructs are tagged with 3X-HA tag and a linker sequence. The constructs were sequence verified and injected into VK37 (BDSC #24872) docking site by phiC31-mediated transgenesis.46
Imaging of fly wings and eyes and of whole animals
For imaging of wings, fly wings were plucked and placed on a white filter paper. For imaging of eyes, whole flies were placed on a white filter paper in an angle so that the whole eye can be viewed. For imaging of whole larvae and adults, animals were placed on a black filter paper. Images were acquired using a digital camera (MicroFire; Olympus) mounted on a stereomicroscope (MZ16; Leica) with ImagePro Plus 5.0 acquisition software (Media Cybernetics). z stacks were taken by the “extend depth of field” function of the AxioVision software (web resources). Selected range of z stack images were max-projected using Imaris (web resources).
Real-time qPCR
Total RNA was extracted from larvae and adult flies using TRIzol (Invitrogen) according to manufacturer’s protocol. Biological triplicate samples of whole animals were processed per condition. qPCR was performed as previously described.44 High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems), SYBR Green Fast Master Mix (Thermo Fisher), and a BioRad C1000 Touch Cycler were used. A housekeeping gene, RpL32, was included for normalization. qPCR primers are listed in Table S5.
Immunostaining and confocal microscopy
Immunostaining of larval brains and imaginal discs, and adult brains were done as previously described.47 Briefly, dissected tissues were fixed in 4% paraformaldehyde (PFA) and blocked in 5% normal goat serum in 0.1% Triton X-100 in 1× PBS. Primary antibodies and dilutions used as follows: anti-Elav (DSHB #7E8A10; 1:250), anti-Repo (DSHB #8D12; 1:50). Donkey-derived secondary antibodies were used at 1:250 (Jackson ImmunoResearch Laboratories). A Leica Sp8x with lightning deconvolution was used for confocal microscopy. Images were taken with either a 20× or 40× Leica objective and processed using Imaris.
Expression of human DOT1L in Drosophila
We overexpressed reference and variant DOT1L cDNA molecules in flies in gpp expression domains by crossing the UAS-DOT1L males to virgin female flies from gppTG4 driver stock. The progeny of these crosses was cultured at 25°C. More than 100 flies were assessed for the expected Mendelian ratios. Percent viabilities (o/e ratios) from three independent experiments were plotted as mean ± SEM, and statistical significance was determined by one-way ANOVA for multiple groups.
For wing phenotypic distribution, the surviving overexpression flies were collected. For each genotype, more than 100 wings (>50 flies) were randomly selected and separated into different groups based on the severity of their wing phenotypes.
Detection of DOT1L and H3K79 methylation levels using western blot
We performed western blot analysis to detect DOT1L and H3K79 methylation status from larva and adult flies. For larval lysates, 10 animals were used. For adult lysates, 5 female and 5 male flies were used. Animals were lysed using a pestle with a cordless motor (VWR) in a buffer (30 μL per animal) containing 150 mM NaCl, 50 mM Tris-Cl (pH 8.0), 1.0% NP-40, 0.1% Triton X-100, 0.4% Na- Deoxycholate, 0.1% SDS in the presence of EDTA-free protease inhibitor (Roche), followed by rotation at 4°C for 20 min. After addition 1× lysis buffer without SDS and Na-Deoxycholate, lysates were sonicated at 20 kHz for 10 cycles of 10 s “on” and 10 s “off” and centrifuged at 1,000 rpm for 2 min. Supernatants were collected and treated with 50U Benzonase (EMD Milipore), 20 μg RNase A (QIAGEN), and 4 mM MgCl2 (final concentration) at 4°C for 30 min. Samples were centrifuged at 5,000 rpm at 4°C for 10 min and the cleared supernatant was collected in fresh 1.5 mL Eppendorf tube. Total of 50 μg protein for each sample was boiled with 2× Laemmli sample buffer. For detection of histones, samples were resolved on a 12% Bis-Tris SDS-polyacrylamide gel (Invitrogen) and immunoblotted with Rabbit anti-H3K79Me (Ab177185, Abcam, 1:1,000) and Mouse anti-H3 (05-499, EMD-Milipore, 1:1,000) antibodies. For detection of DOT1L, samples were resolved on a 3%–8% Tris-Acetate SDS-polyacrylamide gel (Invitrogen) and immunoblotted with Mouse anti-HA.11 (16B12, Biolegend, 1:1,000) and Rabbit anti-α-Tubulin (11H10, Cell signaling, 1:5,000) antibodies. Secondary antibodies Goat anti-Rabbit IRDye680 (Licor) and Goat anti-Mouse IRDye800 (Licor) were used in 1:5,000 dilution.
Recombinant plasmid construction for human and mouse cell culture assays
The full-length human reference DOT1L coding sequence (GenBank: NM_032482.3) was downloaded from NCBI and inserted into pcDNA5 vector, which was added a Flag exogenous tag (DYDDDK) at N-terminal of DOT1L. Meanwhile, two variants of DOT1L were constructed into the same vector pcDNA5 via site directed mutagenesis. The knockdown efficiency of the Dot1l-shRNA plasmid was verified through N2A cell line. The shRNA sequence: 5′-CCGGTCGCCAACACGAGTGTTATATCTCGAGATATAACACTCGTGTTGGCGATTTTTT-3′.
HEK293 cell culture
HEK293 FLP-In cell lines were bought from Thermo Fisher (Invitrogen, K6010–01), which was cultured with DMEM basic (GIBCO, C11995500BT), 10% fetal bovine serum (FBS, GIBCO, 12483020), and penicillin and streptomycin (50 μg/mL) (Invitrogen) and placed into 37°C and 5% CO2 incubator. We used Lipofectamine 3000 Reagent (Invitrogen) to transfect DOT1L reference and variant plasmids into cell lines according to the manual.
Mouse primary neural cell culture
The pregnant mice were sacrificed at day 16.5 and the embryos were dissected out in 1× pre-cooled HBSS. The olfactory bulb and vascular membrane of embryo brains was removed and discarded. The remaining part of the brains were cut it into pieces with scissors and digested in papain at 37°C for 30 min shaking gently every 10 min. After addition of 2 volumes of pre-warmed FBS medium, the neurons were separated gently and filtered with a 40 μm cell strainer. The cells were seeded (150,000 per well) into 24-well plates which contained a poly-lysine-coated coverslip at the bottom. After cells adhered to the cover slips, medium was replaced with pre-warmed B27 medium. We used Lipofectamine 3000 Reagent (Invitrogen) to transfect Dot1l-shRNA plasmids and control plasmids into the primary neurons according to the manual.
Western blot from human cells
The total protein was extracted using RIPA cell lysis buffer (Solarbio #R0010) containg 1% Phenylmethylsulfonyl fluoride (PMSF, Solarbio). The BioRad Mini-PROTEANTM Tetra System was used to electrophorese with ExpressPlus 8% precast gel (GenScript). 25 μg of total protein per sample were run and transferred to a nitrocellulose membrane and immunoblotted with Rabbit anti-H3K79Me3 (A2369, Abclonal), anti-Flag and anti-β-Tubulin antibodies. The protein levels were detected by ECL chemiluminescence.
Results
Summary of clinical findings
Through the UDN,32,33 SickKids Complex Care genomics project,34,35 and GeneMatcher,36,37 we identified nine unrelated individuals (probands 1–9) in whom exome or genome sequencing highlighted candidate variants in DOT1L. All seven distinct variants are missense variants, and in all but one family (with the recurrent p.Glu123Lys variant, where the child was deceased and parents declined further testing), the variant was confirmed to be de novo. The comprehensive clinical information of the probands can be found in Tables 1 and S6. Throughout this report, all sequence variants are described based on reference sequence GenBank: NM_032482.3.
Table 1.
Clinical features of individuals with DOT1L variants
| Proband | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 |
|---|---|---|---|---|---|---|---|---|---|
| DOT1L variant (NM_032482.3) | c.133T>G (p.Cys45Gly) | c.299C>T (p.Thr100Met) | c.367G>A (p.Glu123Lys) | c.367G>A (p.Glu123Lys) | c.367G>A (p.Glu123Lys) | c.385G>A (p.Glu129Lys) | c.1876C>G (p.Leu626Val) | c.2557C>T (p.Arg853Cys) | c.3074A>T (p.Lys1025Met) |
| Inheritance | de novo | de novo | unknown | de novo | de novo | de novo | de novo | de novo | de novo |
| Sex | male | female | male | male | female | male | male | male | male |
| Age at last assessment | 5 years | 4 years 4 months (age at death) | 4 months (later died in infancy) | 5 years | 3 years 11 months | 6 years | 7 years | 4 years | 21 years |
| Medical history | |||||||||
| Brain anomalies (MRI/CT) | cortical dysplasia, periventricular heterotopia | no | HIE | bilateral brain atrophy, bilateral frontotemporal extra brain space widening, multiple small cysts in the corpus callosum | ONH/tract truncated corpus callosum | enlarged VRS, tentorial herniation | normal brain MRI | bilateral brain atrophy, focal cortical dysplasia | cerebellar atrophy, megalencephaly |
| Cardiac anomalies | ASD, PLSVC | myocarditis with cardiomyopathy; resolved by 1 year old | large VSD, moderate size ASD/PDA | ASD | no | no | VSD | no | no |
| Hypotonia | yes | yes | yes | no | yes | yes | no | no | no |
| Musculoskeletal anomalies | hip subluxation | hyperflexible hip joints | tall vertebral bodies, anomalous ribs (first 3), non ossified patella | N/D | hip laxity | no | pain after physical exercise, low endurance | N/D | no |
| Urogenital anomalies | hypospadias with chordee | no | hypospadias | cryptorchidism | grade 1 VUR | no | small penis | no | no |
| Hearing loss | sensorineural and conductive; stenosis external auditory canal | lack of reaction to loud stimuli or startle responses at 9 months | no | mild hearing loss in both ears | no | no | sensorineural w/ bilateral SCC dysplasia | no | no |
| Growth and development | |||||||||
| Global developmental delay | severe | yes | severe | severe | severe | severe | yes | mild | yes |
| Language | non-verbal | non-verbal | N/A (died in infancy) | only a few words | non-verbal | only a few words | only a few words | only a few words | only a few words |
| Stature (percentile) | short (2.94%) | short (4.95%) | N/D | N/D | normal (93.3%) | short (0.64%) | short (0.40%) | N/D | normal (39.7%) |
| Microcephaly (percentile) | no (97%) | no (60%) | yes (<3.0%) | N/D | yes (4.0%) | yes (3.0%) | yes (<1.0%) | yes (N/D) | no (>99%) |
Abbreviations are as follows: HIE, hypoxic ischemic encephalopathy; ONH, optic nerve hypoplasia; VRS, Virchow Robin spaces; ASD, atrial septal defect; PLSVC, persistent left supervior vena cava; VSD, ventricular septal defect; PDA, patent ductus arteriosus; VUR, vesicoureteral reflux; SCC, semi-circular canal; N/D, no data.
All identified probands had some degree of global developmental delay (9/9) and most (5/9) were found to have hypotonia. Whereas motor delay was usually relatively mild, significant speech and language delay was observed in all individuals in which details were provided (seven individuals): two were non-verbal and five were able to speak only individual words. All nine probands were found to have various congenital anomalies although there was considerable heterogeneity across the cohort. Seven had brain structural anomalies, five had cardiac defects (four with septation defects), and five presented with varied urogenital features (of which two had hypospadias). Other common features included hip laxity (3/9), variable ophthalmological features (3/9), and sensorineural hearing loss (3/9). Growth restriction was also a feature seen in the cohort, with five individuals reported to have post-natal microcephaly (<3rd centile for occipital-frontal circumference [OFC]) and four with short stature (<3rd centile for height). While distinctive facial features were described for all probands (9/9), no recognizable pattern was appreciated in this cohort.
In addition to the nine probands described above, we also identified two families with missense variants in DOT1L that were not suspected to be diagnostic by the referring providers. One was heterozygous for a missense variant (c.1352A>G [p.Asp451Gly]) inherited from an unaffected father that had an allele frequency inconsistent with a high-penetrance Mendelian developmental disorder (Table S8), and the other was heterozygous for a de novo missense variant (c.874C>T [p.Arg292Cys]) reported previously5,38 where an alternative genetic diagnosis likely explains the entirety of her clinical features. Clinical features for both individuals can be found in Table S7, and they were included in aspects of our functional studies as negative controls (see below).
Bioinformatic analyses of DOT1L de novo variants are inconclusive
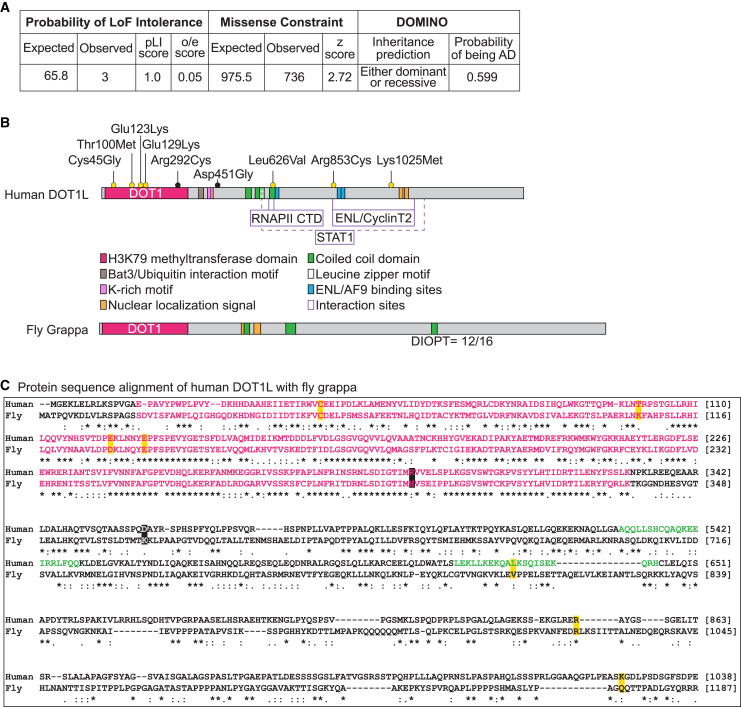
We performed in silico analyses using MARRVEL (Model organism Aggregated resources for Rare Variant ExpLoration)48,49 to collect information about the human DOT1L and its orthologous genes in genetic model organisms. DOT1L is intolerant LoF with a probability LoF intolerant (pLI) score of 1.0 and a LoF variant observed/expected (o/e) ratio of 0.046 (0.02–0.12). Variants in DOT1L are predicted to have a high “probability of being autosomal dominant” based on DOMINO50 (Figure 1A).
Figure 1.
DOT1L is loss of function constrained and conserved in flies
(A) DOT1L is variant constrained based upon the control population database, gnomAD.51 Variants in DOT1L are predicted to be inherited “either recessive or dominant” and have a score of 0.599 for “probability of being autosomal dominant” by DOMINO.
(B) Protein domain organization of human DOT1L and its fly (gpp) ortholog. Domains of DOT1L and the region they span specified with amino acid numbers are as follows: catalytic domain DOT1 (16–330),9,10 Bat3/Ubiquitin interaction motif (361–380),17 K-rich motif (390–407)9 with a partially overlapping nuclear localization signal (NLS) (395–417),52 three coiled-coil domains (529–549, 564–595, 616–636),53 leucine zipper motif (576–594),54 CTD binding patch (618–627),25 three ENL/AF9 binding sites (628–653, 863–878, 877–900),55,56,57 two more NLSs (1,088–1,111 and 1,164–1,171),52 and STAT1 interaction region (580–1,138).58 Variants suspected to be diagnostic are indicated above the protein as yellow dots and the variants suspected to be non-diagnostic are indicated as black dots. DOT1L’s ortholog in fly, gpp, has a DOT1 domain, three coiled-coil domains, and two NLSs. Gpp has a homology score 12/16 (DIOPT, v.8.5).59
(C) Protein sequence alignment of human DOT1L (Uniprot: Q8TEK3) and fly gpp (UniProt: Q8INR6). The variants are highlighted with yellow or black boxes. Pink letters correspond to the DOT1 domain and green letters correspond to a coiled-coil domain. Symbols in the protein alignment: fully conserved (∗), strongly similar (:), weakly similar (.), absent (−).
Based on gnomAD,51 DOT1L is missense constrained with a Z score of 2.72 and a missense variant o/e ratio of 0.754 (0.709–0.802). Of the seven missense variants in our cohort (including one variant that was recurrent in three unrelated families), four variants map to the amino acid residues in the enzymatic domain (DOT1, residues 16–330) of DOT1L (Figure 1B).9,10 The remaining three variants map to residues that are important for interaction of DOT1L with its interaction partners. The variant p.Leu626Val is found in a coiled-coil domain which overlaps with RNA Polymerase II C-Terminal Domain (RNAPII-CTD) binding patch (residues 618–627).20,25,53 The variants p.Arg853Cys and p.Lys1025Met are located in disordered regions of DOT1L. While seeming nonfunctional, these regions have been previously shown to be involved in DOT1L’s interaction with Eleven Nineteen Leukemia (ENL), Cyclin T2, and STAT1.53,55,56,57
Variant allele frequencies and outputs from in silico analysis tools60 are found in Table S8. All proband variants were extremely rare (n = 3) or absent (n = 4) in population-scale databases of genomic variation from approximately 430,000 total individuals,51,61,62 consistent with the variants potentially causing a developmental disorder characterized by variable expressivity. In silico predictions were often conflicting, inconclusive, and/or suggestive of a benign impact (e.g., REVEL scores of <0.5 for six of the seven variants),63 emphasizing the need for additional functional studies. We initially considered the possibility of haploinsufficiency. However, based on the control population data in Database of Genomic Variants (DGV),64 there are forty control individuals that harbor deletions or copy number loss containing DOT1L. There are also two individuals with stop gain variants (c.4064C>A [p.Ser1355∗]) in gnomAD51 control population. Together these data indicate that unlike other KMTs, haploinsufficiency of DOT1L is unlikely to be associated with a disease. Therefore, we hypothesize that DOT1L variants harbored by the individuals may be gain-of-function mutations that correspond to hypermorphs that are more active than the wild-type protein, or they correspond to antimorphs that act in a dominant-negative fashion or to neomorphs that exhibit novel functions.
DNA methylation profiling of proband blood samples
About half of the KMT-associated diseases have unique DNA methylation (DNAm) signatures (Table S1). Even though the molecular basis for these DNAm signatures remains to be determined, they can be used as one indicator to classify variants of uncertain significance (VUSs) as pathogenic or benign.65 To determine whether DOT1L variants impact DNAm patterns, we assessed genome-wide DNAm in peripheral blood from seven individuals with DOT1L variants (p.Cys45Gly, p.Thr100Met, p.Glu123Lys, p.Glu129Lys, p.Leu626Val, p.Lys1025Met, and the suspected non-diagnostic variant p.Arg292Cys) as well as twelve sex- and age-matched controls. We identified 184 differentially methylated CpG sites that meet thresholds of false discovery rate (FDR) < 0.3 and |Δβ| > 0.10 (10% difference in DNAm), using linear regression modeling. We visualized DNAm data at profile sites using principal component analysis (PCA) and hierarchal clustering. Two of the variants, p.Cys45Gly and p.Lys1025Met, were excluded from the discovery cohort as they are age outliers. As seen in Figures S1A and S1B, the significant CpGs could be used to segregate four individuals with suspected diagnostic variants (p.Thr100Met, p.Glu123Lys, p.Glu129Lys, and p.Leu626Val) from control subjects. The suspected non-diagnostic DOT1L variant, p.Arg292Cys, clustered with control individuals. Despite the small sample size, these results suggest that there is a distinct DOT1L-specific methylation signature.
Loss of gpp in Drosophila causes developmental delay, loss of H3K79 methylation, and lethality
The ortholog of DOT1L in the fruit fly Drosophila melanogaster is grappa (Flybase: FBgn0264495). Based on the DRSC Integrative Ortholog Prediction Tool (DIOPT, v.8.5),59 human DOT1L and fly Gpp are highly conserved with a DIOPT score of 12/16. The two proteins exhibit 24% identity and 35% similarity with 28% gaps. There is greater conservation in the DOT1 enzymatic domain (65% identity), to which most variants (5/9, including the suspected non-diagnostic variants) map. Three variants map to disordered regions and the other variant map to a coiled-coil domain (Figure 1B). Three of the nine variants affect fully conserved amino acid residues between DOT1L and Gpp. Four variants occur at highly similar amino acid residues and the last two variants occur at weakly similar amino acid residues between human and fly (Figure 1C).
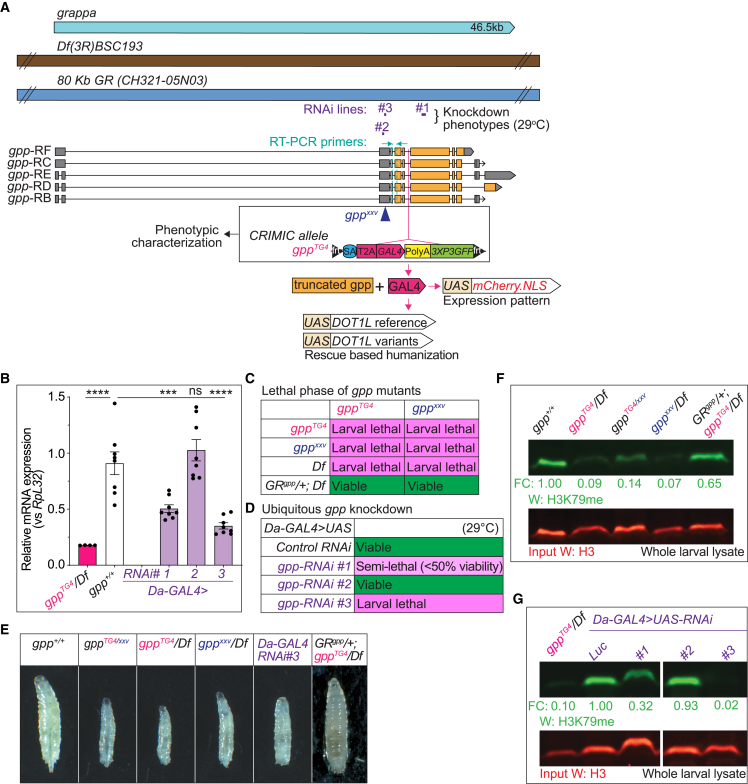
Previous studies have shown that LoF mutants of gpp exhibit slow growth and larval lethality.28 To further assess the phenotypes associated with loss of gpp in flies, we generated a mutant allele, gppTG4, by insertion of a Splice acceptor-T2A-GAL4-polyA cassette into a coding intron (i.e., an intron that is flanked by two coding exons) of gpp shared by all transcripts via CRISPR-Cas9. The gppTG4 allele is expected to behave as a severe LoF allele since the splice acceptor allows the artificial exon to be integrated into the mRNA and the polyA-tail leads to early termination of transcription.42,43 Real-time qPCR data confirm that the gpp transcript levels are severely decreased (<17%) in gppTG4/Df(3R)BSC738 when compared to wild-type larvae (Figure 2B). We also obtained a deletion allele, gppxxv, which was generated by X-ray mutagenesis and one of its breakpoints is in the coding region of gpp.28 We also used three publicly available UAS-RNAi lines targeting gpp (Figure 2A). RNAi #1 leads to a 50% ± 0.03% reduction and RNAi #3 leads to a 65% ± 0.02% reduction of expression of gpp based on real-time qPCR data, while RNAi #2 does not affect the levels of gpp serving as a negative control in our experiments (Figure 2B).
Figure 2.
Fly grappa is essential for development and loss of gpp leads to developmental delay and a reduction in H3K79 methylation
(A) Gene structure of fly gpp, fly reagents, and assays used in this study. The gene region is shown in light blue. A deficiency and genomic rescue (GR) construct are shown in brown and dark blue, respectively. Three independent RNAi lines (#1, P{TRiP.JF01284}attP2; #2, P{TRiP.GL01325}attP2; and #3, P{TRiP.HMJ02129}attP40) tested in this study and their target regions are shown in purple. Different isoforms of gpp (RB, RC, RD, RE, and RF) are shown in the middle panel and exons are indicated in orange, introns as black lines, and untranslated regions (UTR) in gray. gpp mutants used in phenotypic characterization studies include gppxxv (X-ray mutagenesis)28 and gppTG4 (CRIMIC allele).
(B) Relative gpp mRNA expression levels are lower than 20% in gppTG4/Df mutant larvae when compared to controls (gpp+/+) based on real-time qPCR using primers shown in (A). Real-time qPCR analysis of gpp mRNA levels in ubiquitous gpp knockdown larvae shows that Da-GAL4 > gpp-RNAi reduces gpp expression to different levels. Da-GAL4 > gpp-RNAi #1 decreases gpp mRNA levels by ∼50% and RNAi #3 by 70%. RNAi #2 does not have any effect on gpp levels. Normalized gpp levels from three independent experiments were plotted as mean ± SEM, and statistical significance was determined by one-way ANOVA for multiple groups (∗∗∗p < 0.001, ∗∗∗∗p < 0.0001).
(C) Animals homozygous for gpp mutant alleles are larval lethal. None of the alleles or a large deficiency allele which lacks gpp, Df(3R)BSC193, can complement each other. One copy of a genomic rescue construct inserted in second chromosome (VK37), GRgpp/+, rescues lethality of trans-heterozygous allelic combinations.
(D) Ubiquitous knockdown of gpp results in larval lethality. While Da-GAL4 > RNAi #3 flies are larval lethal, -RNAi #1 flies are semi-lethal, as shown by lower-than-expected genotypic ratios of survival into adulthood. RNAi #2 flies are completely viable. Da-GAL4 > UAS-gpp-RNAi flies were compared to Da-GAL4 > control-RNAi (control-RNAi = UAS-luciferase-RNAi). All the crosses were performed at 29°C.
(E) Complete loss or ubiquitous knockdown (<50%) of gpp leads to severe developmental delay. Images of age-matched larvae at third instar larval stage is shown. One copy of a genomic rescue construct inserted in second chromosome (VK37), GRgpp/+, can rescue the developmental delay phenotype.
(F) Gpp mutant animals have a drastic decrease in H3K79 methylation levels. Protein lysate from 10 larvae was prepared for each sample. H3K79 methylation levels were normalized with loading control, H3, and fold change (FC) for each sample were calculated by comparing normalized H3K79 methylation levels to wild-type larvae (gpp+/+). One copy of the genomic rescue construct, GRgpp/+, can increase the H3K79 methylation levels to ∼65%.
(G) Ubiquitous knockdown (<50%) of gpp causes a severe decrease in H3K79 methylation levels. Da-GAL4 > gpp-RNAi #1 decreases H3K79 methylation levels to ∼30% and RNAi #3 to ∼2%. RNAi #2 does not have any effect on methylation levels. Protein lysate from 10 larvae was prepared for each sample. H3K79 methylation levels were normalized with loading control, H3, and FC for each sample were calculated by comparing normalized H3K79 methylation levels to control-RNAi (control-RNAi = UAS-luciferase-RNAi). All the crosses were performed at 29°C. A sample which is excluded from this study is cropped out from the gel image.
The two gpp mutant alleles over a molecularly defined deletion line [DF(3R)BSC193] covering gpp are recessive lethal. Notably, lethality is observed starting from first instar larval stage and animals do not survive beyond the third instar larval stage (Figure 2C). Indeed, a comparison of age-matched mutants, gppTG4/Df or gppTG4/gppxxv, with wild-type third-instar larvae shows severe developmental delay in mutant animals (Figure 2E), consistent with previous findings.28 Mutant larvae also show a severe decrease in total H3K79 methylation levels compared to wild-type animals (Figure 2F). The lethality, larval developmental phenotype and the decrease in H3K79 methylation levels are rescued by a genomic rescue (GR) construct, GRgpp, which carries an independent copy of the entire gpp locus [P[acman]CH321-05N03)].66 Consistently, ubiquitous expression of RNAi #1 using da-GAL4 causes a decrease in viability (<50%) (Figure 2D), and ubiquitous expression of RNAi #3 causes lethality at larval stages with developmental delay (Figures 2D and 2E). Further, ubiquitous knockdown of gpp with different RNAi lines also leads to reduction in H3K79 methylation levels (Figure 2G). Altogether, these data provide compelling evidence that gpp is essential for fly development and is responsible for H3K79 methylation in flies.
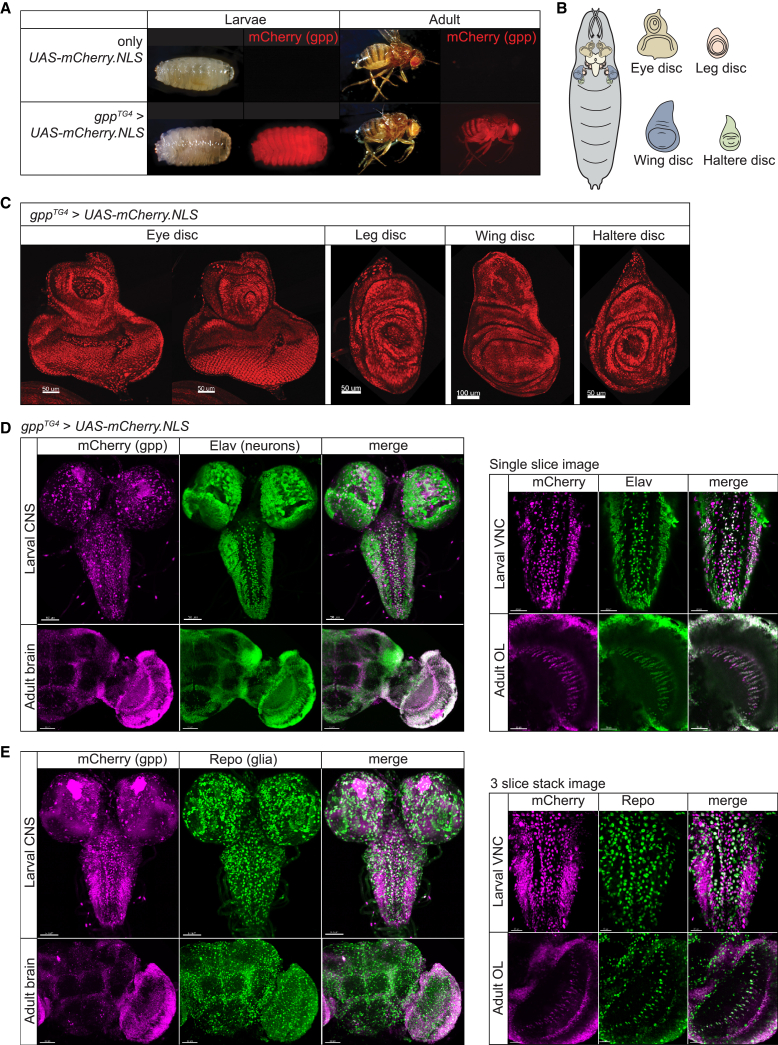
gpp is broadly expressed in flies and primarily expressed in neurons in CNS
Next, we determined the expression pattern of gpp in flies. We crossed gppTG4 animals to UAS-mCherry.NLS (nuclear localized mCherry fluorescent protein) animals to label the nuclei of the cells that express gpp. We find that gpp is expressed broadly in most tissues in both larvae and adult flies (Figure 3A). In the third-instar larvae, gpp is expressed in all imaginal discs including wing, eye, leg, and haltere discs, which gives rise to respective adult structures (Figures 3B and 3C). To determine the cell types in the CNS that express gpp, we counterstained the larval and adult brains with the nuclear pan-neuronal marker Elav67 or nuclear pan-glial marker Repo.68 In both larval and adult brain, many but not all Elav-positive cells express gpp based on whole-mount z stacked confocal microscopy images (Figure 3D, left). In agreement, single slice images also show that gpp is expressed in most Elav-positive cells, indicating that gpp is expressed in a large subset of neurons (Figure 3D, right). Whole-mount z stacked images and 3-slice stack images of Repo staining in both larval and adult brain show a small group of Repo-positive cells that express gpp (Figure 3E). This suggests that gpp is expressed in a small subset of glial cells in addition to its broad neuronal expression.
Figure 3.
Fly grappa is broadly expressed in both larvae and adult flies
The gppTG4 allele was used to drive expression of UAS-fluorescent reporter transgene (UAS-mCherry.NLS).
(A) gpp is expressed broadly and highly throughout the body in both larvae and adult flies. Nuclear mCherry was present and whole larvae or adult was imaged.
(B) Schematic of different tissues of larvae.
(C) gpp is expressed in most tissues in larvae. Nuclear mCherry was present and tissue were dissected from L3 larvae including eye, leg, wing, and haltere discs.
(D and E) gpp is broadly expressed in the nervous system. Nuclear mCherry was driven by gppTG4/+ and tissues were dissected from L3 larvae (CNS, includes central brain and VNC) or adults (brain). Shown is half of the adult brain. Tissue was counterstained with markers for neurons (Elav) (D) or glia (Repo) (E). Z stacked images of gpp expression pattern compared to neurons or glia are shown in the left panels in (D) and (E), respectively. Single slice images (right panels) were used to better visualize cellular co-localization of Nuclear mCherry signal with neurons (D) or glia (E). Note that gpp is expressed in a large subset of neurons and in a small subset of glia. VNC, ventral nerve cord; OL, optic lobe.
To assess whether knockdown of gpp in the nervous system can cause an obvious defect, we performed partial knockdown of gpp in neurons using a neuronal (Elav-GAL4) or a glial (Repo-GAL4) driver. We found that neuronal knockdown of gpp using RNAi #3 causes lethality while glial knockdown does not cause any observable phenotype. Moreover, knockdown of gpp in the developing eye (ey-GAL4) causes a rough eye phenotype (Figure S2A), and knockdown in developing tissues (en-GAL4) leads to lethality and morphological phenotypes of the wing (Figure S2B). These data suggest that gpp have important roles in nervous system function as well as in imaginal disc development.
Functional assays in flies indicate that DOT1L variants are gain-of-function alleles
We next attempted to rescue the lethality observed in gppTG4 flies by expressing human DOT1L in flies. We took advantage of GAL4 expression in gppTG4 flies. In these flies, even though the transcription is terminated early, the produced transcript is translated into a short protein and the presence of the viral T2A sequence allows ribosomal skipping and re-initiation of translation to produce a functional GAL4. Therefore, GAL4 is produced under the control of gpp’s regulatory elements in the same spatial and temporal pattern as gpp.42,43 To use “UASxGAL4” system, we also created transgenic lines expressing C-terminal 3XHA-tagged human reference or variant DOT1L under the control of the UAS promoter (Figure 1A). In addition to modeling the variants seen in our cohort, we also modeled the p.Arg292Cys variant that was suspected to be nonpathogenic as a negative control. Unfortunately, expression of reference or variant DOT1L in gppTG4/Df flies failed to rescue the lethality at all temperatures (18°C, 25°C, 29°C) tested (Figure S3).
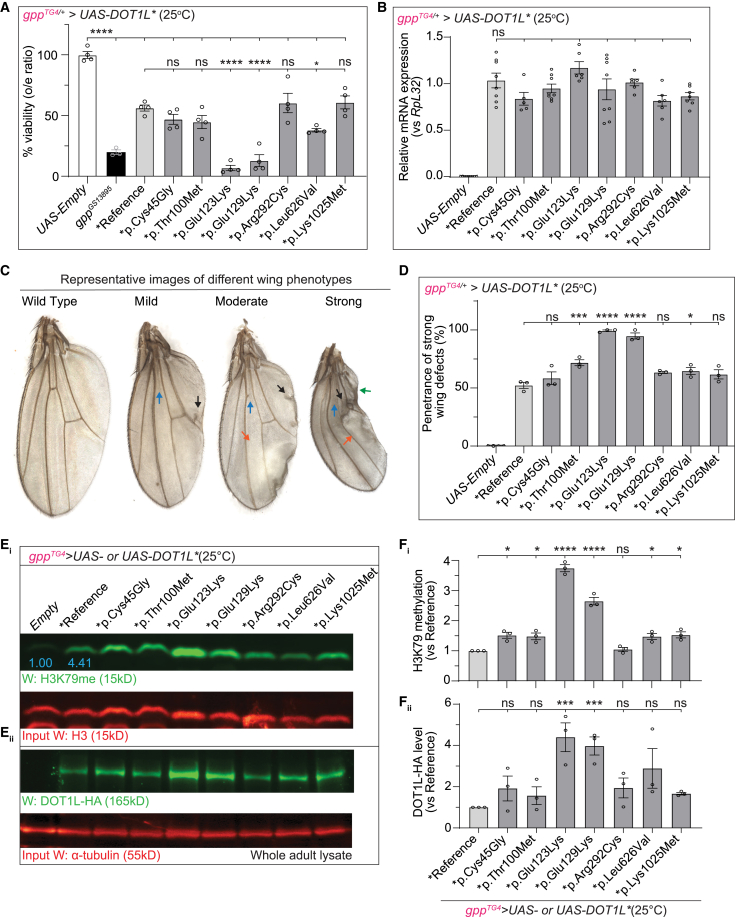
Intriguingly, heterozygous mutant flies, gppTG4/+, expressing reference or variant DOT1L cDNA or fly gpp (gppGS13895, a fly line containing UAS regulatory sequence at 5′ UTR of fly gpp) have reduced viability compared to control flies (gppTG4/+>UAS-Empty) as shown by lower-than-expected Mendelian ratios of eclosion (percent o/e ratios ≤ 60%), indicating that overexpression of human DOT1L in the pattern of the fly gpp causes toxicity. Moreover, the variants p.Glu123Lys and p.Glu129Lys shows a severe reduction, and the variant p.Leu626Val shows a moderate reduction in viability compared to flies expressing reference DOT1L (Figure 4A). A real-time qPCR analysis for mRNA levels of DOT1L in gppTG4/+ flies expressing variant DOT1L cDNAs show no significant difference compared to flies expressing reference DOT1L (Figure 4B). Expression of reference or variant human cDNAs also cause morphological wing defects with high penetrance (>95%) (Figure S4). We classified the wing phenotypes of these flies into three different groups based on their severity. Mild phenotypes include loss of cross-veins and extra vein branching; moderate phenotype includes blistering in addition to mild phenotypes; and strong phenotype includes necrosis in addition to moderate phenotypes (Figure 4C). The variants p.Glu123Lys and p.Glu129Lys cause a more severe wing phenotype as none of the animals have wild-type wings and strong wing phenotypes in these flies are associated with a significantly higher penetrance compared to reference (Figures S4B and 4D). In addition, the variants p.Thr100Met and p.Leu626Val cause significantly more strong phenotypes compared to reference (Figure 4D). Furthermore, expression of human reference or variant DOT1L in developing tissues (en-GAL4) also causes morphological wing defects with full penetrance, and variants p.Glu123Lys and p.Glu129Lys again lead to more severe phenotypes (Figure S5).
Figure 4.
Expression human DOT1L disrupts wing morphology and different variants display different amounts of toxicity and H3K79 methylation
(A) Heterozygous mutant flies, gppTG4/+, expressing reference or variant DOT1L cDNA have reduced viability compared to control flies (gppTG4/+> UAS-Empty) as shown by lower-than-expected genotypic ratios of survival into adulthood. The variants p.Glu123Lys and p.Glu129Lys show significant reduction in viability compared to flies expressing reference DOT1L. All the crosses were performed at 25°C. Percent viabilities (o/e ratios) from three independent experiments were plotted as mean ± SEM, and statistical significance was determined by one-way ANOVA for multiple groups (∗∗∗∗p < 0.0001).
(B) Relative DOT1L mRNA expression levels in gppTG4/+ flies expressing reference or variant DOT1L. Normalized DOT1L levels from three independent experiments were plotted as mean ± SEM, and statistical significance was determined by one-way ANOVA for multiple groups.
(C) Representative images of different wing phenotypes observed in in survivors of heterozygous mutant flies, gppTG4/+, expressing each DOT1L cDNA. Blue arrow, loss of cross-vein; black arrow, extra vein branching; orange arrow, blistered areas; green arrow, necrotic areas.
(D) Penetrance of strong wing phenotypes in survivors of heterozygous mutant flies, gppTG4/+, expressing each DOT1L cDNA. Percent penetrance for strong phenotype from three independent experiments were plotted as mean ± SEM, and statistical significance was determined by one-way ANOVA for multiple groups (∗p < 0.05, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001).
(E) All the DOT1L variants, except p.Arg292Cys, behave as gain-of-function (GoF) mutations. Survivors of heterozygous mutant flies, gppTG4/+, expressing reference or variant DOT1L cDNA show increased H3K79 methylation compared to control flies (gppTG4/+> UAS-Empty). Flies expressing variant DOT1L cDNAs, corresponding to p.Cys45Gly, p.Thr100Met p.Glu123Lys, p.Glu129Lys, p.Leu626Val, and p.Lys1025Met, show higher H3K79 methylation levels when compared to reference (Ei). Flies expressing variant DOT1L cDNAs p.Glu123Lys and p.Glu129Lys show higher DOT1L levels when compared to reference (Eii). Protein lysate from 10 adult flies were prepared for each sample. H3K79 methylation levels were normalized with loading control, H3, and fold change for each sample were calculated by comparing normalized H3K79 methylation levels to reference DOT1L-expressing flies. DOT1L levels were normalized with loading control, α-tubulin, and fold change for each sample were calculated by comparing normalized DOT1L levels to reference DOT1L-expressing flies. All the crosses were performed at 25°C. Blue numbers indicate the fold change of H3K79 methylation level in reference when compared to control (UAS-Empty).
(Fi) Normalized H3K79 methylation band intensities for each group from three independent experiments were plotted as mean ± SEM, and statistical significance was determined by one-way ANOVA for multiple groups (∗p < 0.05, ∗∗∗∗p < 0.0001).
(Fii) Normalized DOT1L band intensities for each group from three independent experiments were plotted as mean ± SEM, and statistical significance was determined by one-way ANOVA for multiple groups (∗∗∗p < 0.001).
To assess the effect of the variants on enzymatic function of DOT1L, we next measured H3K79 methylation levels in heterozygous mutant flies, gppTG4/+, expressing reference or variant DOT1L cDNA. We prepared total protein lysate from 5- to 7-day-old flies to extract histones and histone-bound DOT1L. Previously it was shown that the overall level of methylation at H3K79 residues rather than the level of one specific methylation state, i.e., mono-, di-, or tri-methylation, determines the downstream effect of this particular modification.11 Therefore, we detected the methylation status of our samples using an antibody that recognizes all methylation states at H3K79.69 We found the flies expressing human reference DOT1L have a 4.41-fold increase in H3K79 methylation compared to control flies (UAS-empty) suggesting that human DOT1L methylates H3K79 in Drosophila (Figure 4Ei). Flies expressing all the DOT1L variants, except p.Arg292Cys, show significantly higher H3K79 methylation levels when compared to the reference DOT1L-expressing flies (Figures 4Ei and 4Fi). The variants p.Glu123Lys and p.Glu129Lys cause the highest increase in H3K79 methylation levels possibly due to the higher DOT1L levels in these flies (Figures 4Eii and 4Fii). DOT1L levels in the other variants do not show a statistically significant increase based on one-way ANOVA multiple group comparison analysis. When we ubiquitously expressed reference or variant DOT1L using the Actin-GAL4 driver, we found that the flies expressing human reference DOT1L have a 1.7-fold increase in H3K79 methylation compared to control flies (UAS-empty). We also observed similarly higher H3K79 methylation levels with variants p.Thr100Met, p.Glu123Lys, and p.Glu129Lys when compared to the reference cDNA. DOT1L levels in the variant-expressing flies are not significantly different from the reference-expressing flies (Figures S6A and S6B). Furthermore, there is also no significant change in the viability of flies expressing reference or variant DOT1L ubiquitously when compared to control flies (UAS-Empty) (Figure S6C). To understand the reason underlying the discrepancy in methylation levels and viability in flies expressing DOT1L cDNAs ubiquitously and cDNAs expressed in the endogenous pattern of gpp, we compared the levels of DOT1L reference in these flies. As shown in Figure S6D, the DOT1L level is ∼2-fold higher in gppTG4/+ flies when compared to ubiquitously expressing flies (Figure S6D). Consistent with these observations, ubiquitous expression of fly gpp is less toxic than its expression in the endogenous pattern (Figure S6E).
In summary, our data suggest that all variants tested in flies except p.Arg292Cys (the suspected non-diagnostic variant) alter DOT1L activity or levels and cause higher H3K79 methylation. Moreover, the severity of the phenotypes expressing variant DOT1L correlate with the level of H3K79 methylation.
DOT1L p.Glu123Lys is a gain-of-function variant in vertebrate cells
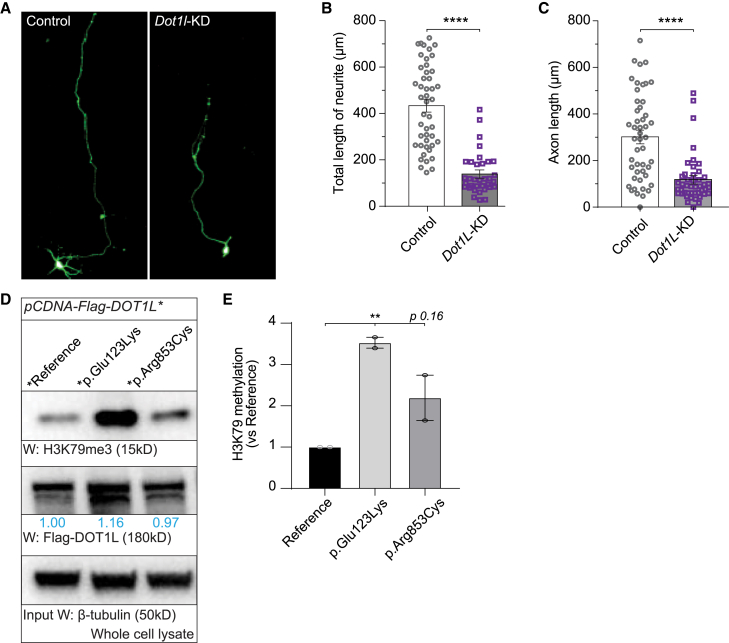
DOT1L is a conserved H3K79 methyltransferase in both invertebrates and vertebrates. Previous mouse studies indicate that germline Dot1L deletion causes embryonic lethality and defects in heart and yolk sac.26 Conditional knockout of Dot1l during embryonic development impairs cortical development and decreases neural progenitor cells.70 To assess the effect of Dot1l loss at the cellular level in nervous system, we performed RNAi assays in mouse primary neuronal cells. Knockdown of Dot1l in primary neuronal cells causes a significant reduction in axon length (Figures 5A and 5B). It also decreases the length of all neurites significantly (Figure 5C).
Figure 5.
Knockdown of Dot1l decreases axonal length and human DOT1L variants increase H3K79 methylation levels
(A) Representative images of different primary neuronal cells. Left panel shows mouse primary neurons transfected with control plasmid and right panel shows mouse primary neurons transfected with Dot1l-shRNA plasmid.
(B and C) Total neurite length (B) and total axonal length (C) was measured using ImageJ (n = 50 per condition). Results were plotted as mean ± SEM, and statistical significance was determined by unpaired t test (∗∗∗∗p < 0.0001).
(D) DOT1L variants behave as gain-of-function (GoF) mutations. HEK293T cells were transfected with N-terminal flag tagged reference or variant (p.Glu123Lys and p.Arg853Cys) DOT1L cDNAs. Whole cell lysate was prepared for each sample. H3K79 methylation levels were normalized with loading control, β-tubulin, and fold change for each sample were calculated by comparing normalized H3K79 methylation levels to reference DOT1L-expressing cells. Blue numbers indicate the fold change of DOT1L levels in cells expressing variant compared to cells expressing reference DOT1L.
(E) Normalized H3K79 methylation band intensities for each group from two independent experiments were plotted as mean ± SEM, and statistical significance was determined by one-way ANOVA for multiple groups (∗∗p < 0.01).
To determine how DOT1L variants affect histone methylation, we transfected HEK293T cells with cDNA from a reference and two variants. The p.Glu123Lys variant, which causes the most severe fly phenotypes, leads to an almost 4-fold increase in H3K79 methylation level compared to reference DOT1L, similar to what was observed in flies. The p.Arg853Cys variant also increases H3K79 methylation level but not to a statistically significant level (Figures 5D and 5E).
Together, these data confirm the conserved roles of DOT1L in neuronal development and show that the DOT1L variants are gain-of-function alleles.
Discussion
We identified nine individuals with rare heterozygous missense variants in DOT1L, encoding a KMT that has not been associated with a Mendelian genetic disorder in humans in OMIM previously. The core clinical features in this cohort includes a range of congenital anomalies and CNS dysfunction (global developmental delay, feeding difficulties, hypotonia, and/or intellectual disability), similar to other KMT disorders (Table S1). We confirm that the fly ortholog gpp is an essential gene and that it is responsible for H3K79 methylation in flies. We also show that it is required for proper development of eyes, wings, and neurons. Expression of the human reference DOT1L increases H3K79 methylation in flies, showing that the human protein can recognize and act on H3K79 in flies. Interestingly, expression of some DOT1L variants decrease the viability of flies when compared to animals expressing the reference DOT1L. All variants, except p.Arg853Cys, increases H3K79 methylation levels compared to reference, consistent with a gain-of-function mechanism. Moreover, the variants associated with the most severe phenotypes in flies also show evidence of a distinct blood-derived DNAm profile in humans. In summary, the clinical, molecular, and biochemical data presented here indicate that select heterozygous missense variants in human DOT1L cause an autosomal-dominant KMT disorder through a gain-of-function mechanism.
The number of Mendelian disorders resulting from dysregulation of epigenetic machinery, including KMTs, have greatly expanded in the past decade.71,72 So far, around 50% of all genes encoding KMTs have been associated with a Mendelian disorder and all these disorders have an autosomal-dominant inheritance pattern. Heterozygous deletions or protein-truncating variants underlie most of these KMT-associated disorders indicating haploinsufficiency as the predominant mechanism (Table S1). Even though missense variants in several genes encoding KMTs have also been reported for individuals with other symptoms including autism, ID, bipolar disorder, and congenital anomalies,5 their causal relationship has not been extensively investigated through functional studies. Here, our functional analysis in flies and human cells show that missense variants in a KMT, DOT1L, correspond to gain-of-function alleles.
DOT1L is the only KMT known to lack SET domain but contains a DOT1 enzymatic domain.7,8 Besides its structure, it is also mechanistically different from SET domain containing KMTs, which has direct consequences for its regulation and function. Compared to the processive mechanism of all SET domain KMTs, DOT1L acts in a distributive manner such that it can only sequentially add methyl groups to each H3K79 residue. For each methylation cycle, it needs to dissociate from and reassociate with the same lysine residue to achieve the higher methylation states (i.e., di- or tri-).10,11 In contrast to the other methylated lysines, H3K79 methylation states (i.e., mono-, di-, or tri-) are similarly distributed over a gene and have redundant roles. Indeed, the overall level of methylation at H3K79, rather than the level of one state, determines the outcome of this histone modification.11 In addition to our knowledge, so far no protein has been identified that can specifically bind to methylated H3K79 in vivo, and there is no known demethylase that can remove H3K79 methylation. Hence, the cells may not be able to easily compensate for the downstream effects of excess methylation due to elevated DOT1L enzymatic activity or elevated levels. Consistent with this hypothesis, we observed H3K79 hypermethylation when DOT1L is expressed in flies that endogenously express gpp. These flies also exhibit reduced viability and wing deformities showing a dominant effect due to H3K79 hypermethylation. Finally, the observed wing phenotypes are consistent with the previous observation of DOT1L-dependent regulation of Wnt target gene expression.29,30,31
Since DOT1L reference and variant transgenes are inserted in the same genomic site in Drosophila and heterozygous mutant flies express similar levels of reference or variant DOT1L mRNAs, we can perform comparative functional analyses of the variants with the reference cDNA. We find that the severity of the phenotypes of the flies expressing DOT1L variants correlates with the levels of DOT1L and H3K79 methylation. The p.Glu123Lys and p.Glu129Lys variants show the highest H3K79 methylation as well as DOT1L levels. This causes a very severe reduction in viability and a more severe wing phenotype as no animals have wild-type wings. The p.Glu123Lys variant causes a drastic increase in H3K79 methylation in human cells as well. These two variants are located in the predicted S-adenosylmethionine (SAM, methyl group donor) binding pocket in the DOT1 domain.73 They might either increase the DOT1L stability or levels or alter SAM binding and lead to enhanced DOT1L activity observed in these flies. The variant p.Leu626Val causes a moderately higher reduction in viability and higher penetrance of strong wing phenotypes compared to reference despite a relatively mild increase in H3K79 methylation. This variant is in a coiled-coil domain that is important for direct interaction of DOT1L with elongating RNAPII on actively transcribed genes.25 Therefore, it might have a direct effect on expression of the target genes leading to a milder but a different phenotype than the reference. Although the other variants (p.Cys45Gly, p.Thr100Met, and p.Lys1025Met) have similar viability and they (except p.Thr100Met) cause similar wing phenotypes as the reference, they show a mild increase in H3K79 methylation. Even though p.Cys45Gly and p.Thr100Met map to the DOT1 domain, which includes regions that are important not only for biochemical activity but also for SAM and histone binding of DOT1L, it is unknown what specific role they may play in DOT1L activity. Any alterations in these bindings, in addition to the modest increase in DOT1L levels in flies expressing these two variants, may lead to the observed increase in H3K79 methylation. The variant p.Lys1025Met is located in a disordered region which is known to be involved in DOT1L’s interaction with its partners.55,56,57 Alterations in the interaction of DOT1L with its partners could lead to higher H3K79 methylation seen in p.Lys1025Met flies. The nondiagnostic variant p.Arg292Cys shows a similar phenotypic profile as p.Cys45Gly and p.Lys1025Met except for H3K79 methylation levels. Further biochemical and structural studies will be needed to understand the specific impact of these variants on DOT1L activity and the interaction with its substrate or interaction partners. Taken together, our data suggest that the DOT1L variants reported here result in a gain of function, are associated with increased activity of DOT1L, and are associated with developmental phenotypes. Understanding the precise molecular mechanisms as to how these variants lead to increased activity in vivo may allow for the use of specific DOT1L inhibitors as a potential therapeutic strategy.
Our study emphasizes the value of functional studies and non-human model systems for rare variant interpretation. Limitations of in silico prediction tools for missense variants are well established,74 and these limitations are exacerbated in situations where the mechanism is not LoF.75 Classification of novel missense variation in DOT1L in the clinical laboratory setting may remain challenging in the short term. Although the number of useable human DNA samples was too low in this study to confidently assign and validate a DNAm signature, this remains a promising area of research that would support clinical diagnostics. We also emphasize that it remains possible that other variants in DOT1L are associated with distinct Mendelian disorders through pathogenic mechanisms beyond gain of function. Furthermore, it should be highlighted that there is some degree of variability with respect to clinical features across the cohort. For example, five individuals have microcephaly, including one with brain atrophy shown by MRI, whereas two others (with p.Cys45Gly and p.Lys1025Met) had large head circumferences, including one with megalencephaly. Further study will be required in order to better understand the specific impact of each missense variant on DOT1L function and the specific phenotypes associated with each.76
In summary, we find that de novo pathogenic DOT1L variants are associated with a Mendelian disorder in humans. We provide functional analysis in flies that supports a gain-of-function model for DOT1L variants. Further studies of the underlying mechanism will be necessary to provide a better understanding of the pathological mechanisms and may provide therapeutic strategies.
Data and code availability
There are restrictions to the availability of the genome-wide (exome and genome) sequencing data because the sequencing was performed as a clinical diagnostic test and/or because of country-specific factors. Fly reagents generated in this study will be deposited to Bloomington Stock Center and Drosophila Genomics Research Center. Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contacts Gregory Costain (gregory.costain@sickkids.ca) and Hugo J. Bellen (hbellen@bcm.edu).
Acknowledgments
We thank all participating individuals and their family members for supporting this study. We thank Hongling Pan for injections to create transgenic flies and Burak Tepe and Lindsey Goodman for suggestions. We thank the Bloomington Drosophila Stock Center (BDSC) for fly stocks and the Developmental Studies Hybridoma Bank for antibodies. This work was supported by a grant from the NIH Commonfund to the Model Organisms Screening Center of the UDN through U54 NS093793 (NINDS), the Office of Research Infrastructure Programs of the NIH (awards R24 OD022005 and R24 OD031447), the Huffington Foundation, the Jan and Dan Duncan Neurological Research Institute at Texas Children’s Hospital to H.J.B., and the Baylor College of Medicine IDDRC P50HD103555 from the Eunice Kennedy Shriver National Institute of Child Health and Human Development for use of the Microscopy Core facilities. S. Banka acknowledges the support by the NIHR Manchester Biomedical Research Centre (NIHR203308). This work was supported by a Canadian Institutes of Health Research (CIHR) grant to R.W. (PJT-178315).
Declaration of interests
The authors declare no competing interests.
Published: October 11, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2023.09.009.
Contributor Information
Gregory Costain, Email: gregory.costain@sickkids.ca.
Hugo J. Bellen, Email: hbellen@bcm.edu.
Web resources
Alamut, https://www.sophiagenetics.com/platform/alamut-visual-plus/
AxioVision, https://www.micro-shop.zeiss.com/en/us/system/software-axiovision+software-products/1007/
deCAF, https://decaf.decode.com/
Ensembl Variant Effect Predictor, http://useast.ensembl.org/Tools/VEP
GeneDx ClinVar submission page, https://www.ncbi.nlm.nih.gov/clinvar/submitters/26957/
GeneMatcher, https://genematcher.org/statistics/
Imaris, https://imaris.oxinst.com/
MARRVEL, http://www.marrvel.org/
OMIM, https://www.omim.org/
Uniprot, https://www.uniprot.org/
Supplemental information
References
- 1.Kouzarides T. Chromatin Modifications and Their Function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 2.Greer E.L., Shi Y. Histone methylation: A dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 2012;13:343–357. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Husmann D., Gozani O. Histone lysine methyltransferases in biology and disease. Nat. Struct. Mol. Biol. 2019;26:880–889. doi: 10.1038/s41594-019-0298-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allis C.D., Berger S.L., Cote J., Dent S., Jenuwien T., Kouzarides T., Pillus L., Reinberg D., Shi Y., Shiekhattar R., et al. New Nomenclature for Chromatin-Modifying Enzymes. Cell. 2007;131:633–636. doi: 10.1016/j.cell.2007.10.039. [DOI] [PubMed] [Google Scholar]
- 5.Faundes V., Newman W.G., Bernardini L., Canham N., Clayton-Smith J., Dallapiccola B., Davies S.J., Demos M.K., Goldman A., Gill H., et al. Histone Lysine Methylases and Demethylases in the Landscape of Human Developmental Disorders. Am. J. Hum. Genet. 2018;102:175–187. doi: 10.1016/j.ajhg.2017.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wood K., Tellier M., Murphy S. DOT1L and H3K79 methylation in transcription and genomic stability. Biomolecules. 2018;8 doi: 10.3390/biom8010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van Leeuwen F., Gafken P.R., Gottschling D.E. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell. 2002;109:745–756. doi: 10.1016/S0092-8674(02)00759-6. [DOI] [PubMed] [Google Scholar]
- 8.Singer M.S., Kahana A., Wolf A.J., Meisinger L.L., Peterson S.E., Goggin C., Mahowald M., Gottschling D.E. Identification of high-copy disruptors of telomeric silencing in Saccharomyces cerevisiae. Genetics. 1998;150:613–632. doi: 10.1093/genetics/150.2.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Min J., Feng Q., Li Z., Zhang Y., Xu R.M. Structure of the catalytic domain of human Dot1L, a non-SET domain nucleosomal histone methyltransferase. Cell. 2003;112:711–723. doi: 10.1016/S0092-8674(03)00114-4. [DOI] [PubMed] [Google Scholar]
- 10.Vlaming H., van Leeuwen F. The upstreams and downstreams of H3K79 methylation by DOT1L. Chromosoma. 2016;125:593–605. doi: 10.1007/s00412-015-0570-5. [DOI] [PubMed] [Google Scholar]
- 11.Frederiks F., Tzouros M., Oudgenoeg G., Van Welsem T., Fornerod M., Krijgsveld J., Van Leeuwen F. Nonprocessive methylation by Dot1 leads to functional redundancy of histone H3K79 methylation states. Nat. Struct. Mol. Biol. 2008;15:550–557. doi: 10.1038/nsmb.1432. [DOI] [PubMed] [Google Scholar]
- 12.Ng H.H., Xu R.M., Zhang Y., Struhl K. Ubiquitination of histone H2B by Rad6 is required for efficient Dot1-mediated methylation of histone H3 lysine 79. J. Biol. Chem. 2002;277:34655–34657. doi: 10.1074/jbc.C200433200. [DOI] [PubMed] [Google Scholar]
- 13.McGinty R.K., Kim J., Chatterjee C., Roeder R.G., Muir T.W. Chemically ubiquitylated histone H2B stimulates hDot1L-mediated intranucleosomal methylation. Nature. 2008;453:812–816. doi: 10.1038/nature06906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Briggs S.D., Xiao T., Sun Z.W., Caldwell J.A., Shabanowitz J., Hunt D.F., Allis C.D., Strahl B.D. Trans-histone regulatory pathway in chromatin. Nature. 2002;418:498. doi: 10.1038/nature00970. [DOI] [PubMed] [Google Scholar]
- 15.Worden E.J., Hoffmann N.A., Hicks C.W., Wolberger C. Mechanism of Cross-talk between H2B Ubiquitination and H3 Methylation by Dot1L. Cell. 2019;176:1490–1501.e12. doi: 10.1016/j.cell.2019.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barry E.R., Krueger W., Jakuba C.M., Veilleux E., Ambrosi D.J., Nelson C.E., Rasmussen T.P. ES cell cycle progression and differentiation require the action of the histone methyltransferase Dot1L. Stem Cell. 2009;27:1538–1547. doi: 10.1002/stem.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wakeman T.P., Wang Q., Feng J., Wang X.-F. Bat3 facilitates H3K79 dimethylation by DOT1L and promotes DNA damage-induced 53BP1 foci at G1/G2 cell-cycle phases. EMBO J. 2012;31:2169–2181. doi: 10.1038/emboj.2012.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ng H.H., Feng Q., Wang H., Erdjument-Bromage H., Tempst P., Zhang Y., Struhl K. Lysine methylation within the globular domain of histone H3 by Dot1 is important for telomeric silencing and Sir protein association. Genes Dev. 2002;16:1518–1527. doi: 10.1101/gad.1001502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim W., Kim R., Park G., Park J.W., Kim J.E. Deficiency of H3K79 histone methyltransferase Dot1-like protein (DOT1L) inhibits cell proliferation. J. Biol. Chem. 2012;287:5588–5599. doi: 10.1074/jbc.M111.328138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim W., Choi M., Kim J.E. The histone methyltransferase Dot1/DOT1L as a critical regulator of the cell cycle. Cell Cycle. 2014;13:726–738. doi: 10.4161/cc.28104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Z., Zang C., Rosenfeld J.A., Schones D.E., Barski A., Cuddapah S., Cui K., Roh T.Y., Peng W., Zhang M.Q., Zhao K. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat. Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Steger D.J., Lefterova M.I., Ying L., Stonestrom A.J., Schupp M., Zhuo D., Vakoc A.L., Kim J.-E., Chen J., Lazar M.A., et al. DOT1L/KMT4 Recruitment and H3K79 Methylation Are Ubiquitously Coupled with Gene Transcription in Mammalian Cells. Mol. Cell Biol. 2008;28:2825–2839. doi: 10.1128/mcb.02076-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vakoc C.R., Sachdeva M.M., Wang H., Blobel G.A. Profile of Histone Lysine Methylation across Transcribed Mammalian Chromatin. Mol. Cell Biol. 2006;26:9185–9195. doi: 10.1128/mcb.01529-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schübeler D., MacAlpine D.M., Scalzo D., Wirbelauer C., Kooperberg C., Van Leeuwen F., Gottschling D.E., O’Neill L.P., Turner B.M., Delrow J., et al. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev. 2004;18:1263–1271. doi: 10.1101/gad.1198204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim S.K., Jung I., Lee H., Kang K., Kim M., Jeong K., Kwon C.S., Han Y.M., Kim Y.S., Kim D., Lee D. Human histone H3K79 methyltransferase DOT1L methyltransferase binds actively transcribing RNA polymerase II to regulate gene expression. J. Biol. Chem. 2012;287:39698–39709. doi: 10.1074/jbc.M112.384057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones B., Su H., Bhat A., Lei H., Bajko J., Hevi S., Baltus G.A., Kadam S., Zhai H., Valdez R., et al. The histone H3K79 methyltransferase Dot1L is essential for mammalian development and heterochromatin structure. PLoS Genet. 2008;4 doi: 10.1371/journal.pgen.1000190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morello G., Porazzi P., Moro E., Argenton F., Basso G., Felix C.A., Germano G. Zebrafish Ortholog of Human DOT1L Regulates Primitive and Transient Definitive Hematopoiesis and Controls hoxa9 and meis1 Expression. Blood. 2012;120:849. doi: 10.1182/blood.v120.21.849.849. [DOI] [Google Scholar]
- 28.Shanower G.A., Muller M., Blanton J.L., Honti V., Gyurkovics H., Schedl P. Characterization of the grappa gene, the Drosophila Histone H3 lysine 79 methyltransferase. Genetics. 2005;169:173–184. doi: 10.1534/genetics.104.033191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Castaño Betancourt M.C., Cailotto F., Kerkhof H.J., Cornelis F.M.F., Doherty S.A., Hart D.J., Hofman A., Luyten F.P., Maciewicz R.A., Mangino M., et al. Genome-wide association and functional studies identify the DOT1L gene to be involved in cartilage thickness and hip osteoarthritis. Proc. Natl. Acad. Sci. USA. 2012;109:8218–8223. doi: 10.1073/pnas.1119899109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mahmoudi T., Boj S.F., Hatzis P., Li V.S.W., Taouatas N., Vries R.G.J., Teunissen H., Begthel H., Korving J., Mohammed S., et al. The leukemia-associated Mllt10/Af10-Dot1l are Tcf4/β-catenin coactivators essential for intestinal homeostasis. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mohan M., Herz H.M., Takahashi Y.H., Lin C., Lai K.C., Zhang Y., Washburn M.P., Florens L., Shilatifard A. Linking H3K79 trimethylation to Wnt signaling through a novel Dot1-containing complex (DotCom) Genes Dev. 2010;24:574–589. doi: 10.1101/gad.1898410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Splinter K., Adams D.R., Bacino C.A., Bellen H.J., Bernstein J.A., Cheatle-Jarvela A.M., Eng C.M., Esteves C., Gahl W.A., Hamid R., et al. Effect of Genetic Diagnosis on Patients with Previously Undiagnosed Disease. N. Engl. J. Med. 2018;379:2131–2139. doi: 10.1056/nejmoa1714458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ramoni R.B., Mulvihill J.J., Adams D.R., Allard P., Ashley E.A., Bernstein J.A., Gahl W.A., Hamid R., Loscalzo J., McCray A.T., et al. The Undiagnosed Diseases Network: Accelerating Discovery about Health and Disease. Am. J. Hum. Genet. 2017;100:185–192. doi: 10.1016/J.AJHG.2017.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Costain G., Walker S., Marano M., Veenma D., Snell M., Curtis M., Luca S., Buera J., Arje D., Reuter M.S., et al. Genome Sequencing as a Diagnostic Test in Children with Unexplained Medical Complexity. JAMA Netw. Open. 2020;3 doi: 10.1001/jamanetworkopen.2020.18109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deshwar A.R., Yuki K.E., Hou H., Liang Y., Khan T., Celik A., Ramani A., Mendoza-Londono R., Marshall C.R., Brudno M., et al. Trio RNA sequencing in a cohort of medically complex children. Am. J. Hum. Genet. 2023;110:895–900. doi: 10.1016/j.ajhg.2023.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: A Matching Tool for Connecting Investigators with an Interest in the Same Gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/HUMU.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hamosh A., Wohler E., Martin R., Griffith S., Rodrigues E.d.S., Antonescu C., Doheny K.F., Valle D., Sobreira N. The impact of GeneMatcher on international data sharing and collaboration. Hum. Mutat. 2022;43:668–673. doi: 10.1002/HUMU.24350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deciphering Developmental Disorders Study. Clayton S., Fitzgerald T.W., Kaplanis J., Prigmore E., Rajan D., Sifrim A., Aitken S., Akawi N., Alvi M., et al. Prevalence and architecture of de novo mutations in developmental disorders. Nature. 2017;542:433–438. doi: 10.1038/nature21062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aryee M.J., Jaffe A.E., Corrada-Bravo H., Ladd-Acosta C., Feinberg A.P., Hansen K.D., Irizarry R.A. Minfi: A flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30:1363–1369. doi: 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Salas L.A., Koestler D.C., Butler R.A., Hansen H.M., Wiencke J.K., Kelsey K.T., Christensen B.C. An optimized library for reference-based deconvolution of whole-blood biospecimens assayed using the Illumina HumanMethylationEPIC BeadArray. Genome Biol. 2018;19:64. doi: 10.1186/s13059-018-1448-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kanca O., Zirin J., Garcia-Marques J., Knight S.M., Yang-Zhou D., Amador G., Chung H., Zuo Z., Ma L., He Y., et al. An efficient CRISPR-based strategy to insert small and large fragments of DNA using short homology arms. Elife. 2019;8 doi: 10.7554/eLife.51539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee P.-T., Zirin J., Kanca O., Lin W.-W., Schulze K.L., Li-Kroeger D., Tao R., Devereaux C., Hu Y., Chung V., et al. A gene-specific T2A-GAL4 library for Drosophila. Elife. 2018;7 doi: 10.7554/eLife.35574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goodman L.D., Cope H., Nil Z., Ravenscroft T.A., Charng W.L., Lu S., Tien A.C., Pfundt R., Koolen D.A., Haaxma C.A., et al. TNPO2 variants associate with human developmental delays, neurologic deficits, and dysmorphic features and alter TNPO2 activity in Drosophila. Am. J. Hum. Genet. 2021;108:1669–1691. doi: 10.1016/j.ajhg.2021.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bischof J., Björklund M., Furger E., Schertel C., Taipale J., Basler K. A versatile platform for creating a comprehensive UAS-ORFeome library in Drosophila. Development. 2013;140:2434–2442. doi: 10.1242/DEV.088757. [DOI] [PubMed] [Google Scholar]
- 46.Venken K.J.T., He Y., Hoskins R.A., Bellen H.J. P[acman]: A BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science. 2006;314:1747–1751. doi: 10.1126/science.1134426. [DOI] [PubMed] [Google Scholar]
- 47.Wang L., Lin G., Zuo Z., Li Y., Byeon S.K., Pandey A., Bellen H.J. Neuronal activity induces glucosylceramide that is secreted via exosomes for lysosomal degradation in glia. Sci. Adv. 2022;8:3326. doi: 10.1126/sciadv.abn3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang J., Al-Ouran R., Hu Y., Kim S.Y., Wan Y.W., Wangler M.F., Yamamoto S., Chao H.T., Comjean A., Mohr S.E., et al. MARRVEL: Integration of Human and Model Organism Genetic Resources to Facilitate Functional Annotation of the Human Genome. Am. J. Hum. Genet. 2017;100:843–853. doi: 10.1016/j.ajhg.2017.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang J., Liu Z., Bellen H.J., Yamamoto S. Navigating MARRVEL, a Web-Based Tool that Integrates Human Genomics and Model Organism Genetics Information. J. Vis. Exp. 2019 doi: 10.3791/59542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Quinodoz M., Royer-Bertrand B., Cisarova K., Di Gioia S.A., Superti-Furga A., Rivolta C. DOMINO: Using Machine Learning to Predict Genes Associated with Dominant Disorders. Am. J. Hum. Genet. 2017;101:623–629. doi: 10.1016/j.ajhg.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P., et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nat. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reisenauer M.R., Wang S.W., Xia Y., Zhang W. Dot1a contains three nuclear localization signals and regulates the epithelial Na+ channel (ENaC) at multiple levels. Am. J. Physiol. Renal Physiol. 2010;299:63–76. doi: 10.1152/ajprenal.00105.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mistry J., Chuguransky S., Williams L., Qureshi M., Salazar G.A., Sonnhammer E.L.L., Tosatto S.C.E., Paladin L., Raj S., Richardson L.J., et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021;49:D412–D419. doi: 10.1093/NAR/GKAA913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang W., Hayashizaki Y., Kone B.C. Structure and regulation of the mDot1 gene, a mouse histone H3 methyltransferase. Biochem. J. 2004;377:641–651. doi: 10.1042/bj20030839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mueller D., García-Cuéllar M.P., Bach C., Buhl S., Maethner E., Slany R.K. Misguided Transcriptional Elongation Causes Mixed Lineage Leukemia. PLoS Biol. 2009;7 doi: 10.1371/journal.pbio.1000249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mueller D., Bach C., Zeisig D., Garcia-Cuellar M.P., Monroe S., Sreekumar A., Zhou R., Nesvizhskii A., Chinnaiyan A., Hess J.L., Slany R.K. A role for the MLL fusion partner ENL in transcriptional elongation and chromatin modification. Blood. 2007;110:4445–4454. doi: 10.1182/blood-2007-05-090514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kuntimaddi A., Achille N.J., Thorpe J., Lokken A.A., Singh R., Hemenway C.S., Adli M., Zeleznik-Le N.J., Bushweller J.H. Degree of Recruitment of DOT1L to MLL-AF9 Defines Level of H3K79 Di- and Tri-methylation on Target Genes and Transformation Potential. Cell Rep. 2015;11:808–820. doi: 10.1016/j.celrep.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shah S., Henriksen M.A. A novel disrupter of telomere silencing 1-like (DOT1L) interaction is required for signal transducer and activator of transcription 1 (STAT1)-activated gene expression. J. Biol. Chem. 2011;286:41195–41204. doi: 10.1074/jbc.M111.284190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hu Y., Flockhart I., Vinayagam A., Bergwitz C., Berger B., Perrimon N., Mohr S.E. An integrative approach to ortholog prediction for disease-focused and other functional studies. BMC Bioinf. 2011;12:357. doi: 10.1186/1471-2105-12-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McLaren W., Gil L., Hunt S.E., Riat H.S., Ritchie G.R.S., Thormann A., Flicek P., Cunningham F. The Ensembl Variant Effect Predictor. Genome Biol. 2016;17:122–214. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Halldorsson B.V., Eggertsson H.P., Moore K.H.S., Hauswedell H., Eiriksson O., Ulfarsson M.O., Palsson G., Hardarson M.T., Oddsson A., Jensson B.O., et al. The sequences of 150,119 genomes in the UK Biobank. Nature. 2022;607:732–740. doi: 10.1038/s41586-022-04965-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Taliun D., Harris D.N., Kessler M.D., Carlson J., Szpiech Z.A., Torres R., Taliun S.A.G., Corvelo A., Gogarten S.M., Kang H.M., et al. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. Nature. 2021;590:290–299. doi: 10.1038/s41586-021-03205-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ioannidis N.M., Rothstein J.H., Pejaver V., Middha S., McDonnell S.K., Baheti S., Musolf A., Li Q., Holzinger E., Karyadi D., et al. REVEL: An Ensemble Method for Predicting the Pathogenicity of Rare Missense Variants. Am. J. Hum. Genet. 2016;99:877–885. doi: 10.1016/j.ajhg.2016.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.MacDonald J.R., Ziman R., Yuen R.K.C., Feuk L., Scherer S.W. The Database of Genomic Variants: A curated collection of structural variation in the human genome. Nucleic Acids Res. 2014;42:D986–D992. doi: 10.1093/nar/gkt958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chater-Diehl E., Goodman S.J., Cytrynbaum C., Turinsky A.L., Choufani S., Weksberg R. Anatomy of DNA methylation signatures: Emerging insights and applications. Am. J. Hum. Genet. 2021;108:1359–1366. doi: 10.1016/j.ajhg.2021.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Venken K.J.T., Carlson J.W., Schulze K.L., Pan H., He Y., Spokony R., Wan K.H., Koriabine M., de Jong P.J., White K.P., et al. Versatile P[acman] BAC libraries for transgenesis studies in Drosophila melanogaster. Nat. Methods. 2009;6:431–434. doi: 10.1038/nmeth.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Soller M., White K. Curr. Biol. 2004;14:R53. doi: 10.1016/j.cub.2003.12.041. [DOI] [PubMed] [Google Scholar]
- 68.Xiong W.C., Okano H., Patel N.H., Blendy J.A., Montell C. repo encodes a glial-specific homeo domain protein required in the Drosophila nervous system. Genes Dev. 1994;8:981–994. doi: 10.1101/gad.8.8.981. [DOI] [PubMed] [Google Scholar]
- 69.Park P.H., Yamamoto T.M., Li H., Alcivar A.L., Xia B., Wang Y., Bernhardy A.J., Turner K.M., Kossenkov A.V., Watson Z.L., et al. Amplification of the mutation-carrying BRCA2 allele promotes RAD51 loading and PARP inhibitor resistance in the absence of reversion mutations. Mol. Cancer Ther. 2020;19:602–613. doi: 10.1158/1535-7163.MCT-17-0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Franz H., Villarreal A., Heidrich S., Videm P., Kilpert F., Mestres I., Calegari F., Backofen R., Manke T., Vogel T. DOT1L promotes progenitor proliferation and primes neuronal layer identity in the developing cerebral cortex. Nucleic Acids Res. 2019;47:168–183. doi: 10.1093/nar/gky953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fahrner J.A., Bjornsson H.T. Mendelian disorders of the epigenetic machinery: postnatal malleability and therapeutic prospects. Hum. Mol. Genet. 2019;28:R254–R264. doi: 10.1093/hmg/ddz174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Luo X., Schoch K., Jangam S.V., Bhavana V.H., Graves H.K., Kansagra S., Jasien J.M., Stong N., Keren B., Mignot C., et al. Rare deleterious de novo missense variants in Rnf2/Ring2 are associated with a neurodevelopmental disorder with unique clinical features. Hum. Mol. Genet. 2021;30:1283–1292. doi: 10.1093/hmg/ddab110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Feng Q., Wang H., Ng H.H., Erdjument-Bromage H., Tempst P., Struhl K., Zhang Y. Methylation of H3-Lysine 79 Is Mediated by a New Family of HMTases without a SET Domain. Curr. Biol. 2002;12:1052–1058. doi: 10.1016/S0960-9822(02)00901-6. [DOI] [PubMed] [Google Scholar]
- 74.Costain G., Andrade D.M. Third-generation computational approaches for genetic variant interpretation. Brain. 2023;146:411–412. doi: 10.1093/brain/awad011. [DOI] [PubMed] [Google Scholar]
- 75.Gerasimavicius L., Livesey B.J., Marsh J.A. Loss-of-function, gain-of-function and dominant-negative mutations have profoundly different effects on protein structure. Nat. Commun. 2022;13:3895–3915. doi: 10.1038/s41467-022-31686-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hou S.M., Hsia C.W., Tsai C.L., Hsia C.H., Jayakumar T., Velusamy M., Sheu J.R. Modulation of human platelet activation and in vivo vascular thrombosis by columbianadin: Regulation by integrin αiIbβ3 inside-out but not outside-in signals. J. Biomed. Sci. 2020;27:60. doi: 10.1186/s12929-020-0619-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
There are restrictions to the availability of the genome-wide (exome and genome) sequencing data because the sequencing was performed as a clinical diagnostic test and/or because of country-specific factors. Fly reagents generated in this study will be deposited to Bloomington Stock Center and Drosophila Genomics Research Center. Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contacts Gregory Costain (gregory.costain@sickkids.ca) and Hugo J. Bellen (hbellen@bcm.edu).