Summary
Polygenic risk scores (PRSs) hold promise for disease risk assessment and prevention. The Genomic Medicine at Veterans Affairs (GenoVA) Study is addressing three main challenges to the clinical implementation of PRSs in preventive care: defining and determining their clinical utility, implementing them in time-constrained primary care settings, and countering their potential to exacerbate healthcare disparities. The study processes used to test patients, report their PRS results to them and their primary care providers (PCPs), and promote the use of those results in clinical decision-making are modeled on common practices in primary care. The following diseases were chosen for their prevalence and familiarity to PCPs: coronary artery disease; type 2 diabetes; atrial fibrillation; and breast, colorectal, and prostate cancers. A randomized clinical trial (RCT) design and primary outcome of time-to-new-diagnosis of a target disease bring methodological rigor to the question of the clinical utility of PRS implementation. The study’s pragmatic RCT design enhances its relevance to how PRS might reasonably be implemented in primary care. Steps the study has taken to promote health equity include the thoughtful handling of genetic ancestry in PRS construction and reporting and enhanced recruitment strategies to address underrepresentation in research participation. To date, enhanced recruitment efforts have been both necessary and successful: participants of underrepresented race and ethnicity groups have been less likely to enroll in the study than expected but ultimately achieved proportional representation through targeted efforts. The GenoVA Study experience to date offers insights for evaluating the clinical utility of equitable PRS implementation in adult primary care.
The Genomic Medicine at Veterans Affairs (GenoVA) Study is addressing three main challenges to the clinical implementation of PRS in preventive care: defining and determining their clinical utility, implementing them in time-constrained primary care settings, and countering their potential to exacerbate healthcare disparities.
Introduction
A pressing question in genomics today is the clinical utility of polygenic risk scores (PRSs).1,2,3,4 PRSs combine information from hundreds to millions of genetic loci, each with a very small association with the risk of common complex disease. The result is a continuous and quantitative measure of an individual’s genetic susceptibility to conditions such as coronary artery disease and type 2 diabetes. Compared to rarer monogenic disease variants, PRSs might have greater transformative potential for public health and preventive medicine in their ability to identify larger proportions of the population at significantly elevated risk for disease, potentially facilitating evidence-based prevention and management.
Although the associations between PRSs and dozens of common diseases have been firmly established, at least three primary challenges impede the ability of PRSs to improve healthcare and health outcomes. First, how to define and determine the clinical utility of PRSs remains uncertain, although there is some consensus that prospectively collected patient outcomes data are needed to demonstrate their clinical utility and yet are lacking.2,5,6 Second, because most preventive care is discussed and delivered in primary-care settings, PRS-based prevention strategies will need to be implemented within this time- and resource-constrained context. Third, despite increasingly large and more diverse discovery and validation cohorts and methodological improvements in trans-ancestry analysis,7,8,9,10 concerns remain that PRS-based prediction models are less valid in underrepresented populations and that their clinical implementation might exacerbate existing healthcare disparities.3,11
Addressing these three overarching challenges to the evidence-based, equitable implementation of PRSs in preventive care, the Genomic Medicine at Veterans Affairs (GenoVA) Study is a pragmatic randomized controlled trial (RCT) of PRS testing and reporting for six common diseases screened for by primary-care providers (PCPs).12 Here we describe how the design, processes, and lessons learned in the study illustrate potential solutions for the equitable implementation of PRSs and for informing their clinical utility in adult primary care.
Study overview and conceptual model
The goal of the GenoVA Study is to model how PRSs might be equitably integrated into the busy primary-care context while using a randomized trial design to rigorously compare the impact of PRS implementation versus usual care on patient outcomes. In the conceptual model for the study (adapted from Vassy 201813), polygenic-risk information acts through both patients and providers to improve preventive health outcomes when linked to specific actionable recommendations, such as tailored screening strategies or targeted preventive therapy.
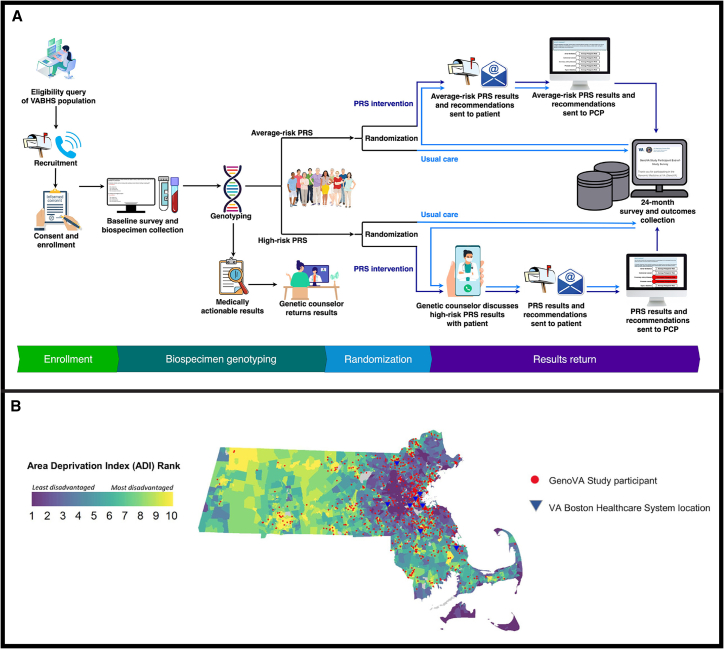
The trial protocol is registered on Clinicaltrials.gov (identifier NCT04331535) and is fully described in Note S1. The study is conducted at the VA Boston Healthcare System (VABHS), an integrated healthcare system within the U.S. Department of Veterans Affairs (VA) comprising eight facilities in eastern Massachusetts. Figure 1 illustrates the GenoVA Study processes from recruitment through results reporting. In brief, patients aged 50 to 70 years without known diagnoses of six target diseases (atrial fibrillation, coronary artery disease, type 2 diabetes, colorectal cancer, breast cancer, and prostate cancer; see full eligibility criteria in Table S1) are recruited and complete genotyping and a baseline survey on enrollment (Note S2). Genotyping categorizes each participant into one of three groups: participants with an actionable monogenic disease result, as defined by the American College of Medicine Genetics and Genomics (ACMG),14 participants with at least one high-risk PRS result, and participants with no high-risk PRS results, a group we term “average risk.” Participants with an ACMG finding receive their monogenic and PRS results from a genetic counselor. All other participants undergo randomization to the PRS intervention or usual-care arm and are stratified by sex and either high-risk or average-risk status. The PRS intervention consists of a PRS laboratory report, targeted genetic counseling for high-risk individuals, communication with each participant’s PCP, and patient- and provider-oriented materials to support decision-making around high-risk PRS results. End-of-study data collection from the EHR and surveys (Note S3) occurs 24 months after randomization, after which participants in the usual-care arm receive their results. The primary outcome compares the time to new diagnosis of the six target diseases for high-risk participants randomized to the PRS intervention or usual-care arms. The GenoVA Study has been approved by the VABHS institutional review board (IRB #3241), and all participants provide informed consent to participate.
Figure 1.
Processes and geographic catchment in the GenoVA Study
A flow diagram of the GenoVA Study process (A) illustrates how eligible participants are identified from an electronic health record (EHR) query and recruited via mailings, emails, and phone calls. Enhanced recruitment efforts target underrepresented gender, race, and ethnicity patient populations. Consent documents are signed remotely prior to baseline survey completion and collection of either blood or saliva. Genotyping identifies the subset of participants with an actionable monogenic disease result, as defined by the American College of Medicine Genetics and Genomics. These participants receive their monogenic and PRS results from a genetic counselor, who refers them to appropriate care. All other participants undergo randomization to the PRS intervention or usual-care arm and are stratified by risk status (at least one high-risk PRS versus only average-risk PRS results). In the intervention arm, any participant with at least one high-risk PRS result receives their results by phone or video from a genetic counselor; participants with only average-risk PRS results receive their results by mail or e-mail. PCPs are notified of results and recommendations by e-mail and through the EHR. End-of-study data collection from the EHR and surveys occurs 24 months after randomization. Participants in the usual-care arm receive their PRS results and recommendations through the same procedures as the intervention arm at the end of study. (B) State-level area deprivation index (ADI) values and relative geolocation of GenoVA Study participants across Massachusetts. ADI is a census block group level neighborhood disadvantage measure composed of 17 factors, including income, education, employment, and housing quality factors derived from American Community Survey data; values range from 1 to 10. Also shown are the eight healthcare facility locations of the VA Boston Health System. Abbreviations: PCP, primary care provider; PRS, polygenic risk score; VABHS, Veterans Affairs Boston Healthcare System.
Modeling clinical polygenic-risk-score testing and reporting
We have previously described our development of the GenoVA Study clinical PRS assay and reporting workflows.12 In brief, we accessed publicly available genetic loci and weights for PRSs for the six target diseases, developed an assay and bioinformatics pipeline to calculate these PRSs from Illumina Global Diversity Array genotype data in a CLIA-certified laboratory, and confirmed the disease associations of the six PRSs from this assay in an independent cohort, the Mass General Brigham Biobank. Figure S1 illustrates the format and content of the resulting PRS report.
The choices made in how to report PRSs to participants and PCPs reflect the study’s conceptual model and focus on actionability. That is, the study chose to report dichotomous PRS categories (“high risk” vs. “average risk”) instead of continuous scores to simplify interpretability for patients and providers.12,15 In the GenoVA Study, we defined a high-risk PRS as one associated with a published odds ratio (OR) of >2 for the target disease, as compared to the median PRS value. Although estimating absolute risk may be considered the gold standard for risk stratification for certain diseases (e.g., the pooled cohort equations for atherosclerotic cardiovascular disease and the BOADICEA model for breast cancer),16,17 validated absolute risk models are not available for most diseases screened for in primary care. The OR > 2 threshold approximates the effect sizes considered significant in Mendelian genetics18 and of other risk factors, such as family history or body-mass index, considered clinically important for risk stratification for the target diseases.19,20,21,22,23 We also chose to report ACMG actionable monogenic findings separately from the PRS results. Despite evidence that PRS might modulate the penetrance of monogenic variants associated with the same diseases,24 integrated models are not yet robustly validated for clinical use in diverse populations and should not be used to lessen the significance of the monogenic findings, which have more established guidelines governing their management.25,26,27 The PRS report itself models the format and content of a traditional laboratory report (Figure S1). That is, it reports the individual patient’s PRS results but does not contextualize those results amidst any other clinical risk factors (e.g., smoking status for cardiovascular disease) or protective factors (e.g., recent negative colonoscopy for colorectal cancer) the patient might have, information often unavailable to a clinical laboratory. On the other hand, the overall delivery of the PRS report back to the primary care context was designed to support its use in clinical decision-making, as described below.
Modeling the primary preventive-care context
Given that most preventive care is delivered in primary-care settings, we aimed to model the GenoVA Study PRS intervention and processes on how they might plausibly be introduced within this constrained clinical context. The following elements of the GenoVA Study reflect this goal. First, eligible patients are 50–70 years of age, a window during which many common preventive screenings occur in adult primary care.19,20,21,22 Second, eligible patients have no known diagnoses of the target diseases, presenting the opportunity for primary prevention or early detection. The study does not, therefore, model the scenario where patients receive PRS results for diseases they are already known to have, an increasingly likely occurrence if PRSs for multiple diseases are more widely implemented.28 Third, we chose six target diseases that are seen commonly in adult primary care and for which PCPs have established guidelines or practice for their prevention, screening, and diagnosis. These choices allow an examination of PRSs as a complementary tool for PCPs’ preventive practices, as opposed to as an isolated technology without familiar clinical anchoring. A high-risk PRS result for a disease is presented as an additional risk factor for the PCP to consider. The choice of familiar diseases also lessens the concern that unprepared PCPs will overinterpret PRS results and order unnecessary, costly, or even harmful follow-up tests or procedures; any test a PCP might recommend upon learning of a high-risk PRS result is likely to fall within guideline-recommended care (e.g., hemoglobin A1c testing for diabetes screening or mammography for breast cancer screening) and simply be an addition to currently used risk-stratification tools. Fourth, the GenoVA Study supports PRS test ordering by removing PRS consent and order entry from the PCP’s responsibilities but still reports the PRS results back to them, similar to other clinical-decision support or population-management programs healthcare systems often use to promote the systematic use of established preventive-care interventions, such as EHR alerts to prompt cholesterol test ordering and nurse-led lung cancer screening programs to identify and consent eligible primary-care patients for computed tomography.29
While the content of GenoVA Study PRS report itself focuses on the technical and laboratory aspects of the results, how the PRS reports and accompanying supportive information are delivered to the patient and PCP supported the interpretation of the individual patient’s PRS results, contextualization amidst other clinical factors, and clinical decision-making. The PRS results are sent to both the PCP and the patient, in addition to being entered in the EHR, both as a portable document format (pdf) report and as structured data in the laboratory information-management system (Figure S2).12 Any patient with at least one high-risk PRS is also contacted by a genetic counselor to discuss the results, potential implications for the patient’s health and health care, and recommendations for next steps in talking about the results with their PCPs. A disease-specific layperson information sheet is provided to the patient and outlines potential clinical management options.12 Similarly, the PCP is sent a provider-oriented disease-specific information sheet with details about the PRS, its limitations, recommendations for contextualizing the results among other patient characteristics and risk factors, and management suggestions, including information about current screening guidelines. In contrast, any patient with no high-risk results simply receives their report and a brief letter stating that none of their PRS results indicated high risk. This delivery was modeled on common primary-care practices in lab-results reporting: patients with abnormal results often receive phone calls or have follow-up visits to discuss the results and next steps in management, whereas patients with normal results often receive their results via brief letters or notifications via patient portal.
Determining clinical utility
There is no clear agreement on how to define the clinical utility of PRSs in the preventive medicine setting, nor how to measure that utility. Proponents of PRSs argue that, because scores in the upper tail of the normal distribution can indicate risk comparable to rare variants associated with monogenic forms of disease, PRSs could similarly influence clinical screening, prevention, and management strategies.1,2,30 Critics argue that PRSs achieve similar discrimination for disease risk as other risk factors already used in clinical care (e.g., body-mass index, family history, and smoking) or readily available without additional testing (e.g., socioeconomic status).31,32 Thus, for some diseases, it is not clear whether PRSs improve current clinical standards of care in disease prediction and prevention. As a concept, the clinical utility of genetic testing in general and PRSs specifically have been variably defined on a spectrum from narrow to expansive, depending on the context and purpose of the definition. The clinical utility of PRSs most often refers to their ability to improve patient outcomes, often through the prevention or amelioration of mortality, morbidity, or disability through the adoption of effective interventions based on the test results.1,3,4,33
In the GenoVA Study context, we define clinical utility as the ability of a PRS test to result in earlier diagnosis of clinically significant cases of the target diseases; earlier diagnosis and treatment are associated with the improved outcomes of lower morbidity and mortality.34,35,36,37 The trial’s primary outcome operationalizes the measurement of that utility: time to diagnosis of both undiagnosed prevalent cases of the six diseases and incident cases during the 24-month observation period after randomization. Despite the apparent paradox that the use of PRSs in preventive medicine might accelerate the diagnosis of disease instead of preventing disease onset, the trial’s primary hypothesis is that, among participants with at least one high-risk PRS result, the time to diagnosis will be shorter for participants receiving PRS results than for those receiving usual care. This choice of outcome best fits the population of 50- to 70-year-old adults, given their baseline annual diagnosis rate of 6.2% (Note S1) and likely high proportion of undiagnosed prevalent cases. On the basis of a priori assumptions that 33% of participants would have at least one high-risk PRS result and that 12% of high-risk patients in the usual-care arm will receive a new target diagnosis during the 24-month observation period, the GenoVA Study has 80% power at a two-tailed ɑ = 0.05 to detect a 2-fold increase in new diagnoses among high-risk patients receiving the PRS intervention.
Participants and their PCPs are not blinded to participant allocation to the intervention or study arms. Although essential for drug trials, blinding is impossible for a trial whose aim is to determine whether participants and their PCPs act on PRS results in a way that improves health outcomes. Indeed, the study hypotheses assume that this unblinded, differential knowledge about disease risk will lead to increased surveillance and disease diagnosis among high-risk patients. This does not represent a detection bias but, rather, the intention of the PRS intervention. The primary threat to study validity from this lack of blinding, then, would be differential outcomes assessment at the end of the trial. The GenoVA Study minimizes the risk of detection bias by including in the primary outcome only strictly defined clinically significant cases (e.g., only prostate cancer cases classified as intermediate risk or higher by National Comprehensive Cancer Network guidelines38 and only cases of atrial fibrillation meeting guidelines for clinical management39), as adjudicated by expert reviewers blinded to randomization status or PRS results. This measure minimizes the overdetection of clinically insignificant disease, the diagnosis of which is less certain to be associated with improved outcomes.
In choosing new diagnoses of clinically significant disease as its primary outcome, the GenoVA Study has adopted a narrow definition of clinical utility. An even narrower definition might require demonstration of reduced mortality or morbidity, often measured with quality-adjusted life years; rigorous trials powered to detect meaningful differences in these outcomes will require longer follow-up. Although narrow definitions of clinical utility are most often used by evidence-based guidelines and healthcare payers,40,41 broader definitions of the clinical utility of PRS include their ability to inform clinical decision-making, a type of clinical actionability.42 Absent evidence for narrower-sense clinical utility, actionability is an admittedly subjective term, despite efforts to generate expert-informed consensus around its definition and quantification.43,44 In the GenoVA Study, the secondary outcome of diagnostic testing ordered by treating clinicians reflects a measurement of actionability. In its broadest sense, clinical utility can also refer to the ability of a test to improve any outcomes considered important to individuals and families; such outcomes might include psychosocial wellbeing and reproductive decision-making and are often termed personal utility.1,45 Additional GenoVA Study outcomes including patient activation in their healthcare, self-assessed health status, and quality of life capture this broader scope of the utility patients might derive from PRS testing. Table S2 shows the secondary and exploratory outcomes, including follow-up diagnostic testing, patient medication adherence, healthcare costs, and quality of life, each representing process and implementation outcomes relevant to determining the value and costs of integrating PRS into adult preventive medicine. Note S4 includes the GenoVA Study statistical analysis plan, which provides greater detail about outcomes measurement and statistical approach.
The choice of a pragmatic randomized clinical trial
There is also no agreement on the type of study design and evidence needed to demonstrate the clinical utility of PRSs. Although observational studies provide valuable evidence when RCTs are not feasible, RCTs remain at the top of the evidence hierarchy, given their ability to minimize bias and confounding.46 However, RCTs have their own limitations, namely that controlled experimental conditions limit the generalizability of the findings to real-world contexts. Whether RCT evidence is needed to demonstrate the clinical utility of PRSs is an unsettled question. On one hand, RCTs are generally accepted as the gold standard for determining the effectiveness of interventions. On the other hand, most laboratory tests used routinely in clinical medicine, such as kidney function testing and complete blood counts, are not supported by RCT evidence.47,48 The question of appropriate study design, then, might hinge on whether a PRS is considered a laboratory test or as one component of a preventive genomics intervention. The GenoVA Study models PRS testing as the latter, the first step of an intervention that also includes interpreted PRS reporting, targeted genetic counseling for high-risk individuals, communication with each participant’s PCP, and patient- and provider-oriented materials to support decision-making around PRS results. An RCT design is thus appropriate for the GenoVA Study in this context. It is worth noting that the ultimate outcomes of the GenoVA Study RCT will need to be interpreted in the setting of this overall intervention, not in terms of a PRS in isolation.
Moreover, a pragmatic design is appropriate, given the study aim to determine the clinical effectiveness of PRS testing in a real-world primary-care context and the multifaceted, preventive nature of the intervention.49 This contrasts with a treatment trial warranting a more explanatory trial design to demonstrate biological impact.49 The pragmatic design also affords the opportunity to collect implementation outcomes relevant to stakeholders interested in the adoption of PRSs in clinical care; such outcomes include healthcare costs and participant- and provider-reported outcomes.49,50 Pragmatic design elements of the GenoVA Study include embedding into existing clinical workflows the PRS test ordering, the send-out to a reference laboratory, and results reporting. Another pragmatic element is the collection of trial-outcome EHR data, supplemented with end-of-study survey data. Figure S3 displays a pragmatic explanatory continuum indicator summary 2 (PRECIS-2) wheel illustrating the degree to which the GenoVA Study design is considered pragmatic versus explanatory.50 Table S3 further elucidates each of the trial’s design features and pragmatic elements. Our intention is that the pragmatic design will increase the likelihood that the RCT results are relevant to how PRSs might reasonably be implemented into routine primary care.
Promoting health equity in clinical PRS implementation
The GenoVA Study affords the opportunity to address a pressing ethical challenge to the clinical implementation of PRS: the risk of exacerbating health disparities among populations already at higher risk of poor health outcomes. The associations between PRSs and disease risk are most robustly validated for populations descended from European continental ancestry groups.11,51 Despite advances in dataset diversity, statistical methods, and trans-ancestry PRS development and validation, this disparity in PRS performance is reduced, but not eliminated.7,8,9,10
Challenges addressed
We have taken several specific actions to leverage the GenoVA Study as an opportunity to promote health equity in the clinical implementation of PRSs. First, recognizing that most data from genome-wide association studues are derived from European-ancestry populations, we paid significant attention to the handling of genetic ancestry in constructing the PRSs and in validating our proposed PRS in the multiracial Mass General Brigham Biobank.12 As described previously, instead of developing multiple population-specific PRSs (e.g., by continental genetic ancestry group or self-reported racial or ethnic group), we chose to validate a single, genetic principal-components-adjusted PRS for each disease for application across populations. At the same time, we transparently include in our PRS laboratory report a description of the populations in which the PRSs were developed and validated, highlighting the limited population diversity for some PRSs. In these efforts, we are intentional in how we use population descriptors such as racial categories and genetic ancestry groups so as not to conflate biological and social constructs or suggest that racial categories have biological meaning.52
Second, we developed recruitment strategies to address underrepresentation in biomedical research. Even if PRSs of equal accuracy across all populations are developed, existing healthcare inequities, including disparate access to care and legacies of untrustworthy healthcare systems, are still likely to impede equitable implementation. The learning healthcare system of the Veterans Health Administration offers a unique setting in which to address these challenges. Although racial and ethnic disparities in healthcare and health outcomes persist, VA as an “equal-access” healthcare system outperforms other systems on several disparity measures.53,54 Even within this setting, the GenoVA Study implemented enhanced recruitment measures to increase representation of patient populations less likely to participate in biomedical research. That is, we preferentially directed recruitment efforts (e.g., mailings and phone calls) to VABHS patients identified as non-white, Hispanic or Latino, or female, all minority populations at VABHS.
Third, we promote gender identity equity in allowing participants to describe their sex assigned at birth and gender identity and use inclusive, specific language to describe which participants may receive PRS results for prostate cancer risk (i.e., those born with a prostate) and breast cancer risk (i.e., those born of natal female sex, given that the validation of breast cancer PRSs has been limited to this population).
Equitable implementation outcomes
Analysis of recruitment and enrollment data to date illustrates these efforts. From June 17, 2020 to May 10, 2023, a total of 10,036 patients across VABHS were deemed eligible for study participation by a computable eligibility classifier described previously (Table 1).55 Among this eligible population, VABHS administrative data categorize 15.5% as having a race other than white, 3.4% as having Hispanic or Latino ethnicity, and 12.2% as female. Figure S4 shows the GenoVA Study recruitment and enrollment efforts as of May 10, 2023. Among the 2,083 participants who actively declined participation, 1,165 (56%) individuals offered a reason. Common reasons included time constraints (n = 407, 35%); ethical, legal, and social considerations (n = 173, 15%); lack of interest (n = 162, 14%); health reasons (n = 114, 9.8%); dislike of research (n = 60, 5.2%); and VA- or government-related reasons (n = 38, 3.2%).
Table 1.
Baseline demographic and clinical characteristics of first 966 GenoVA Study participants with collected biospecimens and the remaining 9,070 eligible members of the VABHS patient population
|
GenoVA patients with collected biospecimens (n = 966) |
GenoVA eligible patients (n = 9,070) |
|||
|---|---|---|---|---|
| Mean | SD | Mean | SD | |
| Age (50–70 years) | 60.47 | 5.65 | 60.68 | 5.83 |
| Body mass index (BMI) | 30.37 | 6.30 | 29.65 | 5.95 |
| Systolic blood pressure (SBP) | 130.1 | 14.45 | 131.03 | 16.22 |
| Diastolic blood pressure (DBP) | 79.74 | 8.73 | 80.00 | 9.49 |
| Low-density lipoprotein cholesterol (LDL-C) | 110.65 | 33.09 | 112.69 | 34.67 |
| n | % | N | % | |
| Gender | ||||
| Femalea | 182 | 18.5 | 1,106 | 12.2 |
| Male | 784 | 81.5 | 7,964 | 87.8 |
| Race | ||||
| American Indian/Alaskan Native | 3 | 0.3 | 35 | 0.4 |
| Asian | 9 | 0.9 | 46 | 0.5 |
| Black/African American | 112 | 11.6 | 1,120 | 12.4 |
| Native Hawaiian/Pacific Islander | 5 | 0.5 | 34 | 0.4 |
| White | 766 | 79.3 | 6,965 | 76.8 |
| Multiracial | 15 | 1.6 | 162 | 1.8 |
| Unknown or declined | 56 | 5.8 | 708 | 7.8 |
| Ethnicity | ||||
| Hispanic of Latino | 53 | 5.5 | 306 | 3.4 |
| Not Hispanic or Latino | 824 | 85.3 | 7,616 | 84.0 |
| Unknown or declined | 89 | 9.2 | 1,148 | 12.7 |
| Ruralityb | ||||
| Rural | 48 | 5.0 | 796 | 8.8 |
| Urban | 917 | 95.0 | 8,269 | 91.2 |
| Area deprivation indexc category (ADI; state rank) | ||||
| Least disadvantaged (ADI 1–3) | 228 | 23.6 | 2,109 | 23.3 |
| Moderately disadvantaged (ADI 4–6) | 359 | 37.2 | 3,363 | 37.1 |
| Most disadvantaged (ADI 7–10) | 364 | 37.7 | 3,466 | 38.2 |
| Unknown or suppressed | 15 | 1.5 | 132 | 1.5 |
Including three transgender women with male biological sex.
Rural or urban designation is attributed to geocoded patient location data as validated by the VA Geospatial Service Support Center. Rural status includes “rural” and “highly rural” designations from VA data. Six individuals included in the total cohort (n = 10,036) have undesignated rurality status.
Area deprivation index (ADI) derived via 2020 FIPS-level ADI, 2020 US Census Block Group shapefile boundaries, and VA Boston Healthcare System (station 523) geocoded patient location data as validated by the VA Geospatial Service Support Center. State rank is based on validated geolocated state of residence. 84.16% of the total cohort (n = 8,446) and 95.24% of participants with collected biospecimens (n = 920) were designated as Massachusetts residents. 147 individuals in the total cohort have missing geolocation data or suppressed ADI designations of high group quarters, low population or housing, both high group quarters and low population or housing, or questionable data integrity.
We observed that our enhanced recruitment efforts were necessary—as we did observe that participants of non-white race or Hispanic ethnicity were less likely to enroll than expected—and successful, in that they ultimately yielded proportional or higher representation of women and non-white or Hispanic participants from the VABHS population overall. Table S4 shows overall observed and expected rates of study acceptance (defined as agreement to receive a consent packet among eligible phone call respondents, n = 3,855) and enrollment (defined as return of consent documents among those who agreed to receive a consent packet, n = 2,107). Overall, women accepted study participation (p < 0.001, Cramer’s V 0.072) and enrolled (p < 0.046, Cramer’s V 0.039) in slightly greater proportions than would be expected if acceptance and enrollment were proportional across demographic categories.56 Non-white or Hispanic participants accepted study participation (p < 0.001, Cramer’s V 0.087) in slightly greater proportions than expected but were less likely to enroll (p < 0.001, Cramer’s V 0.104) by returning consent documents than expected. Table 1 shows baseline demographic and clinical characteristics of the 966 participants who have enrolled and provided a viable biospecimen, in comparison to the overall eligible VABHS patient population who did not enroll. The study achieved overrepresentation of women and Hispanic/Latino enrollees. Among enrollees, 182 (19%) identify as women, including three transgender women, compared to 12.8% of the overall eligible VABHS population (p < 0.001, Cramer’s V 0.058), and 53 (5.5%) report Hispanic/Latino ethnicity (p < 0.001, Cramer’s V 0.042), in comparison to 3.6% of the overall eligible VABHS population. Racial representation was achieved: 144 (14.9%) of enrollees report one or more racial identities other than white, in comparison to 15.4% of the overall eligible population (p = 0.149, Cramer’s V 0.019).
We also examined differences in expected and observed recruitment and enrollment outcomes by neighborhood disadvantage, as measured with the state-level area deprivation index (ADI; 1 = least deprived to 10 = most deprived). ADI is a census block group-level neighborhood disadvantage measure composed of 17 factors, including income, education, employment, and housing quality factors derived from American Community Survey (ACS) data.57 Similar proportions of participants within each ADI category were as likely to accept study participation (p = 0.241, Cramer’s V 0.015) and enroll (p = 0.982, Cramer’s V < 0.001) as expected (Table S4). Among enrollees with available data (n = 951), large proportions reside in highly disadvantaged (ADI 7–10, 38.7%) and moderately disadvantaged (ADI 4–6, 37.6%) areas. Figure 1 maps the ADI and relative geolocation of GenoVA Study enrollees across Massachusetts. In contrast, fewer rural participants have enrolled, in comparison to the remainder of the eligible patient cohort (5.0% versus 8.8%, respectively); rural status was defined by the VA Geospatial Service Support Center.58 We did not implement enhanced efforts to recruit participants from rural areas and did in fact observe lower representation of rural enrollees. Although rural participants were as likely to accept study participation as expected (p = 0.243, Cramer’s V 0.012), they were ultimately slightly less likely to enroll than expected (p = 0.031, Cramer’s V 0.044). Recruitment and retention of socioeconomically deprived and rural populations is a well-known challenge in clinical research, including among Veterans,59,60 but our success in targeted recruitment from other underrepresented populations gives hope that similar efforts would help reach these groups.
As of May 10, 2023, study staff have received interpreted PRS reports for 840 participants. Thirteen (2%) of these had at least one positive result from the current ACMG actionable secondary findings gene list (Table S5).14 Table S6 shows the distribution of all 840 PRS results received as of May 10, 2023; 307 (37.1%) participants have at least one high-risk PRS result, and 54 (6.5%) of these have two or more high-risk PRS results, consistent with expected results.12 Of these 307 high-risk participants with available demographic data, 238 are white (77.5%) and 60 are non-white or Hispanic (19.5%); this racial and ethnic demographic composition is consistent with overall recruitment percentages and is not significantly different from participants with average-risk results (p = 0.483, Cramer’s V < 0.001).
Enrolled participants are now being followed for 24 months for the study outcomes, including disease diagnoses (primary outcome) and diagnostic testing (secondary outcome). We will report these outcomes in aggregate and stratified by gender; race and ethnicity; neighborhood deprivation; and rural status.61 Planned analyses of primary and secondary outcomes will include participant sex as a covariate because of the study’s sex-stratified randomization (Note S4). We will also perform exploratory analyses to investigate whether heterogeneous effects of the PRS intervention exist among different demographic groups. For example, we will include participant race, ethnicity, ADI, and rurality separately and in combination as factor variables in our statistical models for the pre-specified outcomes to identify between-group differences and generate hypotheses for how the introduction of PRSs in primary care might differentially impact certain groups. These analyses of GenoVA Study process and outcome measures will facilitate the identification of points in the PRS clinical-implementation pathway where disparities might exist and should be addressed.
Conclusions
The clinical implementation of PRSs is moving forward through clinical programs, research projects, and commercial laboratory and direct-to-consumer offerings,28,62,63,64 and a limited number of important RCTs have or will inform the clinical utility of PRS in single-disease settings.65,66,67,68,69,70 As a pragmatic RCT implementing a multi-disease PRS intervention, the GenoVA Study makes a unique contribution to informing the equitable implementation of PRSs for preventive medicine in the time-constrained primary-care context. Its design as a pragmatic trial enhances the generalizability of its ultimate findings, and its RCT design adds rigor to hypothesis-testing about the impact of PRS testing on preventive-medicine processes and outcomes. The VA is the largest healthcare system in the United States. Although this setting limits the generalizability of the GenoVA Study’s findings to other settings in some respects, lessons learned from the study still offer potential solutions for assessing the clinical utility of implementing PRSs into adult primary care while attending to the potential of that implementation to hinder or promote health equity.
Acknowledgments
This work was supported by NIH (National Institutes of Health) National Human Genome Research Institute (NHGRI) grant R35HG010706. P.N. is supported by NIH NHGRI grant U01HG011719. S.A.L. previously received support from NIH grants R01HL139731 and R01HL157635, and American Heart Association 18SFRN34250007 during this project. K.D.C. was previously supported by a research grant from Sanford Health. R.C.G.’s research is supported by grant funding from the National Institutes of Health, the Department of Defense, and the Franca Sozzani Fund for Preventive Genomics.
Author contributions
J.L.V., C.A.B., M.S.L., N.V.J.A., M.G., R.C.G., P.K., S.A.L., and P.N. conceived the study and contributed to its design. M.S.L. analyzed the data from the MGBB and GenoVA samples. A.A.A., C.A.B., M.D., N.E.J., P.K., E.H., K.M., M.P.C., and J.L.V. collected GenoVA data. J.L.V., C.A.B., K.M., N.E.J., and A.A.A. drafted the manuscript, and all authors reviewed the scientific content of the manuscript prior to submission.
Declaration of interests
P.N. reports research grants from Allelica, Apple, Amgen, Boston Scientific, Genentech/Roche, and Novartis; and personal fees from Allelica, Apple, AstraZeneca, Blackstone Life Sciences, Eli Lilly & Co, Foresite Labs, Genentech/Roche, GV, HeartFlow, Magnet Biomedicine, and Novartis; scientific advisory board membership of Esperion Therapeutics, Preciseli, and TenSixteen Bio; status as a scientific co-founder of TenSixteen Bio; equity in MyOme, Preciseli, and TenSixteen Bio; and spousal employment at Vertex Pharmaceuticals, all unrelated to the present work. S.A.L. is an employee of Novartis as of July 18, 2022, has previously received sponsored research support from Bristol Myers Squibb, Pfizer, Boehringer Ingelheim, Fitbit, Medtronic, Premier, and IBM, and has consulted for Bristol Myers Squibb, Pfizer, Blackstone Life Sciences, and Invitae. A.C.F.L. reports owning stock in Fabric Genomics. R.C.G. has received compensation for advising Allelica, Atria, Fabric, Genome Web, Genomic Life, and Juniper Genomics and is co-founder of Genome Medical and Nurture Genomics. The other authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2023.10.001.
Web resources
2020 12-Digit FIPS code block group area deprivation index (ADI) data, https://www.neighborhoodatlas.medicine.wisc.edu/
2020 United States Census Bureau cartographic boundary files, https://www.census.gov/geographies/mapping-files/time-series/geo/cartographic-boundary.2020.html#list-tab-1883739534
Supplemental information
References
- 1.Torkamani A., Wineinger N.E., Topol E.J. The personal and clinical utility of polygenic risk scores. Nat. Rev. Genet. 2018;19:581–590. doi: 10.1038/s41576-018-0018-x. [DOI] [PubMed] [Google Scholar]
- 2.Lambert S.A., Abraham G., Inouye M. Towards clinical utility of polygenic risk scores. Hum. Mol. Genet. 2019;28:R133–R142. doi: 10.1093/hmg/ddz187. [DOI] [PubMed] [Google Scholar]
- 3.Lewis C.M., Vassos E. Polygenic risk scores: from research tools to clinical instruments. Genome Med. 2020;12:44. doi: 10.1186/s13073-020-00742-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abu-El-Haija A., Reddi H.V., Wand H., Rose N.C., Mori M., Qian E., Murray M.F., ACMG Professional Practice and Guidelines Committee. Electronic address: documents@acmg.net The clinical application of polygenic risk scores: A points to consider statement of the American College of Medical Genetics and Genomics (ACMG) Genet. Med. 2023;25:100803. doi: 10.1016/j.gim.2023.100803. [DOI] [PubMed] [Google Scholar]
- 5.Huntley C., Torr B., Sud A., Rowlands C.F., Way R., Snape K., Hanson H., Swanton C., Broggio J., Lucassen A., et al. Utility of polygenic risk scores in UK cancer screening: a modelling analysis. Lancet Oncol. 2023;24:658–668. doi: 10.1016/S1470-2045(23)00156-0. [DOI] [PubMed] [Google Scholar]
- 6.Grosse S.D., Khoury M.J. What is the clinical utility of genetic testing? Genet. Med. 2006;8:448–450. doi: 10.1097/01.gim.0000227935.26763.c6. [DOI] [PubMed] [Google Scholar]
- 7.Wang Y., Tsuo K., Kanai M., Neale B.M., Martin A.R. Challenges and Opportunities for Developing More Generalizable Polygenic Risk Scores. Annu. Rev. Biomed. Data Sci. 2022;5:293–320. doi: 10.1146/annurev-biodatasci-111721-074830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ge T., Irvin M.R., Patki A., Srinivasasainagendra V., Lin Y.-F., Tiwari H.K., Armstrong N.D., Benoit B., Chen C.-Y., Choi K.W., et al. Development and validation of a trans-ancestry polygenic risk score for type 2 diabetes in diverse populations. Genome Med. 2022;14:70. doi: 10.1186/s13073-022-01074-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weissbrod O., Kanai M., Shi H., Gazal S., Peyrot W.J., Khera A.V., Okada Y., Biobank Japan Project. Martin A.R., Finucane H.K., Price A.L. Leveraging fine-mapping and multipopulation training data to improve cross-population polygenic risk scores. Nat. Genet. 2022;54:450–458. doi: 10.1038/s41588-022-01036-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ding Y., Hou K., Xu Z., Pimplaskar A., Petter E., Boulier K., Privé F., Vilhjálmsson B.J., Olde Loohuis L.M., Pasaniuc B. Polygenic scoring accuracy varies across the genetic ancestry continuum. Nature. 2023;618:774–781. doi: 10.1038/s41586-023-06079-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martin A.R., Kanai M., Kamatani Y., Okada Y., Neale B.M., Daly M.J. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat. Genet. 2019;51:584–591. doi: 10.1038/s41588-019-0379-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hao L., Kraft P., Berriz G.F., Hynes E.D., Koch C., Korategere V Kumar P., Parpattedar S.S., Steeves M., Yu W., Antwi A.A., et al. Development of a clinical polygenic risk score assay and reporting workflow. Nat. Med. 2022;28:1006–1013. doi: 10.1038/s41591-022-01767-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vassy J.L., Chun S., Advani S., Ludin S.A., Smith J.G., Alligood E.C. Impact of SLCO1B1 Pharmacogenetic Testing on Patient and Healthcare Outcomes: A Systematic Review. Clin. Pharmacol. Ther. 2019;106:360–373. doi: 10.1002/cpt.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller D.T., Lee K., Abul-Husn N.S., Amendola L.M., Brothers K., Chung W.K., Gollob M.H., Gordon A.S., Harrison S.M., Hershberger R.E., et al. ACMG SF v3.1 list for reporting of secondary findings in clinical exome and genome sequencing: A policy statement of the American College of Medical Genetics and Genomics (ACMG) Genet. Med. 2022;24:1407–1414. doi: 10.1016/j.gim.2022.04.006. [DOI] [PubMed] [Google Scholar]
- 15.Lewis A.C.F., Green R.C., Vassy J.L. Polygenic risk scores in the clinic: Translating risk into action. HGG Adv. 2021;2 doi: 10.1016/j.xhgg.2021.100047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee A., Mavaddat N., Wilcox A.N., Cunningham A.P., Carver T., Hartley S., Babb de Villiers C., Izquierdo A., Simard J., Schmidt M.K., et al. BOADICEA: a comprehensive breast cancer risk prediction model incorporating genetic and nongenetic risk factors. Genet. Med. 2019;21:1708–1718. doi: 10.1038/s41436-018-0406-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stone N.J., Robinson J.G., Lichtenstein A.H., Bairey Merz C.N., Blum C.B., Eckel R.H., Goldberg A.C., Gordon D., Levy D., Lloyd-Jones D.M., et al. 2013 ACC/AHA Guideline on the Treatment of Blood Cholesterol to Reduce Atherosclerotic Cardiovascular Risk in Adults: A Report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines. Circulation. 2014;129:S1–S45. doi: 10.1161/01.cir.0000437738.63853.7a. [DOI] [PubMed] [Google Scholar]
- 18.Senol-Cosar O., Schmidt R.J., Qian E., Hoskinson D., Mason-Suares H., Funke B., Lebo M.S. Considerations for clinical curation, classification, and reporting of low-penetrance and low effect size variants associated with disease risk. Genet. Med. 2019;21:2765–2773. doi: 10.1038/s41436-019-0560-8. [DOI] [PubMed] [Google Scholar]
- 19.US Preventive Services Task Force. Grossman D.C., Curry S.J., Owens D.K., Bibbins-Domingo K., Caughey A.B., Davidson K.W., Doubeni C.A., Ebell M., Epling J.W., Jr., et al. Screening for Prostate Cancer: US Preventive Services Task Force Recommendation Statement. JAMA. 2018;319:1901–1913. doi: 10.1001/jama.2018.3710. [DOI] [PubMed] [Google Scholar]
- 20.US Preventive Services Task Force. Davidson K.W., Barry M.J., Mangione C.M., Cabana M., Caughey A.B., Davis E.M., Donahue K.E., Doubeni C.A., Krist A.H., et al. Screening for Colorectal Cancer: US Preventive Services Task Force Recommendation Statement. JAMA. 2021;325:1965–1977. doi: 10.1001/jama.2021.6238. [DOI] [PubMed] [Google Scholar]
- 21.Siu A.L., U.S. Preventive Services Task Force Screening for Breast Cancer: U.S. Preventive Services Task Force Recommendation Statement. Ann. Intern. Med. 2016;164:279–296. doi: 10.7326/M15-2886. [DOI] [PubMed] [Google Scholar]
- 22.ElSayed N.A., Aleppo G., Aroda V.R., Bannuru R.R., Brown F.M., Bruemmer D., Collins B.S., Hilliard M.E., Isaacs D., Johnson E.L., et al. 2. Classification and Diagnosis of Diabetes: Standards of Care in Diabetes-2023. Diabetes Care. 2023;46:S19–S40. doi: 10.2337/dc23-S002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arnett D.K., Blumenthal R.S., Albert M.A., Buroker A.B., Goldberger Z.D., Hahn E.J., Himmelfarb C.D., Khera A., Lloyd-Jones D., McEvoy J.W., et al. 2019 ACC/AHA Guideline on the Primary Prevention of Cardiovascular Disease: A Report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. Circulation. 2019;140:e596–e646. doi: 10.1161/CIR.0000000000000678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fahed A.C., Wang M., Homburger J.R., Patel A.P., Bick A.G., Neben C.L., Lai C., Brockman D., Philippakis A., Ellinor P.T., et al. Polygenic background modifies penetrance of monogenic variants for tier 1 genomic conditions. Nat. Commun. 2020;11:3635. doi: 10.1038/s41467-020-17374-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bevers T.B., Niell B.L., Baker J.L., Bennett D.L., Bonaccio E., Camp M.S., Chikarmane S., Conant E.F., Eghtedari M., Flanagan M.R., et al. NCCN Guidelines® Insights: Breast Cancer Screening and Diagnosis, Version 1.2023: Featured Updates to the NCCN Guidelines. J. Natl. Compr. Canc. Netw. 2023;21:900–909. doi: 10.6004/jnccn.2023.0046. [DOI] [PubMed] [Google Scholar]
- 26.Weiss J.M., Gupta S., Burke C.A., Axell L., Chen L.-M., Chung D.C., Clayback K.M., Dallas S., Felder S., Gbolahan O., et al. NCCN Guidelines® Insights: Genetic/Familial High-Risk Assessment: Colorectal, Version 1.2021. J. Natl. Compr. Canc. Netw. 2021;19:1122–1132. doi: 10.1164/jnccn.2021.0048. [DOI] [PubMed] [Google Scholar]
- 27.Watts G.F., Gidding S.S., Hegele R.A., Raal F.J., Sturm A.C., Jones L.K., Sarkies M.N., Al-Rasadi K., Blom D.J., Daccord M., et al. International Atherosclerosis Society guidance for implementing best practice in the care of familial hypercholesterolaemia. Nat. Rev. Cardiol. 2023:1–25. doi: 10.1038/s41569-023-00892-0. [DOI] [PubMed] [Google Scholar]
- 28.Linder J.E., Allworth A., Bland H.T., Caraballo P.J., Chisholm R.L., Clayton E.W., Crosslin D.R., Dikilitas O., DiVietro A., Esplin E.D., et al. Returning integrated genomic risk and clinical recommendations: The eMERGE study. Genet. Med. 2023;25 doi: 10.1016/j.gim.2023.100006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saab M.M., O’Driscoll M., Sahm L.J., Leahy-Warren P., Noonan B., FitzGerald S., Kilty C., O’Malley M., Lyons N., Hegarty J. Referring high-risk individuals for lung cancer screening: A systematic review of interventions with healthcare professionals. Eur. J. Cancer Prev. 2022;31:540–550. doi: 10.1097/CEJ.0000000000000755. [DOI] [PubMed] [Google Scholar]
- 30.Khera A.V., Chaffin M., Aragam K.G., Haas M.E., Roselli C., Choi S.H., Natarajan P., Lander E.S., Lubitz S.A., Ellinor P.T., Kathiresan S. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat. Genet. 2018;50:1219–1224. doi: 10.1038/s41588-018-0183-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khoury M.J., Janssens A.C.J.W., Ransohoff D.F. How can polygenic inheritance be used in population screening for common diseases? Genet. Med. 2013;15:437–443. doi: 10.1038/gim.2012.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Inouye M., Abraham G., Nelson C.P., Wood A.M., Sweeting M.J., Dudbridge F., Lai F.Y., Kaptoge S., Brozynska M., Wang T., et al. Genomic Risk Prediction of Coronary Artery Disease in 480,000 Adults: Implications for Primary Prevention. J. Am. Coll. Cardiol. 2018;72:1883–1893. doi: 10.1016/j.jacc.2018.07.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reddi H.V., Wand H., Funke B., Zimmermann M.T., Lebo M.S., Qian E., Shirts B.H., Zou Y.S., Zhang B.M., Rose N.C., et al. Laboratory perspectives in the development of polygenic risk scores for disease: A points to consider statement of the American College of Medical Genetics and Genomics (ACMG) Genet. Med. 2023;25:100804. doi: 10.1016/j.gim.2023.100804. [DOI] [PubMed] [Google Scholar]
- 34.Crosby D., Bhatia S., Brindle K.M., Coussens L.M., Dive C., Emberton M., Esener S., Fitzgerald R.C., Gambhir S.S., Kuhn P., et al. Early detection of cancer. Science. 2022;375 doi: 10.1126/science.aay9040. [DOI] [PubMed] [Google Scholar]
- 35.Chatterjee S., Khunti K., Davies M.J. Type 2 diabetes. Lancet. 2017;389:2239–2251. doi: 10.1016/S0140-6736(17)30058-2. [DOI] [PubMed] [Google Scholar]
- 36.Kirchhof P., Camm A.J., Goette A., Brandes A., Eckardt L., Elvan A., Fetsch T., van Gelder I.C., Haase D., Haegeli L.M., et al. Early Rhythm-Control Therapy in Patients with Atrial Fibrillation. N. Engl. J. Med. 2020;383:1305–1316. doi: 10.1056/NEJMoa2019422. [DOI] [PubMed] [Google Scholar]
- 37.Fihn S.D., Gardin J.M., Abrams J., Berra K., Blankenship J.C., Dallas A.P., Douglas P.S., Foody J.M., Gerber T.C., Hinderliter A.L., et al. 2012 ACCF/AHA/ACP/AATS/PCNA/SCAI/STS Guideline for the Diagnosis and Management of Patients With Stable Ischemic Heart Disease: Executive Summary: A Report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines, and the American College of Physicians, American Association for Thoracic Surgery, Preventive Cardiovascular Nurses Association, Society for Cardiovascular Angiography and Interventions, and Society of Thoracic Surgeons. J. Am. Coll. Cardiol. 2012;60:2564–2603. doi: 10.1016/j.jacc.2012.07.012. [DOI] [PubMed] [Google Scholar]
- 38.Moses K.A., Sprenkle P.C., Bahler C., Box G., Carlsson S.V., Catalona W.J., Dahl D.M., Dall’Era M., Davis J.W., Drake B.F., et al. NCCN Guidelines® Insights: Prostate Cancer Early Detection, Version 1.2023: Featured Updates to the NCCN Guidelines. J. Natl. Compr. Canc. Netw. 2023;21:236–246. doi: 10.6004/jnccn.2023.0014. [DOI] [PubMed] [Google Scholar]
- 39.Noseworthy P.A., Kaufman E.S., Chen L.Y., Chung M.K., Elkind M.S.V., Joglar J.A., Leal M.A., McCabe P.J., Pokorney S.D., Yao X., American Heart Association Council on Clinical Cardiology Electrocardiography and Arrhythmias Committee; Council on Arteriosclerosis, Thrombosis and Vascular Biology; Council on Cardiovascular and Stroke Nursing; and Stroke Council Subclinical and Device-Detected Atrial Fibrillation: Pondering the Knowledge Gap: A Scientific Statement From the American Heart Association. Circulation. 2019;140:e944–e963. doi: 10.1161/CIR.0000000000000740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pitini E., De Vito C., Marzuillo C., D’Andrea E., Rosso A., Federici A., Di Maria E., Villari P. How is genetic testing evaluated? A systematic review of the literature. Eur. J. Hum. Genet. 2018;26:605–615. doi: 10.1038/s41431-018-0095-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walcott S.E., Miller F.A., Dunsmore K., Lazor T., Feldman B.M., Hayeems R.Z. Measuring clinical utility in the context of genetic testing: a scoping review. Eur. J. Hum. Genet. 2021;29:378–386. doi: 10.1038/s41431-020-00744-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goddard K.A.B., Lee K., Buchanan A.H., Powell B.C., Hunter J.E. Establishing the Medical Actionability of Genomic Variants. Annu. Rev. Genomics Hum. Genet. 2022;23:173–192. doi: 10.1146/annurev-genom-111021-032401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Berg J.S., Foreman A.K.M., O’Daniel J.M., Booker J.K., Boshe L., Carey T., Crooks K.R., Jensen B.C., Juengst E.T., Lee K., et al. A semiquantitative metric for evaluating clinical actionability of incidental or secondary findings from genome-scale sequencing. Genet. Med. 2016;18:467–475. doi: 10.1038/gim.2015.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hunter J.E., Irving S.A., Biesecker L.G., Buchanan A., Jensen B., Lee K., Martin C.L., Milko L., Muessig K., Niehaus A.D., et al. A standardized, evidence-based protocol to assess clinical actionability of genetic disorders associated with genomic variation. Genet. Med. 2016;18:1258–1268. doi: 10.1038/gim.2016.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.ACMG Board of Directors Clinical utility of genetic and genomic services: a position statement of the American College of Medical Genetics and Genomics. Genet. Med. 2015;17:505–507. doi: 10.1038/gim.2015.41. [DOI] [PubMed] [Google Scholar]
- 46.Burke W. Genetic tests: clinical validity and clinical utility. Curr. Protoc. Hum. Genet. 2014;81:9.15.1–9.15.8. doi: 10.1002/0471142905.hg0915s81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sackett D.L., Rosenberg W.M., Gray J.A., Haynes R.B., Richardson W.S. Evidence based medicine: what it is and what it isn’t. BMJ. 1996;312:71–72. doi: 10.1136/bmj.312.7023.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Elwenspoek M.M.C., Scott L.J., Alsop K., Patel R., Watson J.C., Mann E., Whiting P. What methods are being used to create an evidence base on the use of laboratory tests to monitor long-term conditions in primary care? A scoping review. Fam. Pract. 2020;37:845–853. doi: 10.1093/fampra/cmaa074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ford I., Norrie J. Pragmatic Trials. N. Engl. J. Med. 2016;375:454–463. doi: 10.1056/NEJMra1510059. [DOI] [PubMed] [Google Scholar]
- 50.Loudon K., Treweek S., Sullivan F., Donnan P., Thorpe K.E., Zwarenstein M. The PRECIS-2 tool: designing trials that are fit for purpose. BMJ. 2015;350:h2147. doi: 10.1136/bmj.h2147. [DOI] [PubMed] [Google Scholar]
- 51.Duncan L., Shen H., Gelaye B., Meijsen J., Ressler K., Feldman M., Peterson R., Domingue B. Analysis of polygenic risk score usage and performance in diverse human populations. Nat. Commun. 2019;10:3328. doi: 10.1038/s41467-019-11112-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Using Population Descriptors in Genetics and Genomics Research: A New Framework for an Evolving Field (2023). (National Academies Press) 10.17226/26902. [DOI] [PubMed]
- 53.Peterson K., Anderson J., Boundy E., Ferguson L., McCleery E., Waldrip K. Mortality Disparities in Racial/Ethnic Minority Groups in the Veterans Health Administration: An Evidence Review and Map. Am. J. Public Health. 2018;108:e1–e11. doi: 10.2105/AJPH.2017.304246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wong M.S., Hoggatt K.J., Steers W.N., Frayne S.M., Huynh A.K., Yano E.M., Saechao F.S., Ziaeian B., Washington D.L. Racial/Ethnic Disparities in Mortality Across the Veterans Health Administration. Health Equity. 2019;3:99–108. doi: 10.1089/heq.2018.0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alexander N.V.J., Brunette C.A., Guardino E.T., Yi T., Kerman B.J., MacIsaac K., Harris E.J., Antwi A.A., Vassy J.L. Performance of EHR classifiers for patient eligibility in a clinical trial of precision screening. Contemp. Clin. Trials. 2022;121 doi: 10.1016/j.cct.2022.106926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kamiran F., Calders T. Data preprocessing techniques for classification without discrimination. Knowl. Inf. Syst. 2012;33:1–33. doi: 10.1007/s10115-011-0463-8. [DOI] [Google Scholar]
- 57.Kind A.J.H., Buckingham W.R. Making Neighborhood-Disadvantage Metrics Accessible - The Neighborhood Atlas. N. Engl. J. Med. 2018;378:2456–2458. doi: 10.1056/NEJMp1802313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gonsoulin M. US Department of Veteran Affairs; 2018. VIReC Factbook: Corporate Data Warehouse (CDW) Patient 3.1 Domain. [Google Scholar]
- 59.Brockman T.A., Shaw O., Wiepert L., Nguyen Q.A., Kelpin S.S., West I., Albertie M., Williams S., Abbenyi A., Stephenson N., et al. Community engagement strategies to promote recruitment and participation in clinical research among rural communities: A narrative review. J. Clin. Transl. Sci. 2023;7:e84. doi: 10.1017/cts.2023.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weeks W.B., Wallace A.E., West A.N., Heady H.R., Hawthorne K. Research on Rural Veterans: An Analysis of the Literature. J. Rural Health. 2008;24:337–344. doi: 10.1111/j.1748-0361.2008.00179.x. [DOI] [PubMed] [Google Scholar]
- 61.Fain K.M., Nelson J.T., Tse T., Williams R.J. Race and ethnicity reporting for clinical trials in ClinicalTrials.gov and publications. Contemp. Clin. Trials. 2021;101 doi: 10.1016/j.cct.2020.106237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Homburger J.R., Neben C.L., Mishne G., Zhou A.Y., Kathiresan S., Khera A.V. Low coverage whole genome sequencing enables accurate assessment of common variants and calculation of genome-wide polygenic scores. Genome Med. 2019;11:74. doi: 10.1186/s13073-019-0682-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen S.-F., Dias R., Evans D., Salfati E.L., Liu S., Wineinger N.E., Torkamani A. Genotype imputation and variability in polygenic risk score estimation. Genome Med. 2020;12:100. doi: 10.1186/s13073-020-00801-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hughes E., Tshiaba P., Gallagher S., Wagner S., Judkins T., Roa B., Rosenthal E., Domchek S., Garber J., Lancaster J., et al. Development and Validation of a Clinical Polygenic Risk Score to Predict Breast Cancer Risk. JCO Precis. Oncol. 2020;4:585–592. doi: 10.1200/PO.19.00360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Grant R.W., O’Brien K.E., Waxler J.L., Vassy J.L., Delahanty L.M., Bissett L.G., Green R.C., Stember K.G., Guiducci C., Park E.R., et al. Personalized genetic risk counseling to motivate diabetes prevention: a randomized trial. Diabetes Care. 2013;36:13–19. doi: 10.2337/dc12-0884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kullo I.J., Jouni H., Austin E.E., Brown S.A., Kruisselbrink T.M., Isseh I.N., Haddad R.A., Marroush T.S., Shameer K., Olson J.E., et al. Incorporating a Genetic Risk Score into Coronary Heart Disease Risk Estimates: Effect on LDL Cholesterol Levels (the MIGENES Clinical Trial) Circularion. 2016;133:1181–1188. doi: 10.1161/CIRCULATIONAHA.115.020109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Turner A.R., Lane B.R., Rogers D., Lipkus I., Weaver K., Danhauer S.C., Zhang Z., Hsu F.C., Noyes S.L., Adams T., et al. Randomized trial finds that prostate cancer genetic risk score feedback targets prostate-specific antigen screening among at-risk men. Cancer. 2016;122:3564–3575. doi: 10.1002/cncr.30162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Smit A.K., Allen M., Beswick B., Butow P., Dawkins H., Dobbinson S.J., Dunlop K.L., Espinoza D., Fenton G., Kanetsky P.A., et al. Impact of personal genomic risk information on melanoma prevention behaviors and psychological outcomes: a randomized controlled trial. Genet. Med. 2021;23:2394–2403. doi: 10.1038/s41436-021-01292-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gray M.P., Berman Y., Bottà G., Grieve S.M., Ho A., Hu J., Hyun K., Ingles J., Jennings G., Kilov G., et al. Incorporating a polygenic risk score-triaged coronary calcium score into cardiovascular disease examinations to identify subclinical coronary artery disease (ESCALATE): Protocol for a prospective, nonrandomized implementation trial. Am. Heart J. 2023;264:163–173. doi: 10.1016/j.ahj.2023.06.009. [DOI] [PubMed] [Google Scholar]
- 70.Esserman L.J., WISDOM Study and Athena Investigators The WISDOM Study: breaking the deadlock in the breast cancer screening debate. NPJ Breast Cancer. 2017;3:34. doi: 10.1038/s41523-017-0035-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.