Summary
As large-scale genomic screening becomes increasingly prevalent, understanding the influence of actionable results on healthcare utilization is key to estimating the potential long-term clinical impact. The eMERGE network sequenced individuals for actionable genes in multiple genetic conditions and returned results to individuals, providers, and the electronic health record. Differences in recommended health services (laboratory, imaging, and procedural testing) delivered within 12 months of return were compared among individuals with pathogenic or likely pathogenic (P/LP) findings to matched individuals with negative findings before and after return of results. Of 16,218 adults, 477 unselected individuals were found to have a monogenic risk for arrhythmia (n = 95), breast cancer (n = 96), cardiomyopathy (n = 95), colorectal cancer (n = 105), or familial hypercholesterolemia (n = 86). Individuals with P/LP results more frequently received services after return (43.8%) compared to before return (25.6%) of results and compared to individuals with negative findings (24.9%; p < 0.0001). The annual cost of qualifying healthcare services increased from an average of $162 before return to $343 after return of results among the P/LP group (p < 0.0001); differences in the negative group were non-significant. The mean difference-in-differences was $149 (p < 0.0001), which describes the increased cost within the P/LP group corrected for cost changes in the negative group. When stratified by individual conditions, significant cost differences were observed for arrhythmia, breast cancer, and cardiomyopathy. In conclusion, less than half of individuals received billed health services after monogenic return, which modestly increased healthcare costs for payors in the year following return.
Keywords: clinical outcomes, electronic health records, translational, monogenic sequencing, return of results, actionable genetic findings, healthcare utilization, healthcare costs
The eMERGE network sequenced and returned actionable genetic findings and then analyzed changes in provision of healthcare services. We found that individuals with pathogenic or likely pathogenic results more frequently received guideline-indicated services and had modestly higher annual costs after return compared to before return of results and compared to individuals with negative results.
Introduction
Population genomic screening has a variable yet potentially profound impact on routine clinical care such as risk assessment, diagnosis, and therapeutic decision making. Genomic screening more reliably identifies individuals at monogenic high risk of disease compared to testing based on family history alone1 and identifies individuals eligible for tailored preventive care according to widely disseminated clinical guidelines.2,3,4,5,6 Over individuals’ lifetimes, screening is projected to prevent or ameliorate cancers and cardiovascular disease to justify the initial investment in testing.7 While genomic screening tests are increasingly affordable and popular using direct-to-consumer and health-system-based testing models, they are not currently standard of care for primary prevention in clinical practice nor are they reimbursable. One barrier to adoption is the lack of data on the cost to manage these risks and adherence to recommended follow up when such screening is deployed within health systems.
To improve clinical outcomes after return of genomic risk, patients at increased risk should receive genetic counseling, education, and follow-up evaluations that may consist of laboratory, imaging, or procedural screening and diagnostic testing. The initial evaluations and follow up may be considered process outcomes that mediate changes in health. For example, hypertrophic cardiomyopathy risks may be evaluated with electrocardiography, echocardiography, Holter monitor, or cardiac magnetic resonance imaging (MRI) in order to assess the need for an implantable cardiac defibrillator designed to treat potentially fatal ventricular arrhythmias. Utilization of EHR data and analytic methods that isolate outcomes attributable to return of results (RoR) provide a practical approach.
This manuscript details the short-term impact of RoR in a cohort of 16,218 adult individuals recruited from 10 health systems by the Electronic Medical Records and Genomics (eMERGE) network. The laboratory, imaging, and procedural services delivered in the year following return were assessed from administrative data. Among the genetic results returned by eMERGE, outcomes for the five most common conditions—arrhythmia (long QT [MIM: PS192500]), breast cancer (BCa [MIM: 114480]), cardiomyopathy (cardiomyopathy, familial hypertrophic [MIM: PS115200, PS192600]), colorectal cancer (CRC [MIM: PS120435]), and familial hypercholesterolemia (FH [MIM: PS143890])—were analyzed. Frequency of health care utilization as well as impact on costs incurred by payors after return of genomic results were examined to determine the impact of genomic sequencing and return on an unselected population.
Subjects and methods
Setting and population
The eMERGE consortium was established in 2007 and focuses on translational research utilizing large-scale genome wide- and phenome wide-association analyses.8,9,10,11,12,13,14,15 The third phase of the network, which was conducted from 2015 to 2019, consisted of 10 institutions that enrolled a large prospective cohort in order to return results from a targeted next-generation sequencing (NGS) panel. The “eMERGEseq” panel content was designed to include genes with known actionability.11,16 While all individuals received the same panel test and were required to have a longitudinal care provider, sites varied in cohort enrollment ascertainment (indications for enrollment), age (pediatric and adult), return method, ethnicity/race distributions, and management of genetic risk. As described in Fossey et al.,17 seven of the sites used genetic counselors to return P/LP variants, two sites additionally used primary care providers or a clinical genomics team, and two sites used only primary care physicians. Leppig et al.18 found that participants utilized genetic counseling services when offered (38.4% of the time) or embedded (42.8% of the time) in the return process. Approximately 4.2% of the eMERGE III cohort were found to have pathogenic or likely pathogenic (P/LP) variants. To better represent the general population, individuals from eMERGE sites using specific enrollment ascertainments (e.g., selecting from populations of individuals with cancer diagnosis or high cholesterol) were excluded from the analyses presented in this paper. As we focused on conditions with actionable findings in adults, individuals <18 years of age at time of return were excluded; outcomes for pediatric patients with P/LP variants are reported elsewhere.19 Additionally, we excluded those who did not have any administrative codes available or were missing the timing of the return of results to the electronic health record. All individuals in the cohort that were included in the analysis had data indicating whether they were male or female. For the analyses centered around breast cancer, only individuals who were indicated as female in EHR records (57.6% of study population) were included. Due to small sample size and lack of standard screening guidelines, males were not included in breast cancer-related analyses. Variants of unknown significance were returned by only a subset of sites, and due to low sample sizes and pre-existing indication for sequencing, they were excluded from analysis.
Outcome assessment
The eMERGE consortium used chart review to assess health care utilization for those individuals with a positive P/LP result. However, this approach was impractical to assess the large population of individuals with negative findings. Thus, we aimed to create a high-throughput way to accurately capture the linkage of clinical evaluation and interventions attributable to return of genetic results. This is a current gap in large-scale research studies and influences the ability to accurately attribute clinical outcomes to genetic testing across large populations over time.5 To this end we created The High Throughput Assessment of Genomic outcomes (HI-TAG; Table S1) knowledge base to assess outcomes based on administrative data representing health care services billed to payors. HI-TAG consists of current procedural knowledge (CPT)20 codes that would be expected to be ordered after RoR for five categories of heritable conditions: arrhythmia, female breast cancer (BCa), cardiomyopathy, colorectal cancer (CRC), and familial hypercholesterolemia (FH). HI-TAG was focused on CPT codes associated with procedures that would be ordered after return of genomic results, and codes that were unrelated to this return were not included. Details on creation and validation of the HI-TAG knowledge base can be found in the supplemental methods and Figure S1.
Assessment of costs
Costs, which were assigned to the individual clinical services based on CPT, were obtained from a claims database consisting of de-identified patient-level health data for 30 million employees, retirees, and dependents of more than 250 medium and large employers and health plans in the United States (IBM Marketscan).21 Only individuals covered under private insurance plans are included in the database. The average of total gross payments to a provider for a specific service for a given CPT code was linked to HI-TAG for analysis. Codes and costs were examined one year before and after return of results. All the codes included in our analyses that contained participant data were found to have corresponding cost data in the claims database.
Study design
We utilized the HI-TAG knowledge base to assess outcomes across individuals with P/LP and negative results one year before and after return of results. Results are given for the health care service-related codes included in the HI-TAG knowledge base (Table S1) and CPT data collected through June of 2021. Any occurrence of International Classification of Disease (ICD) codes with dates were utilized to determine the length of time participants interacted with the health care system.
Time zero for return of results was defined as the date reports were uploaded to the EHR. We selected this date as it represented the earliest date at which the provider could initiate changes to clinical care based on the genetic results. All sites returned P/LP results to individuals using genetic counselors and provided the reports to providers and the EHR. Negative reports were returned to the provider, EHR and (for five sites) to individuals directly. Sites encouraged individuals to contact the study genetic counselors if there were questions, even when negative reports were returned. We used the following four groups to generate analyses: (1) pre-RoR with P/LP findings, (2) post-RoR with P/LP findings, (3) pre-RoR with negative findings, and (4) post-RoR with negative findings. The negative report group was 1:1 matched to the P/LP group and controlled for age, sex, and enrollment site. The matching by site aimed to control for site effects associated with different return methods across the study. The analysis could not be stratified by site due to insufficient sample size at many of the sites. To assess differences in health care utilization, we conducted several comparisons. (1) We examined the number of individuals interacting with the healthcare system in the P/LP and negative report groups both before and after return of results. (2) We conducted a prespecified subgroup analysis limiting the analysis dataset to those individuals within the recommended age ranges of testing based on clinical findings (Table S2) for the P/LP group before and after return of results. (3) We examined the number of unique HI-TAG codes associated with each individual in the P/LP and negative report group, both before and after return of results. (4) We conducted a difference-in-difference analysis on both the frequencies of services in the P/LP group (after and before RoR) compared to those receiving negative reports and (5) we conducted a cost analysis difference using Marketscan data. Chi-square statistics and p values are given for the analyses involving comparisons between the number of unique individuals receiving services. Means, medians, interquartile range (IQR), and p values based on Wilcoxon-Pratt signed-rank test are given for the difference-in-difference analyses.
Ethical considerations
All procedures for the de-identified data analysis were followed in accordance with the ethical standards of the institutional review board. Outcomes and cost analysis were covered under Vanderbilt University Medical Center IRB # 150209 and # 221360, respectively. Informed consent for the eMERGE cohort was obtained by each site prior to aggregating de-identified data.
Results
Study population
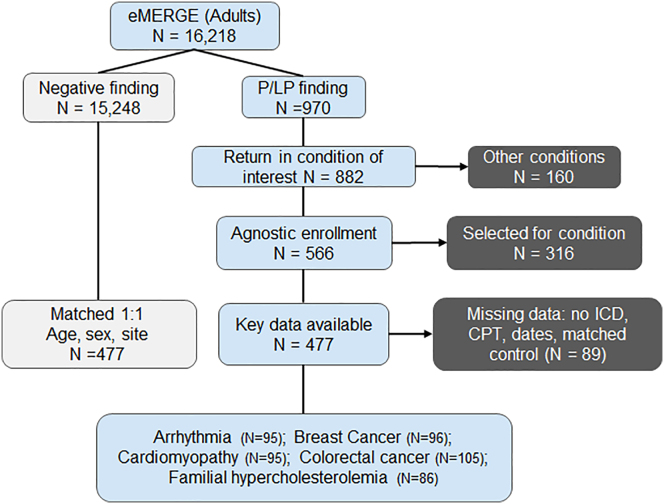
The eMERGE III cohort enrolled 25,068 participants across 10 sites, of whom 16,218 had data indicating they received a return of result in the EHR as an adult; 8,785 of these were females. Individuals who were under 18 years (n = 1,818) as well as those missing return date were excluded. Of the 16,218 adult individuals, 970 had P/LP findings and 882 had findings in at least one of the 49 actionable genes in the five conditions of interest. A list of genes associated with each condition can be found in Table S3. Individuals who were enrolled by sites with condition-specific criteria (selected for one of the conditions) and those without at least one ICD or CPT code were also removed from analysis. 477 individuals were included in the final analysis and were matched 1:1 to an individual with a negative (no variant) report (Figure 1). The final cohort included in the study was distributed across six eMERGE sites: Cincinnati Children’s Hospital Medical Center, Columbia, Geisinger, Northwestern, Massachusetts General Brigham, and Vanderbilt University Medical Center. The other four sites could not be included due to age-specific enrollments (pediatrics), cohorts specifically enrolled for a given condition, and lack of CPT data. Demographics of the included cohort are described in Table S4. The time interval in years between return of results and last administrative code (CPT or ICD; censored at one year) for the four main analysis groups were as follows (mean, standard error): P/LP pre-RoR (0.99, ±0.003); P/LP post-RoR (0.83, ±0.02); negative pre-RoR (0.99, ±0.0001); negative post-RoR (0.87, ±0.02). There was no significant difference in the time of follow-up post-RoR in the P/LP compared to the negative group (p > 0.05). Table 1 shows instances of CPT groupings and number of individuals in the P/LP group before and after return of results in our dataset.
Figure 1.
eMERGE cohort inclusion criteria
There were 16,218 adults in the eMERGE III cohort that had return of result data, of which 970 had pathogenic or likely pathogenic (P/LP) findings. After removing participants that had been enrolled for specific conditions, had P/LP finding in other conditions, did not have adequate code data (at least 1 CPT or ICD code), or were missing return age, the individuals were 1:1 matched on age, sex, and enrollment site to individuals receiving negative reports. The final analysis cohort consisted of 477 adults in the P/LP and negative report category each.
Table 1.
Frequency of healthcare services before and after return of results
|
# Instances |
# Individuals |
|||
|---|---|---|---|---|
| Pre-RoR | Post-RoR | Pre-RoR | Post-RoR | |
| Arrhythmia | ||||
| Electrocardiography | 68 | 163 | 21 | 54 |
| Cardiac MRI | 3 | 4 | 2 | 2 |
| Electrophysiology procedure | 2 | 3 | 2 | 2 |
| Stress ecg testing | 2 | 17 | 1 | 8 |
| Myocardial perfusion imaging | 1 | 6 | 1 | 6 |
| Breast cancer | ||||
| Mammography | 13 | 38 | 10 | 24 |
| Repair & reconstruction | 8 | 4 | 2 | 2 |
| Diagnostic imaging | 2 | 3 | 1 | 2 |
| Surgical pathology | 2 | 3 | 2 | 2 |
| Ultrasound | 2 | 7 | 2 | 6 |
| MRI breast | 1 | 11 | 1 | 9 |
| Breast biopsy | 0 | 6 | 0 | 6 |
| Cardiomyopathy | ||||
| Electrocardiogram | 115 | 168 | 30 | 49 |
| Transthoracic echocardiogram | 30 | 44 | 19 | 32 |
| Cardiac catheterization | 6 | 7 | 5 | 4 |
| Electrophysiology procedure | 5 | 5 | 2 | 2 |
| Therapeutic cardioversion | 5 | 3 | 2 | 2 |
| Transesophageal echocardiogram | 4 | 3 | 1 | 2 |
| CT angiography | 4 | 3 | 4 | 2 |
| Stress echocardiogram | 3 | 3 | 2 | 2 |
| Cardiac MRI | 1 | 26 | 1 | 20 |
| Implantable cardiac defibrillator | 0 | 4 | 0 | 2 |
| Stress ecg | 0 | 15 | 0 | 7 |
| Myocardial perfusion imaging | 0 | 5 | 0 | 5 |
| Colorectal cancer | ||||
| Colonoscopy | 8 | 43 | 6 | 24 |
| FH | ||||
| Lipid panel or components of lipid panel | 59 | 70 | 41 | 40 |
Data are not shown for instances of CPT groupings where the post-RoR group counts were <3. RoR, return of results; CT, computed tomography; ECG, electrocardiogram; MRI, magnetic resonance imaging; FH, familial hypercholesterolemia.
Health care service utilization
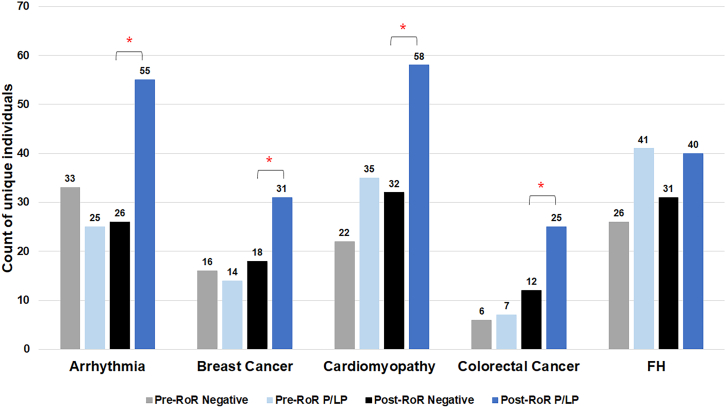
We examined the number of unique individuals across the cohort that obtained health care services. In the P/LP group, 43.8% (n = 209) of individuals had at least one prespecified qualifying health care service across the study period after return of results, while only 24.9% (n = 119) of individuals received a qualifying health care service after a negative result (p < 0.0002). Furthermore, 25.6% (n = 122) in the P/LP group had at least one service prior to return. When comparisons were stratified by condition, four of the five conditions showed significant differences (Figure 2) comparing individuals with P/LP versus those with negative reports in the post-return period: arrhythmia (p = 0.0002); BCa (p = 0.03), cardiomyopathy (p = 0.0002), CRC (p = 0.02), and FH (p > 0.05).
Figure 2.
Number of individuals receiving at least one healthcare service differs based on actionable genetic finding
More individuals received at least one qualifying health care service in the P/LP (dark blue) group after RoR compared with those with negative reports (black) in four of the five conditions examined. Number of unique individuals shown in the pre-RoR period (P/LP) are shown in light blue and (negative) gray. P/LP, pathogenic or likely pathogenic, RoR, return of results; FH, familial hypercholesterolemia; ∗p < 0.05 when post-RoR P/LP and post-RoR negative reports are compared.
A subgroup analysis examining qualifying health care services within recommended testing age ranges was also conducted. Results were similar to the analysis containing all individuals with 29.0% (n = 115) of individuals pre-RoR P/LP; 46.4% (n = 184) post-RoR P/LP; and 26.7% (n = 106) post-RoR negative group.
Frequency of individual’s health care utilization
The frequency of health care utilization per individual post- and pre-RoR for those receiving P/LP reports compared to negative reports was examined. Significantly larger differences were found in the P/LP group compared to negative group one year after return in three of the five conditions (median [25% IQR, 75% IQR]; p value), i.e., arrhythmia (1 [0, 3.5]; p < 0.0001), BCa (0 [0, 1]; p = 0.006), and cardiomyopathy (1 [−2, 3]; p = 0.02); a marginally significant difference was found in CRC (0 [0, 1]; p = 0.052); and no change was found in FH (0 [−1, 1]; p = 0.82) (Figure S2). IQRs of all the comparison groups are shown in Table S5.
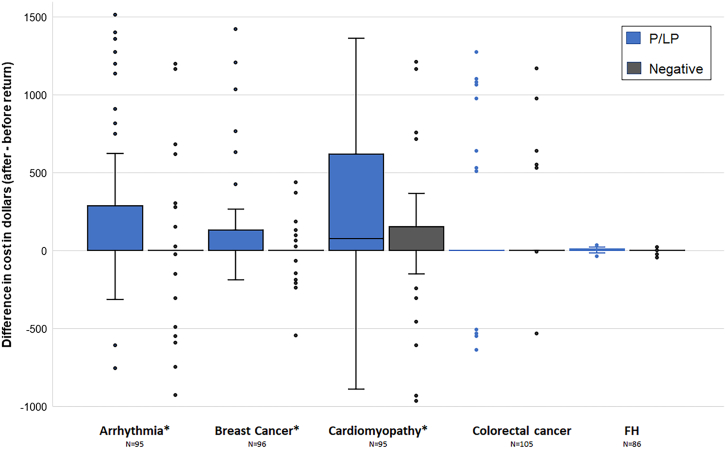
Impact of RoR on healthcare cost
The difference in cost after and before RoR was compared in the P/LP and negative report groups (Figure 3). Mean average (median [25% IQR, 75% IQR]) and p value are given. Individuals receiving P/LP reports had a post-return annual cost of $343.04 and those pre-return had a cost of $162.32 on average; the difference of $180.72 (0 [0, 151.61]); p < 0.0001 was significant. However, individuals receiving negative reports did not show a significant difference in spending post- and pre-return: $31.50 (0 [0, 0]); p > 0.05. The difference in differences in P/LP compared to the negative group was significant and showed an average cost difference of $149.22 (0 [0, 262.32]); p < 0.0001.
Figure 3.
Cost differences are observed after the return of arrhythmia, breast cancer, and cardiomyopathy genomic results
Difference in spending after return of results minus before return for individuals receiving P/LP findings (blue) and negative (black) findings. There were significantly higher costs in three conditions (arrhythmia, p < 0.0001; breast cancer, p = 0.0013; and cardiomyopathy, p = 0.001), and a marginally significant difference in colorectal cancer (p = 0.059). No significant difference was observed in FH (p > 0.05). Box represents interquartile range (IQR), with whiskers representing variability outside the quartiles, and outliers represented as dots. Data shown are limited to 5 to 95 percentiles of costs. IQR for colorectal cancer and FH are 0. FH, familial hypercholesterolemia; ∗p < 0.05.
In stratified analyses, significant differences were observed after P/LP reports were returned compared to those receiving negative reports in arrhythmia ($288.14; p < 0.0001), BCa ($65.28; p = 0.0013), and cardiomyopathy ($241.95; p = 0.001), and a marginally significant difference was observed in CRC ($137.51; p = 0.059). No significant difference was observed in FH ($1.31; p > 0.05). Table S6 shows the median and IQR of the comparisons between all analysis groups.
Discussion
Within a large, sequenced cohort, less than half of adults (43.8%) with a monogenic risk result received a qualifying health service at an eMERGE site as determined by CPT codes within a year of return, which led to a modest increase in average health care costs. Changes in health care services were largely attributable to completing recommended follow-up testing of arrhythmia risks (electrocardiograms), hereditary breast cancer risks (mammograms and breast magnetic resonance imaging), cardiomyopathy (echocardiograms and cardiac magnetic resonance imaging), and colorectal cancer (colonoscopy), whereas lipid testing rates for familial hypercholesterolemia were similar to the control populations. The mean increase in cost attributable to return of P/LP results was $149 per individual; as expected, costs were highly skewed so that a small number of individuals received higher cost care. Limiting the analysis to age groups that would be expected to receive services based on clinical guidelines slightly increased the proportion (between 1.8% and 3.4%) receiving those services. As the costs described here are from the payor (health insurance) to the provider; there may be additional out-of-pocket costs to individuals that are not captured. Additionally, as HI-TAG focused on codes associated with common procedures after the return of genomic results, these findings do not capture all potential healthcare services and costs associated with return, such as additional office visits, referrals, or services that are not routinely billed.
In addition to examining the pragmatic immediate effect of a large-scale return, the study was unique in development and use of claims data to measure follow-up testing and procedures for five conditions across multiple health systems. Chart review requires significant time and effort and cannot be conducted on public databases and cross-institutional biobanks that contain abstracted, de-identified data without full narrative text. HI-TAG or similar knowledge bases that compile standard codes and administrative data for outcomes analysis close a gap in current outcomes research. The ability to update and evolve these knowledge bases to improve sensitivity and predictive value is also needed as the field progresses. While this study demonstrates the health services impact of returning results across multiple US health systems, other studies have documented the impact of returning genomic results. The MyCode program implemented within Geisinger reported that 70% of individuals receiving P/LP CDC tier 1 results—hereditary breast and ovarian cancer (HBOC), Lynch syndrome, and familial hypercholesterolemia—received at least one recommended service after multiple years of follow up.22 The MyCode program analyzed the cost consequences of returning HBOC results to 59 of 50,726 participants and found that total health care costs were not significantly different between pre- and post-return periods after HBOC disclosure.23 Finally, more specific investigation of outcomes for the eMERGE cohort were previously published, including breast cancer,24 familial hypercholesterolemia,25 and arrhythmia26; however, these analyses used manually abstracted data and were not able to compare the P/LP cohort to a matched cohort with negative reports.
In this study, approximately half of participants receiving P/LP results did not receive a qualifying health care service related to the monogenic risk at their site. While we were not able to ascertain the reason for lack of utilization, a number of explanations are possible. First, individuals may have received testing prior to the study observation period for screening purposes or symptoms and be considered still up to date after return; an example would be an individual with identified Lynch syndrome who had already been screened with a routine colonoscopy. Secondly, individuals may have declined or deferred additional testing due to health care access challenges or increased out-of-pocket costs related to insurance coverage. The study did not systematically obtain insurance status of enrolled participants; however, the majority were likely insured as they were required to have longitudinal care at large academic or integrated healthcare institutions. Lack of follow-up testing could also be due to prevalent disease in the individual. While individuals who were enrolled for specific conditions were excluded from our analysis, it is possible that some individuals had prevalent disease. For these individuals, the return of genomics results may not have increased health care utilization and may be an explanatory factor in the approximately 50% of individuals with P/LP findings that did not receive additional care. Provider factors, such as knowledge of clinical guidelines for genomic risks, could also have reduced follow-up testing, although high-risk individuals received genetic counseling with recommendations communicated to providers. Finally, individuals may have received care elsewhere, and these services were not coded in the eMERGE site’s EHR such as breast MRIs conducted at partner screening facilities. For individuals with identified FH, the lack of difference in costs or frequency of testing between affected individuals and control subjects may be related to the routine and frequent measurement of lipid panels at baseline such that return of FH did not prompt more frequent measurement. Additionally, modest increase in lipid testing frequency is unlikely to alter average or median costs, since individual lipid tests are relatively inexpensive.
There are several limitations to our study that should be considered for future work. Approximately 14% of the eMERGE cohort was made up of individuals typically underrepresented in biomedical research. As health care access and utilization may differ in those communities due to trust, availability of care, and financial constraints, more study cohort diversity is needed to understand the full range of healthcare utilization after return of results. Secondly, we selected de-identified CPT data to capture a broad range of process outcomes that could be readily collected and reconciled across healthcare institutions. Though CPT-based measurement of process outcomes shows reasonable sensitivity and positive predictive value for most conditions and test types, validation with chart review indicated discrepancies with both false positives and false negatives. Low sensitivity was particularly evident for echocardiograms and colonoscopies which may reflect the varied locations those tests are obtained, as billing data reported outside the EHR were not captured by the study. Likewise, lower positive predictive value for some of the tests, such as lipid studies, may reflect the heterogeneous ways those tests are billed. Future iterations of this process outcome assessment method could add other structured data elements from the EHR, such as presence of a laboratory or procedure report to improve the fidelity of the test measurements. The current phase of eMERGE is also surveying study participants about receipt of healthcare services after return of results, which could help improve understanding of the gaps inherent in single-institution EHRs.27 We anticipate expanding the knowledge base (HI-TAG) to support measurement of additional genomic conditions and applying it to biobank cohorts that feature return of results. While the study matched individuals with P/LP to those with negative results by age, sex, and enrollment site, there could be other clinical factors unrelated to the return of results that explain the study findings such as increased high-risk family history and social determinants of health. As the de-identified databases utilized for this study did not contain family history data, future work could benefit from incorporating additional clinical and family history covariates.
As genomic screening of unselected populations becomes more common through large-scale research studies such as the All of Us Research Program28,29 and direct-to-consumer testing,1 understanding the impact of return of actionable results is critical for determining success of these initiatives. Further work should examine downstream costs of additional screening, the impact of genomic risks on longer-term screening behavior, and overall cost effectiveness of genomic screening, as was recently reported for tier 1 conditions.7 This study informs other national and international efforts to collect outcomes and informs the potential cost of screening immediately following return.
Data and code availability
The accession number for the genetic data reported in this study are available on AnVIL and dbGaP: phs001616.v2.p2. HI-TAG knowledge base is included as Table S1.
Acknowledgments
The authors would like to thank the eMERGE III participants and network for their contributions of data. The third phase of the eMERGE Network was initiated and funded by the NHGRI through the following grants: U01HG008657 (Group Health Cooperative/University of Washington); U01HG008685 (Brigham and Women's Hospital); U01HG008672 (Vanderbilt University Medical Center); U01HG008666 (Cincinnati Children's Hospital Medical Center); U01HG006379 (Mayo Clinic); U01HG008679 (Geisinger Clinic); U01HG008680 (Columbia University Health Sciences); U01HG008684 (Children's Hospital of Philadelphia); U01HG008673 (Northwestern University); U01HG008701 (Vanderbilt University Medical Center serving as the Coordinating Center); U01HG008676 (Partners Healthcare/Broad Institute); U01HG008664 (Baylor College of Medicine); and U54MD007593 (Meharry Medical College).
Author contributions
The following authors were involved in the concept, data, method, manuscript draft writing review, and editing: R.L.C., W.K.C., N.K.W., B.C., H.H., M.H., C.H., G.P.J., K.K., I.J.K., K.A.L., C. Liu, C.A.P., A.K.R., L.J.R.-T., M.E.S., D.L.V., C.W., J.L.W., and M.S.W. The following authors were involved in the concept, method, manuscript draft review, and editing: S.A., J.J.C., O.E., A.S.G., I.A.H., S.J., C. Lee, T.A.M., E.M.M., R.R., W.-Q.W., and G.L.W. The following authors were involved in the data, analysis, manuscript draft review, and editing: N.A.P.-A. and R.T. The following authors were involved in the concept, data, method, analysis, and original manuscript draft writing, review, and editing: J.E.L. and J.F.P.
Declaration of interests
W.K.C. is on the board of directors of Prime Medicine and D.L.V. is a consultant for Illumina and has a funded project from GeneDx.
Published: October 25, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2023.10.006.
Supplemental information
First tab displays a summary of services examined for each condition including counts of CPT code types included in a given service and how many codes were considered primarily evaluation screening. Subsequent tabs list the codes and descriptions for each condition and service.
References
- 1.Grzymski J.J., Elhanan G., Morales Rosado J.A., Smith E., Schlauch K.A., Read R., Rowan C., Slotnick N., Dabe S., Metcalf W.J., et al. Population genetic screening efficiently identifies carriers of autosomal dominant diseases. Nat. Med. 2020;26:1235–1239. doi: 10.1038/s41591-020-0982-5. [DOI] [PubMed] [Google Scholar]
- 2.Daly M.B., Pilarski R., Yurgelun M.B., Berry M.P., Buys S.S., Dickson P., Domchek S.M., Elkhanany A., Friedman S., Garber J.E., et al. NCCN Guidelines Insights: Genetic/Familial High-Risk Assessment: Breast, Ovarian, and Pancreatic, Version 1.2020: Featured Updates to the NCCN Guidelines. J. Natl. Compr. Canc. Netw. 2020;18:380–391. doi: 10.6004/jnccn.2020.0017. [DOI] [PubMed] [Google Scholar]
- 3.Grundy S.M., Stone N.J., Bailey A.L., Beam C., Birtcher K.K., Blumenthal R.S., Braun L.T., de Ferranti S., Faiella-Tommasino J., Forman D.E., et al. AHA/ACC/AACVPR/AAPA/ABC/ACPM/ADA/AGS/APhA/ASPC/NLA/PCNA Guideline on the Management of Blood Cholesterol: A Report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. Circulation. 2019;139:e1082–e1143. doi: 10.1161/CIR.0000000000000625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Landstrom A.P., Chahal A.A., Ackerman M.J., Cresci S., Milewicz D.M., Morris A.A., Sarquella-Brugada G., Semsarian C., Shah S.H., Sturm A.C., American Heart Association Data Science and Precision Medicine Committee of the Council on Genomic and Precision Medicine and Council on Clinical Cardiology; Council on Cardiovascular and Stroke Nursing; Council on Hypertension; Council on Lifelong Congenital Heart Disease and Heart Health in the Young; Council on Peripheral Vascular Disease; and Stroke Council Interpreting Incidentally Identified Variants in Genes Associated With Heritable Cardiovascular Disease: A Scientific Statement From the American Heart Association. Circ. Genom. Precis. Med. 2023;16 doi: 10.1161/HCG.0000000000000092. [DOI] [PubMed] [Google Scholar]
- 5.Ommen S.R., Mital S., Burke M.A., Day S.M., Deswal A., Elliott P., Evanovich L.L., Hung J., Joglar J.A., Kantor P., et al. AHA/ACC Guideline for the Diagnosis and Treatment of Patients With Hypertrophic Cardiomyopathy. Circulation. 2020;142:e558–e631. doi: 10.1161/CIR.0000000000000937. [DOI] [PubMed] [Google Scholar]
- 6.Weiss J.M., Gupta S., Burke C.A., Axell L., Chen L.M., Chung D.C., Clayback K.M., Dallas S., Felder S., Gbolahan O., et al. NCCN Guidelines® Insights: Genetic/Familial High-Risk Assessment: Colorectal, Version 1.2021. J. Natl. Compr. Canc. Netw. 2021;19:1122–1132. doi: 10.1164/jnccn.2021.0048. [DOI] [PubMed] [Google Scholar]
- 7.Guzauskas G.F., Garbett S., Zhou Z., Schildcrout J.S., Graves J.A., Williams M.S., Hao J., Jones L.K., Spencer S.J., Jiang S., et al. Population Genomic Screening for Three Common Hereditary Conditions. Ann. Intern. Med. 2023;176:585–595. doi: 10.7326/M22-0846. [DOI] [PubMed] [Google Scholar]
- 8.Kho A.N., Pacheco J.A., Peissig P.L., Rasmussen L., Newton K.M., Weston N., Crane P.K., Pathak J., Chute C.G., Bielinski S.J., et al. Electronic medical records for genetic research: results of the eMERGE consortium. Sci. Transl. Med. 2011;3:79re1. doi: 10.1126/scitranslmed.3001807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCarty C.A., Chisholm R.L., Chute C.G., Kullo I.J., Jarvik G.P., Larson E.B., Li R., Masys D.R., Ritchie M.D., Roden D.M., et al. The eMERGE Network: A consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med. Genomics. 2011;4:13. doi: 10.1186/1755-8794-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pathak J., Wang J., Kashyap S., Basford M., Li R., Masys D.R., Chute C.G. Mapping clinical phenotype data elements to standardized metadata repositories and controlled terminologies: the eMERGE Network experience. J. Am. Med. Inform. Assoc. 2011;18:376–386. doi: 10.1136/amiajnl-2010-000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.eMERGE Consortium Lessons learned from the eMERGE Network: balancing genomics in discovery and practice. HGG Adv. 2021;2 doi: 10.1016/j.xhgg.2020.100018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Denny J.C., Ritchie M.D., Basford M.A., Pulley J.M., Bastarache L., Brown-Gentry K., Wang D., Masys D.R., Roden D.M., Crawford D.C. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene–disease associations. Bioinformatics. 2010;26:1205–1210. doi: 10.1093/bioinformatics/btq126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ritchie M.D., Verma S.S., Hall M.A., Goodloe R.J., Berg R.L., Carrell D.S., Carlson C.S., Chen L., Crosslin D.R., Denny J.C., et al. Electronic medical records and genomics (eMERGE) network exploration in cataract: several new potential susceptibility loci. Mol. Vis. 2014;20:1281–1295. [PMC free article] [PubMed] [Google Scholar]
- 14.Dumitrescu L., Ritchie M.D., Denny J.C., El Rouby N.M., McDonough C.W., Bradford Y., Ramirez A.H., Bielinski S.J., Basford M.A., Chai H.S., et al. Genome-wide study of resistant hypertension identified from electronic health records. PLoS One. 2017;12 doi: 10.1371/journal.pone.0171745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hellwege J.N., Stallings S., Torstenson E.S., Carroll R., Borthwick K.M., Brilliant M.H., Crosslin D., Gordon A., Hripcsak G., Jarvik G.P., et al. Heritability and genome-wide association study of benign prostatic hyperplasia (BPH) in the eMERGE network. Sci. Rep. 2019;9:6077. doi: 10.1038/s41598-019-42427-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.eMERGE Consortium Harmonizing Clinical Sequencing and Interpretation for the eMERGE III Network. Am. J. Hum. Genet. 2019;105:588–605. doi: 10.1016/j.ajhg.2019.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fossey R., Kochan D., Winkler E., Pacyna J.E., Olson J., Thibodeau S., Connolly J.J., Harr M., Behr M.A., Prows C.A., et al. Ethical Considerations Related to Return of Results from Genomic Medicine Projects: The eMERGE Network (Phase III) Experience. J. Pers. Med. 2018;8 doi: 10.3390/jpm8010002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leppig K.A., Kulchak Rahm A., Appelbaum P., Aufox S., Bland H.T., Buchanan A., Christensen K.D., Chung W.K., Clayton E.W., Crosslin D., et al. The reckoning: The return of genomic results to 1444 participants across the eMERGE3 Network. Genet. Med. 2022;24:1130–1138. doi: 10.1016/j.gim.2022.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blumling A.A., Prows C.A., Harr M.H., Chung W.K., Clayton E.W., Holm I.A., Wiesner G.L., Connolly J.J., Harley J.B., Hakonarson H., et al. Outcomes of Returning Medically Actionable Genomic Results in Pediatric Research. J. Pers. Med. 2022;12:1910. doi: 10.3390/jpm12111910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.American Medical Association Current Procedural Knowledge (CPT) Am Med Assoc. 2017 [Google Scholar]
- 21.Roberson M.L., Padi-Adjirackor N.A., Hooker G., Pal T. Evaluating Costs Associated With Genetic Counseling Among Commercially Insured US Patients With Cancer From 2013 to 2019. JAMA Health Forum. 2022;3 doi: 10.1001/jamahealthforum.2022.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buchanan A.H., Lester Kirchner H., Schwartz M.L.B., Kelly M.A., Schmidlen T., Jones L.K., Hallquist M.L.G., Rocha H., Betts M., Schwiter R., et al. Clinical outcomes of a genomic screening program for actionable genetic conditions. Genet. Med. 2020;22:1874–1882. doi: 10.1038/s41436-020-0876-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hao J., Hassen D., Manickam K., Murray M.F., Hartzel D.N., Hu Y., Liu K., Rahm A.K., Williams M.S., Lazzeri A., et al. Healthcare Utilization and Costs after Receiving a Positive BRCA1/2 Result from a Genomic Screening Program. J. Pers. Med. 2020;10:7. doi: 10.3390/jpm10010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fan X., Wynn J., Shang N., Liu C., Fedotov A., Hallquist M.L.G., Buchanan A.H., Williams M.S., Smith M.E., Hoell C., et al. Penetrance of Breast Cancer Susceptibility Genes From the eMERGE III Network. JNCI Cancer Spectr. 2021;5 doi: 10.1093/jncics/pkab044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dikilitas O., Sherafati A., Saadatagah S., Satterfield B.A., Kochan D.C., Anderson K.C., Chung W.K., Hebbring S.J., Salvati Z.M., Sharp R.R., et al. Familial Hypercholesterolemia in the Electronic Medical Records and Genomics Network: Prevalence, Penetrance, Cardiovascular Risk, and Outcomes After Return of Results. Circ. Genom. Precis. Med. 2023;16 doi: 10.1161/CIRCGEN.122.003816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Glazer A.M., Davogustto G., Shaffer C.M., Vanoye C.G., Desai R.R., Farber-Eger E.H., Dikilitas O., Shang N., Pacheco J.A., Yang T., et al. Arrhythmia Variant Associations and Reclassifications in the eMERGE-III Sequencing Study. Circulation. 2022;145:877–891. doi: 10.1161/CIRCULATIONAHA.121.055562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Linder J.E., Allworth A., Bland H.T., Caraballo P.J., Chisholm R.L., Clayton E.W., Crosslin D.R., Dikilitas O., DiVietro A., Esplin E.D., et al. Returning integrated genomic risk and clinical recommendations: The eMERGE study. Genet. Med. 2023;25 doi: 10.1016/j.gim.2023.100006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramirez A.H., Sulieman L., Schlueter D.J., Halvorson A., Qian J., Ratsimbazafy F., Loperena R., Mayo K., Basford M., Deflaux N., et al. The All of Us Research Program: Data quality, utility, and diversity. Patterns. 2022;3 doi: 10.1016/j.patter.2022.100570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.All of Us Research Program Investigators. Denny J.C., Rutter J.L., Goldstein D.B., Philippakis A., Smoller J.W., Jenkins G., Dishman E. The “All of Us” Research Program. N. Engl. J. Med. 2019;381:668–676. doi: 10.1056/NEJMsr1809937. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
First tab displays a summary of services examined for each condition including counts of CPT code types included in a given service and how many codes were considered primarily evaluation screening. Subsequent tabs list the codes and descriptions for each condition and service.
Data Availability Statement
The accession number for the genetic data reported in this study are available on AnVIL and dbGaP: phs001616.v2.p2. HI-TAG knowledge base is included as Table S1.