Abstract
The impact of preozonation and filter contact time (depth) on microbial communities was examined in drinking water biofilters treating Ohio River water which had undergone conventional treatment (coagulation, flocculation, sedimentation) or solutions of natural organic matter isolated from groundwater (both ozonated and nonozonated). With respect to filter depth, compared to filters treating nonozonated waters, preozonation of treated water led to greater differences in community phospholipid fatty acid (PLFA) profiles, utilization of sole carbon sources (Biolog), and arbitrarily primed PCR fingerprints. PLFA profiles indicated that there was a shift toward anaerobic bacteria in the communities found in the filter treating ozonated water compared to the communities found in the filter treating nonozonated settled water, which had a greater abundance of eukaryotic markers.
The presence of natural organic matter (NOM) in drinking water leads to conditions that promote the regrowth of microorganisms in distribution systems. In addition, NOM reacts with disinfectants to form disinfection by-products which present health concerns. Biofiltration can remove much of the biodegradable NOM, which results in decreases in the regrowth potential of water and in significant reductions in the formation of disinfection by-products. Recent studies of microbial biomass present in drinking water biofilters have indicated that there is a marked decrease in biomass with increasing filter depth (and corresponding contact time) (11, 17). This change occurs concomitantly with the utilization of NOM and its constituents, such as the surrogate parameters dissolved organic carbon (DOC) and assimilable organic carbon (8, 11). Preozonation of the NOM has been shown to increase the concentration of assimilable compounds, which leads to higher biomass contents in filters (15, 17). These differences in the nature and quantity of substrates present in biofilters, as well as in filter operational conditions, may cause shifts in the microbial communities present in biofilters and in the substrate utilization capabilities of biofilters. The objective of this study was to determine microbial community structure depth profiles in drinking water biofilters by using phospholipid fatty acid (PLFA) analysis, arbitrarily primed PCR (AP-PCR) fingerprinting, and carbon source utilization pattern assessment (Biolog, Inc., Hayward, Calif.). Biofilters treating settled Ohio River water (ORW) and ozonated and nonozonated NOM isolated from groundwater were evaluated.
Filter operation and sampling.
A full-scale rapid sand filter that had been in operation for several years at a local drinking water treatment plant (Cincinnati Water Works Richard Miller Plant) was studied. The filter received conventionally treated (coagulation, flocculation, sedimentation), nondisinfected ORW at a DOC concentration of 2.1 mg/liter and a hydraulic loading rate of 6.1 m/h and was backwashed as needed with disinfectant-free water, which rendered the filter biologically active. Two bench scale filters that were fed solutions of ozonated or nonozonated NOM isolated from groundwater by using an anion-exchange resin were also studied. The filters were seeded by recirculating raw ORW through them for 2 weeks. The filters were then acclimated with the NOM solutions in single-pass mode at a hydraulic loading rate of 5.0 m/h for 3 to 5 months before sampling. The nonozonated NOM solution had a DOC concentration of 4.0 mg/liter, and the ozonated solution, which was ozonated at a dose of 0.35 mg of O3/mg of DOC, had a DOC concentration of 3.5 mg/liter. The bench scale filters were operated without backwashing.
Sand samples were taken from the top, middle, and bottom 2 cm of a 3- by 75-cm vertical core extracted from the full-scale filter immediately prior to backwashing and from the segmented bench scale sand filters packed in three glass chromatography columns (2.54 by 30 cm; Ace Glass, Louisville, Ky.).
Community structure and substrate utilization analyses.
Samples for PLFA and Biolog analyses were taken from the filters, homogenized, and subsampled in duplicate to assess sampling and analytical error. Samples for AP-PCR analysis were analyzed without replication.
PLFA profiles were determined with 10-g (wet weight) sand samples by the method of Guckert et al. (7). Solvent-only controls were treated exactly like samples were treated. The lipids were fractionated by silicic acid column chromatography (13), and polar lipid fatty acids were released as fatty acid methyl esters by mild alkaline transesterification. Nonadecanoic acid methyl ester was used as an internal standard to determine recovery efficiency. PLFAs were quantified and identified with a Hewlett-Packard model 5890 gas chromatograph equipped with a flame ionization detector linked to an HP 5972 series MS detector (14) by Microbial Insights, Inc., Knoxville, Tenn. The relative abundance of each individual fatty acid was expressed as a percentage of the total molar concentration of fatty acids in each sample.
AP-PCR was used to profile the total-community DNA isolated from 1-g (wet weight) sand samples. The DNA was isolated by the method of Tsai and Olson (12) and was purified with a Geneclean II kit (Bio 101 Inc., La Jolla, Calif.). Amplification was performed by using an eight-nucleotide DNA oligomer [86-D; d(GTAACGCC)] for random priming of the extracted DNA (3), a thermostable DNA polymerase, and a programmable thermocycler (M.J. Research, Watertown, Mass.). Each PCR mixture consisted of 65 μl of sterile water, 10.5 μl of 10× Stoeffel buffer, 10 μl of 25 mM MgCl2, 8 μl of a deoxynucleoside triphosphate mixture, 5 μl of primer, 2 μl of template, and 1 μl of the Stoeffel fragment of Taq polymerase (Perkin-Elmer, Foster City, Calif.). The temperature program used was as follows: four cycles consisting of 94°C for 5 min, 36°C for 5 min, and 72°C for 5 min, followed by 30 cycles consisting of 94°C for 1 min, 36°C for 1 min, and 72°C for 2 min, and then one cycle consisting of 72°C for 10 min. As contamination can be a problem, controls containing no template DNA were routinely run. The AP-PCR amplification products were resolved by electrophoresis on 2% Metaphor agarose (FMC Bioproducts, Rockland, Maine) gels. The molecular weights of the PCR products were determined by comparing the positions of the bands to the positions of a series of DNA molecular weight markers (123 bp). Similarity indices were calculated based on the number of bands shared between samples relative to the total number of bands observed for each sample (10).
Carbon source utilization patterns (Biolog) were determined with 20- to 30-g (wet weight) samples. All glassware and solutions were sterilized by autoclaving. The microbial community attached to the sand medium was removed by shaking the samples for 30 min in 100 ml of a 0.3% sodium pyrophosphate solution in a 250-ml plastic bottle, followed by sonication of the suspension in a bath sonicator (model FS3; Fisher Scientific, Pittsburgh, Pa.) for 30 s. The supernatant liquid was decanted into a 250-ml centrifuge bottle, and the procedure was repeated. Negative controls were prepared by processing sterile bottles with 0.3% sodium pyrophosphate by using identical procedures. The two supernatant fractions were pooled and centrifuged for 10 min at 10,000 × g to concentrate the cells. The cells were diluted with sterile saline (0.85% NaCl) to a concentration of approximately 3.0 × 108 cells/ml, as determined by comparing the solution transmittance to the transmittance of high and low turbidity standards (Biolog). Diluted cell suspensions were inoculated into Biolog GN microplates (Biolog) according to the manufacturer’s instructions. Color development was quantified by determining absorbance at 590 nm with a microplate reader after 48 h of incubation at room temperature (22 to 25°C). A control well-corrected absorbance of 0.25 was used as a threshold for defining a positive reaction for all substrates.
Statistical analyses.
The relationships among the PLFA profiles of the biofilters were assessed by using the principal components analysis (PCA) option of the Factor module in SYSTAT (SYSTAT version 5.03 for Windows; Systat, Inc., Evanston, Ill.). The data used for PCA were row centered and standardized to give a constant variance. Component loadings are the covariances of the original variables with the components, and the signs of the loadings are arbitrary. Component loadings quantify the relationship between the original variables and the components.
Community PLFA profiles.
PLFA profile analysis identified 90 fatty acids obtained from the biofilters treating settled ORW and the ozonated NOM solution. The individual fatty acids ranged in abundance from undetectable levels to 28% of the total fatty acids detected in a sample. The median coefficient of variation of the average mole percent abundance values for all fatty acids in all samples was 0.09. The 29 most abundant fatty acids, which accounted for at least 89% of the total PLFAs present in each sample, were assigned to functional groups of microorganisms (4–6, 16), and their mole percent abundance values were compared by PCA. Ester-linked fatty acids are essential cell constituents of members of the Bacteria and Eucarya but not of members of the Archaea, so members of the Archaea are not represented in PLFA analyses. A study of drinking water distribution system populations performed with rRNA probes, however, indicated that members of the Archaea may not make significant contributions to planktonic and biofilm drinking water microbial communities (9). Fatty acids characteristic of prokaryotes (branched chain, odd chain, and cyclopropyl derivatives) accounted for 70 to 78% of all PLFAs in each of the samples. Of the prokaryotic markers, 76 to 81% were contributed by aerobic eubacteria, and 10 to 13% were contributed by gram-positive anaerobic bacteria. Polyenoics, which are indicative of microeukaryotes, accounted for 6 to 10% of all PLFAs in the filters and were evenly divided between markers for photoautotrophs and heterotrophs.
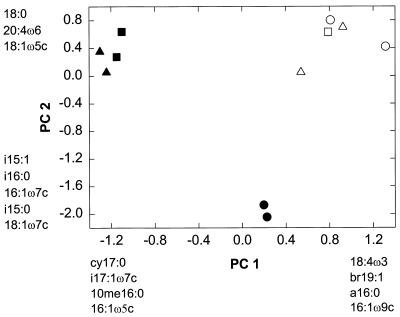
PCA revealed differences in the microbial communities present in the biofilters treating different water sources and with filter depth (Fig. 1). Principal components 1 and 2 explained 41 and 34% of the total variance, respectively. Component 1 differentiated samples taken from the filter treating ORW from samples from the filter treating the ozonated NOM solution. PLFAs carrying negative loadings with respect to component 1 (cy17:0, i17:1ω7, 10me16:0, and i17:0), which were indicative of sulfate-reducing bacteria and other anaerobes, were present in greater abundance in the bottom and middle of the filter treating ozonated NOM. The data for polyenoic fatty acids, including 18:2ω6, 18:4ω3, and 20:5ω3, which carried positive loadings on component 1, indicated that eukaryotes were present in greater abundance in the filter treating settled ORW. Component 2 separated samples taken from the top of the filter treating ozonated NOM from samples taken from the middle and bottom of that filter and from all depths of the filter treating ORW. PLFAs characteristic of gram-positive bacteria (i15:0 and i16:0) and aerobic eubacteria (16:1ω7c and 18:1ω7c), which carried high negative loadings on component 2, were enriched at the top of the filter treating ozonated NOM.
FIG. 1.
PCA of PLFA profiles for microbial communities from drinking water biofilters. Symbols: ○, □, and ▵, top, middle, and bottom, respectively, of a filter treating settled ORW; •, ▪, and ▴, top, middle, and bottom, respectively, of a filter treating ozonated NOM. Fatty acids with high loadings on each principal component (PC) are indicated next to each axis.
Community sole carbon source utilization patterns.
After 48 h of incubation, the control well-corrected absorbance values for the substrate-containing wells ranged from 0 to 2. The median coefficient of variation of the average absorbance values for all substrates in all samples was 0.40, with most of the variation attributed to values below the threshold value. The communities from all levels of the biofilter treating the nonozonated NOM solution used more compounds as sole carbon sources than the communities from all levels of the biofilter treating ozonated NOM (Table 1). The total number of carbon sources utilized by the biofilter communities decreased as a function of filter depth, regardless of the type of influent NOM. Although the substrate utilization diversity changed for the different communities, the specific compounds used as sole carbon sources did not shift drastically as a function of filter depth or type of water treated (Table 1).
TABLE 1.
Sole carbon source utilization by drinking water biofilter communities
| Compound | Sole carbon source utilization by filter microbial communities treating:
|
Compound | Sole carbon source utilization by filter microbial communities treating:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ozonated NOM
|
Nonozonated NOM
|
Ozonated NOM
|
Nonozonated NOM
|
|||||||||||
| 0 cma | 40 cm | 90 cm | 0 cm | 40 cm | 90 cm | 0 cm | 40 cm | 90 cm | 0 cm | 40 cm | 90 cm | |||
| Polymers | α-Keto valeric acid | − | − | − | − | − | − | |||||||
| α-Cyclodextrin | − | − | − | − | − | − | dl-Lactic acid | + | + | + | + | + | + | |
| α-Keto glutaric acid | + | + | − | + | + | + | Malonic acid | + | − | − | − | − | − | |
| Dextrin | − | − | − | + | + | − | Propionic acid | + | + | − | + | + | + | |
| Glycogen | − | − | − | + | + | − | Quinic acid | + | − | − | + | + | + | |
| Tween 40 | + | + | + | + | + | + | Quinic acid | + | − | − | + | + | + | |
| Tween 80 | + | + | + | + | + | + | d-Saccharic acid | + | − | − | + | + | + | |
| Sebacic acid | + | − | − | − | + | + | ||||||||
| Carbohydrates | Succinic acid | + | + | + | + | + | + | |||||||
| N-Acetyl-d-galactosamine | − | − | − | − | + | + | ||||||||
| N-Acetyl-d-glucosamine | − | − | − | + | + | − | Bromosuccinic acid | + | + | + | − | + | + | |
| Adonitol | − | − | − | − | − | − | ||||||||
| l-Arabinose | − | − | − | + | + | + | Amides | |||||||
| d-Arabitol | + | + | − | + | − | − | Succinamic acid | − | − | − | + | − | − | |
| Cellobiose | + | − | − | + | + | − | Glucuronamide | − | − | − | − | − | − | |
| i-Erythritol | − | − | − | − | − | − | Alaninamide | − | − | − | − | − | − | |
| d-Fructose | − | − | − | + | + | − | d-Alanine | − | − | − | + | + | − | |
| l-Fucose | − | − | − | + | − | − | l-Alanine | + | − | − | + | + | + | |
| d-Galactose | − | − | − | + | + | − | l-Alanyl-glycine | − | − | − | + | + | + | |
| Gentiobiose | − | − | − | − | − | − | l-Asparagine | + | + | − | + | + | + | |
| α-d-Glucose | + | − | − | + | + | + | l-Glutamic acid | + | + | + | + | + | + | |
| m-Inositol | + | − | − | − | − | − | Glycyl-l-aspartic acid | − | − | − | − | − | − | |
| α-d-Lactose | − | − | − | − | + | − | Glycyl-l-glutamic acid | − | − | − | − | − | − | |
| Lactulose | − | − | − | − | + | − | l-Histidine | + | + | − | + | + | + | |
| Maltose | − | − | − | + | + | + | Hydroxy-l-proline | + | − | − | + | + | + | |
| d-Mannitol | + | + | − | + | + | + | l-Leucine | − | − | − | − | − | − | |
| d-Mannose | − | − | − | + | + | + | l-Ornithine | − | − | − | + | + | + | |
| d-Melibiose | − | − | − | + | − | + | l-Phenylalanine | − | − | − | − | − | − | |
| β-Methyl-d-glucoside | − | − | − | + | + | − | l-Proline | + | − | − | + | + | + | |
| d-Psicose | − | − | − | − | − | + | l-Pyroglutamic acid | + | + | + | + | + | + | |
| d-Raffinose | − | − | − | + | − | − | d-Serine | + | − | − | + | + | + | |
| l-Rhamnose | − | − | − | − | − | − | l-Serine | + | − | − | + | + | + | |
| d-Sorbitol | − | − | − | + | − | − | l-Threonine | − | − | − | − | − | − | |
| Sucrose | + | − | − | + | + | + | l-Threonine | − | − | − | − | − | − | |
| d-Trehalose | + | − | − | + | + | − | dl-Carnitine | + | − | − | + | + | + | |
| Turanose | + | − | − | + | + | + | γ-Aminobutyric acid | + | + | − | + | + | + | |
| Xylitol | − | − | − | + | − | − | Urocanic acid | + | − | − | + | + | + | |
| Esters | Aromatic compounds | |||||||||||||
| Methyl pyruvate | + | − | − | + | + | + | Inosine | + | − | − | + | + | + | |
| Mono-methyl succinate | + | − | − | − | + | + | Uridine | − | − | − | − | + | − | |
| Thymidine | − | − | − | − | − | − | ||||||||
| Carboxylic acids | ||||||||||||||
| Acetic acid | − | − | − | − | − | + | Amines | |||||||
| cis-Aconitic acid | + | + | − | + | + | + | Phenylethylamine | + | − | − | + | − | − | |
| Citric acid | + | + | − | + | + | + | Putrescine | + | − | − | + | + | + | |
| Formic acid | − | − | − | − | − | − | ||||||||
| d-Galactonic acid lactone | − | − | − | + | + | − | Alcohols | |||||||
| d-Galacturonic acid | + | − | − | + | + | + | 2-Aminoethanol | − | − | − | + | + | − | |
| d-Gluconic acid | + | − | − | + | + | + | 2,3-Butanediol | − | − | − | − | − | − | |
| d-Glucosaminic acid | − | − | − | − | − | − | Glycerol | − | − | − | + | + | + | |
| d-Glucuronic acid | + | + | − | + | + | + | ||||||||
| α-Hydroxybutyric acid | − | − | − | − | − | − | Phosphate-containing compounds | |||||||
| β-Hydroxybutyric acid | + | + | − | + | + | + | ||||||||
| γ-Hydroxybutyric acid | + | − | − | + | − | − | dl-α-Glycerol phosphate | − | − | − | + | + | − | |
| p-Hydroxyphenylacetic acid | + | − | − | + | + | + | Glucose-1-phosphate | + | − | − | + | + | + | |
| Itaconic acid | + | − | − | − | − | − | Glucose-6-phosphate | + | − | − | + | + | + | |
| α-Keto butyric acid | − | − | − | − | − | − | ||||||||
Filter depth. The total numbers of compounds used at filter depths of 0, 40, and 90 cm were 48, 19, and 8, respectively, for ozonated NOM and 64, 62, and 50, respectively, for nonozonated NOM.
The patterns of utilization of the groups of related compounds by the communities isolated from the different filter medium samples generally revealed the same trend, with the communities from all depths of the filter treating nonozonated water and the community from the top of the filter treating ozonated water displaying the highest percentages of substrates used for each substrate class. Of the substrate classes represented by the highest number of compounds, the amino acids had the highest utilization extent for all communities, followed by the carboxylic acids. These classes of substrates were well utilized by the communities from all depths of the filter treating nonozonated NOM and by the community from the top of the filter treating ozonated NOM. A lower percentage of carbohydrates than of the other substrate classes was utilized, especially by the communities from the filter treating ozonated NOM.
Community AP-PCR fingerprints.
DNA fingerprinting methods for analysis of complex genomes, which originally were developed by Welsh and McClelland (18), have been applied to complex microbial systems. In this study we employed AP-PCR due to the ease and rapidity of sample processing. As with any PCR-based method, a limiting factor in the analysis can be preferential amplification of certain sequences. Unlike many currently used DNA-based methods, AP-PCR is amenable to sample-intensive monitoring and therefore is useful for reactor work or field studies.
AP-PCR fingerprints were determined for the filters treating ozonated NOM and settled ORW. Many more bands were present in all of the fingerprints from the filter treating ORW than in the fingerprints from the ozonated NOM filter, indicating that the communities were genetically distinct in these two filters (data not shown). Only slight differences were seen in the band patterns obtained for samples from different depths of the filter treating ORW, indicating that there was a high degree of genetic similarity in the communities present throughout this filter. The fingerprint of the community from the top of the filter treating ozonated NOM had fewer bands than the fingerprints of the communities from the middle and bottom of the filter, indicating that the communities in the top of this filter were genetically distinct from the communities in the middle and the bottom of the filter.
The relationships among the microbial communities in the different filter samples were elucidated by calculating similarity indices for the AP-PCR band patterns (10). Table 2 shows that the largest differences among the communities were the differences between the community from the top of the filter treating ozonated NOM and the communities from the middle and bottom of that filter. Samples from each filter generally exhibited lower similarity indices with samples from another filter than with samples from different depths of the same filter (Table 2).
TABLE 2.
Similarity indices for AP-PCR patterns of microbial communities from drinking water biofilters
| Community | Proportion of bands shared bya:
|
||||
|---|---|---|---|---|---|
| Ozonated NOM community
|
ORW community
|
||||
| Top | Middle | Bottom | Middle | Bottom | |
| Ozonated NOM | |||||
| Middle | 0.36 | ||||
| Bottom | 0.36 | 1.00 | |||
| ORW | |||||
| Top | 0.19 | 0.42 | 0.42 | 0.52 | |
| Middle | 0.00 | 0.35 | 0.35 | 0.48 | 0.62 |
| Bottom | 0.20 | 0.35 | 0.35 | ||
Similarity indices for DNA fingerprints (F) were calculated as follows: F = 2 × nxy/(nx + ny), where nx is the number of bands in fingerprint x, ny is the number of bands in fingerprint y, and nxy is the number of bands shared by the two fingerprints.
Preozonation.
Preozonation and source water characteristics significantly affected community PLFA profiles, substrate utilization patterns, and AP-PCR fingerprints. The results observed are consistent with results of previous studies of biofilters in which pure culture techniques were used (1, 2). PLFA profiles showed that sulfate-reducing bacteria and other anaerobes were more prevalent in the filter treating ozonated NOM than in the filter treating settled ORW. Sulfate-reducing bacteria may have been present in higher numbers in anaerobic microniches, which may have been more prevalent deep within the filter treating ozonated water due to the 3.4-fold-higher total biomass in that filter (data not shown). Eukaryotic markers made up a greater proportion of the community in the filter treating settled ORW. This difference could have been due to the continuous input of fresh surface water into the full-scale filter, whereas the bench scale filter was used to treat groundwater NOM diluted with tap water.
Substrate utilization patterns also differentiated the populations in the filters treating ozonated and nonozonated NOM. The populations in the filter treating nonozonated NOM utilized more substrates at all filter depths than the populations in the filter treating ozonated NOM (Table 1). While amino acids and carboxylic acids were fairly well utilized by all communities in the filter treating nonozonated NOM and by the community from the top of the filter treating ozonated NOM, these substrates were not well utilized by communities from the middle and bottom of the filter treating ozonated water. Carbohydrates were better utilized by communities from all depths of the filter treating nonozonated NOM than by communities from all depths of the filter treating ozonated NOM.
AP-PCR fingerprints showed that the communities in the filters treating ozonated NOM and settled ORW were genetically different (Table 2), suggesting that preozonation and the subsequent biodegradation of labile substrates select for different populations, not simply for different phenotypic expression of the same genotypic communities.
Filter depth.
Biofilter microbial communities were also differentiated as a function of filter depth, particularly for the filter treating ozonated water. Ozonation increased the amount of quickly biodegradable DOC, which was fully utilized after a very short contact time, corresponding to the top of the biofilter (data not shown). Thus, the microbial community present at the top of the filter was adapted for efficient utilization of the labile biodegradable DOC fraction. Sole carbon source substrate utilization patterns indicated that the community at the top of the biofilter treating ozonated NOM could utilize 50% of the compounds as sole carbon sources, compared to the less than 20% of the compounds utilized by the communities isolated from the middle and bottom of that filter (Table 1). This may indicate that ozonation did not select for a metabolically different community but that depletion of substrates during biofiltration was an important selection pressure. In addition, PLFA markers for aerobic prokaryotes (mainly gram-negative bacteria) and gram-positive bacteria made up a greater portion of the community at the top of the filter than at the middle and bottom. AP-PCR results also showed that the communities in the biofilter treating ozonated NOM differed genetically as a function of filter depth (Table 2), probably due to the availability of only relatively recalcitrant substrates for microbial growth and maintenance.
Little change with depth was seen in the community structure profiles of the biofilters treating settled ORW and nonozonated NOM. Samples from all depths of the filter treating ORW were closely grouped on the PCA plot of the PLFA profiles, indicating that the communities populating various depths of the filter treating settled ORW were closely related (Fig. 1). Biolog profiles (Table 1) and AP-PCR similarity indices (Table 2) also demonstrated that the communities present at various filter depths in the filters treating ORW and nonozonated NOM were more similar genetically and in their substrate utilization patterns than the communities from various depths of the filter treating ozonated water. The high degrees of similarity in PLFA profiles, DNA fingerprints, and substrate utilization patterns may have been due to the homogeneity of substrates in the water sources and the relatively low concentration of labile substrates in these waters.
The information provided by PLFA profiles, sole carbon source utilization patterns, and DNA fingerprinting differentiated the microbial communities in drinking water biofilters based on preozonation and filter depth. Each of the methods contributes different types of information about the communities, and when taken together, the data provide an overview of microbial community structure. Thus, PLFA profiles, sole carbon source utilization profiles, and DNA fingerprinting may be used to monitor changes in drinking water biofilter communities when different pretreatment processes and/or operational changes in the treatment plant are being evaluated.
Acknowledgments
This work was supported by the U.S. EPA under cooperative agreement CR-821891.
We thank Cincinnati Water Works for providing the full-scale biofilter core.
REFERENCES
- 1.Becke J, Bonde G J. Organics in Danish drinking water. Part 3. Bacteriological examinations of samples from a specialised water treatment method for lake water using ozone and biologically-active carbon, but without the use of chlorine. Aqua. 1984;6:375–386. [Google Scholar]
- 2.Burlingame G A, Suffet I H, Pipes W O. Predominant bacterial genera in granular activated carbon water treatment systems. Can J Microbiol. 1986;32:225–230. doi: 10.1139/m86-045. [DOI] [PubMed] [Google Scholar]
- 3.Caetano-Anollés G, Bassam B J, Gresshoff P M. DNA amplification fingerprinting using very short arbitrary oligonucleotide primers. Bio/Technology. 1994;9:554–556. doi: 10.1038/nbt0691-553. [DOI] [PubMed] [Google Scholar]
- 4.Dobbs F C, Guckert J B. Microbial food resources of the macrofaunal-deposit feeder Ptychodera bahamensis (Hemichordata: Enteropneusta) Mar Ecol Prog Ser. 1988;45:127. [Google Scholar]
- 5.Findlay R H, Dobbs F C. Quantitative description of microbial communities using lipid analysis. In: Kemp P F, Sherr B F, Sherr E B, Cole J C, editors. Handbook of methods in aquatic microbial ecology. Boca Raton, Fla: Lewis Publishers; 1993. pp. 271–284. [Google Scholar]
- 6.Findlay R H, Trexler M B, White D C. Response of a benthic microbial community to biotic disturbance. Mar Ecol Prog Ser. 1990;62:135. [Google Scholar]
- 7.Guckert J B, Anworth C P, Nichols P D, White D C. Phospholipid ester-linked fatty acid profiles as reproducible assays for changes in prokaryotic community structure in estuarine sediments. FEMS Microbiol Ecol. 1985;31:147–158. [Google Scholar]
- 8.LeChevallier M W, Becker W C, Schorr P, Lee R G. Evaluating the performance of biologically active rapid filters. Am Water Works Assoc J. 1992;84(4):136–146. [Google Scholar]
- 9.Manz W, Szewzyk U, Ericsson P, Amann R, Schleifer K-H, Stenström T-A. In situ identification of bacteria in drinking water and adjoining biofilms by hybridization with 16S and 23S rRNA-directed oligonucleotide probes. Appl Environ Microbiol. 1993;59:2293–2298. doi: 10.1128/aem.59.7.2293-2298.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nei M, Li W H. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA. 1979;76:5265–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Servais P, Billen G, Ventresque C, Bablon G P. Microbial activity in GAC filters at the Choisy-le-Roi treatment plant. Am Water Works Assoc J. 1991;83(2):62–68. [Google Scholar]
- 12.Tsai Y-L, Olson B H. Rapid method for direct extraction of DNA from soil and sediments. Appl Environ Microbiol. 1991;57:1070–1074. doi: 10.1128/aem.57.4.1070-1074.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tunlid A, Baird B H, Drexler M B, Ollson S, Findlay R H, Odham G, White D C. Determination of PLFAs and poly-beta-hydroxybutyrate for the estimation of bacterial biomass and activity in the rhizosphere of the rape plant Brassica napus. Can J Microbiol. 1985;31:1113–1119. [Google Scholar]
- 14.Tunlid A, Hoitink H A, Low C, White D C. Characterization of bacteria that suppress Rhizoctonia damping-off in bark compost media by analysis of fatty acid biomarkers. Appl Environ Microbiol. 1989;55:1368–1374. doi: 10.1128/aem.55.6.1368-1374.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Urfer D, Huck P M, Booth S D J, Coffey B M. Biological filtration for BOM and particle removal: a critical review. Am Water Works Assoc J. 1997;89(12):83–98. [Google Scholar]
- 16.Vestal J R, White D C. Lipid analysis in microbial ecology: quantitative approaches to the study of microbial communities. BioScience. 1989;39:535–541. [PubMed] [Google Scholar]
- 17.Wang J Z, Summers R S, Miltner R J. Biofiltration performance. Part 1. Relationship to biomass. Am Water Works Assoc J. 1995;87(12):55–63. [Google Scholar]
- 18.Welsh J, McClelland M. Fingerprinting genomes using PCR using arbitrary primers. Nucleic Acids Res. 1990;18:7213–7218. doi: 10.1093/nar/18.24.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]