Summary
Valosin-containing protein (VCP) is an AAA+ ATPase that plays critical roles in multiple ubiquitin-dependent cellular processes. Dominant pathogenic variants in VCP are associated with adult-onset multisystem proteinopathy (MSP), which manifests as myopathy, bone disease, dementia, and/or motor neuron disease. Through GeneMatcher, we identified 13 unrelated individuals who harbor heterozygous VCP variants (12 de novo and 1 inherited) associated with a childhood-onset disorder characterized by developmental delay, intellectual disability, hypotonia, and macrocephaly. Trio exome sequencing or a multigene panel identified nine missense variants, two in-frame deletions, one frameshift, and one splicing variant. We performed in vitro functional studies and in silico modeling to investigate the impact of these variants on protein function. In contrast to MSP variants, most missense variants had decreased ATPase activity, and one caused hyperactivation. Other variants were predicted to cause haploinsufficiency, suggesting a loss-of-function mechanism. This cohort expands the spectrum of VCP-related disease to include neurodevelopmental disease presenting in childhood.
Valosin-containing protein (VCP) is an ATPase that assists in cellular recycling pathways. Pathogenic variants in VCP are associated with adult-onset multisystem proteinopathy (MSP), affecting the brain, muscle, and bone. This article describes 13 individuals with VCP variants associated with childhood-onset neurodevelopmental disorder, possibly mediated by a loss-of-function mechanism.
Introduction
Valosin-containing protein (VCP; MIM: 601023), also known as p97, Cdc48, and Ter94 in other organisms, is a ubiquitous AAA+ protein (ATPase associated with other activities) that facilitates protein degradation through the ubiquitin-proteasome and autophagy pathways.1,2 By controlling the degradation of key signaling molecules and misfolded proteins, it plays a critical role in multiple cellular functions, including autophagy and lysosomal degradation,3,4,5 maintenance of mitochondria,6,7,8 DNA replication and repair,9,10,11 and stress response.12,13 Cryo-electron microscopy (cryo-EM) shows that it forms a homohexamer14 in which each identical monomer is composed of three major domains (N, D1, and D2). The N-terminal domain regulates co-factor/adaptor protein binding, controlling the localization of the hexamer and its functionality. D1 and D2 are ATPase domains; ATP hydrolysis leads to a conformational change in VCP’s N domain, inducing substrate binding and release, and unfolding of the substrate through a central pore to facilitate degradation or remodeling.1,15,16,17,18,19,20
Heterozygous pathogenic variants in VCP are associated with multisystem proteinopathy (MSP), or inclusion body myopathy with early-onset Paget disease and frontotemporal dementia-1 (IBMPFD1; MIM: 167320). This autosomal-dominant disorder manifests in adulthood with incomplete penetrance of several phenotypes: inclusion body myopathy (proximal and distal muscle weakness), Paget disease of bone (localized bone deformity and pain), frontotemporal dementia (executive function deficits), and motor neuron disease (spasticity and weakness).21,22,23,24 Over 250 individuals have been reported in the literature. Most pathogenic variants that cause MSP are localized at the N-D1 interface in three-dimensional space (Figure 1) and are hypothesized to alter co-factor binding or unfolding kinetics, which causes disease by altering the stability of key signaling factors or causing accumulation of proteins that form inclusion bodies.25,26
Figure 1.
Variants in VCP in this cohort versus published cases
(A) Displayed is the canonical VCP protein and the N, D1, and D2 domains.27 The theoretical protein products for probands 1 and 3 are shown above; the novel sequence is in light blue, and both products are predicted to undergo nonsense-mediated mRNA decay before protein production. In-frame deletions are noted by yellow circles, and missense mutations in our cohort are denoted by orange squares, followed by the proband ID in parentheses. Below the bar, 44 previously described missense mutation sites are noted in blue (classic MSP with features of IBMPFD1), red (any other phenotype), or purple (classic MSP and other phenotype overlap).21,28,29
(B) Cryo-EM structures with a single subunit of VCP14; the N, D1, and D2 domains are in white, blue, and purple, respectively. On the left, marked in red are several classic MSP-associated variants, clustered in the N-D1 interface. On the right are the sites of variants described in this cohort, which are spread across the protein.
A growing number of pathogenic variants in VCP, some outside of the N-D1 interface, have been associated with other neurological conditions, including isolated frontotemporal dementia (FTD; MIM: 613954),30 familial amyotrophic lateral sclerosis (ALS; MIM: 613954),31 Parkinson disease,32,33 hereditary spastic paraplegia,34 Charcot-Marie-Tooth (CMT; MIM: 616687),35 and autism.36 How these variants affect VCP function is not fully understood, but they might represent a neuron-predominant form of MSP.
Here, we describe a cohort of 13 individuals with heterozygous VCP pathogenic variants, including missense, in-frame deletion, frameshift, and splice variants, associated with a childhood-onset disorder characterized by developmental delay (DD), intellectual disability (ID), hypotonia, and macrocephaly.
Material and methods
Standard protocol approvals, registrations, and proband consents
GeneMatcher37 facilitated the identification of probands with VCP variants. Written informed consent and authorization for publication were received from probands’ guardians according to institution-specific protocols. Probands 1–3 and 6 signed a consent form approved by Washington University’s institutional review board (IRB) (“Media Authorization for the Use and Disclosure of Protected Health Information”). Proband 4 signed a consent form approved by Universitetssykehuset Nord-Norge (“Samtykke Til Publisering Av Resultater Genetisk Utredning”). Proband 5 signed a consent form approved by Centre Hospitalier Universitaire de Brest (“Consentement du Patient”). Proband 7 signed a consent form approved by Charité Universitätsmedizin Berlin (“Einwilligungserklärung zur Durchführung genetischer Analysen”). Proband 8 signed a consent form approved by Medizinische Universität Graz (“Consent Form for Publication”). Proband 9 signed a consent form approved by Seattle Children’s Hospital’s IRB (“Media Authorization for the Use and Disclosure of Protected Health Information”). Proband 10 signed a consent form approved by Cincinnati Children’s Hospital Medical Center (“Authorization and Release for Use and/or Disclosure of Protected Health Information for Scientific Presentation and/or Publication”). Proband 11 signed a consent form approved by the Western Institutional Review Board-Copernicus Group IRB (protocol no. 20120789), which included consent for the publication of photographs. Proband 12 was considered exempt from IRB requirements at the University of Hawaii. Proband 13 signed a consent form approved by the Hôpital Trousseau (protocol no. 2010-A00715-34). No interventions were performed on probands, and no biological specimens were collected from participants for this research study.
Genetic testing
Next-generation sequencing was performed at each institution on a clinical basis for the evaluation of specific clinical findings in all probands. See the supplemental note for details on the testing on each proband. In brief, VCP variants were identified on trio exome sequencing (ES) for all probands except proband 5, who had an ID gene panel run at Brest University Hospital, and proband 10, whose variant was identified via GeneDx’s Autism/ID Xpanded panel (Gaithersburg, MD); both panels were run with parental samples as a comparison and included parentage analysis. ES was analyzed by the following laboratories: GeneDx (probands 2, 3, 9, 11, and 12), Greenwood Genetic Center (proband 1), Telemark Hospital (proband 4), the Institute of Human Genetics Hamburg (proband 6), Charité Universitätsmedizin Berlin (proband 7), and the D&R Institute of Human Genetics Graz (proband 8). All VCP variants were de novo except in proband 12, whose variant was paternally inherited. All were reported by the performing laboratories as variants of uncertain significance (VUSs).
Clinical history
Demographic data, clinical and developmental histories, and the results of additional diagnostic workups were obtained from evaluations from the probands’ geneticists and/or neurologists, chart review, and/or information from the probands’ parents (available in the supplemental note). All probands were examined by a clinical geneticist, and their genetic results were discussed by a geneticist or genetic counselor.
In silico analysis
Variants were analyzed in silico with CADD and REVEL scoring38,39 and classified on the basis of current American College of Medical Genetics and Genomics (ACMG) criteria (https://clinicalgenome.org/working-groups/sequence-variant-interpretation/).40,41,42,43 mRNA and genomic sequences were acquired from NCBI Gene (https://www.ncbi.nlm.nih.gov/gene/7415), and the reference sequences were GenBank: NG_007887.1 (GRCh38.p13 Primary Assembly), GenBank: NM_007126.5 (transitional endoplasmic reticulum ATPase isoform 1), and GenBank: NP_009057.1. ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) and GnomAD v2.1.1 (GRCh37/hg19) and v3.1.2 (GRch38/hg38) (https://gnomad.broadinstitute.org/), last accessed in August 2023, were examined for variants. NCBI BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) was used for assessing conservation at the affected residues in Homo sapiens, Pan troglodytes, Mus musculus, Danio rerio, Drosophila melanogaster, and Saccharomyces cerevisiae. Sequences were manually annotated and manipulated in A plasmid Editor (ApE) v3.0.8 by M. Wayne Davis (https://jorgensen.biology.utah.edu/wayned/ape/). UnitProt (https://www.uniprot.org/) was used for visualizing protein structures; Figure S1 features PDB: 7BP9.44 The RaptorX Contact Prediction Server was used for analyzing the folding of the in-frame deletion variants (https://raptorx.uchicago.edu/).45 Figure 1A was made with Illustrator for Biological Sequences.46 Figure 1B was made with ChimeraX 1.4 using PDB: 5FTM, although variants were also examined with PDB: 5FTN (down vs. up configuration of the N domain).14
In vitro assays of ATPase activity
We subjected human VCP plasmid (TCB197) to site-directed mutagenesis with primers containing mutations to create each of the indicated variants. Proteins were purified as described from E. coli.47 Hexamer formation was confirmed by gel filtration using Superose 6 Increase 10/300 GL columns (Sigma-Aldrich). We diluted purified VCP (12.5 μL of 50 μM; the final concentration in the reaction was 25 nM) in 20 mL of assay buffer (5 mL of 5× assay buffer A [1× = 50 mM Tris (pH 7.4), 20 mM MgCl2, 1 mM EDTA], mixed with 15 mL of water, 25 μL of 0.5 M TCEP, and 25 μL of 10% Triton) to make the enzyme solution. 40 μL of enzyme solution was dispensed into each well of a 96-well plate. We carried out the ATPase assay by adding 10 μL of 1,000 mM ATP (pH 7.5; Roche) to each well and incubating the reaction at room temperature for 25 min. We stopped the reactions by adding 50 μL of BIOMOL Green reagent (Enzo Life Sciences). Absorbance at 635 nm was measured after 4 min on the Synergy Neo Microplate Reader (BioTek). All assays were performed with two to four biological replicates, and the results were consistent between independent experiments.
Statistical analysis
Data were expressed as numbers and percentages for categorical variables and as means ± standard deviation (SD) or Z scores for quantitative variables. Statistical analysis was performed with GraphPad Prism v9.5.1. For ATPase assays, statistical significance was assessed with a one-way ANOVA. Bartlett’s test showed a significant difference in standard deviation across samples (p < 0.01), so Brown-Forsythe and Welch’s ANOVA tests were also run; they showed the same results as an ordinary one-way ANOVA. Dunnett’s multiple-comparison correction was used for comparing each variant against the wild-type control. For genotype-phenotype correlations, an ordinal score was given for severity (0–3 on a scale of normal to severe), and Prism was used for calculating Spearman’s rank correlation for individual probands and Fisher’s exact test for groups of probands.
Cell culture and immunoblotting
U2OS cells were transfected with pcDNA3.1 with mouse Vcp (the protein of which is 100% homologous to the human protein at the amino acid level) containing a sequence encoding an in-frame C-terminal myc/his tag. Variants were made via site-directed mutagenesis with primers containing mutations that would create each of the indicated variants. Cell transfection was performed with Lipofectamine 2000 (ThermoFisher) according to the manufacturer’s instructions with 1 μg of plasmid. The transfection complex was removed, and the cells were replaced with new media after 24 h. After 48 h, U2OS cells were suspended in radioimmunoprecipitation assay lysis buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1% NP-40 [Sigma, I3021], 0.25% Na deoxycholate [Sigma-Aldrich, 30970], and 1 mM EDTA) supplemented with a protease inhibitor cocktail (Sigma-Aldrich, S8820). Lysates were centrifuged at 14,000 × g for 10 min. Aliquots of the supernatant were solubilized in Laemmli sample buffer, and equal amounts were separated on 10% SDS-PAGE gels, transferred to nitrocellulose, and blocked with 5% nonfat dry milk in TBST (Tris-buffered saline and 0.1% Tween 20 [Sigma-Aldrich, 30970]). Membrane was incubated with primary antibody at 1:500 dilution overnight and then with secondary antibody conjugated with horseradish peroxidase at a 1:5,000 dilution. The Amersham ECL Western Blotting Detection Reagents Kit (GE Healthcare, RPN3244) was used for protein detection, and immunoblots were visualized with G:Box Chemi XT4 Genesys v1.1.2.0 (Syngene, Cambridge, UK). Antibodies used included anti-GAPDH (Cell Signaling Technology, 2118), anti-VCP (Fitzgerald, 10R-P104A), anti-MYC (Cell Signaling Technology, 2276).
Results
We describe 13 probands with heterozygous VCP variants associated with childhood-onset neurodevelopmental disease. The probands ranged from 2 to 22 years old (mean = 11 ± 6), came from multiple ethnic backgrounds, and shared phenotypic features that have not been previously associated with VCP-related disease in OMIM, including DD, ID, hypotonia, neurobehavioral abnormalities, dysmorphic features, and macrocephaly. Although their variants were classified as VUSs by the diagnostic laboratories, our in vitro and in silico analyses support that these variants are damaging and that VCP variants might have a different disease mechanism than adult-onset VCP-related syndromes.
VCP variants
Thirteen VCP variants were identified on a trio ES or gene panel: nine missense variants, two in-frame deletions, one frameshift leading to early termination, and one splice variant (Table 1). All variants arose de novo except for that in proband 12, who inherited the variant from his father. These variants contribute significant diversity to previously reported VCP genotypes. Although most VCP variants linked to MSP are missense variants located at 37 residues at the N-D1 interface in the protein’s tertiary structure,24,28,29 the variants in our cohort spanned the length of the protein, including residues within the D2 domain (Figure 1). One variant, c.801_803del (GenBank: NM_007126.5) (p.Phe267del), had been described in two relatives with adult-onset FTD,48 but no other variants had been previously reported as pathogenic or benign in ClinVar or gnomAD.
Table 1.
13 VCP variants associated with childhood-onset disease
| Proband |
Varianta |
ACMG criteriab |
|||||
|---|---|---|---|---|---|---|---|
| mRNA | Protein | Inheritance modec | Population frequencyd | In silico analysise | Functional dataf | Classificationg | |
| 1 | c.265del | p.Arg89Glyfs∗8 | de novo (PS2-P) | PM2-P | PVS1-M | N/A | VUS |
| 2 | c.685C>T | p.Leu229Phe | de novo (PS2-P) | PM2-P | PP3 | PP2, PS3-P | VUS |
| 3 | c.709-2A>G | p.? | de novo (PS2-P) | PM2-P | PVS1-M | N/A | VUS |
| 4 | c.753G>T | p.Lys251Asn | de novo (PS2-P) | PM2-P | PP3-M | PP2, PS3-P | LP |
| 5 | c.766C>G | p.Arg256Gly | de novo (PS2-P) | PM2-P | PP3-M | PP2, PS3-P | LP |
| 6 | c.801_803del | p.Phe267del | de novo (PS2-P) | PM2-P | PM4-P | PS3-P | VUS |
| 7 | c.812G>A | p.Gly271Asp | de novo (PS2-P) | PM2-P | PP3-S | PP2, PS3-P | LP |
| 8 | c.901_903del | p.Ile301del | de novo (PS2-P) | PM2-P | PM4-P | PS3-P | VUS |
| 9 | c.1084C>T | p.Arg362Cys | de novo (PS2-P) | PM2-P | PP3-S | PP2, PS3-P | LP |
| 10 | c.1622C>A | p.Ser541Tyr | de novo (PS2-P) | PM2-P | PP3-M | PP2, PS3-P | LP |
| 11 | c.1874G>C | p.Arg625Pro | de novo (PS2-P) | PM2-P | PP3-S | PP2, PS3-P | LP |
| 12 | c.2257C>T | p.Arg753Trp | paternal (BS4-P) | PM2-P | PP3-M | PP2, PS3-P | VUS |
| 13 | c.892C>T | p.Pro298Ser | de novo (PS2-P) | PM2-P | PP3 | PP2, PS3-P | VUS |
The mRNA and protein coordinates are mapped to isoform 1 of VCP (GenBank: NM_007126.5, NP_009057.1).
The ACMG criteria are based on the latest guidelines,40,41,42,43 although these criteria are not meant to be applied without a validated gene-disease association. Modifications of the strength of evidence are given as supporting (-P), moderate (-M), and strong (-S).
De novo inheritance with paternity or maternity confirmed by trio sequencing is scored as supporting (PS2-P) given that the phenotype is not highly specific and has significant genetic heterogeneity. Paternal inheritance in proband 12, i.e., nonsegregation with disease, is adjusted to benign supporting (BS4-P) given the uncertain penetrance of our phenotype.
Population frequency is scored as pathogenic supporting (PM2-P) because all variants were absent from gnomAD and other databases.
Frameshift and splice variants predicted to undergo nonsense-mediated decay were scored as pathogenic moderate (PVS1-M) given that haploinsufficiency is not a known mechanism of disease for VCP. For missense variants, in silico algorithms predicted a deleterious effect and were weighted (PP3-P, PP3-M, or PP3-S) according to the REVEL score. Pathogenic supporting (PM4-P) was applied for a protein-length-changing variant of −1 amino acid.
For missense variants, pathogenic supporting (PP2) was applied because missense variants in VCP are rare and a known mechanism of disease. ATPase activity is not considered a definitive assay to confirm a pathogenic mechanism in VCP-related disease, so we considered our in vitro data to be a supporting level of evidence (PS3-P). N/A, not available.
The final ACMG classification proposed for each variant is based on the current point-system criteria (+1 for supporting, +2 for moderate, +4 for strong evidence) and given as likely pathogenic (LP; score 6–9) or variant of uncertain significance (VUS; score 0–5).
Most probands underwent extensive genetic and metabolic testing prior to the discovery of these variants (Table S2), but no proband met the diagnostic criteria for an alternative genetic condition. More importantly, our probands did not have clinical or laboratory features suggestive of MSP. The similar phenotypes and recurring clinical features, as well as the lack of other genetic abnormalities among the probands in this cohort, strongly support a pathogenic role for heterozygous VCP variants in childhood-onset neurodevelopmental disease.
In silico characterization of VCP variants
All variants were classified by their reporting laboratories as VUSs. Although the ACMG criteria are not technically valid for describing novel diseases, the application of these criteria,40,41,42,43 inheritance mode, and in vitro ATPase assay results upgraded six variants to “likely pathogenic” (Tables 1 and S1).
Two variants were predicted to lead to nonsense-mediated mRNA decay: a frameshift causing early termination (c.265del [p.Arg89Glyfs∗8]) in exon 3 in proband 1 and a splice variant (c.709-2A>G [p.?]) leading to the inclusion of intron 6 and early termination in proband 3 (Figure 1A). No transcript isoforms containing this intron were found by RNA sequencing in the HAVANA/Ensembl database (https://useast.ensembl.org/Homo_sapiens/Location/View?g=ENSG00000165280;r=9:35053928-35072668). Interestingly, proband 7’s c.812G>A (p.Gly271Asp) variant is at the 3′ splice acceptor site of intron 7-exon 8, but splicing predictions were mixed (Table S1).49,50 The presence of variants that are predicted to cause nonsense-mediated decay highly suggests that haploinsufficiency of VCP is a pathomechanism for this disease.
It is unclear what effect the in-frame deletions in probands 6 and 8 would have on protein function. Proband 6’s p.Phe267del variant was recently reported in two relatives with FTD or aphasia diagnosed around 60 years of age, which suggests pathogenicity for adult-onset disease.48 Proband 8’s c.901_903del (p.Ile301del) variant had not been previously reported as pathogenic or benign in gnomAD or ClinVar. According to the cryo-EM structure, p.Phe267del and p.Ile301del remove residues from the center of a β strand in the same pleated sheet in D1 (Figure 1B), relatively near the ATP binding site. Contact prediction using the deep-learning software RaptorX45 predicts that the β sheet secondary structure would be retained in both cases without striking changes in the orientation of nearby α helices (Figure S1). However, minor conformational changes could still interfere with ATP binding, hexamerization, protein dynamics, or other functions.
In silico algorithms predicted deleterious effects for all nine missense variants in this cohort (Tables 1 and S1). Variants at these sites had not been reported in association with human disease in ClinVar. The residues involved are highly conserved between species, from human to S. cerevisiae (Figure S2). Proband 4’s c.753G>T (p.Lys251Asn) variant affects the “Walker A motif,” required for ATP interaction, and should therefore interfere with ATPase activity.51,52 VCP has dual arginine fingers formed by Arg359 and Arg362 (replaced by cysteine in proband 9), which are necessary for stabilizing the leaving group.20,53 Other notable variants include c.685C>T (p.Leu229Phe), in an α helix near the N-D1 interface, and two arginine variants (c.1874G>C [p.Arg625Pro] and c.2257C>T [p.Arg753Trp]) that sit at the exposed bottom surface of the D2 ring (Figure 1B). Interestingly, four out of the nine missense variants affect arginine, a bulky and highly charged residue often involved in ionic interactions.
In vitro ATPase activity of variants
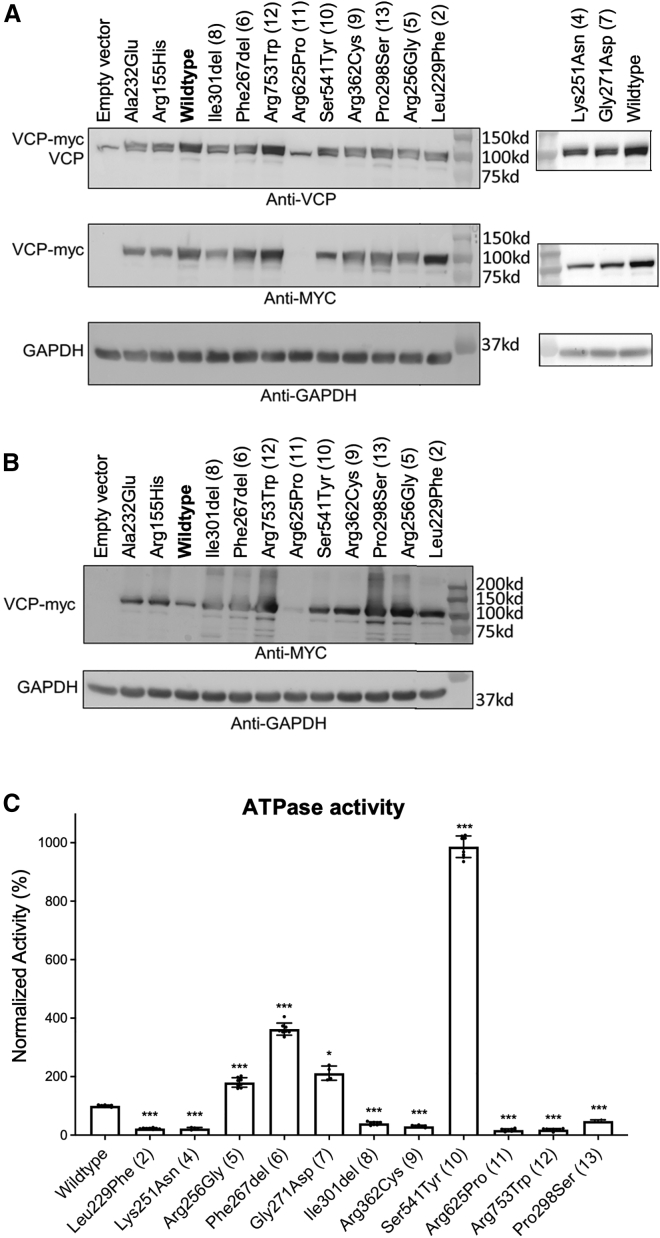
The frameshift and splice VCP variants, which were predicted to lead to haploinsufficiency, suggest that the other variants could similarly behave as loss-of-function alleles given our cohort’s shared phenotype. We first examined the protein stability of each variant in U2OS cells. Most variants were produced at levels similar to that of wild-type VCP, except for p.Arg625Pro (proband 11), which showed significantly decreased protein abundance on western blotting (Figure 2A). There appeared to be an accumulation of higher-molecular-weight species and smaller degradation products in this cohort’s variants, but not in the wild-type protein or MSP-associated variants c.695C>A (p.Ala232Glu) and c.464G>A (p.Arg155His) (Figure 2B). These data suggest that the missense variants reported here could destabilize VCP.
Figure 2.
ATPase activity of VCP variants
(A) Immunoblots of lysates from U2OS cells expressing an empty vector or expression plasmids with VCP variants and a C-terminal Myc tag with an anti-VCP antibody (top), anti-Myc antibody (middle), or anti-GAPDH as the loading control (bottom). Variants include classic MSP-associated variants p.Ala232Glu and p.Arg155His and variants from all probands except 1 and 3, whose variants are predicted to undergo nonsense-mediated decay. On the right, data were run separately for p.Lys251Asn (proband 4) and p.Gly271Asp (proband 7). Note that a small doublet is present with anti-VCP antibody, demonstrating the endogenous VCP (lower band) and the recombinant myc-tagged VCP (upper band). The p.Arg625Pro variant fails to be produced.
(B) Longer exposure, using the anti-myc antibody, of a similar set of lysates shows a faint band for the p.Arg625Pro variant, as well as a high-molecular-weight smear and multiple degradation products for the variants.
(C) Recombinant purified VCP was obtained from bacteria, and in vitro ATPase activity was assessed by a standard colorimetric assay and normalized to that of wild-type VCP. A comparison of these variants against classic MSP mutations can be found in Figure S3. n = 2–4 biological replicates with two technical replicates each. Error bars display the SD. p values from one-way ANOVA with correction for multiple comparisons with the wild type are denoted by ∗p < 0.05 and ∗∗∗p < 0.001.
We reasoned that these variants could cause loss of function by disrupting VCP’s intrinsic ATPase activity. We measured in vitro ATPase activity by using recombinant VCP purified from bacteria. These formed stable homo-hexamers, including the p.Arg625Pro variant (Figure S3B). As shown in previous publications, MSP-associated VCP variants increase ATPase activity, supporting a gain of function (Figure S3C).54 Three of our variants fit this pattern: c.801_803del (p.Phe267del), which was previously reported in adult-onset FTD48 and had ATPase activity similar to that of classic MSP variants, and c.766C>G (p.Arg256Gly) and c.812G>A (p.Gly271Asp), which had intermediately elevated ATPase activity. In contrast, the other variants in this cohort exhibited significantly decreased ATPase activity: c.685C>T (p.Leu229Phe), c.753G>T (p.Lys251Asn), c.892C>T (p.Pro298Ser), c.901_903del (p.Ile301del), c.1084C>T (p.Arg362Cys), c.1874G>C (p.Arg625Pro), and c.2257C>T (p.Arg753Trp) had <50% of wild-type activity (Figure 2C). One hyperactivating variant, c.1622C>A (p.Ser541Tyr), had >1,000% of wild-type ATP hydrolysis and around twice that of MSP variants. This degree of ATPase activity could significantly affect VCP’s ability to coordinate ATP hydrolysis with co-factor association and protein unfolding. All missense variants disrupted normal VCP ATPase function, suggesting that they are potentially pathogenic.
Clinical phenotypes
In our cohort of 13 individuals (seven males and six females), ranging from 2 to 22 years of age at the time of last evaluation, the most striking phenotypic features were DD, ID, hypotonia, and macrocephaly. A significant number of probands also had musculoskeletal abnormalities, abnormal but nonspecific brain MRI findings, psychiatric or behavioral disorders, ophthalmological abnormalities, and dysmorphic features (Figure 3). These data are summarized in Tables 2 and 3 and provided in more detail in Tables S2–S5. A detailed clinical history and physical examination of each individual are provided in the supplemental note.
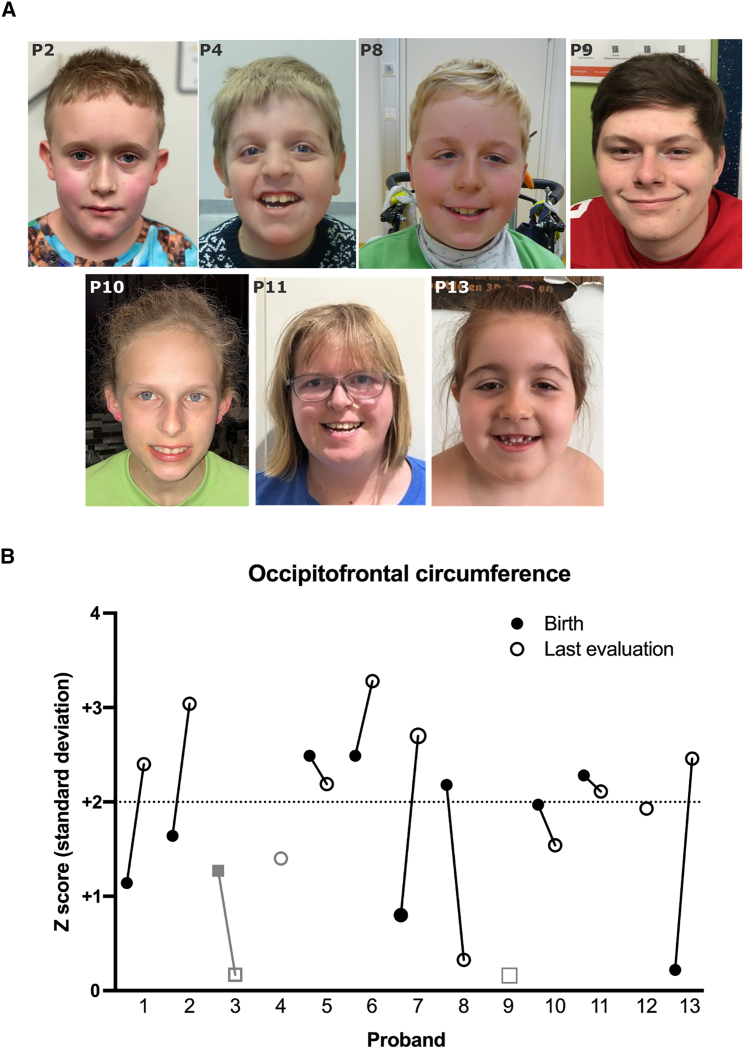
Figure 3.
Facial features and head circumference
(A) Photos of probands (from left to right): 2, 4, 8, 9, 10, 11, and 13. There was no recognizable facial gestalt for this syndrome, although some probands share features such as a broad forehead and down-slanting palpebral fissures (in probands 4, 8, 11, and 13).
(B) Macrocephaly is defined as a head circumference > 2 SD above the mean. The number of SD (Z score) was plotted for the head circumference of each proband at birth (filled circle or square) and at last evaluation (open circle or square). Proband 4 (gray circle) had relative macrocephaly, as defined by a standardized head circumference > 2 SD above standardized height.55 Probands 3 and 9 (gray squares) were never macrocephalic. Proband 10 was evaluated at the 98th percentile (Z + 1.97), and proband 12 was evaluated at Z + 1.93, which we consider within the margin of measuring error for macrocephaly.
Table 2.
Proband phenotypes and molecular data
|
Proband |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | |
| Sexa | M | M | F | M | M | F | F | M | M | F | F | M | F |
| Age (years) | 2 | 9 | 8 | 13 | 11 | 4 | 3 | 11 | 18 | 13 | 22 | 18 | 13 |
| Ethnicityb | NE | NE | NE | Arab | NE | NE + A | NE | NE | NE | NE | NE | NE + H | NE |
| Molecular data | |||||||||||||
| Variant | p.Arg89 Glyfs∗8 |
p.Leu229Phe | p.? | p.Lys251 Asn |
p.Arg256Gly | p.Phe 267del |
p.Gly271 Asp |
p.Ile301 del |
p.Arg362 Cys |
p.Ser541 Tyr |
p.Arg625 Pro |
p.Arg753 Trp |
p.Pro298 Ser |
| Variant typec | FS | D1 | splice | D1 | D1 | D1Δ | D1 | D1Δ | D1 | D2 | D2 | D2 | D1 |
| ATPase functiond | N/A | − | N/A | − | + | + | + | − | − | ++ | − | − | − |
| DD and IDe | |||||||||||||
| Gross motor delay | mild | mod | mild | sev | mod | mod | sev | sev | mild | mild | mild | absent-mild | mild |
| Fine motor delay | mild | mod | + | + | + | − | + | + | + | + | + | + | + |
| Speech delay | − | mild-mod | sev | sev | mild | mod | sev | sev | mod | mod | mild | − | mod |
| ID | − | − | mod | sev | − | mild-mod | sev | sev | mild | mild | mild-mod | − (↓IQ) | mod |
| Neurological and psychiatric disorders | |||||||||||||
| ADHD | − | + | − | − | + | − | − | − | + | + | − | + | − |
| Autism | − | + | −f | − | − | − | − | + | − | − | − | + | − |
| Anxiety | − | + | − | − | + | − | − | − | − | + | − | + | − |
| Hypotonia | + | + | + | + | + | + | + | + | + | + | + | − | + |
| Epilepsy | − | − | − | −g | + | − | − | − | − | + | − | + | − |
| Tremor | + | − | − | − | − | − | − | − | − | − | + | + | + |
| Macrocephalyh | + | + | − | rel | + | + | + | + | − | + | + | + | + |
| Abnormal MRI | − | + | + | + | + | + | + | + | + | + | + | − | + |
| Congenital defects and other featuresi | |||||||||||||
| Dysmorphic | + | + | − | + | − | + | + | + | − | − | + | − | + |
| MSK | + | + | + | + | − | − | − | + | + | + | + | + | − |
| Ophthalmic | − | − | − | + | + | + | + | + | + | − | + | + | − |
| GI | + | + | + | − | + | − | − | + | − | + | + | + | − |
| GU | + | + | + | − | − | − | − | + | − | − | − | − | + |
| Cardiac | − | − | − | − | − | − | + | − | − | − | − | + | + |
Aggregated phenotypic data for the 13 probands in our cohort. Unless otherwise noted, a plus sign (+) indicates the presence of the symptom or finding, and a minus sign (−) indicates the absence of the symptom or finding.
Sex assigned at birth: M, male; F, female.
NE, northern European; A, Asian; H, Hispanic; +, proband shares both ancestries.
FS, frameshift; D1Δ, in-frame deletion in D1; D1 or D2, missense in this domain.
ATPase function was higher (+), much higher (++), or lower (−) than wild-type function (see Figure 2C). N/A, not available.
DD is qualified as mild (≤2× months to milestone), moderate (mod; >2× normal), or severe (sev; milestone never reached), and ID is based on an IQ score < 70 and/or clinical impression: mild IQ = 55–70, moderate IQ = 35–55, and severe IQ < 35 according to DSM-IV criteria. Absent-mild, absent to mild; mild-mod, mild to moderate.
Some features of autism.
EEG with epileptic activity but no clinical seizures.
Macrocephaly was absolute (+) or relative (rel).
Table 3.
Summary of proband phenotypes
| Characteristic or phenotype | Proband statistics |
|---|---|
| Demographics | |
| Age at last clinical update | average: 11 ± 6 years (range: 2–22 years) |
| Sex | 7 males, 6 females |
| Ethnicity and ancestry | 10 northern European, 1 NE + Thai, 1 NE + Hispanic, 1 Arab |
| DD and ID | |
| Gross motor delay | 13/13 affected |
| Fine motor delay | 12/13 affected |
| Language delay | 11/13 affected |
| ID | 9/13 affected + 1 drop in IQ |
| Behavioral disorders | |
| ADHD | 5/13 affected |
| Anxiety | 4/13 affected |
| Sleep abnormalities | 4/13 affected |
| Autism spectrum | 3/13 affected |
| Neurological findings | |
| Abnormal brain MRI | 11/13 affected |
| Macrocephaly | 10/13 absolute, 1/13 relative |
| Hypotonia | 12/13 affected |
| Tremor | 4/13 affected |
| Epilepsy | 3/13 affected |
| Other features | |
| Dysmorphic facies | 11/13 affecteda |
| Musculoskeletal | 9/13 affected |
| Ophthalmologic | 8/13 affected |
| Gastrointestinal | 8/13 affected |
| Other congenital defects | 5/13 GU, 3/13 cardiac |
Phenotypes shared across probands with heterozygous VCP variants causing childhood-onset disease. Musculoskeletal abnormalities include kyphosis, scoliosis, hammer toes, pes cavus (including acquired), long-bone deformities, torticollis, and hip dysplasia or misalignments; five probands required surgery, splinting, or orthotics. Ophthalmologic abnormalities include corneal clouding, astigmatism, strabismus, and hyperopia or myopia. Gastrointestinal features include pyloric stenosis, gastroesophageal reflux, constipation, and poor feeding. Genitourinary (GU) abnormalities include inguinal hernia, cryptorchidism, hydrocele, hypospadias, and a duplicated collecting system. Cardiac abnormalities include ventricular septal defects, atrial septal defects, patent foramen ovalia, and patent ductus arteriosus.
Including three probands with minor features.
DD and ID
The most striking shared phenotypes in this cohort were DD and ID, which had not been previously described in individuals with VCP variants.21,28,29 The severity of DD and ID among the probands is summarized in Table 2, and specifics for each proband can be found in Table S3. None of the probands had significant developmental regression.
All individuals in our cohort had either gross or fine motor delays, and most experienced both. Most gross motor delays were mild to moderate such that the majority learned to walk between 18 months and 3 years old. Two probands had earlier delays but walked on time. Gross motor delays were associated with hypotonia, which was present in all but proband 12. For example, probands 4 and 8 required orthoses to walk, and proband 4 used a wheelchair. Hypotonia might have also affected fine motor development in our probands. For instance, proband 2 was noted to develop his pincer grasp around 3 years of age and used a keyboard because generalized hypotonia made handwriting difficult for him. Probands 11–13 had tremor involving their hands, which limited their fine motor skills. Additional findings that affected motor development included poor coordination or balance in probands 5, 9, and 10 and hemiparesis with hemiatrophy in proband 11.
Most individuals, except for probands 1 and 12, exhibited language delay, which was variable in severity. Five individuals were mostly nonverbal or spoke few words. Two had delayed speech onset (the first word around 2 years of age) but produced a reasonable vocabulary as they grew older. Two others had speech onset within the normal range, but the formation of sentences was delayed to 4 or 5 years old. Proband 13 had delayed onset of both her first word and sentence formation, and her vocabulary remains limited. Other associated speech findings included dysarthria, apraxia, and expressive aphasia in one proband each.
9 of 13 individuals in this cohort exhibited ID, ranging from mild to severe. Few probands underwent formal IQ testing; three had IQs around 50, and two others were diagnosed with severe ID. Proband 12’s IQ was above the range for ID (<70) but had regressed from 105 at 8 years of age to 76 only 6 years later. This could have been related to the development of seizures, although cognitive decline due to the natural history of the condition cannot be excluded.
Behavioral and psychiatric phenotypes
6 of 13 individuals in this cohort were diagnosed with behavioral or psychiatric conditions, most with multiple diagnoses (Tables 2, 3, and S4). This includes five diagnoses of attention-deficit/hyperactivity disorder (ADHD), three formal diagnoses of autism, and four diagnoses of anxiety. Note that an individual with a frameshift variant in VCP was previously described to have autism; no further information was given, but he might share features with this cohort.36 Several individuals had abnormal behaviors without a formal diagnosis, including head-banging behavior, hand flapping, stereotypic hand movements, and aggressive outbursts (Table S4). Four individuals had sleep difficulties.
Neurological and neuroimaging findings
12 out of 13 probands evaluated were noted to have hypotonia, making this a central phenotype in the cohort. The reporting radiologist noted that 11 had some abnormalities on brain MRI (Table S4). However, the findings were generally mild and nonspecific, and no major brain malformations or structural anomalies were noted. Shared features included mild cerebral atrophy or generalized volume loss with prominence of cerebrospinal fluid spaces (4/13), hydrocephalus with more significant cerebral atrophy (2/13), decreased white matter (3/13), and a thin corpus callosum (3/13). With focal dysplasia in the left parietal cortex, proband 13 had the most abnormalities. Note that for most individuals, cerebral atrophy was generalized; however, proband 11 had some mild frontal lobe atrophy that developed between 10 and 21 years of age, reminiscent of adult-onset FTD. Probands 1 and 12, two of our least developmentally affected probands, had normal MRIs.
Proband 12 had multiple seizures and was on antiepileptic medications for years. Two other individuals (probands 5 and 10) also had seizures, and proband 4 had an epileptiform electroencephalogram (EEG) but no clinical evidence of seizures. These three individuals (probands 4, 5, and 10) were noted to have brain MRI abnormalities, but other probands with similar MRI changes did not have known seizures.
Ten individuals in our cohort had absolute macrocephaly, as defined by an occipitofrontal circumference (OFC) in the ≥98th percentile at any point in their development, and one (proband 4) had relative macrocephaly, as defined by a standardized head circumference > 2 SD above standardized height (Table 2).55 The mechanisms of development of macrocephaly in our cohort appear to be varied. Six probands were macrocephalic at birth, whereas four others had head circumferences in the normal range at birth with progressive macrocephaly as they aged (Figure 3B). In probands 5 and 7, hydrocephalus might have contributed to pre- or postnatal enlargement of the OFC. Proband 12’s mother also had a head circumference in the 98th percentile, which could suggest familial factors for his macrocephaly.
Evaluation for features of MSP
Few probands had testing that could rule out MSP given that this is not a childhood-onset condition. Generally, there were no concerns for myopathy or Paget disease of bone. All eight probands who had creatinine kinase (CK) measured were in the normal range. Nine probands had alkaline phosphatase (ALP) measured, which was generally normal, although three had mild elevations during acute illness (Table S2). The only proband to have a muscle biopsy was proband 3, in whom no cytoplasmic inclusions were found. Proband 2 had a muscle ultrasound that was normal. As noted above, only proband 11 showed MRI findings that could have been consistent with FTD, but clinical suspicion for the condition was low.
MSP is also associated with motor neuron disease, which can manifest with limb weakness, fasciculations, spasticity, and/or hyperreflexia.23 Although proband 12 was hyperreflexic, most of our probands had low muscle tone. Testing for neuropathy or myopathy was performed in four probands on the basis of their symptoms, and three had abnormal findings on electromyography (EMG) or nerve conduction velocity (NCV) testing. Proband 4 had a predominantly demyelinating sensorimotor polyneuropathy in the upper and lower extremities on NCV studies; his EMG was normal. Proband 9 initially had normal EMG and NCV at 14 years of age; however repeat EMG at 18 years showed a mild sensorimotor polyneuropathy, and review of prior EMG showed similar findings at both ages. At age 10, proband 11 had an EMG that showed a mixed myopathic-neuronopathic picture, but repeat study at age 16 was normal (Table S2). Overall, these results were not consistent with the motor neuron disease seen in adults with MSP but could suggest CMT.23 Only one additional proband had EMG testing (proband 2), which was normal at 8 years old.
Facial features
8 of 13 individuals in our cohort had dysmorphic facial features, although there was no obvious characteristic gestalt for this cohort (Figure 3A). Three others had minor features but were not judged to be overtly dysmorphic. The most common facial features included frontal bossing or a prominent or broad forehead (9/13); a thin upper lip (7/13); down-slanting palpebral fissures (5/13); low-set, posteriorly rotated, or dysplastic ears (4/13); nasal anomalies (4/13); deep-set eyes (3/13); a high or arched palate (3/13); and a smooth philtrum (3/13) (Table S5).
Congenital anomalies
There were no common birth defects in our cohort, but most probands had at least one congenital anomaly (Table S5). These included urogenital abnormalities, such as cryptorchidism, hypospadias, and a duplicated collecting system (5/13); cardiac malformations, such as ventricular septal defects, patent ductus arteriosus, and patent foramen ovalia (3/13); and musculoskeletal findings, such as hip dysplasia and underdeveloped epiphyses (3/13). No cardiac findings required repair. Other findings included congenital torticollis, an inguinal hernia, and an umbilical hernia (each in one proband). Minor defects included a palmar transverse crease, a high foot arch, and hammer toes.
Miscellaneous findings
8 out of the 13 individuals in our cohort had ophthalmological abnormalities in infancy or childhood. Two developed strabismus, and one was noted to have punctate corneal clouding. Five probands had refractive errors, including myopia, hyperopia, and/or astigmatism. Eight probands had gastrointestinal abnormalities, including five individuals with constipation and four with gastroesophageal reflux. Six probands had acquired musculoskeletal abnormalities, such as scoliosis (Table S5).
Discussion
We describe 13 individuals with heterozygous VCP variants (12 de novo and 1 inherited) associated with a childhood-onset syndrome distinct from adult-onset presentations and characterized by DD, ID, hypotonia, and macrocephaly, as well as some behavioral and psychiatric disease and dysmorphic features. In silico analysis and in vitro assays of ATPase activity supported a pathogenic classification of these variants, and six would reach likely pathogenic status if a gene-disease association were established. Additional evidence supporting VCP pathogenic variants as the cause of this phenotype includes shared neurological and developmental features, an extensive workup (including genetic studies and brain imaging) excluding other etiologies, and no alternative compelling diagnoses. The alterations in ATPase activity and the identification of variants predicted to lead to nonsense-mediated decay suggest that this childhood-onset VCP-related disease has a different pathomechanism than MSP, which could explain the earlier presentation of disease.
There is no clear genotype-phenotype correlation within our cohort, although statistical comparisons were underpowered given the small number of probands, each with a unique variant. There was no clear relationship between the severity of presentation, imaging findings, dysmorphic features, and the molecular consequence of the variant. For example, it does not appear that variants in the same region of the protein have similar effects. Probands 6 and 8 have in-frame deletions affecting residues in the same β-pleated sheet but have very different ATPase activity levels and exhibit differences in their MRI findings, facial features, and ID severity. The p.Leu229Phe variant in proband 2 is located close to the classic MSP variant p.Ala232Glu, indicating that nearby variants could have disparate effects on VCP function and subsequent phenotypes. There was also no correlation between ATPase activity and in silico predictions of deleteriousness or severity of DD (Figure S4A), indicating that these tools cannot be used for predicting alterations in ATPase activity or phenotype severity. Even though their variants have similar increases in ATPase activity, probands 5–7 are discordant in DD severity, head size, brain imaging, seizure history, and dysmorphic features. Proband 10, who has a hyperactivating variant, had a relatively mild ID or DD phenotype, although he did have epilepsy and brain MRI changes. When grouped together, the four probands with increased ATPase activity had milder motor delays and possibly more severe language and ID than the seven probands with variants that decreased ATPase activity (Figure S4B), but these observations were not significant by Spearman’s correlation (p = 0.5–0.99) or by Fisher’s exact test when severity was grouped as normal or mild vs. moderate or severe (p = 0.5–0.99). These data imply that there is not a straightforward correlation between ATPase function and pathological consequence. Additionally, probands 1 and 3, whose variants are predicted to lead to nonsense-mediated decay, do not have a more or less severe phenotype than the probands with missense variants. However, there are clear differences in both the genotype and phenotype between our cohort and variants that cause MSP and other adult-onset VCP-related conditions, as we discuss later.
In silico predictions and in vitro data suggest a VCP loss-of-function mechanism via haploinsufficiency or decreased protein activity for some of these variants. Haploinsufficiency is suggested by probands 1 and 3, whose variants are predicted to undergo nonsense-mediated mRNA decay, and proband 11’s p.Arg625Pro variant, which was poorly produced in cells. VCP is predicted to be intolerant to loss-of-function variants (pLI ∼ 1, LOUEF = 0.03).56 However, in vivo evidence supporting haploinsufficiency as the disease mechanism of VCP is limited given that knockout mice heterozygous for Vcp were initially described as indistinguishable from their wild-type littermates.57 Studies focusing on neuron-specific knockout of Vcp show more promising data: knockdown of Vcp by ∼60% decreased dendritic spine formation in cultured rodent neurons,58,59 and neuron-specific knockout mice had reduced brain volume, hyperactivity, and poor spatial learning, which could correlate to our probands’ MRI and behavioral phenotypes.60 Assessing cognition and behaviors in heterozygous mice will require further studies given that the developmental phenotypes seen in our cohort were not fully evaluated in earlier mouse studies. In humans, 9p13 microdeletion syndrome leads to haploinsufficiency of VCP and is associated with DD, ID, and tremor, along with other findings, such as short stature, genital anomalies, and precocious puberty.61,62,63 Although many other genes are present in the ∼2–5 Mb deletions, VCP should be considered a candidate gene for the 9p13 deletion neurological-developmental phenotype.
Most missense and in-frame deletion variants in our cohort caused VCP loss of function via decreased ATPase activity. This is opposed to MSP-causing variants, which leave ATPase function intact or elevated (Figure S3).64,65,66 However, four variants (p.Arg256Gly, p.Phe267del, p.Gly271Asp, and p.Ser541Tyr) in our cohort increased ATPase activity. With its super-rapid ATPase activity, proband 10’s p.Ser541Tyr hyperactivating variant might not allow enough time for coordinated protein unfolding or co-factor binding and release. Other hyperactivating mutations, for example, in UBE3A (MIM: 601623) and KIF1A (MIM: 601255), have been described to cause disease when loss of function of these genes is the usual mechanism.67,68 Variants p.Arg256Gly and p.Phe267del in probands 5 and 6, respectively, are near the N-D1 interface of VCP and thus might alter adaptor or substrate binding similarly to MSP-causing variants. In fact, the c.801_803del (p.Phe267del) variant has been described in two adults with FTD or progressive primary aphasia.48 We initially speculated that all variants in our cohort cause earlier or more profound disruption of cellular function than the variants that cause MSP, leading to earlier disease onset. Although this could be true for most variants in our cohort, the p.Phe267del variant suggests that the syndrome we describe can exist in a spectrum with other VCP-related diseases. It would be interesting to know whether the adults with this variant had features of the childhood-onset syndrome we describe or whether proband 6, who harbors this variant, will go on to develop FTD. In fact, all individuals with deleterious VCP variants should be prospectively monitored for the development of findings associated with MSP as they age given that no one in our cohort has reached the normal age of onset of MSP symptoms (the third to fourth decade of life).29 We hypothesize that individuals in this cohort have additional risk factors that contribute to early disease development, which might also explain the development of proband 12’s behavioral phenotypes, which were not found in his father, from whom he inherited his VCP variant.
The variants and phenotypes we describe are distinct from MSP and show no signs of myopathy or bone disease; however, there might be some overlap with other VCP-associated neurological conditions. Three probands had abnormal EMG or NCV studies with evidence of sensorimotor polyneuropathy; these are reminiscent of CMT type 2Y, which is caused by VCP variants c.290G>A (p.Gly97Glu) and c.553G>A (p.Glu185Lys).35,69 Neuropathy appears to have reduced penetrance in our cohort given that not all older probands had a noticeable neuropathy; formal EMG and NCV studies of more individuals could clarify this association further. Another potential overlapping phenotype is FTD; the oldest individual in our study, proband 11, showed atrophy of the frontal lobes on brain MRI. It is possible that specific VCP variants primarily affect neurons and spare other cell types given that not all VCP variants are associated with neuropathy or dementia. The neurodegenerative effects of VCP loss of function could be due to the accumulation of toxic proteins, such as TDP-43 or Tau, as has been shown in knockout mice59 and VCP hypomorph variant c.1184A>G (p.Asp395Gly).70 Additional work is needed for understanding whether the VCP variants in our cohort also produce inclusion bodies or whether a novel mechanism leads to early-onset neurological disease. Ongoing evaluation will be important for understanding the potential overlap between this syndrome and other VCP-associated syndromes, such as MSP, CMT, and FTD.
There are several limitations of this study. Compiling this cohort of individuals required international collaboration, which meant that each proband was evaluated by different healthcare professionals, and analysis was performed with non-standardized sequencing techniques and algorithms. Additionally, there was no uniform measurement of the degree of DD or ID, making it more challenging to compare phenotypic severity. Although 13 individuals is a relatively large cohort for an ultra-rare disease, this number of probands did not allow for artificial-intelligence-based analysis of facial features or statistical analysis for genotype-phenotype correlations. Under current ACMG criteria, establishing a gene-disease association and upgrading variant classifications to likely pathogenic or pathogenic will require additional cases for each variant and/or established functional studies. Despite demonstrating alterations from the wild-type allele, our in vitro ATPase studies are necessarily limited given that ATPase activity is indirectly related to disease pathogenesis and decreased levels of ATPase activity have not yet been correlated with disease. Ideally, these assays would be performed with mixed hexamers containing both variant and wild-type subunits to simulate heterozygous individuals; however, this has been done for only one variant so far because several technical challenges limit the feasibility of mixing assays.66 In order to efficiently mimic heterozygous VCP variants, new techniques will be needed. VCP variants might also cause differences in intracellular localization, chaperone binding, and unfolding activity that are not analyzed by this assay but that can drastically affect cellular function. Therefore, haploinsufficiency might not be the mechanism of pathogenesis for some of the variants in this cohort; there might be a change in function or dominant negative effects. For the variants predicted to cause nonsense-mediated decay, functional studies assessing mRNA and protein production, as well as the study of behavioral and neurological phenotypes in model organisms, would ideally be performed. Although assessing ATPase function and protein stability provides a point of comparison against MSP-causing variants, evaluating the impact of the variants in this cohort will require more extensive molecular studies. We hope the variants and phenotype described here will provide rich fuel for future studies, clarify the pathogenicity of VUSs in VCP, and assist in the diagnosis of children with DD and/or ID.
Conclusion
We present 13 individuals with a childhood-onset VCP-related disorder, characterized by DD, ID, hypotonia, and macrocephaly, that is distinct from adult-onset MSP. We provide evidence that this disease is caused by damaging heterozygous variants in VCP. The 13 variants include missense variants spread throughout the protein, in-frame deletions, and a frameshift and a splicing variant that could lead to haploinsufficiency. Additionally, we provide evidence for a loss-of-function pathomechanism given that many of these VCP variants caused decreased ATPase activity, and we identify one variant that results in hyperactivation of ATPase activity. This contrasts with the studied MSP-associated variants, which moderately increase ATPase activity. Although there was no clear overlap in symptomatology with MSP, one of the variants in this cohort was seen in adult-onset FTD, and some probands have a neuropathy reminiscent of CMT, which suggests possible overlap between this neurodevelopmental disorder and MSP and other VCP-related diseases. Overall, this cohort provides direction for further research and expands our understanding of the functions of VCP in neurologic disease.
Data and code availability
We, or the sequencing laboratory, have submitted these variants and associated phenotypes to ClinVar under accession numbers 1331681, 2582677, 1303410, 2582678, 2582679, 1913089, 2429745, 2582680, 2582681, 2582682, 2444456, 1303647, and 2575609. Anonymized data will be shared by request from any qualified investigator.
Acknowledgments
This research would be impossible without the generosity of our probands and their families, and we thank them from the bottom of our hearts. We were not able to recognize as authors all clinical and research team members who helped care for these individuals, but their contributions were invaluable. We would also like to thank Kathryn Russell and Moriel Singer-Berk of the Broad Institute for their assistance with variant classification. E.B.L. is supported by National Institutes of Health grant RF1AG065341. T.-F.C. is supported by National Institute of Neurological Disorders and Stroke grant R01NS102279. C.C.W. is supported by National Institute on Aging grant R01AG031867 and National Institute of Arthritis and Musculoskeletal and Skin Diseases grant K24AR073317.
Author contributions
Conceptualization, M.S.S.; writing – original draft, A.Y.M. with proband supplements contributed by W.B., M.F.S., K.U., T.S., H.L.S., H.T., S.V., J.T., C.G.S., K.R., L.H.S., and A.A.; writing – review & editing, A.Y.M.-S., B.C.C., L.P., E.B.L., C.C.W., and M.S.S.; investigation – clinical & variant analysis, W.B., S.A.S., Ø.L.H., M.F.S., M.P., K.U., S.R., T.B., T.S., M.A.M, H.L.S., H.T., S.V., J.B., E.S., J.T., C.L., C.G.S, L.P., K.R., V.N., L.H.S, D.R., A.A., L.B., and M.S.S.; supervision – clinical, C.L., J.D., and M.S.S.; investigation – in vitro, J.D., D.H., and M.W. with supervision by T.-F.C. and C.C.W.; visualization, A.Y.M.-S. and B.C.C.
Declaration of interests
The authors declare no competing interests.
Published: October 25, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2023.10.007.
Contributor Information
Conrad C. Weihl, Email: weihlc@wustl.edu.
Marwan S. Shinawi, Email: mshinawi@wustl.edu.
Web resources
Supplemental information
References
- 1.Olszewski M.M., Williams C., Dong K.C., Martin A. The Cdc48 unfoldase prepares well-folded proteins substrates for degradation by the 26S proteasome. Commun. Biol. 2019;2:29. doi: 10.1038/s42003-019-0283-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beskow A., Grimberg K.B., Bott L.C., Salomons F.A., Dantuma N.P., Young P. A conserved unfoldase activity for the p97 AAA-ATPase in proteasomal degradation. J. Mol. Biol. 2009;394:732–746. doi: 10.1016/j.jmb.2009.09.050. [DOI] [PubMed] [Google Scholar]
- 3.Ju J.S., Fuentealba R.A., Miller S.E., Jackson E., Piwnica-Worms D., Baloh R.H., Weihl C.C. Valosin-containing protein (VCP) is required for autophagy and is disrupted in VCP disease. J. Cell Biol. 2009;187:875–888. doi: 10.1083/jcb.200908115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tresse E., Salomons F.A., Vesa J., Bott L.C., Kimonis V., Yao T.-P., Dantuma N.P., Taylor J.P. VCP/p97 is essential for maturation of ubiquitin-containing autophagosomes and this function is impaired by mutations that cause IBMPFD. Autophagy. 2010;6:217–227. doi: 10.4161/auto.6.2.11014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson A.E., Shu H., Hauswirth A.G., Tong A., Davis G.W. VCP-dependent muscle degeneration is linked to defects in a dynamic tubular lysosomal network in vivo. Elife. 2015;4 doi: 10.7554/eLife.07366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Custer S.K., Neumann M., Lu H., Wright A.C., Taylor J.P. Transgenic mice expressing mutant forms VCP/p97 recapitulate the full spectrum of IBMPFD including degeneration in muscle, brain and bone. Hum. Mol. Genet. 2010;19:1741–1755. doi: 10.1093/hmg/ddq050. [DOI] [PubMed] [Google Scholar]
- 7.Kim N.C., Tresse E., Kolaitis R.M., Molliex A., Thomas R.E., Alami N.H., Wang B., Joshi A., Smith R.B., Ritson G.P., et al. VCP is essential for mitochondrial quality control by PINK1/Parkin and this function is impaired by VCP mutations. Neuron. 2013;78:65–80. doi: 10.1016/j.neuron.2013.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang T., Mishra P., Hay B.A., Chan D., Guo M. Valosin-containing protein (VCP/p97) inhibitors relieve Mitofusin-dependent mitochondrial defects due to VCP disease mutants. Elife. 2017;6 doi: 10.7554/eLife.17834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mouysset J., Deichsel A., Moser S., Hoege C., Hyman A.A., Gartner A., Hoppe T. Cell cycle progression requires the CDC-48UFD-1/NPL-4 complex for efficient DNA replication. Proc Natl Acad Sci USA. 2008;105:12879–12884. doi: 10.1073/pnas.0805944105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meerang M., Ritz D., Paliwal S., Garajova Z., Bosshard M., Mailand N., Janscak P., Hübscher U., Meyer H., Ramadan K. The ubiquitin-selective segregase VCP/p97 orchestrates the response to DNA double-strand breaks. Nat. Cell Biol. 2011;13:1376–1382. doi: 10.1038/ncb2367. [DOI] [PubMed] [Google Scholar]
- 11.Vaz B., Halder S., Ramadan K. Role of p97/VCP (Cdc48) in genome stability. Front. Genet. 2013;4:60. doi: 10.3389/fgene.2013/00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ye Y., Meyer H.H., Rapoport T.A. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature. 2001;414:652–656. doi: 10.1038/414652a. [DOI] [PubMed] [Google Scholar]
- 13.Wang T., Xu W., Qin M., Yang Y., Bao P., Shen F., Zhang Z., Xu J. Pathogenic mutations in the valosin-containing protein/p97(VCP) N-domain inhibit the SUMOylation of VCP and lead to impaired stress response. J. Biol. Chem. 2016;291:14373–14384. doi: 10.1074/jbc.M116.729343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Banerjee S., Bartesaghi A., Merk A., Rao P., Bulfer S.L., Yan Y., Green N., Mroczkowski B., Neitz R.J., Wipf P., et al. 2.3 Å resolution cryo-EM structure of human p97 and mechanism of allosteric inhibition. Science. 2016;351:871–875. doi: 10.1126/science.aad7974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y., Ballar P., Zhong Y., Zhang X., Liu C., Zhang Y.-J., Monteiro M.J., Li J., Fang S. SVIP induces localization of p97/VCP to the plasma and lysosomal membranes and regulates autophagy. PLoS One. 2011;6 doi: 10.1371/journal.pone.0024478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahlstedt B.A., Ganji R., Raman M. The functional importance of VCP to maintaining cellular protein homeostasis. Biochem. Soc. Trans. 2022;50:1457–1469. doi: 10.1042/BST20220648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Twomey E.C., Ji Z., Wales T.E., Bodnar N.O., Ficarro S.B., Marto J.A., Engen J.R., Rapoport T.A. Substrate processing by the Cdc48 ATPase complex is initiated by ubiquitin unfolding. Science. 2019;365 doi: 10.1126/science.aax1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cooney I., Han H., Stewart M.G., Carson R.H., Hansen D.T., Iwasa J.H., Price J.C., Hill C.P., Shen P.S. Structure of the Cdc48 segregase in the act of unfolding an authentic substrate. Science. 2019;365:502–505. doi: 10.1126/science.aax0486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pan M., Yu Y., Ai H., Zheng Q., Xie Y., Liu L., Zhao M. Mechanistic insight into substrate processing and allosteric inhibition of human p97. Nat. Struct. Mol. Biol. 2021;28:614–625. doi: 10.1038/s41594-021-00617-2. [DOI] [PubMed] [Google Scholar]
- 20.Xu Y., Han H., Cooney I., Guo Y., Moran N.G., Zuniga N.R., Price J.C., Hill C.P., Shen P.S. Active conformation of the p97-p47 unfoldase complex. Nat. Commun. 2022;13:2640. doi: 10.1038/s41467-022-30318-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pfeffer G., Lee G., Pontifex C.S., Fanganiello R.D., Peck A., Weihl C.C., Kimonis V. Multisystem proteinopathy due to VCP mutations: a review of clinical heterogeneity and genetic diagnosis. Genes. 2022;13:963. doi: 10.3390/genes13060963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kimonis V.E., Fulchiero E., Vesa J., Watts G. VCP disease associated with myopathy, Paget disease of bone and frontotemporal dementia: review of a unique disorder. Biochim. Biophys. Acta. 2008;1782:744–748. doi: 10.1016/j.bbadis.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 23.Benatar M., Wuu J., Fernandez C., Weihl C.C., Katzen H., Steele J., Oskarsson B., Taylor J.P. Motor neuron involvement in multisystem proteinopathy: implications for ALS. Neurology. 2013;80:1874–1880. doi: 10.1212/WNL.0b013e3182929fc3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schiava M., Ikenaga C., Villar-Quiles R.N., Caballero-Ávila M., Topf A., Nishino I., Kimonis V., Udd B., Schoser B., Zanoteli E., et al. Genotype-phenotype correlations in valosin-containing protein disease: a retrospective multicentre study. J. Neurol. Neurosurg. Psychiatry. 2022;93:1099–1111. doi: 10.1136/jnnp-2022-328921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fernandez-Saiz V., Buchberger A. Imbalances in p97 co-factor interactions in human proteinopathy. EMBO Rep. 2010;11 doi: 10.1038/embor.2010.49. 479-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Manno A., Noguchi M., Fukushi J., Motohashi Y., Kakizuka A. Enhanced ATPase activities as a primary defect of mutant valosin-containing proteins that cause inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia. Gene Cell. 2010;15:911–922. doi: 10.1111/j.1365-2443.2010.01428.x. [DOI] [PubMed] [Google Scholar]
- 27.DeLaBarre B., Brunger A.T. Complete structure of p97/valosin-containing protein reveals communication between nucleotide domains. Nat. Struct. Biol. 2003;10:856–863. doi: 10.1038/nsb972. [DOI] [PubMed] [Google Scholar]
- 28.Tang W.K., Xia D. Mutations in the human AAA+ chaperone p97 and related diseases. Front. Mol. Biosci. 2016;3:79. doi: 10.3389/fmolb.2016.00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Al-Obeidi E., Al-Tahan S., Surampalli A., Goyal N., Wang A.K., Hermann A., Omizo M., Smith C., Mozaffar T., Kimonis V. Genotype-phenotype study in patients with VCP valosin-containing protein mutations associated with multisystem proteinopathy. Clin. Genet. 2018;93:119–125. doi: 10.1111/cge.13095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wong T.H., Pottier C., Hondius D.C., Meeter L.H.H., van Rooij J.G.J., Melhem S., Netherlands Brain Bank. van Minkelen R., van Duijn C.M., Rozemuller A.J.M., et al. Three VCP mutations in patients with frontotemporal dementia. J. Alzheimers Dis. 2018;65:1139–1146. doi: 10.3233/JAD-180301. [DOI] [PubMed] [Google Scholar]
- 31.Johnson J.O., Mandrioli J., Benatar M., Abramzon Y., Van Deerlin V.M., Trojanowski J.Q., Gibbs J.R., Brunetti M., Gronka S., Wuu J., et al. Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron. 2010;68:857–864. doi: 10.1016/j.neuron.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Majounie E., Traynor B.J., Chiò A., Restagno G., Mandrioli J., Benatar M., Taylor J.P., Singleton A.B. Mutational analysis of the VCP gene in Parkinson’s disease. Neurobiol. Aging. 2012;33:209.e1–209.e2. doi: 10.1016/j.neurobiolaging.2011.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Regensburger M., Türk M., Pagenstecher A., Schröder R., Winkler J. VCP-related multisystem proteinopathy presenting as early-onset Parkinson disease. Neurology. 2017;89:746–748. doi: 10.1212/WNL.0000000000004240. [DOI] [PubMed] [Google Scholar]
- 34.van de Warrenburg B.P., Schouten M.I., de Bot S.T., Vermeer S., Meijer R., Pennings M., Gilissen C., Willemsen M.A., Scheffer H., Kamsteeg E.J. Clinical exome sequencing for cerebellar ataxia and spastic paraplegia uncovers novel gene-disease associations and unanticipated rare disorders. Eur. J. Hum. Genet. 2016;24:1460–1466. doi: 10.1038/ejhg.2016.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gonzalez M.A., Feely S.M., Speziani F., Strickland A.V., Danzi M., Bacon C., Lee Y., Chou T.F., Blanton S.H., Weihl C.C., et al. A novel mutation in VCP causes Charcot-Marie-Tooth type 2 disease. Brain. 2014;137:2897–2902. doi: 10.1093/brain/awu224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iossifov I., Ronemus M., Levy D., Wang Z., Hakker I., Rosenbaum J., Yamrom B., Lee Y.H., Narzisi G., Leotta A., et al. De novo gene disruptions in children on the autistic spectrum. Neuron. 2012;74:285–299. doi: 10.1016/j.neuron.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sobreira N., Schiettecatte F., Valle D., Hamosh A. A matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rentzsch P., Schubach M., Shendure J., Kircher M. CADD-Splice—improving genome-wide variant effect prediction using deep learning-derived splice scores. Genome Med. 2021;13:31. doi: 10.1186/s13073-021-00835-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ioannidis N.M., Rothstein J.H., Pejaver V., Middha S., McDonnell S.K., Baheti S., Musolf A., Li Q., Holzinger E., Karyadi D., et al. REVEL: an ensemble method for predicting the pathogeneicity of rare missense variants. Am. J. Hum. Genet. 2016;99:877–885. doi: 10.1016/j.ajhg.2016.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tavtigian S.V., Harrison S.M., Boucher K.M., Biesecker L.G. Fitting a naturally scaled point system to the ACMG/AMP variant classification guidelines. Hum. Mutat. 2020;41:1734–1737. doi: 10.1002/humu.24088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abou Tayoun A.N., Pesaran T., DiStefano M.T., Oza A., Rehm H.L., Biesecker L.G., Harrison S.M., ClinGen Sequence Variant Interpretation Working Group (ClinGen SVI) Recommendations for interpreting the loss of function PVS1 ACMG/AMP variant criterion. Hum. Mutat. 2018;39:1517–1524. doi: 10.1002/humu.23626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pejaver V., Byrne A.B., Feng B.-J., Pagel K.A., Mooney S.D., Karchin R., O’Donnell-Luria A., Harrison S.M., Tavtigian S.V., Greenblatt M.S., et al. Calibration of computational tools for missense variant pathogenicity classification and ClinGen recommendations for use of PP3/BP4 criteria. Am. J. Hum. Genet. 2022;109:2163–2177. doi: 10.1016/j.ajhg.2022.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu K., Cai Y., Si X., Ye Z., Gao Y., Liu C., Wang R., Ma Z., Zhu H., Zhang L., et al. The phosphorylation and dephosphorylation switch of VCP/p97 regulates the architecture of centrosome and spindle. Cell Death Differ. 2022;29:2070–2088. doi: 10.1038/s41418-022-01000-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Källberg M., Wang H., Wang S., Peng J., Wang Z., Lu H., Xu J. Template-based protein structure modeling using the RaptorX web server. Nat. Protoc. 2012;7:1511–1522. doi: 10.1038/nprot.2012.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu W., Xie Y., Ma J., Luo X., Nie P., Zuo Z., Lahrmann U., Zhao Q., Zheng Y., Zhao Y., et al. IBS: an illustrator for the presentation and visualization of biological sequences. Bioinformatics. 2015;31:3359–3361. doi: 10.1093/bioinformatics/btv362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chou T.F., Bulfer S.L., Weihl C.C., Li K., Lis L.G., Walters M.A., Schoenen F.J., Lin H.J., Deshaies R.J., Arkin M.R. Specific inhibition of p97/VCP ATPase and kinetic analysis demonstrate interaction between D1 and D2 ATPase domains. J. Mol. Biol. 2014;426:2886–2899. doi: 10.1016/j.jmb.2014.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sassi C., Capozzo R., Hammer M., Zecca C., Federoff M., Blauwendraat C., Bernstein N., Ding J., Gibbs J.R., Price T., et al. Exploring dementia and neuronal ceroid lipofuscinosis genes in 100 FTD-like patients from 6 towns and rural villages on the Adriatic Sea cost of Apulia. Sci. Rep. 2021;11:6353. doi: 10.1038/s41598-021-85494-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jaganathan K., Kyriazopoulou Panagiotopoulou S., McRae J.F., Darbandi S.F., Knowles D., Li Y.I., Kosmicki J.A., Arbelaez J., Cui W., Schwartz G.B., et al. Predicting splicing from primary sequence with deep learning. Cell. 2019;176:535–548.e24. doi: 10.1016/j.cell.2018.12.015. [DOI] [PubMed] [Google Scholar]
- 50.Jian X., Boerwinkle E., Liu X. In silico prediction of splice-altering single nucleotide variants in the human genome. Nucleic Acids Res. 2014;42:13534–13544. doi: 10.1093/nar/gku1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Song C., Wang Q., Li C.C.H. ATPase activity of p97-valosin-containing protein (VCP): D2 mediates the major enzyme activity, and D1 contributes to the heat-induced activity. J. Biol. Chem. 2003;278:3648–3655. doi: 10.1074/jbc.M208422200. [DOI] [PubMed] [Google Scholar]
- 52.Bodnar N.O., Rapoport T.A. Molecular mechanism of substrate processing by the Cdc48 ATPase complex. Cell. 2017;169:722–735.e9. doi: 10.1016/j.cell.2017.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Davies J.M., Brunger A.T., Weis W.I. Improved structures of full-length p97, an AAA ATPase: implications for mechanisms of nucleotide-dependent conformational change. Structure. 2008;16:715–726. doi: 10.1016/j.str.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 54.Schiava M., Ikenaga C., Topf A., Caballero-Ávila M., Chou T.-F., Li S., Wang F., Daw J., Stojkovic T., Villar-Quiles R., et al. Clinical classification of variants in the valosin-containing protein gene associated with multisystem proteinopathy. Neurol. Genet. 2023;9 doi: 10.1212/NXG.0000000000200093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lainhart J.E., Bigler E.D., Bocian M., coon H., Dinh E., Dawson G., Deutsch C.K., Dunn M., Estes A., Tager-Flusberg H., et al. Head circumference and height in autism: a study by the Collaborative Program of Excellence in Autism. Am. J. Med. Genet. 2006;140:2257–2274. doi: 10.1002/ajmg.a.31465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P., et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Müller J.M.M., Deinhardt K., Rosewell I., Warren G., Shima D.T. Targeted deletion of p97 (VCP/CDC48) in mouse results in early embryonic lethality. Biochem. Biophys. Res. Commun. 2007;354:459–465. doi: 10.1016/j.bbrc.2006.12.206. [DOI] [PubMed] [Google Scholar]
- 58.Wang H.F., Shih Y.T., Chen C.Y., Chao H.W., Lee M.J., Hsueh Y.P. Valosin-containing protein and neurofibromin interact to regulate dendritic spine density. J. Clin. Invest. 2011;121:4820–4837. doi: 10.1172/JCI45677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shih Y.T., Hsueh Y.P. VCP and ATL1 regulate endoplasmic reticulum and protein synthesis for dendritic spine formation. Nat. Commun. 2016;7 doi: 10.1038/ncomms11020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wani A., Zhu J., Ulrich J.D., Eteleeb A., Sauerbeck A.D., Reitz S.J., Arhzaouy K., Ikenaga C., Yuede C.M., Pittman S.K., et al. Neuronal VCP loss of function recapitulates FTLD-TDP pathology. Cell Rep. 2021;36 doi: 10.1016/j.celrep.2021.109399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Niemi A.K., Kwan A., Hudgins L., Cherry A.M., Manning M.A. Report of two patients and further characterization of interstitial 9p13 deletion – a rare but recurrent microdeletion syndrome? Am. J. Med. Genet. 2012;158A:2328–2335. doi: 10.1002/ajmg.a.35536. [DOI] [PubMed] [Google Scholar]
- 62.Crone M., Thomas M.A. 9p13.1p13.3 interstitial deletion: a case report and further delineation of a rare condition. Am. J. Med. Genet. 2016;170A:1095–1098. doi: 10.1002/ajmg.a.37534. [DOI] [PubMed] [Google Scholar]
- 63.Ferreira S.I., Cinnirella G., Ramos L., Suppa A., Pires L.M., Nardone A.M., Camerota L., Lanciotti S., Galasso C., De Maio F., et al. Tremor is a major feature of 9p13 deletion syndrome. Am. J. Med. Genet. 2020;182:2694–2698. doi: 10.1002/ajmg.a.61807. [DOI] [PubMed] [Google Scholar]
- 64.Halawani D., LeBlanc A.C., Rouiller I., Michnick S.W., Servant M.J., Latterich M. Hereditary inclusion body myopathy-linked p97/VCP mutations in the NH2 domain and the D1 ring modulate p97/VCP ATPase activity and D2 ring conformation. Mol. Cell Biol. 2009;29:4484–4494. doi: 10.1128/MCB.00252-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Blythe E.E., Olson K.C., Chau V., Deshaies R.J. Ubiquitin- and ATP-dependent unfoldase activity of P97/VCP-NPLOC4-UFD1L is enhanced by a mutation that causes multisystem proteinopathy. Proc Natl Acad Sci. 2017;114:E4380–E4388. doi: 10.1073/pnas.1706205114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Blythe E.E., Gates S.N., Deshaies R.J., Martin A. Multisystem proteinopathy mutations in VCP/p97 increase NPLOC4-UFD1L binding and substrate processing. Structure. 2019;27:1820–1829.e4. doi: 10.1016/j.str.2019.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Weston K.P., Gao X., Zhao J., Kim K.-S., Maloney S.E., Gotoff J., Parikh S., Leu Y.-C., Wu K.-P., Shinawi M., et al. Identification of disease-linked hyperactivating mutations in UBE3A through large-scale functional variant analysis. Nat. Commun. 2021;12:6809. doi: 10.1038/s41467-021-27156-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chiba K., Takahashi H., Chen M., Obinata H., Arai S., Hashimoto K., Oda T., McKenney R.J., Niwa S. Disease-associated mutations hyperactivate KIF1A motility and anterograde axonal transport of synaptic vesicle precursors. Proc Nat Acad Sci. 2019;116:18429–18434. doi: 10.1073/pnas.1905690116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jerath N.U., Crockett C.D., Moore S.A., Shy M.E., Weihl C.C., Chou T.-F., Grider T., Gonzalez M.A., Zuchner S., Swenson A. Rare manifestation of a c.290 C>T, p.gly97glu VCP mutation. Case Rep. Genet. 2015;2015 doi: 10.1155/2015/239167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Darwich N.F., Phan J.M., Kim B., Suh E., Papatriantafyllou J.D., Changolkar L., Nguyen A.T., O’Rourke C., He Z., Porta S., et al. Autosomal dominant VCP hypomorph mutation impairs disaggregation of PHF-tau. Science. 2020;370 doi: 10.1126/science.aay8826. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
We, or the sequencing laboratory, have submitted these variants and associated phenotypes to ClinVar under accession numbers 1331681, 2582677, 1303410, 2582678, 2582679, 1913089, 2429745, 2582680, 2582681, 2582682, 2444456, 1303647, and 2575609. Anonymized data will be shared by request from any qualified investigator.