Abstract
Lung cancer is the leading cause of cancer-related deaths. Early detection and diagnosis are critical, as survival decreases with advanced stages. Approximately 1.6 million nodules are incidentally detected every year on chest CT scan images in the United States. This number of nodules identified is likely much larger after accounting for screening-detected nodules. Most of these nodules, whether incidentally or screening detected, are benign. Despite this, many patients undergo unnecessary invasive procedures to rule out cancer because our current stratification approaches are suboptimal, particularly for intermediate probability nodules. Thus, noninvasive strategies are urgently needed. Biomarkers have been developed to assist through the continuum of lung cancer care and include blood protein-based biomarkers, liquid biopsies, quantitative imaging analysis (radiomics), exhaled volatile organic compounds, and bronchial or nasal epithelium genomic classifiers, among others. Although many biomarkers have been developed, few have been integrated into clinical practice as they lack clinical utility studies showing improved patient-centered outcomes. Rapid technologic advances and large network collaborative efforts will continue to drive the discovery and validation of many novel biomarkers. Ultimately, however, randomized clinical utility studies showing improved patient outcomes will be required to bring biomarkers into clinical practice.
Key Words: biomarkers, early detection, indeterminate pulmonary nodules, lung cancer
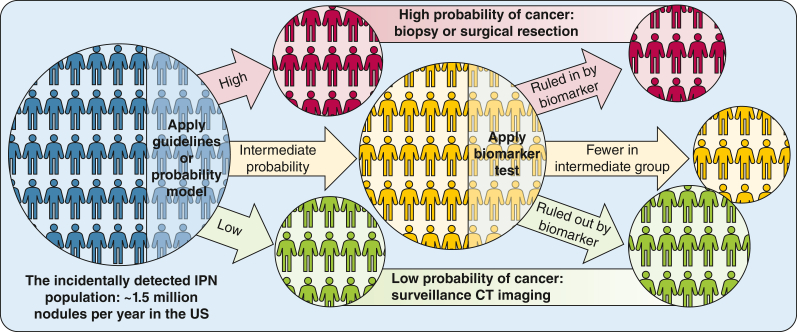
An estimated 4,000 indeterminate pulmonary nodules (IPNs) are identified every day in the United States alone1 leading to follow-up imaging, biopsy, or surgery, depending on the estimated likelihood of malignancy or “pretest probability.” The intermediate pretest probability category, for which additional diagnostic imaging or biopsies are indicated, remains challenging from a management standpoint, with heterogeneity in guideline recommendations and implementation,2 exposing patients to attendant morbidity and mortality. In addition, Medicare data suggest that > 40% of the overall cost of lung cancer management can be attributed to benign nodules managed with an invasive procedure.3 Thus, improving the reclassification from intermediate probability to low or high probability remains a research priority to improve the early diagnosis and treatment of lung cancer (Fig. 1).
Figure 1.
New stratification algorithm based on validated and clinically useful biomarkers. US = United States.
Many biomarkers aimed at reclassifying intermediate pretest probability IPNs have been developed in the past decade. Few have been commercialized, and even fewer are routinely used in clinical practice. These include, among others, blood protein-based biomarkers, liquid biopsies, quantitative imaging analysis (radiomics), exhaled volatile organic compounds, bronchial or nasal epithelium genomic classifiers, and combinations thereof.4 Despite evidence of biological validity, they are underutilized due to accessibility, cost, clinical practice inertia, and, more importantly, the absence of clinical utility studies showing improved patient outcomes. The current review provides a comprehensive overview of the available literature on emerging and validated biomarkers for IPNs, discusses their potential clinical role, and considers the barriers currently limiting their implementation in practice.
Methods
We conducted a comprehensive review of the literature related to lung cancer biomarkers. PubMed and the Cochrane Library were searched using the terms “lung cancer” AND “biomarker” plus terms specific to the respective sections (eg, “radiomics” or “quantitative imaging” for radiomic biomarkers) from January 2011 through December 2022. Studies were selected that included > 100 subjects and reported both internal and external validation. Only manuscripts written in English were considered. Studies that did not meet these criteria but were believed to be of particular relevance were also included.
Circulating Blood Protein-Based Biomarkers
Several protein biomarkers have been studied within the context of IPN diagnosis, the most widely studied being carcinoembryonic antigen (CEA) and cytokeratin fragment 21-1 (CYFRA 21-1),4,5 alone or in combination.6 In addition, several panels have been developed. A panel including CEA, CYFRA, epidermal growth factor receptor, prosurfactant protein B, neutrophil activating protein 2, and tissue inhibitor of metalloproteinase-1 has shown potential utility to reclassify intermediate probability nodules.7 Another four-marker panel, consisting of precursor form of surfactant protein B, cancer antigen 125, CYFRA 21-1, and CEA, may be useful for both IPN diagnosis and in the context of lung cancer screening.8,9
Biomarkers that have been studied in a validation cohort or clinical usefulness analysis are listed in Table 1.4, 5, 6, 7, 8,10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22 Based on the available data, performance characteristics are described as follows: the results of clinical impact analysis (I), the results of reclassification analysis (II), or the sensitivity/specificity of the test in the validation cohort (III). The sensitivities and specificities listed are at the thresholds defined by the authors in each article and therefore may not be directly comparable.
Table 1.
Blood Protein-Based Biomarkers for IPN Diagnosis
| Study | Biomarker Candidates | Study Population Size (Cancer/Benign/No Nodule) | Performance Characteristics |
|---|---|---|---|
| Farlow et al,10 2010 | IMPDF, phosphoglycerate mutase, ubiquillin, annexin I, annexin II, HSP70-9B | Training: 117/13/61 | III: 95/91 |
| Ostroff et al,11 2010 | Cadherin-1, CD30 ligand, endostatin, HSP 90α, LRIG3, MIP-4, pleiotrophin, PRKCI, RGM-C, SCF sR, sL-Selectin, YES | Training: 213/420/352 Validation: 78/245/118 |
III: 91/84 |
| Kupert et al,12 2011 | sPLA2-IIa, CEA, CYFRA 21-1 | Training: 96/29/– Validation: 44/0/20 |
III: 63/76 |
| Patz et al,13 2013 | CEA, AAT, SCC | Training: 298/211 Validation: 203/196/– |
III: 88/82 |
| Daly et al,14 2013 | IL-6, IL-10, IL-1ra, sIL-2Rα, SDF-1α+β, TNF-α, MIP-1α | Training: 69/67/– Validation: 20/60/– |
III: 95/23 |
| Okamura et al,15 2013 | CEA, CYFRA 21-1 | Training: 655/237/– | III: 33/95 |
| Fahrmann et al,16 2016 | Phosphatidylethanolamines PE34:2, PE36:2, and PE38:4 | Training: 61/29/– | III: 94/62 |
| Massion et al,17 2017 | Autoantibodies to CAGE, GBU 4–5, p53, MAGE A4, HuD, NY-ES0-1, and SOX-2 (Oncimmune/Biodesix Nodify CDT) | Training: 75/221/– | III: 33/97 |
| Ajona et al,18 2018 | C4d | Training: 59/0/79 Validation: 148/92/– |
III: 44/89 |
| Du et al,19 2018 | Autoantibodies to p53, PGP9.5, SOX2, GAGE7, GBU4-5, CAGE, and MAGEA1 | Training: 352/45/74 | III: 57/92 |
| Yang et al,5 2018 | ProGRP, CEA, SCC, CYFRA 21-1 | Training: 163 Validation: 179 (authors did not specific the patient breakdown by final disease status) |
III: 84/81 |
| Lastwika et al,20 2019 | IgG complexed FCGR2A, EPB41L3, LINGO1, IGM complexed S100A7L2 | Training: 125/125/– | III: 33/90 |
| Kammer et al,4 2021 | hsCYFRA 21-1 | Training: 150/75/– | I: 20% reduction in benign procedures, 39 days faster time-to-diagnosis |
| Ostrin et al,8 2021 | Pro-SFBT, CA125, CEA, CYFRA 21-1 (4MP) | Validation 1: 100/100/– Validation 2: 32/32/– |
III: 26/95 |
| Tanner et al,21 2021 | LG3BP and C163A (Biodesix Nodify XL2) | Validation: 29/132/– | I: 40% fewer procedures on benign nodules |
| Marmor et al,6 2022 | CYFRA 21-1, CEA (Abbott Architect) | Training: 217/121/– | II: cNRI of 0.21 for cancer |
| Marmor et al,22 2023 | Histoplasmosis IgG/IgM | Training: 117/40/– Validation: 75/95/– |
II: cNRI of 0.18 for cancer |
| Vachani et al,7 2023 | CEA, CYFRA, EGFR, ProSB, NAP2, and TIMP (MagArray Reveal) | Training: 186/243/– Validation: 212/277/– |
II: Increase in percentage of patients in the highest and lowest risk groups |
Performance characteristics are described as: I, results of estimation of clinical utility; II, results of a reclassification analysis; and III, sensitivity/specificity of the test in the validation cohort. If there is no validation cohort, the sensitivity/specificity reported is from the training cohort. Sensitivity/specificity reported as Youden Index. For biomarkers/classifiers that have been reported in multiple publications, only the most recent was included in this table. 4MP = 4-marker panel; AAAT = alpha-1-antitrypsin; BNAP2 = neutrophil-activating peptide 2; CA125 = cancer antigen 125; CAGE = cancer/testis antigen 1B; CD30 = cluster of differentiation 30 (also known as TNFRSF8); cNRI = bias-corrected clinical net reclassification index; CYFRA = cytokine fragment; CYFRA 21-1 = cytokeratin fragment 21-1; EGFR = epidermal growth factor receptor; EPB41L3 = erythrocyte membrane protein band 4.1-like 3; FCGR2A = Fc fragment of IgG receptor IIa (also known as CD32); GAGE7 = G antigen 7; GBU 4–5 = gamma-butyrobetaine dioxygenase 4–5 (also known as BBOX1); hsCYFRA 21-1 = high-sensitivity cytokeratin fragment 21-1; HSP = heat shock protein; HuD = Hu antigen D (also known as ELAVL4); IL = interleukin; IPN = indeterminate pulmonary nodule; LINGO1 = leucine-rich repeat and Ig domain-containing nogo receptor-interacting protein 1; LRIG3 = leucine-rich repeats and immunoglobulin-like domains protein 3; MAGEA1 = melanoma-associated antigen 1; MAGE A4 = melanoma-associated antigen 4; MIP-1α = macrophage inflammatory protein 1 alpha (also known as CCL3); MIP-4 = macrophage inflammatory protein 4 (also known as CCL18); MPDF = interferon-induced mitochondrial protein; NAP2 = neutrophil activating protein 2; p53 = tumor protein p53; PGP9.5 = protein gene product 9; PRKCI = protein kinase C iota; ProSB = prosurfactant protein B; Pro-SFBT = pro-surfactant protein B precursor (also known as SFTPB); RGM-C = repulsive guidance molecule C (also known as RGMC); S100A7L2 = S100 calcium-binding protein A7-like 2; SCC = squamous cell carcinoma antigen (also known as SCCA); SCF sR = stem cell factor soluble receptor (also known as c-kit ligand); SDF-1α+β = stromal cell-derived factor 1 alpha and beta; sIL = oluble interleukin; sL-Selectin = soluble L-selectin (also known as CD62L); SOX-2 = SRY (sex determining region Y)-box 2; sPLA2-IIa = secretory phospholipase A2 group II; TIMP1 = tissue inhibitor of metalloproteinase-1; TNF-α = tumor necrosis factor-α; YES = tyrosine kinase substrate with YEDQ motifI.
An alternative to tumor-specific antigens are tumor-associated antibodies, which offer the promise of high specificity at earlier time points in the disease course due to signal amplification by the immune system.20 Several autoantibody panels have been developed, including one panel of seven tumor-associated antibodies (to p53, PGP9.5, SOX2, GAGE7, GBU4-5, CAGE, and MAGEA1).19 When combined with CT scan imaging, this signature yielded a specificity of 91.6% for cancers vs benign nodules, with a sensitivity of 56.5%. Another panel that included CAGE, GBU 4-5, p53, MAGE A4, HuD, NY-ES0-1, and SOX-2 achieved high specificity and positive predictive values.17 As an alternative to diagnosing cancer, in areas endemic for Histoplasma capsulatum, serum antibodies to Histoplasma have been used to diagnose benign disease. In two studies, dual positive IgG/IgM antibodies to Histoplasma were associated with only benign nodules.22 This test is best used in the appropriate clinical context and suggestive radiologic findings.
To date, no blood-based biomarker has been shown to definitely improve patient outcomes. A proteomic classifier that uses two proteins (LG3BP and C163A) was prospectively evaluated as part of an integrated classifier using clinical and imaging features in the Pulmonary Nodule Plasma Proteomic Classifier (PANOPTIC) study, which suggested a 32% relative reduction in unnecessary invasive procedures for benign nodules.21 Although the potential for these biomarkers may be inferred from retrospective and prospective observational studies, clinical utility can only be shown through prospective randomized controlled clinical trials with evidence of improved patient outcomes. Of the biomarkers listed earlier, the integrated classifier Nodify XL2 (Biodesix) is the only currently enrolling randomized controlled study aimed at showing improved patient outcomes.23
Circulating Blood Transcriptomic Biomarkers: MicroRNAs
MicroRNAs (miRNAs) are endogenous short noncoding RNAs of 20 to 22 nucleotides that operate as regulators of gene expression at the posttranscriptional level. Selected miRNAs are located in cancer-associated genomic regions, acting as oncogenes or tumor suppressors.24 An increasing number of investigations suggest that miRNAs profiled from blood samples may be promising adjuncts to existing methods for IPN evaluation and early lung cancer detection. Early data suggest a potential for high sensitivity and specificity even for early-stage (stage I and II) non-small cell lung cancer (NSCLC) detection.
Several studies have explored miRNA signatures alone or in combination with other blood-based biomarkers for IPN evaluation and early detection of lung cancer.25, 26, 27 These studies, however, have several limitations, including small sample size, lack of external validation, inclusion of late-stage cancers, and lack of demographic diversity. The largest miRNA study in early-stage NSCLC detection is a multicenter study that included 744 patients with NSCLC and 944 matched control subjects. A five-miRNA panel (miR-375, miR-1291, miR-214-3p, miR-1-3p, and let-7a-5p) distinguished participants with NSCLC subjects from control participants with areas under the curve (AUCs) of 0.936 and 0.984 in the discovery and verification cohorts, respectively. This panel was then validated in three independent cohorts with AUCs of 0.973, 0.916, and 0.917 and a sensitivity of 81.3% for all stages, 82.9% for stages I and II, and 83% for stage I NSCLC, with a specificity of 90.7%. The accuracy of the five-miRNA panel in detecting NSCLC was significantly higher than that of CEA or CYFRA21-1.28 This study included Asian and White populations, used samples that were not matched for demographic and clinicopathologic characteristics, and was not adjusted for other patient-specific variables.
None of the existing miRNA panels have been prospectively validated in clinical trials of individuals with early-stage NSCLC, although their use as biomarkers for IPNs seems promising. Thus far, none of these signatures are used in clinical practice. Additional research will be needed to confirm these preliminary but encouraging findings and to optimize reproducibility, detection, quantification, and clinical utility of miRNAs.
Circulating Tumor Cells and Tumor DNA: Liquid Biopsies
Circulating tumor cells (CTCs), circulating tumor DNA (ctDNA), and circulating methylated DNA are promising noninvasive biomarkers frequently referred to as “liquid biopsies.” They provide a composite view of tumor burden, avoiding sampling bias and the technical complexity inherent in tissue biopsy while theoretically yielding similar information. Currently, their primary clinical application in thoracic oncology is molecular characterization of known lung cancers (eg, detection of therapeutically targetable mutations). However, there is increasing interest in using these biomarkers for IPN evaluation. The most relevant studies are highlighted in Table 2.29, 30, 31, 32, 33, 34, 35
Table 2.
Characteristics and Performance of Liquid Biopsies (CTCs, ctDNA, and Methylated DNA) Predicting Malignancy in Pulmonary Nodules
| Publication | Population | Technique | Performance | Comments |
|---|---|---|---|---|
| Marquette et al, 29 2020 | AIR prospective cohort (high-risk individuals, n = 614; lung cancers, n = 19) | Blood-based CTC detection via isolation by size of epithelial tumor cell technique | Sens 26%, Spec 96% | LDCT imaging detected 178 pulmonary nodules with Sens 83%, Spec 71% |
| Li et al,30 2019 | Single-center case-control (patients with lung cancer, n = 174; healthy control subjects, n = 90) | Blood-based CTC detection via negative enrichment/fluorescence in situ hybridization | AUC 0.85 (Sens 68%, Spec 100%) Among patients with early-stage (I-II) lung cancer, Sens 64% |
Combination of CTCs and tumor markers (CEA, CA125, CYFRA 21-1, and SCC) improved diagnosis for all patients (Sens 83%) and in those with early-stage disease (Sens 79%) |
| Mathios et al,31 2021 | Training: LUCAS prospective cohort (patients with lung cancer, n = 129; high-risk individuals, n = 236) Validation: multinational cohort (healthy control subjects, n = 385; patients with early-stage lung cancer, n = 46) |
Blood-based cell-free DNA fragment detection and machine learning model | Training: AUC = 0.90, with greater AUCs for stages II, III, and IV (0.89, 0.92, and 0.92) vs stage I disease (0.76) Independent validation: comparable range of Sens and Spec across various stages and histologic subtypes |
Multimodal model of fragmentation profiles with CEA, age, tobacco use history, and presence of COPD increased AUC to 0.93 Use of multimodal model as a prescreen to stratify patients requiring LDCT imaging yielded improved performance with Sens 94% and Spec 80% |
| Leung et al,32 2020 | Single-center prospective cohort (patients with known or suspected early primary or secondary lung cancer, n = 192) | Blood-based ctDNA detection via qRT-PCR | Sens 75%, Spec 89% | Detected mutations in ctDNA and corresponding tissue biopsy results showed concordance of 83% and kappa statistic of 0.63 (P < .001) |
| Kneip et al,33 2011 | Training: single-center case-control (patients with stage IV lung cancer, n = 20; healthy control subjects, n = 20) Validation: two-center case-control (patients with suspected lung cancer, n = 343; healthy control subjects, n = 150) |
Blood-based methylation-specific qRT-PCR for SHOX2 | Training: Sens 75%, 95% Testing: AUC 0.78 (Sens 60%, Spec 90%) |
Small cell lung cancer and squamous cell carcinoma were detected more readily than adenocarcinomas (Sens 80% and 63% vs 39%), likely reflecting underlying differences in tumor biology |
| Hulbert et al,34 2017 | Single-center case-control (stage I or IIA lung cancer, n = 150; control with suspicious pulmonary nodule, n = 60) | Blood and sputum-based methylation-specific qRT-PCR for 6 cancer-specific methylated genes | Three-gene panel (TAC1, HOXA17, and SOX17) yielded Sens 93%, Spec 62% with blood and Sens 98%, Spec 71% and with sputum | Combination of blood-based methylated DNA and age, pack-y, COPD status, and FVC values using blinded random forest prediction models improved AUC to 0.89 (Sens 93%, Spec 67%) |
| Ooki et al,35 2017 | NYU LCBC case-control (patients with stage IA adenocarcinoma, n = 43; patients with stage IA SCC, n = 40; healthy control subjects, n = 42) | Blood-based promoter methylation-specific qRT-PCR for 30 cancer-specific methylated genes | Six-gene panel (CDO1, HOXA9, AJAP1, PTGDR, UNCX, and MARCH11) yielded Sens 72% and 60% for stage IA adenocarcinoma and SCC, respectively; Spec 71% | Risk stratification based on differential methylation significantly stratified prognosis, with 5-y OS of 100.0% for low risk, 96.0% for moderate risk, and 55.6% for high risk (P = .015) |
AIR = AIR Project Study Group; AUC = area under the curve; CTC = circulating tumor cell; ctDNA = circulating tumor DNA; CA-125 = cancer antigen 125; CEA = carcinoembryonic antigen; CYFRA 21-1 = cytokeratin 19 fragment; LDCT = low-dose CT scan; LUCAS = Longitudinal Urban Cohort Aging Study; qRT-PCR = quantitative reverse transcription polymerase chain reaction; NYU LCBC = New York University Lung Cancer Biomarker Center; OS = overall survival; SCC = squamous cell carcinoma; SCCA = squamous cell carcinoma antigen; Sens = sensitivity; Spec = specificity.
CTCs are cells that have left the primary tumor site and entered the bloodstream or lymphatic system and can be detected via size-based or biophysical separation methods. Limited available data in patients with lung cancer have shown relatively high specificity (96%-100%) but unreliable sensitivity (26%-68%).29,30 This performance has been improved using a combination of CTC detection alongside four tumor markers, even among patients with early-stage lung cancer.30
ctDNA is fragmented DNA not associated with tumor cells that can be found in the bloodstream and that may be more readily detected than CTCs.31,32 Although the sensitivity of ctDNA may be higher than that of CTCs, the use of ctDNA alone for lung cancer detection might result in more missed diagnoses compared with low-dose CT scan imaging. The sensitivity of ctDNA assays for lung cancer detection has been improved by using machine learning models that leverage clinical data such as age, tobacco use history, and presence of COPD and other relevant biomarkers such as CEA in addition to ctDNA. The use of such a multimodal screen to stratify patients requiring low-dose CT scan imaging could reduce the number of unnecessary biopsies while maintaining a low false-negative rate.31
Aberrant CpG promoter methylation that occurs early in tumorigenesis presents yet another diagnostic opportunity. Genome-wide DNA methylation profiles in lung cancer have revealed differentially methylated regions that include specific candidate genes for diagnostic assays. Quantitative methylation-specific real-time polymerase chain reaction enables amplification of methylated DNA target sequences of these candidate genes. Several studies investigating candidate genes and gene panels have shown promising sensitivities of 60% to 98% and specificities of 62% to 95%.33, 34, 35 Similar to studies of ctDNA, models incorporating methylated gene panels with clinical data may yield a performance superior to that of methylation biomarkers alone.
These studies highlight the potential of CTCs, ctDNA, and methylated DNA for IPN evaluation and early detection of lung cancer. Although performance metrics, particularly sensitivity, remain suboptimal, several studies suggest that performance could be improved by combining clinical data, imaging, and other serum biomarkers. Further refinement, validation, and prospective demonstration of clinical utility are needed prior to broad clinical application.
Airway Classifiers
The ability to detect cancer-associated molecular differences in relatively accessible normal-appearing airway epithelium36, 37, 38, 39 has served as the foundation for developing several noninvasive and minimally invasive biomarkers. Tissue collection via bronchoscopy is a common approach for diagnosing lung cancer in patients with IPNs with intermediate to high pretest probability. This procedure has high specificity but can have low sensitivity, especially in smaller or more peripheral nodules.40 Augmenting bronchoscopic sampling of the lesion with molecular analysis of more proximal bronchial brushings represents an attractive option. Blomquist et al41 reported the levels of 14 genes involved in antioxidant and DNA repair pathways, measured via reverse transcription polymerase chain reaction, in the normal bronchial airway epithelium that were associated with lung cancer status. This resulted in a biomarker currently being tested in the Lung Cancer Risk Test trial.41, 42, 43 Similarly, using microarray-based transcriptomic profiling, Spira et al44 developed and validated an 80-gene signature for differentiating participants who had ever used tobacco with or without lung cancer using endobronchial brushings from normal-appearing mainstem airways. This signature was later refined into a 23-gene lung cancer biomarker for patients with suspected lung cancer undergoing bronchoscopy.45 This biomarker was extensively validated in independent cohorts consisting of > 600 patients.46 The high sensitivity, either alone (89%) or in combination with bronchoscopy (97%), and the high negative predictive value of this biomarker allow physicians to choose surveillance with imaging for intermediate probability patients with an inconclusive bronchoscopy and negative biomarker result. The potential clinical impact of this biomarker was reported in a registry study at 35 US medical centers.47 This study examined the management of pulmonary nodules in 283 patients with a nondiagnostic bronchoscopy in whom the test was ordered. A negative result down-classified the risk of malignancy in 34% of cases. Of these, 74% had a change in management from invasive procedure to surveillance, without delaying the time to diagnosis.
Although biomarkers based on bronchial airway gene expression have shown clinical promise, biospecimen collection via bronchoscopy limits the intended use population.39 As an alternative, researchers have explored lung cancer-associated gene expression alterations in the nasal epithelium. Studies have shown that bronchial and nasal epithelium share similar transcriptomic alterations associated with tobacco use status48 and lung diseases such as COPD49 and idiopathic pulmonary fibrosis.50 Using nasal brushing samples and microarray expression profiling from participants in the Airway Epithelial Gene Expression in the Diagnosis of Lung Cancer (AEGIS) cohorts, Perez-Rogers et al51 developed a clinical genomic classifier with 30 genes that can identify lung cancer among participants who had ever used tobacco with high sensitivity of 91%. The preliminary results from RNA sequencing of nasal swab samples have been reported; they suggest 95% sensitivity for cancer among individuals classified as low risk and 90% specificity for cancer among individuals classified as high risk.52 Similar high performance was observed among patients who met or did not meet US Preventive Services Task Force eligibility criteria for lung cancer screening.53 These studies highlight the potential clinical promise of nasal epithelium gene expression for early lung cancer detection.
There is evidence for other airway molecular analytes that may aid in the early detection of lung cancer. A four-gene methylation biomarker was developed in samples from bronchial washings and showed high performance (82% sensitivity and 91% specificity) in an independent validation set.54 Pavel et al55 reported that adding miR-146a-5p expression to the bronchial gene biomarker can significantly improve the performance of the lung cancer diagnosis within the samples of AEGIS cohorts. These studies show the feasibility and importance of integrating data from different platforms to build multimodal biomarkers for early lung cancer diagnosis.
Exhaled Volatile Organic Compound-Based Biomarkers
The use of exhaled breath analysis in medicine has been studied since the 1970s.56 Regarding lung cancer detection, gas chromatography with mass spectrometry (GC/MS) has generally been the most used method of detection, with sensitivity and specificity ranging from 70% to 90%.57, 58, 59, 60 A recent study by Smirnova et al61 designed to overcome historical volatile organic compound (VOC) trial limitations and aimed at investigating the predictive performance of VOCs in 225 patients with stage I lung cancer reported an accuracy of 0.61. Additional studies have combined GC/MS with machine learning and discovered that a combination of five VOCs with machine learning assistance resulted in an AUC between 0.89 and 0.98.62, 63, 64 Artificial neural networks have also been used with GC/MS, with a high discriminatory power showing a sensitivity of 80% and a specificity of 91%.65 Another study using artificial neural networks found that a VOC combination was able to distinguish healthy control subjects from patients with lung cancer with an AUC of 0.99, and benign nodules from lung cancer with an AUC of 0.81,66 suggesting potential promise for future clinical usefulness.
The electronic nose device (e-nose) is another measurement device that has been used to analyze VOCs. The device is portable and inexpensive and uses gas sensors to detect various combinations of chemical compounds.67, 68, 69 Studies have reported a sensitivity up to 96% and specificity up to 94%.67,69, 70, 71, 72, 73, 74 Given the limitations of single-strategy approaches, the e-nose combined with machine learning is becoming increasingly popular to improve diagnostic accuracy.67,68,75,76 A recent multicenter study trained and validated a predictive model that combined clinical variables with VOC analysis from an e-nose device to distinguish patients with NSCLC from control participants (training set, n = 376; validation set, n = 199).77 In the validation cohort, the model yielded a sensitivity and specificity of 95% and 49%, respectively, and an AUC of 0.86.
There are numerous methods of both identifying VOCs and analyzing signatures, with respective advantages and disadvantages. However, estimates of diagnostic test performance vary significantly across studies using the same technology with limited reproducibility. Although breath analysis of VOCs holds promise, there has been little overall change in the accuracy of these tests over the last decade. Novel technologic advances, additional validation, and clinical utility studies are needed before VOC analysis can be used in clinical practice.
Imaging-Based Biomarkers: Quantitative Image Analysis or Radiomics
Current clinical guidelines for the management of lung nodules primarily rely on a few discrete imaging and clinical features. Some of these imaging features are subject to individual reader assessment, with substantial variability in clinical practice. The widespread availability of CT scan imaging data at the time of nodule identification positions CT scan-based radiomic approaches as an attractive, accessible, and potentially cost-effective option for the stratification of IPNs.
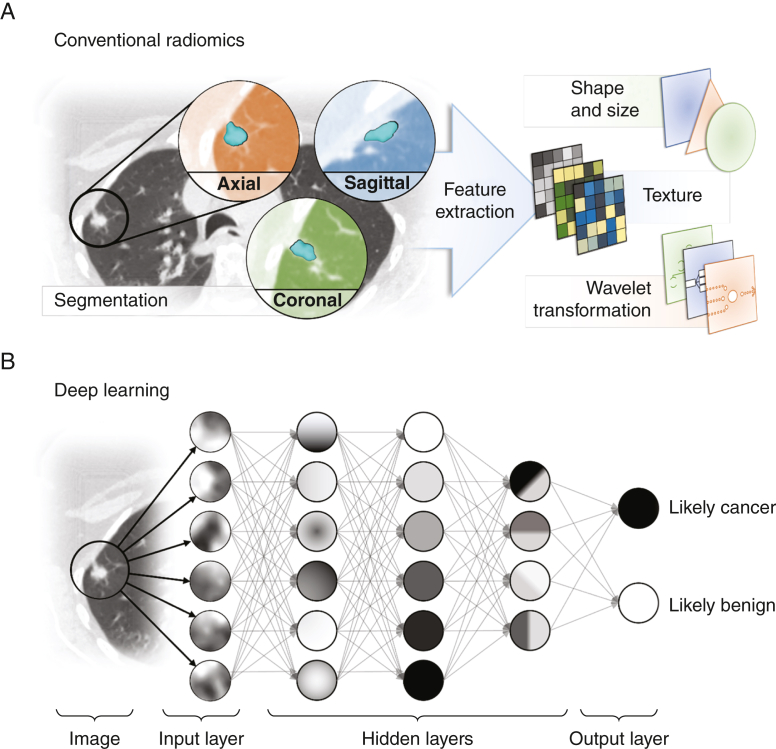
Radiomics is the process of identification, extraction, quantification, and analysis of imaging features from radiologic images that are beyond the capability of the radiologist (Fig 2). Applied clinical radiomics can enhance image interpretation by objective assessment of radiologic features associated with a given outcome or phenotype such as primary lung cancer. Radiomic features consist of semantic (visible) and agnostic features that require mathematical extraction and computation such as tumor heterogeneity or skewness. These features can either be prespecified from discrete conventional variables believed to be associated with particular phenotypes or “learned” through artificial neural networks (“deep learning”) for supervised approaches.
Figure 2.
A, B, Schematic representation of feature extraction and selection with conventional radiomics and deep learning. A, With conventional radiomics, nodules are segmented, and expert-defined features are extracted and analyzed by using traditional statistical regression methods. B, With artificial neural networks, automated learning of relevant features through multilayered networks mitigates expert bias, but extraction and analysis are hidden, and only the output (benign or cancer) is known (black box).
Clinically valuable tools should be externally validated and show consistent performance across different patient populations and sources of CT scan imaging data, which vary by scanner type, image slice thickness, reconstruction methods, or acquisition protocols.
Over the last few years, numerous algorithms have been proposed. Table 378, 79, 80, 81, 82 summarizes the main studies published on this topic. Although these models are promising, there is inconsistency in methodology (deep learning vs conventional radiomics), baseline characteristics and rates of malignancy in the training data sets, and quality of external validation cohorts and studies. For example, some models were developed using only patients with identified IPNs, whereas others use broader training, testing, and tuning sets using scans with and without any pulmonary nodules. The additional value of changes in radiomic features over time (longitudinal changes or “delta radiomics”) has also been studied and best approximates CT scan image interpretation in practice, but this approach adds complexity and requires further validation.83 The most advanced model and the only US Food and Drug Administration-cleared radiomic tool for lung nodule classification is the Lung Cancer Predictor Convolutional Neural Network. This model was trained in > 15,000 CT scans from the National Lung Screening Trial and validated in North America and Europe. Published studies reported improved discrimination between benign and malignant pulmonary nodules over available clinical prediction models, and this model is available commercially for clinical practice.78,84
Table 3.
Characteristics and Performance of Radiomic Models Predicting Malignancy in Pulmonary Nodules
| Publication | Training Population | Radiomics Technique | Feature Descriptions | External Validation |
Performance | Comments |
|---|---|---|---|---|---|---|
| Massion et al,78 2020 | NLST data set 14,761 CT scans (5,972 patients) with benign nodules, 932 CT scans (575 patients) with malignant nodules |
Deep learning- LCP-CNN, developed by Optellum | Unknown/not reported (Dense Convolutional Network) Clinical variables excluded from model |
Vanderbilt: incidentally detected pulmonary nodules: 52 benign, 64 malignant Oxford: incidentally detected pulmonary nodules: 400 benign, 63 malignant |
Internal validation: AUC = 0.921 vs Brock model, AUC = 0.856 External validation: Vanderbilt: LCP-CNN: AUC = 0.835 vs Mayo Clinic model AUC = 0.781 Oxford: LCP-CNN: AUC = 0.919 vs Mayo Clinic model AUC = 0.819 |
Optellum received FDA clearance for commercial use of its Virtual Nodule Clinic LCP-CNN showed superiority in net reclassification of benign and malignant nodules over Mayo Clinic model in both validation sets |
| Ardila et al,79 2019 | NLST data set 42,290 CT scans (14,851 patients) with and without nodules (70% used for training, 15% for tuning, 15% for testing); 638 malignant nodules |
Deep learning- Convolutional Neural Network, developed by Google Inc. | 1,024 learned features | Northwestern: screening CT scans: 1,139 CT scans (907 patients); 27 malignant nodules | Internal (training): AUC = 0.944 (Sens 83%, Spec 95%) External validation: AUC = 0.955 (Sens 84%, Spec 96%) |
Model showed improved performance in predicting malignancy in 1 year over retrospective Lung-RADS criteria when prior imaging was not available Similar performance to applied Lung-RADS when prior imaging was available |
| Peikert et al,80 2018 | NLST data set 318 benign nodules, 408 malignant nodules |
Conventional radiomic model (BRODERS) developed at the Mayo Clinic | Eight features selected by LASSO multivariate modeling from 57 features Notable variable categories include nodule location, nodule shape, nodule surface characteristics, and texture analysis |
Vanderbilt (Maldonado et al, 202181) incidentally detected pulmonary nodules: 79 benign nodules, 91 malignant nodules | Internal validation: BRODERS model AUC = 0.939 External validation: AUC = 0.900 (Sens 92%, Spec 62%) vs Brock model AUC = 0.870 |
Modest improvement over Brock model Brock model (clinical risk calculator developed on a screen-detected population) was applied to an external validation set with high rate of malignancy |
| Lv et al,82 2021 | NLST data set and Jinling Hospital (incidentally detected nodules) 1,078 benign nodules, 1,028 malignant nodules |
Deep learning- Filter-guided pyramid network | Unknown/not reported | Incidentally detected pulmonary nodules from three different hospitals 80 benign nodules, 261 malignant nodules |
External validation: AUC = 0.847 (Sens 85%, Spec 68%) | Performance of filter-guided pyramid network was similar to that of a panel of radiologists Established the utility of coupling deep learning strategies with human review for indeterminate pulmonary nodules |
AUC = area under the curve; BRODERS = Benign Versus Aggressive Nodule Evaluation using Radiomic Stratification; FDA = US Food and Drug Administration; LASSO = Least Absolute Shrinkage and Selection Operator; LCP CNN = Lung Cancer Predictor Convolutional Neural Network; NLST = National Lung Screening Trial.
Currently, the optimal clinical imaging-based biomarker to distinguish benign from malignant lung nodules across different CT scans and patient populations remains elusive, and this task may ultimately require the combination of imaging and non-imaging-based biomarkers.
Future Directions, Clinical Utility, and Biomarker Study Designs
In addition to showing improved accuracy compared with current stratification tools, a clinically useful biomarker must either: (1) reduce the time to diagnosis in patients with lung cancer without increasing the number of procedures in those with benign nodules or (2) reduce the number of procedures in patients with benign nodules without delaying the diagnosis in those with lung cancer.85 In the absence of studies documenting such outcomes, biomarkers for lung cancer are likely to remain underutilized.
Statistical methods such as the net reclassification index have been proposed to estimate the potential clinical usefulness of a biomarker. However, interpretation of the bias-adjusted net reclassification index86 is predicated on the assumption that a movement across threshold-guided decision groups will result in a change in clinical management, which may be flawed. To account for a more clinically relevant model of patient management, the intervention probability curve was recently developed. The intervention probability curve models the decision to perform a diagnostic procedure as a continuous function of the risk of disease and can therefore capture more granular and “real-world” changes in potential clinical usefulness.87
Although many biomarkers have been proposed as standalone diagnostic tools, it is more likely that the future of early detection will require a multifaceted approach combining different platforms to take advantage of their respective strengths. Many of the studies highlighted throughout this article have combined clinical information with one or more biomarkers such as blood-based biomarkers and radiomics.4 However, the use of advanced statistical modeling and multimodality biomarkers will have to be balanced against higher complexity, feasibility, and increased health care costs.
The uptake of biomarkers to guide the management of lung cancer will ultimately require well-designed randomized controlled trials, which are currently lacking despite the existence of multiple commercially available candidate biomarkers (Table 4). This is a problem for two reasons: (1) potentially impactful biomarkers are not currently used despite availability and (2) biomarkers without clinical benefit are nonetheless used by some clinicians. The dearth of clinical utility trials is likely due to their very high cost, large sample size requirement, prolonged accrual and follow-up time, need for multiple clinical sites, and the fact that the US Food and Drug Administration does not typically mandate randomized controlled clinical trials prior to commercialization of biomarkers. In addition, the pace of innovation might hinder the ability to generate clinically relevant data using traditional randomized controlled trial designs. Pragmatic approaches with more flexible and efficient designs that maximize patient accrual and optimize biomarker evaluation, such as cluster randomized clinical trials or platform trials, may represent an appealing potential alternative.
Table 4.
Selected Commercially Available Biomarkers for Lung Cancer in the United States
| Name | Assay/Methods | Assay Measurement/Variables Measured | Biomarker Source |
Company |
|---|---|---|---|---|
| Nodify XL2 | MRM-MS | Two proteins | Blood | Biodesix |
| Nodify CDT | ELISA | Seven autoantibodies | Blood | Biodesix |
| Percepta Genomic Sequencing Classifier | RNA sequencing | Multigene biomarker score | Bronchial epithelial brushing | Veracyte |
| Percepta Nasal Swab | RNA sequencing | Multigene biomarker score | Nasal epithelial brush | Veracyte |
| LCP CNN | Convolutional neural network | Radiomic signature | CT scan image | Optellum |
| REVEAL | MagArray | Protein panel | Blood | MagArray |
ELISA = enzyme-linked immunosorbent assay; LCP CNN = Lung Cancer Predictor Convolutional Neural Network; MRM-MS = multiple reaction monitoring mass spectrometry.
Summary
Biomarkers are urgently needed to reduce lung cancer mortality while also reducing unnecessary procedures, morbidity, and health care costs. Rapid technologic advances and large network collaborative efforts will continue to drive the discovery of many novel biomarkers. These should either improve risk stratification to enable participation in lung cancer screening or improve our ability to predict whether a detected nodule is benign or malignant. Ultimately, integrating biomarkers into patient care hinges on our ability to show clinical utility and improved patient-centered outcomes through rigorously designed clinical trials. Improving our ability to successfully implement and conduct randomized clinical trials in the field of biomarker research is a crucial step and an unmet need that is required to bring biomarkers into clinical practice.
Funding/Support
B. N. was supported by the Moorman-Simon Fellowship in Computational Biomedicine and the National Cancer Institute (NCI [2U01CA152751]). M. E. L. was supported by the NCI [2U01CA152751 and R01CA210360]. F. M. was supported by the NCI [R01 CA253923 and U01 CA196405]. W. T. I. was supported by a National Comprehensive Cancer Network Young Investigator Award. K. M. F. is supported by the National Health and Medical Research Council Medical Research Future Fund (Australia). S. D. and E. L. G. were supported by the NCI [5U01CA152662].
Financial/Nonfinancial Disclosures
The authors have reported to CHEST the following: M. E. L. is an inventor on several patents related to biomarkers for lung cancer detection that are owned by Boston University; and has received payment from Veracyte for consulting services. F. M. and T. P. are inventors on a patent related to radiomic biomarkers for lung cancer. W. T. I. reports consulting for Bristol Myers Squibb, Biodesix, Jazz Pharma, G1 Therapeutics, Mirati, Takeda, OncLive, Clinical Care Options, Chardan, Outcomes Insights, Cello Health, and Curio Science. M. N. K. has received payment from Meru Biotechnologies and Biodesix for consulting services and is an inventor on patents related to technology used for biomarker detection. None declared (R. P., N. T. T., S. S., B. E. H., M. L. B., B. N., C. M., L. Y., K. M. F., S. D., E. L. G.).
Acknowledgments
Role of sponsors: The sponsor had no role in the design of the study, the collection and analysis of the data, or the preparation of the manuscript.
Other contributions: The authors acknowledge their dear friend, mentor, and colleague Pierre P. Massion, who dedicated his career to the discovery of biomarkers to improve the care and quality of life of his patients, and who proposed an early version of this project.
References
- 1.Gould M.K., Tang T., Liu I.L., et al. Recent trends in the identification of incidental pulmonary nodules. Am J Respir Crit Care Med. 2015;192(10):1208–1214. doi: 10.1164/rccm.201505-0990OC. [DOI] [PubMed] [Google Scholar]
- 2.Tanner N.T., Porter A., Gould M.K., Li X.J., Vachani A., Silvestri G.A. Physician assessment of pretest probability of malignancy and adherence with guidelines for pulmonary nodule evaluation. Chest. 2017;152(2):263–270. doi: 10.1016/j.chest.2017.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lokhandwala T., Bittoni M.A., Dann R.A., et al. Costs of diagnostic assessment for lung cancer: a Medicare claims analysis. Clin Lung Cancer. 2017;18(1):e27–e34. doi: 10.1016/j.cllc.2016.07.006. [DOI] [PubMed] [Google Scholar]
- 4.Kammer M.N., Lakhani D.A., Balar A.B., et al. Integrated biomarkers for the management of indeterminate pulmonary nodules. Am J Respir Crit Care Med. 2021;204(11):1306–1316. doi: 10.1164/rccm.202012-4438OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang D., Zhang X., Powell C.A., et al. Probability of cancer in high-risk patients predicted by the protein-based lung cancer biomarker panel in China: LCBP study. Cancer. 2018;124(2):262–270. doi: 10.1002/cncr.31020. [DOI] [PubMed] [Google Scholar]
- 6.Marmor H.N., Jackson L., Gawel S., et al. Improving malignancy risk prediction of indeterminate pulmonary nodules with imaging features and biomarkers. Clin Chim Acta. 2022;534:106–114. doi: 10.1016/j.cca.2022.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vachani A., Lam S., Massion P.P., et al. Development and validation of a risk assessment model for pulmonary nodules using plasma proteins and clinical factors. Chest. 2023;163(4):966–976. doi: 10.1016/j.chest.2022.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ostrin E.J., Bantis L.E., Wilson D.O., et al. Contribution of a blood-based protein biomarker panel to the classification of indeterminate pulmonary nodules. J Thorac Oncol. 2021;16(2):228–236. doi: 10.1016/j.jtho.2020.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guida F., Sun N., Bantis L.E., et al. Integrative Analysis of Lung Cancer Etiology and Risk (INTEGRAL) Consortium for Early Detection of Lung Cancer, Assessment of lung cancer risk on the basis of a biomarker panel of circulating proteins. JAMA Oncol. 2018;4(10) doi: 10.1001/jamaoncol.2018.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farlow E.C., Patel K., Basu S., et al. Development of a multiplexed tumor-associated autoantibody-based blood test for the detection of non-small cell lung cancer. Clin Cancer Res. 2010;16(13):3452–3462. doi: 10.1158/1078-0432.CCR-09-3192. [DOI] [PubMed] [Google Scholar]
- 11.Ostroff R.M., Bigbee W.L., Franklin W., et al. Unlocking biomarker discovery: large scale application of aptamer proteomic technology for early detection of lung cancer. PloS One. 2010;5(12) doi: 10.1371/journal.pone.0015003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kupert E., Anderson M., Liu Y., et al. Plasma secretory phospholipase A2-IIa as a potential biomarker for lung cancer in patients with solitary pulmonary nodules. BMC Cancer. 2011;11:513. doi: 10.1186/1471-2407-11-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patz E., Campa M., Gottlin E., et al. Biomarkers to help guide management of patients with pulmonary nodules. Am J Respir Crit Care Med. 2013;188(4):461–465. doi: 10.1164/rccm.201210-1760OC. [DOI] [PubMed] [Google Scholar]
- 14.Daly S., Rinewalt D., Fhied C., et al. Development and validation of a plasma biomarker panel for discerning clinical significance of indeterminate pulmonary nodules. J Thorac Oncol. 2013;8(1):31–36. doi: 10.1097/JTO.0b013e31827627f8. [DOI] [PubMed] [Google Scholar]
- 15.Okamura K., Takayama K., Izumi M., Harada T., Furuyama K., Nakanishi Y. Diagnostic value of CEA and CYFRA 21-1 tumor markers in primary lung cancer. Lung Cancer. 2013;80(1):45–49. doi: 10.1016/j.lungcan.2013.01.002. [DOI] [PubMed] [Google Scholar]
- 16.Fahrmann J., Grapov D., DeFelice B., et al. Serum phosphatidylethanolamine levels distinguish benign from malignant solitary pulmonary nodules and represent a potential diagnostic biomarker for lung cancer. Cancer Biomarkers. 2016;16(4):609–617. doi: 10.3233/CBM-160602. [DOI] [PubMed] [Google Scholar]
- 17.Massion P.P., Healey G.F., Peek L.J., et al. Autoantibody signature enhances the positive predictive power of computed tomography and nodule-based risk models for detection of lung cancer. J Thorac Oncol. 2017;12(3):578–584. doi: 10.1016/j.jtho.2016.08.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ajona D., Okrój M., Pajares M.J., et al. Complement C4d-specific antibodies for the diagnosis of lung cancer. Oncotarget. 2018;9(5):6346–6355. doi: 10.18632/oncotarget.23690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Du Q., Yu R., Wang H., et al. Significance of tumor-associated autoantibodies in the early diagnosis of lung cancer. Clin Respir J. 2018;12(6):2020–2028. doi: 10.1111/crj.12769. [DOI] [PubMed] [Google Scholar]
- 20.Lastwika K.J., Kargl J., Zhang Y., et al. Tumor-derived autoantibodies identify malignant pulmonary nodules. Am J Respir Crit Care Med. 2019;199(10):1257–1266. doi: 10.1164/rccm.201804-0628OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanner N.T., Springmeyer S.C., Porter A., et al. Assessment of integrated classifier’s ability to distinguish benign from malignant lung nodules: extended analyses and 2-year follow-up results of the PANOPTIC (Pulmonary Nodule Plasma Proteomic Classifier) trial. Chest. 2021;159(3):1283–1287. doi: 10.1016/j.chest.2020.10.069. [DOI] [PubMed] [Google Scholar]
- 22.Marmor H.N., Deppen S.A., Welty V., et al. Improving lung cancer diagnosis with computed tomography radiomics and serum Histoplasmosis testing. Cancer Epidemiol Biomarkers Prev. 2023;32(3):329–336. doi: 10.1158/1055-9965.EPI-22-0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.National Institutes of Health Clinical Center. Nodify XL2 classifier clinical utility study in low to moderate risk lung nodules (ALTITUDE). NCT04171492. ClinicalTrials.gov. National Insitutes of Health; 2020. Updated November 12, 2022. Accessed June 25, 2023. https://www.clinicaltrials.gov/study/NCT04171492
- 24.Barger J.F., Rahman M.A., Jackson D., Acunzo M., Nana-Sinkam S.P. Extracellular miRNAs as biomarkers in cancer. Food Chem Toxicol. 2016;98(pt A):66–72. doi: 10.1016/j.fct.2016.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu W., Zhou K., Zha Y., et al. Diagnostic value of serum miR-182, miR-183, miR-210, and miR-126 levels in patients with early-stage non-small cell lung cancer. PLoS One. 2016;11(4) doi: 10.1371/journal.pone.0153046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leng Q., Lin Y., Jiang F., et al. A plasma miRNA signature for lung cancer early detection. Oncotarget. 2017;8(67):111902–111911. doi: 10.18632/oncotarget.22950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dou Y., Zhu Y., Ai J., et al. Plasma small ncRNA pair panels as novel biomarkers for early-stage lung adenocarcinoma screening. BMC Genomics. 2018;19(1):545. doi: 10.1186/s12864-018-4862-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ying L., Du L., Zou R., et al. Development of a serum miRNA panel for detection of early stage non-small cell lung cancer. Proc Natl Acad Sci U S A. 2020;117(40):25036–25042. doi: 10.1073/pnas.2006212117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marquette C.H., Boutros J., Benzaquen J., et al. Circulating tumour cells as a potential biomarker for lung cancer screening: a prospective cohort study. Lancet Respir Med. 2020;8(7):709–716. doi: 10.1016/S2213-2600(20)30081-3. [DOI] [PubMed] [Google Scholar]
- 30.Li Y., Tian X., Gao L., et al. Clinical significance of circulating tumor cells and tumor markers in the diagnosis of lung cancer. Cancer Med. 2019;8(8):3782–3792. doi: 10.1002/cam4.2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mathios D., Johansen J.S., Cristiano S., et al. Detection and characterization of lung cancer using cell-free DNA fragmentomes. Nat Commun. 2021;12(1):5060. doi: 10.1038/s41467-021-24994-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leung M., Freidin M.B., Freydina D.V., et al. Blood-based circulating tumor DNA mutations as a diagnostic and prognostic biomarker for lung cancer. Cancer. 2020;126(8):1804–1809. doi: 10.1002/cncr.32699. [DOI] [PubMed] [Google Scholar]
- 33.Kneip C., Schmidt B., Seegebarth A., et al. SHOX2 DNA methylation is a biomarker for the diagnosis of lung cancer in plasma. J Thorac Oncol. 2011;6(10):1632–1638. doi: 10.1097/JTO.0b013e318220ef9a. [DOI] [PubMed] [Google Scholar]
- 34.Hulbert A., Jusue-Torres I., Stark A., et al. Early detection of lung cancer using DNA promoter hypermethylation in plasma and sputum. Clin Cancer Res. 2017;23(8):1998–2005. doi: 10.1158/1078-0432.CCR-16-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ooki A., Maleki Z., Tsay J.J., et al. A panel of novel detection and prognostic methylated DNA markers in primary non-small cell lung cancer and serum DNA. Clin Cancer Res. 2017;23(22):7141–7152. doi: 10.1158/1078-0432.CCR-17-1222. [DOI] [PubMed] [Google Scholar]
- 36.Billatos E., Vick J.L., Lenburg M.E., Spira A.E. The airway transcriptome as a biomarker for early lung cancer detection. Clin Cancer Res. 2018;24(13):2984–2992. doi: 10.1158/1078-0432.CCR-16-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hackett N.R., Heguy A., Harvey B.G., et al. Variability of antioxidant-related gene expression in the airway epithelium of cigarette smokers. Am J Respir Cell Mol Biol. 2003;29(3 pt 1):331–343. doi: 10.1165/rcmb.2002-0321OC. [DOI] [PubMed] [Google Scholar]
- 38.Spira A., Beane J., Shah V., et al. Effects of cigarette smoke on the human airway epithelial cell transcriptome. Proc Natl Acad Sci U S A. 2004;101(27):10143–10148. doi: 10.1073/pnas.0401422101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nelson H.H., Christiani D.C., Mark E.J., Wiencke J.K., Wain J.C., Kelsey K.T. Implications and prognostic value of K-ras mutation for early-stage lung cancer in women. J Natl Cancer Inst. 1999;91(23):2032–2038. doi: 10.1093/jnci/91.23.2032. [DOI] [PubMed] [Google Scholar]
- 40.Ost D.E., Ernst A., Lei X., et al. Diagnostic yield and complications of bronchoscopy for peripheral lung lesions. Results of the AQuIRE Registry. Am J Respir Crit Care Med. 2016;193(1):68–77. doi: 10.1164/rccm.201507-1332OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blomquist T., Crawford E.L., Mullins D., et al. Pattern of antioxidant and DNA repair gene expression in normal airway epithelium associated with lung cancer diagnosis. Cancer Res. 2009;69(22):8629–8635. doi: 10.1158/0008-5472.CAN-09-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.National Institutes of Health Clinical Center. Validation of a multi-gene test for lung cancer risk. NCT01130285. National Institutes of Health; 2011. Updated October 19, 2020. Accessed June 25, 2023. https://www.clinicaltrials.gov/study/NCT01130285
- 43.Crawford E.L., Levin A., Safi F., et al. Lung cancer risk test trial: study design, participant baseline characteristics, bronchoscopy safety, and establishment of a biospecimen repository. BMC Pulm Med. 2016;16:16. doi: 10.1186/s12890-016-0178-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spira A., Beane J.E., Shah V., et al. Airway epithelial gene expression in the diagnostic evaluation of smokers with suspect lung cancer. Nat Med. 2007;13(3):361–366. doi: 10.1038/nm1556. [DOI] [PubMed] [Google Scholar]
- 45.Whitney D.H., Elashoff M.R., Porta-Smith K., et al. Derivation of a bronchial genomic classifier for lung cancer in a prospective study of patients undergoing diagnostic bronchoscopy. BMC Med Genomics. 2015;8:18. doi: 10.1186/s12920-015-0091-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Silvestri G.A., Vachani A., Whitney D., et al. A bronchial genomic classifier for the diagnostic evaluation of lung cancer. N Engl J Med. 2015;373(3):243–251. doi: 10.1056/NEJMoa1504601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee H.J., Mazzone P., Feller-Kopman D., et al. Impact of the Percepta Genomic Classifier on clinical management decisions in a multicenter prospective study. Chest. 2021;159(1):401–412. doi: 10.1016/j.chest.2020.07.067. [DOI] [PubMed] [Google Scholar]
- 48.Zhang X., Sebastiani P., Liu G., et al. Similarities and differences between smoking-related gene expression in nasal and bronchial epithelium. Physiol Genomics. 2010;41(1):1–8. doi: 10.1152/physiolgenomics.00167.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boudewijn I.M., Faiz A., Steiling K., et al. Nasal gene expression differentiates COPD from controls and overlaps bronchial gene expression. Respir Res. 2017;18(1):213. doi: 10.1186/s12931-017-0696-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sala M.A., Balderas-Martinez Y.I., Buendia-Roldan I., et al. Inflammatory pathways are upregulated in the nasal epithelium in patients with idiopathic pulmonary fibrosis. Respir Res. 2018;19(1):233. doi: 10.1186/s12931-018-0932-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.AEGIS Study Team Shared gene expression alterations in nasal and bronchial epithelium for lung cancer detection. J Natl Cancer Inst. 2017;109(7):djw327. doi: 10.1093/jnci/djw327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mazzone P.J., Lamb C., Rieger-Christ K.M., et al. Early candidate nasal swab classifiers developed using machine learning and whole transcriptome sequencing may improve early lung cancer detection. J Clin Oncol. 2021;39(suppl 15):8551. [Google Scholar]
- 53.Lamb C.R., Rieger-Christ K., Reddy C., et al. A nasal genomic classifier for assessing risk of malignancy in lung nodules demonstrates similar performance in patients that meet screening criteria for high baseline risk and those who do not. B30 The Quest for the Holy Grail: Modeling and Biomarkers for Nodules and Lung Cancer. Am J Respir Crit Care Med. 2022;205:A5585. [Google Scholar]
- 54.Nikolaidis G., Raji O.Y., Markopoulou S., et al. DNA methylation biomarkers offer improved diagnostic efficiency in lung cancer. Cancer Res. 2012;72(22):5692–5701. doi: 10.1158/0008-5472.CAN-12-2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pavel A.B., Campbell J.D., Liu G., et al. Alterations in bronchial airway miRNA expression for lung cancer detection. Cancer Prev Res (Phila) 2017;10(11):651–659. doi: 10.1158/1940-6207.CAPR-17-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pauling L., Robinson A.B., Teranishi R., Cary P. Quantitative analysis of urine vapor and breath by gas-liquid partition chromatography. Proc Natl Acad Sci U S A. 1971;68(10):2374–2376. doi: 10.1073/pnas.68.10.2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang Y., Hu Y., Wang D., et al. The analysis of volatile organic compounds biomarkers for lung cancer in exhaled breath, tissues and cell lines. Cancer Biomark. 2012;11(4):129–137. doi: 10.3233/CBM-2012-00270. [DOI] [PubMed] [Google Scholar]
- 58.Phillips M., Bauer T.L., Cataneo R.N., et al. Blinded validation of breath biomarkers of lung cancer, a potential ancillary to chest CT screening. PLoS One. 2015;10(12) doi: 10.1371/journal.pone.0142484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Callol-Sanchez L., Munoz-Lucas M.A., Gomez-Martin O., et al. Observation of nonanoic acid and aldehydes in exhaled breath of patients with lung cancer. J Breath Res. 2017;11(2) doi: 10.1088/1752-7163/aa6485. [DOI] [PubMed] [Google Scholar]
- 60.Phillips M., Bauer T.L., Pass H.I. A volatile biomarker in breath predicts lung cancer and pulmonary nodules. J Breath Res. 2019;13(3) doi: 10.1088/1752-7163/ab21aa. [DOI] [PubMed] [Google Scholar]
- 61.Smirnova E., Mallow C., Muschelli J., et al. Predictive performance of selected breath volatile organic carbon compounds in stage 1 lung cancer. Transl Lung Cancer Res. 2022;11(6):1009–1018. doi: 10.21037/tlcr-21-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zou Y., Wang Y., Jiang Z., et al. Breath profile as composite biomarkers for lung cancer diagnosis. Lung Cancer. 2021;154:206–213. doi: 10.1016/j.lungcan.2021.01.020. [DOI] [PubMed] [Google Scholar]
- 63.Koureas M., Kalompatsios D., Amoutzias G.D., Hadjichristodoulou C., Gourgoulianis K., Tsakalof A. Comparison of targeted and untargeted approaches in breath analysis for the discrimination of lung cancer from benign pulmonary diseases and healthy persons. Molecules. 2021;26(9) doi: 10.3390/molecules26092609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tsou P.H., Lin Z.L., Pan Y.C., et al. Exploring volatile organic compounds in breath for high-accuracy prediction of lung cancer. Cancers (Basel) 2021;13(6) doi: 10.3390/cancers13061431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rudnicka J., Kowalkowski T., Buszewski B. Searching for selected VOCs in human breath samples as potential markers of lung cancer. Lung Cancer. 2019;135:123–129. doi: 10.1016/j.lungcan.2019.02.012. [DOI] [PubMed] [Google Scholar]
- 66.Chen X., Muhammad K.G., Madeeha C., et al. Calculated indices of volatile organic compounds (VOCs) in exhalation for lung cancer screening and early detection. Lung Cancer. 2021;154:197–205. doi: 10.1016/j.lungcan.2021.02.006. [DOI] [PubMed] [Google Scholar]
- 67.Liu L., Li W., He Z., et al. Detection of lung cancer with electronic nose using a novel ensemble learning framework. J Breath Res. 2021;15(2) doi: 10.1088/1752-7163/abe5c9. [DOI] [PubMed] [Google Scholar]
- 68.Krauss E., Haberer J., Barreto G., Degen M., Seeger W., Guenther A. Recognition of breathprints of lung cancer and chronic obstructive pulmonary disease using the Aeonose® electronic nose. J Breath Res. 2020;14(4) doi: 10.1088/1752-7163/ab8c50. [DOI] [PubMed] [Google Scholar]
- 69.Shlomi D., Abud M., Liran O., et al. Detection of lung cancer and EGFR mutation by electronic nose system. J Thorac Oncol. 2017;12(10):1544–1551. doi: 10.1016/j.jtho.2017.06.073. [DOI] [PubMed] [Google Scholar]
- 70.McWilliams A., Beigi P., Srinidhi A., Lam S., MacAulay C.E. Sex and smoking status effects on the early detection of early lung cancer in high-risk smokers using an electronic nose. IEEE Trans Biomed Eng. 2015;62(8):2044–2054. doi: 10.1109/TBME.2015.2409092. [DOI] [PubMed] [Google Scholar]
- 71.Gasparri R., Santonico M., Valentini C., et al. Volatile signature for the early diagnosis of lung cancer. J Breath Res. 2016;10(1) doi: 10.1088/1752-7155/10/1/016007. [DOI] [PubMed] [Google Scholar]
- 72.Kort S., Tiggeloven M.M., Brusse-Keizer M., et al. Multi-centre prospective study on diagnosing subtypes of lung cancer by exhaled-breath analysis. Lung Cancer. 2018;125:223–229. doi: 10.1016/j.lungcan.2018.09.022. [DOI] [PubMed] [Google Scholar]
- 73.van de Goor R., van Hooren M., Dingemans A.M., Kremer B., Kross K. Training and validating a portable electronic nose for lung cancer screening. J Thorac Oncol. 2018;13(5):676–681. doi: 10.1016/j.jtho.2018.01.024. [DOI] [PubMed] [Google Scholar]
- 74.Cai X., Chen L., Kang T., et al. A prediction model with a combination of variables for diagnosis of lung cancer. Med Sci Monit. 2017;23:5620–5629. doi: 10.12659/MSM.904738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li W., Jia Z., Xie D., Chen K., Cui J., Liu H. Recognizing lung cancer using a homemade e-nose: a comprehensive study. Comput Biol Med. 2020;120 doi: 10.1016/j.compbiomed.2020.103706. [DOI] [PubMed] [Google Scholar]
- 76.Binson V.A., Subramoniam M., Mathew L. Discrimination of COPD and lung cancer from controls through breath analysis using a self-developed e-nose. J Breath Res. 2021;15(4) doi: 10.1088/1752-7163/ac1326. [DOI] [PubMed] [Google Scholar]
- 77.Kort S., Brusse-Keizer M., Schouwink H., et al. Diagnosing non-small cell lung cancer by exhaled breath profiling using an electronic nose: a multicenter validation study. Chest. 2023;163(3):697–706. doi: 10.1016/j.chest.2022.09.042. [DOI] [PubMed] [Google Scholar]
- 78.Massion P.P., Antic S., Ather S., et al. Assessing the accuracy of a deep learning method to risk stratify indeterminate pulmonary nodules. Am J Respir Crit Care Med. 2020;202(2):241–249. doi: 10.1164/rccm.201903-0505OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ardila D., Kiraly A.P., Bharadwaj S., et al. End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography. Nat Med. 2019;25(6):954–961. doi: 10.1038/s41591-019-0447-x. [DOI] [PubMed] [Google Scholar]
- 80.Peikert T., Duan F., Rajagopalan S., et al. Novel high-resolution computed tomography-based radiomic classifier for screen-identified pulmonary nodules in the National Lung Screening Trial. PLoS One. 2018;13(5) doi: 10.1371/journal.pone.0196910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Maldonado F., Varghese C., Rajagopalan S., et al. Validation of the BRODERS classifier (Benign versus aggRessive nODule Evaluation using Radiomic Stratification), a novel HRCT-based radiomic classifier for indeterminate pulmonary nodules. Eur Respir J. 2021;57(4) doi: 10.1183/13993003.02485-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lv W., Wang Y., Zhou C., et al. Development and validation of a clinically applicable deep learning strategy (HONORS) for pulmonary nodule classification at CT: a retrospective multicentre study. Lung Cancer. 2021;155:78–86. doi: 10.1016/j.lungcan.2021.03.008. [DOI] [PubMed] [Google Scholar]
- 83.Cherezov D., Hawkins S.H., Goldgof D.B., et al. Delta radiomic features improve prediction for lung cancer incidence: a nested case-control analysis of the National Lung Screening Trial. Cancer Med. 2018;7(12):6340–6356. doi: 10.1002/cam4.1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kim R.Y., Oke J.L., Pickup L.C., et al. Artificial intelligence tool for assessment of indeterminate pulmonary nodules detected with CT. Radiology. 2022;304:683–691. doi: 10.1148/radiol.212182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mazzone P.J., Sears C.R., Arenberg D.A., et al. Evaluating molecular biomarkers for the early detection of lung cancer: when is a biomarker ready for clinical use? An official American Thoracic Society Policy Statement. Am J Respir Crit Care Med. 2017;196(7):e15–e29. doi: 10.1164/rccm.201708-1678ST. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Paynter N.P., Cook N.R. A bias-corrected net reclassification improvement for clinical subgroups. Med Decis Making. 2013;33(2):154–162. doi: 10.1177/0272989X12461856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kammer M.N., Rowe D.J., Deppen S.A., et al. The intervention probability curve: modeling the practical application of threshold-guided decision making, evaluated in lung, prostate, and ovarian cancers. Cancer Epidemiol Biomarkers Prev. 2022;31(9):1752–1759. doi: 10.1158/1055-9965.EPI-22-0190. [DOI] [PMC free article] [PubMed] [Google Scholar]