Abstract
We analysed both pooled and individual tick samples collected from four countries in Eastern Europe and the Black Sea region, using metagenome-based nanopore sequencing (NS) and targeted amplification. Initially, 1337 ticks, belonging to 11 species, were screened in 217 pools. Viruses (21 taxa) and human pathogens were detected in 46.5% and 7.3%, respectively. Tick-borne viral pathogens comprised Tacheng Tick Virus 2 (TTV2, 5.9%), Jingmen Tick Virus (JMTV, 0.9%) and Tacheng Tick Virus 1 (TTV1, 0.4%). An association of tick species with individual virus taxa was observed, with the exception of TTV2, which was observed in both Dermacentor and Haemaphysalis species. Individual ticks from pools with pathogen detection were then further screened by targeted amplification and then NS, which provided extensive genome data and revealed probable pathogen Haseki Tick Virus (HTV, 10.2%). Two distinct TTV2 clades were observed in phylogenetic analysis, one of which included closely related Dermacentor reticulatus Uukuviruses. JMTV detection indicated integrated virus sequences. Overall, we observed an expansion of newly documented pathogenic tick-borne viruses into Europe, with TTV1 being identified on the continent for the first time. These viruses should be included in the diagnostic assessment of symptomatic cases associated with tick bites and vector surveillance efforts. NS is shown as a useful tool for monitoring tick-associated pathogens in pooled or individual samples.
Subject terms: Metagenomics, Viral epidemiology, Viral evolution, Viral reservoirs
Introduction
The overall prevalence of tick-borne infections is increasing globally, due to the expansion of tick populations into new geographic regions and increased human exposure as a result of environmental and climatic changes1,2. Accounting for a major portion of vector-borne diseases in many countries, tick-borne infections may produce a significant disease burden on local healthcare providers and constitute an eminent public health threat3. With ticks acting as biological vectors, a wide palette of pathogenic microorganisms including viruses, protozoans and bacteria can be transmitted to humans and other susceptible vertebrates3.
Viruses transmitted by ticks are taxonomically heterogenous and exhibit diverse characteristics4. Those considered significant human or animal pathogens due to disease severity, distribution, potential to emerge and notable impact include Crimean-Congo hemorrhagic fever virus (CCHFV) and Nairobi sheep disease virus (NSDV) (family Nairoviridae genus Orthonairovirus); Tick-borne encephalitis virus (TBEV), Omsk hemorrhagic fever virus (OHFV), Kyasanur forest disease virus (KFDV), Powassan virus (POWV) and Alkhurma hemorrhagic fever virus (AHFV) (family Flaviviridae genus Flavivirus); Severe fever with thrombocytopenia syndrome virus (SFTSV) and Heartland virus (HRTV) (family Phenuiviridae genus Banyangvirus); Colorado tick fever virus (CTFV) (family Reoviridae, genus Coltivirus), Bourbon virus (BRBV) (family Orthomyxoviridae, genus Thogotovirus) and African swine fever virus (ASFV) (family Asfarviridae, genus Asfivirus)4,5. Expanding activity zones and changing epidemiology have been documented for some of these viruses, such as for CCHFV around the Mediterranean Sea region, POWV in North America and AHFV in Africa5. In addition to these established tick-borne pathogens, a number of newly characterized viruses have been associated with human infections, including Jingmen Tick Virus (JMTV) and Alongshan virus (ALSV), (family Flaviviridae), Tacheng Tick Virus 1 (TTV1) and Songling virus (SGLV) (family Nairoviridae, genus Orthonairovirus), Tacheng Tick Virus 2 (TTV2) (family Phenuiviridae genus Uukuvirus) and Haseki Tick Virus (HTV) (family Flaviviridae)6–11. Although cases have been reported from China (JMTV, ALSV, TTV1, TTV2, SGLV), Kosovo (JMTV) and the Russian Federation (HTV), information on pathogenesis, distribution and public health impact of these viruses are currently scarce.
Expansion and spillover of tick-borne viruses may be easily overlooked until the emergence of symptomatic case clusters or local epidemics involving humans or domestic animals. Timely identification of the circulating pathogens and assessment of public health threats rely heavily on surveillance2,4. Metagenome-based screening, enabling analysis of the nucleic acid content of any sample without prior information facilitates bio- or xeno-surveillance, where ticks can be utilized as “blood bags” to screen pathogens over multiple hosts12. Metagenome-based screening by Nanopore Sequencing (NS) has been documented as a powerful tool to detect known and novel tick-borne viruses13. In this study, we analyzed pooled and subsequently individual ticks collected from several locations in Eastern Europe and the Black Sea region using NS and targeted amplification for several recently described tick-borne viruses.
Results
In total, 1337 ticks, belonging to 11 species, were included in the study and screened in 217 pools. Virus sequences were detected in 101 pools (46.5%) and viruses documented as tick-borne human pathogens were observed in 16 (7.3%). In pools with detectable viruses, probable coinfections, evidenced by multiple virus sequences from various families/genera, were present in 31 (30.6%). Co-infections of human pathogens were not detected in pools. Tick species and virus detection prevalences according to the country of collection are provided in Table 1.
Table 1.
Summary of tick species and virus detection.
| Tick species | Poland | Bulgaria | Ukraine | Georgia | ||||
|---|---|---|---|---|---|---|---|---|
| # | Pool | # | Pool | # | Pool | # | Pool | |
| Dermacentor marginatus | – | – | 3 | 1 | – | – | 16 | 9 |
| Dermacentor reticulatus | 66 | 13 | 0 | 0 | 718 | 43 | 65 | 23 |
| Haemaphysalis parva | 0 | 0 | 0 | 0 | – | – | 120 | 27 |
| Ixodes ricinus | 80 | 14 | 124 | 20 | – | – | 32 | 18 |
| Hyalomma marginatum | 0 | 0 | 2 | 1 | – | – | 6 | 6 |
| Haemaphysalis punctata | – | – | 17 | 2 | – | – | 12 | 10 |
| Haemaphysalis innermis | – | – | 19 | 3 | – | – | 29 | 12 |
| Haemaphysalis sulcata | – | – | – | – | – | – | 8 | 2 |
| Rhipicephalus bursa | – | – | – | – | – | – | 10 | 10 |
| Rhipicephalus turanicus | – | – | 8 | 1 | – | – | 1 | 1 |
| Rhipicephalus rossicus | – | – | – | – | – | – | 1 | 1 |
| Total | 146 | 27 (12.4%) | 173 | 28 (12.9%) | 718 | 43 (19.8%) | 300 | 119 (54.8%) |
| Virus (+) | 19 (70.3%) | 17 (60.7%) | 37 (86%) | 28 (23.5%) | ||||
| Pathogen (+) | 5 (18.5%) | – | 1 (2.3%) | 10 (8.4%) | ||||
Viruses belonging to 21 separate taxa were documented in the tick pools (Table 2). Detected tick-borne human pathogens comprising TTV2, JMTV and TTV1 were present in 5.9% (n = 13), 0.9% (n = 2) and 0.4% (n = 1) of the pools, respectively. All three pathogens were present in ticks collected from Poland, whereas none could be identified in samples from Bulgaria. An association with tick species was observed for viruses, regardless of the country of collection. A notable exception was TTV2, which was detected mainly in Dermacentor marginatus and Dermacentor reticulatus ticks, and also in a Haemaphysalis punctata pool from Georgia. Ixodes ricinus harbored the broadest virus diversity (10 virus taxa) and all JMTV detections. Information on individual pools with detectable virus sequences are provided in Supplementary Tables S1 and S2.
Table 2.
Virus detection prevalences in pooled ticks according to country of collection and species.
| Virus | Location | Tick species | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Poland (n = 27, 12.4%) | Bulgaria (n = 28, 12.9%) | Ukraine (n = 43, 19.8%) | Georgia (n = 119, 54.8%) | I. ricinus (n = 52, 23.9%) | D. reticulatus (n = 79, 36.4%) | D. marginatus (n = 10, 4.6%) | H. marginatum (n = 7, 3.2%) | Rhipicephalus spp. (n = 13, 5.9%) | Haemaphysalis spp. (n = 56, 25.8%) | |
| Tacheng tick virus 2 (n = 13; 5.9%) | 3 (11.1%) | – | 1 (2.3%) | 9 (7.5%) | – | 9 (11.3%) | 3 (30%) | – | – | 1 (1.8%) |
| Tacheng tick virus 1 (n = 1, 0.4%) | 1 (3.7%) | – | – | – | – | 1 (1.9%) | – | – | – | – |
| Jingmen tick virus (n = 2, 0.9%) | 1 (3.7%) | – | – | 1 (0.8%) | 2 (3.8%) | – | – | – | – | – |
| Changping tick virus 1 (n = 25, 11.5%) | – | – | 19 (44.1%) | 6 (5%) | – | 25 (31.6%) | – | – | – | – |
| Norwavirus (n = 22, 10.1%) | 7 (25.9%) | 15 (53.5%) | – | – | 22 (42.3%) | – | – | – | – | – |
| D. reticulatus pestivirus-like virus 1 (n = 14, 6.4%) | 9 (33.3%) | – | – | 5 (4.2%) | – | 14 (17.7%) | – | – | – | – |
| D. reticulatus uukuvirus (n = 9, 4.1%) | 5 (18.5%) | – | – | 4 (3.3%) | – | 9 (11.3%) | – | – | – | – |
| Bole tick virus 4 (n = 4, 1.8%) | – | 1 | – | 3 (2.5%) | – | – | – | 4 (57.1%) | – | – |
| Bole tick virus 3 (n = 3, 1.3%) | – | – | – | – | – | – | – | – | 3 (23.1%) | – |
| Iflavirus (n = 3, 1.3%) | 1 (3.7%) | 2 (7.1%) | – | – | 3 (5.7%) | – | – | – | – | – |
| Tick phlebovirus (n = 3, 1.3%) | – | 1 (3.5%) | – | 2 (1.6%) | – | – | – | – | 2 (15.3%) | 1 (1.8%) |
| Hubei toti-like virus 24 (n = 2, 0.9%) | – | – | – | 2 (1.6%) | – | 1 (1.9%) | 2 (20%) | – | – | – |
| I.scapularis bunyavirus (n = 2, 0.9%) | – | 2 (7.1%) | – | – | 2 (3.8%) | – | – | – | – | – |
| Norway luteo-like virus 3 (n = 2, 0.9%) | 2 (7.4%) | – | – | – | 2 (3.8%) | – | – | – | – | – |
| Norway partiti-like virus 1 (n = 2, 0.9%) | – | – | – | 2 (1.6%) | 2 (3.8%) | – | – | – | – | – |
| Bole tick virus 2 (n = 1, 0.4%) | – | – | – | 1 (0.8%) | – | – | – | 1 (14.2%) | – | – |
| Chimay rhabdovirus (n = 1, 0.4%) | – | 1 (3.5%) | – | – | 1 (1.9%) | – | – | – | – | – |
| Norway mononegavirus 1 (n = 1, 0.4%) | 1 (3.7%) | – | – | – | 1 (1.9%) | – | – | – | – | – |
| Serbia narna-like virus (n = 1, 0.4%) | – | 1 (3.5%) | – | – | 1 (1.9%) | – | – | – | – | – |
| Taishun tick virus (n = 1, 0.4%) | – | – | – | 1 (0.8%) | – | – | – | 1 (14.2%) | – | – |
| Uukuniemi virus (n = 1, 0.4%) | – | – | – | 1 (0.8%) | 1 (1.9%) | – | – | – | – | – |
Individual PCR and NS screening
We performed targeted PCR and additional NS on individual ticks from pools with detectable viral pathogens. Fifty-nine individual samples from 15 pools were amplified and subsequently sequenced, including samples with expected band sizes for TTV2 and JMTV observed in electrophoresis (n = 31), selected samples negative in targeted amplification (n = 6) and controls comprising a female and a male I. ricinus specimens (n = 2, 39 in total). A pool with TTV2 detection from Ukraine and JMTV detection from Poland could not be included in PCR screening due to the lack of sufficient material for testing.
TTV2 findings
Using targeted PCR, we screened all individuals from 10 pools from Georgia and 3 pools from Poland, which were NS positive for TTV2 (n = 12) and closely-related Dermacentor reticulatus uukuvirus (DRUV) (n = 1). Subsequently, we sequenced all PCR positive (n = 29) and selected PCR negative (n = 3) individuals. Expected viruses, based on NS sequencing, were observed in 19 individual samples—18 PCR (+) and 1 PCR (−). The largest contigs were characterized as TTV2 (n = 15) and DRUV (n = 4) by BLAST, with mapped read counts of 3–86 and sizes between 332 and 5659 bp on the L and S genome segments. Breakup of pooled and single samples and virus detections are provided in Supplementary Table S2.
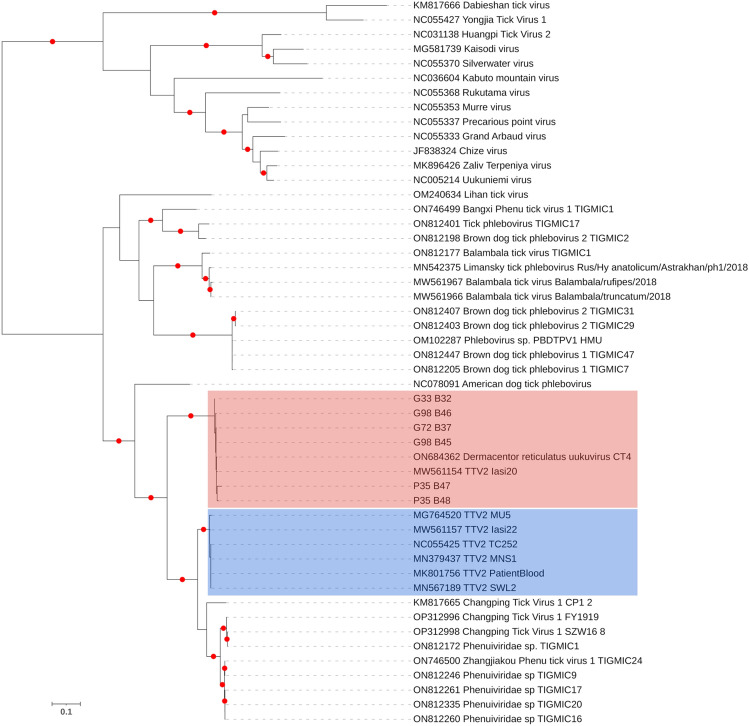
We further analyzed 6 larger TTV2 L segment contigs (> 2330 bp) from different individuals and collection sites. Pairwise comparisons revealed up to 9.9% and 2.5% diversity on nucleotide and amino acid alignments, respectively (Supplementary Table S3). In the phylogenetic tree, two well-demarcated TTV2 clusters were observed, where the sequences identified in this study clustered with a TTV2 and DRUV sequence from Eastern Europe, distinct from other TTV2s from Europe and Asia (Fig. 1).
Figure 1.
The maximum likelihood consensus tree of the phenuivirus polymerase sequences (783 amino acids), constructed using 1000 replicates. Branches achieving ≥ 95% bootstrap support are annotated with red dots. Tacheng Tick Virus 2 (TTV2) lineages are marked. Viruses are indicated by GenBank accession, name and isolate identifier where available. Sequences detected in the study are indicated by sample IDs.
A large portion (1885 of 2189 amino acids) of the TTV2 L segment, encoding for the RNA-dependent RNA polymerase (viral replicase), was available in two individual samples from Georgia (G33-B32, G98-B45, GenBank accessions). The sequences demonstrated 6.6% and 1.7% divergence based on nucleotide and putative amino acid alignments, respectively, which increased to 32.1% nucleotide and 5.7% amino acid divergence in pairwise comparisons with the closest relative (TTV2 strain Iasi20) (Supplementary Table S3). Conserved domain searches revealed the N-terminal endonuclease motif (PFAM15518) required for cap-dependent transcription (amino acids 1–45), Bunyavirus RNA-dependent RNA polymerase (PFAM04196) (amino acids 529–1223) and associated domain (PFAM12603) (amino acids 101–344) in the putative viral proteins. Phylogeny construction based on these sequences confirmed the findings from shorter TTV2 contigs and displayed a separate grouping of these sequences and DRUV (Supplementary Fig. S1). No strong signal for potential recombination was noted among TTV2/DRUV sequences.
TTV1 findings
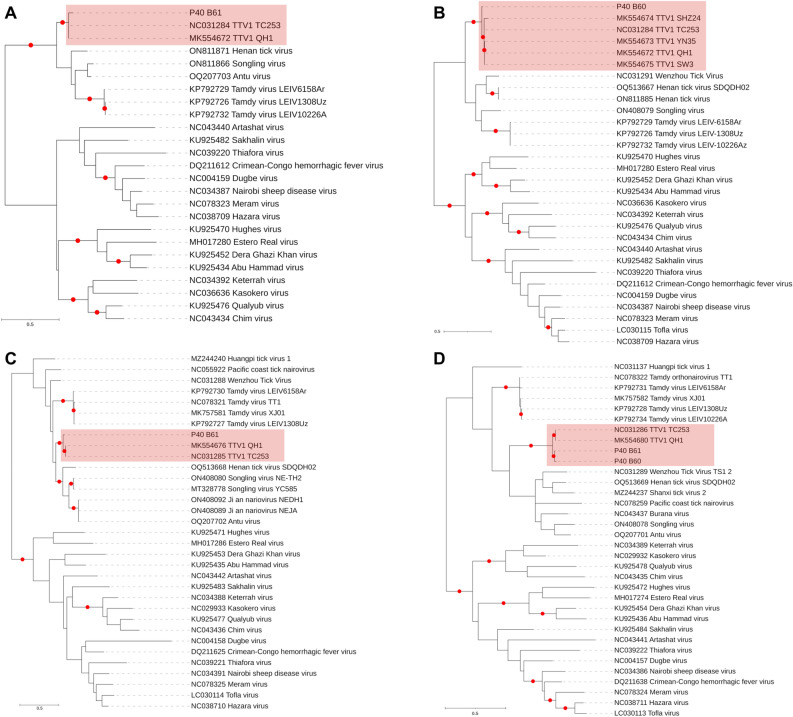
Targeted PCR was carried out in individual D. reticulatus samples from Poland, with TTV1 sequences identified as a single pool (Table 2). Despite all samples being negative in PCR, we selected three samples and performed individuals NS. Interestingly, TTV1 could be detected in two of these individuals with contig sizes of 391–997 bp, representing all three virus genome segments (L, M and S) in one sample and two segments (L and S) in another. Pairwise sequence comparisons with available TTV1 genomes showed 19.4% and 7.3% divergence based on nucleotide and amino acid alignments, respectively (Supplementary Table S3). The partial S segment contigs identified in both samples did not cover the primer binding sites utilized for the screening PCR. We further carried out maximum likelihood phylogenies for each virus genome segment (performed individually for the L segment due to non-aligning contigs). In all trees, TTV1 sequences from Poland grouped with previously reported TTV1 isolates, distinct from other orthonairoviruses (Fig. 2).
Figure 2.
The maximum likelihood consensus tree of the nairovirus polymerase (A: 335 amino acids, B: 359 amino acids), glycoprotein precursor (C: 115 amino acids) and nucleocapsid (D: 135 amino acids) sequences. The trees are constructed using 1000 replicates. Branches achieving ≥ 95% bootstrap support are annotated with red dots. Tacheng Tick Virus 1 (TTV1) sequences in each alignment are marked. Viruses are indicated by GenBank accession, name and isolate identifier where available. Sequences detected in the study are indicated by sample IDs.
JMTV findings
JMTV sequences were detected by NS in two I. ricinus pools from Poland and Georgia (Table 2). In these samples, only virus replicase sequences (located on genome segment 1) with low abundance (2–3 mapped reads) were noted. We could not perform targeted JMTV screening in individual ticks from Georgia, due to the insufficient material available. Two out of 9 individual samples tested were observed as positive. However, subsequent NS performed on both positive samples revealed a single read of 285 bp in one sample, with a BLAST similarity of 90.97% to a JMTV replicase sequence observed as integrated into the I. ricinus chromosome (Supplementary Table S2).
Other viruses
In addition to documented pathogens described above, 18 viral taxa were detected in pooled ticks (Table 2). Changping tick virus 1 (family Phenuiviridae, genus Phlebovirus) was detected in 11.5% of the pools, which were exclusively D. reticulatus ticks. Sequences with highest identities to members of the Norwavirus genus (family Nairoviridae) were also common, detected in 10.1% of the pools and in I. ricinus ticks. Our phylogenetic analyses based on Bunyavirus polymerase and nucleocapsid amino acid alignments revealed diverse virus sequences related to Grotenhout virus and other proposed members, distinct from the recently described congeneric pathogen Beiji nairovirus (Supplementary Fig. S2)7,14,15.
Another frequently observed virus was D. reticulatus Pestivirus-like Virus 1 (DRPV1) (family Flaviviridae, genus Pestivirus), present in 6.4% (14/217) of pooled and 23.1% (9/39) of single D. reticulatus ticks. This virus is related to HTV (unclassified Riboviria), with 91.8% and 94.4% nucleotide and putative amino acid identities, respectively. Despite being absent in pooled ticks, sequences with highest BLAST identities to HTV were observed in four individual D. reticulatus samples from Georgia and Poland, with a prevalence of 10.2% (4/39) (Supplementary Table S2). Alignment and phylogenetic analysis revealed subgrouping of the sequences with particular HTV isolates as well as DRPV1. Further phylogeny construction using contigs of various sizes from pooled samples also confirmed a close relationship between these viruses, probably representing geographically segregated lineages (Supplementary Fig. S3).
Virus taxa observed in pooled NS were also noted in individual ticks. However, additional viral taxa (n = 4) including Tacheng Tick Virus 5 (TTV5; unclassified Riboviria), Tacheng Tick Virus 3 (TTV3; Rhabdoviridae), Dermacentor reticulatus Rhabdovirus 1 (DRR1; Rhabdoviridae) and Betaricinrhavirus chimay (Rhabdoviridae) were identified during analyses of individual ticks, with < 10 mapped reads (Supplementary Table S2).
Discussion
Our pooled and individual metagenome-based tick screening using NS revealed 25 virus taxa including four tick-borne pathogens in samples collected from countries from Eastern Europe and around the Black Sea. Known tick-borne viruses endemic in the region, such as CCHFV or TBEV, were not detected, owing to the low prevalence of these viruses within the tick populations in targeted sites or sampling bias. However, we identified recently described tick-borne pathogens TTV2, TTV1, JMTV (7.2% in pools) and HTV (10.2% in individuals) in the sample cohort and produced abundant virus genome data for further downstream analysis.
Among the tick-borne viral pathogens identified, TTV2 was the most frequent, detected in 5.9% of the tick pools (13/217) from several sites in three of the four countries of collection, including Eastern Europe (Poland and Ukraine) and the Asian Black Sea region (Georgia). Detections were further confirmed in individual targeted PCR and NS (Supplementary Table S2). Classified in the Uukuvirus tachengense species, TTV2 was initially described during a metagenomic investigation of arthropods from China and has since been detected in D. marginatus, Dermacentor nuttalli, Dermacentor silvarum, and Hyalomma asiaticum ticks9,16,17. Subsequently, it has been documented as the causative agent in a patient with tick-associated febrile disease from China, with implications of person-to-person transmission through droplets or direct contact with body fluids9. Further detections of TTV2 in Hyalomma / Dermacentor spp. from Kazakhstan (according to GenBank records) and D. reticulatus from Romania are reported18. TTV2 is also documented in D. marginatus and the prominent CCHFV vector, H. marginatum, from ecologically diverse locations in Asia Minor, with evidence for trans-stadial or horizontal virus transmission among ticks19,20. Interestingly, we observed two distinct TTV2 clades in our phylogenetic analysis, where one of the groups also included DRUV, an unclassified Ribovirus separately detected in 4.1% of the tick pools in the study. DRUV was recently described from Croatia in D. reticulatus pools21, but to date there is scant information. Genomes of both TTV2 and DRUV consist of L and S segments, while seemingly lacking the glycoprotein encoding M segment, even in cell culture grown isolates9,21. The presence of TTV2 and DRUV divergent clades and human exposure in Europe or around the Black Sea region require further investigation.
We further detected TTV1 in 0.4% of the pools, comprising D. reticulatus from Poland. Although no amplification was observed in virus specific targeted PCR, individual NS revealed several contigs of varying sizes, encompassing all virus genome segments. Subsequent phylogenetic analyses placed all contigs with the global TTV1 sequences as a separate group within the orthonairovirus genus (Fig. 2). This is the first documentation of TTV1 in Europe. Currently, TTV1 is classified as the sole taxon in the Tacheng orthonairovirus species (genus Orthonairovirus, family Nairoviridae)15. Initially described during virus discovery efforts in arthropods from China, TTV1 was also detected in febrile diseases associated with tick bites, and as a co-infecting agent in a case with Rickettsial fever and meningitis8,16,22. Virus excretion patterns in infected individuals suggested possible transmission by direct contact with body fluids or droplets, similar to CCHFV, as noted for TTV28,9. Virus exposure was further documented in local sheep, cattle and human populations in China, with Dermacentor spp. suggested as probable vectors9. Virus genomes were detected in D. marginatus, D. silvarum, D. nuttalli, and H. asiaticum ticks in China, and also identified from spleen tissues of great gerbils (Rhombomys opimus)9,23. Recently, the virus has been documented in pools of Hyalomma aegyptium from southern Turkey24. It remains to be determined whether the lack of detection in targeted amplification in our tick cohort reflects genome diversity or low sensitivity of the PCR.
JMTV is the best studied emerging tick-borne pathogen observed in our cohort. It was identified in 0.9% of the samples, comprising I. ricinus samples collected from Poland and Georgia (Table 2). Classified within the Flaviviridae family, JMTV and related viruses (Jingmenviruses) appear to be widely distributed, having been identified in a diverse spectrum of tick species as well as in bats, cattle and rodents from Asia, Africa, Europe and the Americas25. JMTV and Alongshan virus (another Jingmenvirus) have been documented to cause human infections and subsequent seroconversion, with further evidence for exposure in domestic animals and humans6,7. Co-infections with JMTV were reported among individuals with CCHFV in Kosovo, with possible impacts on disease outcome26. In Europe, JMTV-infected ticks were reported from Romania, Serbia and the Thrace region of Turkey, while Alonghsan virus detection is more pronounced in northern Europe25–28. Recently, the integration of the JMTV polymerase gene into I. ricinus genome was documented, in ticks of both sexes and I. ricinus embryo-derived tick cell line IRE/CTVM1929. The integrated fragment of the viral genome was reported to be variable in size and forming several genovariants, with an average of 1.5 copies per cell. Several tick species other than I. ricinus were further suggested to contain the integrated segments. In field collected I. ricinus samples, the integration event was observed with a prevalence of up to 34.3% in various regions of Russia29. The JMTV findings in our study strongly suggest integrated virus sequences being detected, rather than replicating viruses. We also have previously reported NS as capable of detecting all JMTV genome segments, surpassing the sensitivity of screening by nested PCR13. Documentation of virus integration in tick vectors also has ramifications for PCR-based screening, which may require additional genome targets or steps to exclude amplification of integrated fragments.
Our findings on HTV and DRPV1 demand further discussion. We detected DRPV1 in 6.4% and 23.1% of the pooled and individual D. reticulatus samples, respectively. This virus was first described in D. reticulatus from Croatia, sharing significant NS3–NS5 protein homologies with members of the Pestivirus genus21. It is closely related to other recently documented pestivirus-like viruses, Bole Tick Virus 4 (BTV4), Trinbago Virus (TBOV), as well as HTV21. HTV genomes were recently documented in retrospectively screened sera from individuals with febrile disease associated with tick bites, with prevalences of 0.9–9.3% in different regions of the Russian Federation, with and without TBEV or Borrelia spp.11. We detected HTV in 10.2% of the single D. reticulatus samples from Georgia and Poland and observed clustering of the sequences with HTVs as well as DRPV1 in phylogenetic analyses. Given these findings and limited sequence variation between complete HTV and DRPV1 polyprotein coding sequences, these viruses represent sub-lineages of the same taxon, observed in geographically segregated regions. The capacity of either virus to produce human symptomatic infections requires further investigation.
Finally, we detected Norwaviruses, distinct from Beiji nairovirus, in 10.1% of the pools, comprising I. ricinus ticks from Bulgaria and Poland. Norwavirus is a recently described genus in the family Nairoviridae, which currently includes Grotenhout Virus (GRHV) as the sole member of the Grotenhout norwavirus species15. Closely related viruses have been documented in northern Europe and central Asia, such as Pustyn Virus (PTV) and Norway Nairovirus 1 (NWNV1), but these strains currently remain taxonomically unclassified7,14,15. Similar to TTV2, norwaviruses lack the genomic segment M, which encodes for nairovirus glycoproteins. Beiji nairovirus (BJNV), a norwavirus-related virus, was documented as a potential agent causing tick-borne febrile diseases in China, supported by experimental inoculations7. The public health implications of norwaviruses currently remain underexplored.
In conclusion, our findings reveal several tick-borne viral pathogens, recently reported from various regions from Asia, to be present in various locations in Eastern Europe and Asian Black Sea region. In any of the sampling regions, actual prevalences of individual viruses is hard to assess, due to our cross-sectional collection and non-targeted screening strategy employed in the study. Moreover, local virus clades or integrated virus genomes might undermine targeted screening due to sequence variation (TTV1) or amplification of the integrated fragments (JMTV). Currently, the lack of standardized antibody testing further hinders investigation of previous exposures and serological diagnosis. Nevertheless, these viruses should be considered in the diagnostic assessment of symptomatic cases associated with tick bites and tested when possible. NS proves to be a useful tool for monitoring tick-associated pathogens in pooled and individual samples.
Methods
Sample collection, processing and barcoding
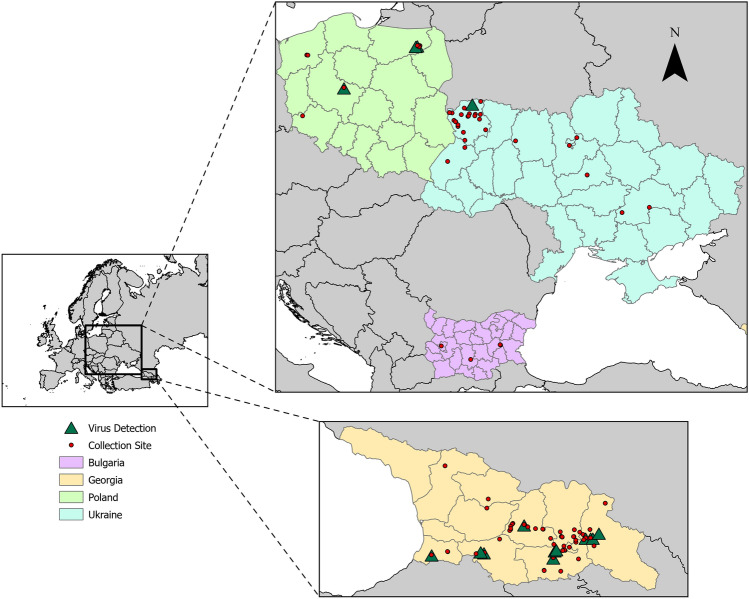
Questing adult ticks were collected via drag/flagging at 75 ad hoc sites in Bulgaria (2021–2022), Poland (2021), Georgia (2021) and Ukraine (2012–2013) (Fig. 3). The samples were morphologically identified using appropriate keys30 and stored at − 80 °C. Subsequently, samples from Bulgaria, Poland and Georgia were shipped to Walter Reed Biosystematics Unit (WRBU), Museum Support Center, Smithsonian Institution, MD, USA, in dry ice. Samples from Ukraine were pooled according to species and collection site, processed locally and nucleic acids were shipped to WRBU for further processing. The dorsal and ventral sides of intact specimens were imaged using IDX machine (Vectech, Baltimore, MD, USA).
Figure 3.
Map indicating the tick sampling locations in the study. Sites with detectable human pathogens are marked by a triangle.
Nucleic acid extraction was carried out individually following homogenization in sterile 1 × PBS (ThermoFisher Scientific, Waltham, MA, USA) and ATL Lysis Buffer with regent DX (Qiagen, Valencia, CA, USA) with 5 mm stainless steel grinding balls (OPS Diagnostics, Lebanon, NJ, USA) using TissueLyser (Qiagen, Valencia, CA, USA). Following centrifugation, the supernatant was purified using the IndiMag Pathogen Kit (Indical Bioscience, Leipzig, Germany) on BioSprint 96 DNA/RNA Purification System (Qiagen, Valencia, CA, USA), according to the manufacturer recommendations.
Morphological identification of the individual ticks was confirmed using DNA barcoding, by amplification of the 658 bp region of the mitochondrial cytochrome c oxidase subunit I (COI) gene, as described previously31,32. Subsequently, 5 µl of the individual tick nucleic acids were pooled according to species, sex and locality of collection (up to a maximum of 9), and stored in − 80 °C.
Nanopore sequencing (NS)
Pooled nucleic acids were subjected to cDNA synthesis using NEBNext Ultra II RNA First Strand and Non-Directional RNA Second Strand Synthesis modules, utilizing a random primer mix (New England Biolabs, Ipswich, MA, USA). Double stranded cDNA was cleaned up using Agencourt AMPure XP reagent (Beckman Coulter Biosciences, Indianapolis, IN, USA) and quantitated by Qubit dsDNA HS Assay Kit (ThermoFisher Scientific, Waltham, MA, USA). The NEBNext Ultra II End repair/dA-tailing and Quick Ligation modules (New England Biolabs, Ipswich, MA, USA), and Ligation Sequencing Kit SQK-LSK109 (Oxford Nanopore Technologies, Oxford, UK) were used to prepare libraries, which were further quantitated by Qubit (ThermoFisher Scientific, Waltham, MA, USA) using 1 × dsDNA HS Assay Kit. Each sample was separately barcoded with the Native Barcoding Expansion 96-EXP-NBD 196 (Oxford Nanopore Technologies, Oxford, UK). An epMotion 5075 workstation (Eppendorf, Hamburg, Germany) was used for automated liquid handling and library preparation, carried out according to the manufacturer recommendations. Sequencing libraries containing 20–24 barcoded pools were loaded on a single ONT R9.4.1 flow cell and run on the GridION sequencing device (Oxford Nanopore Technologies, Oxford, UK) for 48 h.
Targeted viral pathogen detection
Nested polymerase chain reaction (PCR) was used to amplify JMTV, TTV1 and TTV2 in single tick samples with the corresponding virus sequences observed in pooled NS. We employed previously published primer sets and conditions for each assay, in a 30 µl total volume per sample. The assays targeted JMTV NS5-like protein on segment 1 and the nucleocapsid protein on segment S for TTV1 and TTV28,9,33. The amplicons of the nested amplification (JMTV: 394 bp, TTV1: 328 bp, TTV2: 252 bp) were visualized by electrophoresis on 1.5% agarose gels using GelGreen Nucleic Acid Gel Stain (Biotium Inc., Fremont, CA, USA).
Data analysis
Following NS, base-calling and demultiplexing was carried out on the GridION with the MinKNOW operating software v21.11.7 (Oxford Nanopore Technologies) and Guppy v5.1.1334. Raw reads were trimmed with Porechop to remove adapter sequences and then filtered with NanoFilt to remove reads with q-scores ≤ 9 and read lengths ≤ 100 bp34,35. Tick genome data were then removed as hosts using Minimap2 v2.24 and Samtools v1.936,37. Subsequently, the data was aligned to the National Center for Biotechnology Information (NCBI) non-redundant (NR) database using DIAMOND v2.0.1438 and visualized using MEGAN6 (v6.23.2)39.
Sequences were handled using Geneious Prime (v2022.2.1) (Biomatters Ltd., Auckland, New Zealand). BLASTn and BLASTp algorithms were used for similarity searches in the NCBI database40. Read mapping was carried out using Minimap2 plug-in for Geneious Prime, with default settings optimized for nanopore data. Longest reads were used for similarity assessment when contigs were not available. Alignment and pairwise sequence comparisons were carried out using CLUSTAL W41. Potential genetic exchange and recombination were assessed using the RDP4 software in the default settings42. Protein domain and motif searches were performed using the NCBI conserved domain search tool and MOTIF Search in the PFAM database43,44. Phylogenetic analysis was performed on sequences using IQ-TREE 245. Optimal evolutionary models and partitioning schemes were determined for nucleotide and amino acid sequence alignment using the automatic model selection tools (-m MFP + MERGE). The maximum likelihood extended majority-rule consensus trees were constructed using the ultrafast bootstrap approximation approach (UFBoot) for 1000 replicates46.
Supplementary Information
Acknowledgements
The study was financially supported by the Armed Forces Health Surveillance Division—Global Emerging Infections Surveillance (AFHSD-GEIS) awards P0057_22_WR, P0044_23_WR and P0008_21_GA, to the Walter Reed Army Institute of Research (WRAIR) One Health Branch, and US Army Medical Detachment-Georgia (USAMRD-G), respectively. Collections in Ukraine were enabled by funding from the Defense Threat Reduction Agency (DTRA) through the Cooperative Biological Engagement Program in Ukraine (Project UP-2). The authors are grateful to the Area Support Group—Poland (ASG-P), Epidemiological Response Center of Polish Armed Forces (ERC), Bulgarian Military Medical Academy (MMA) and Colonel Associate Professor Andrey Galev, Head of the Scientific Applied Center for Military Epidemiology and Hygiene (NPTSVEH-MMA) for assistance in access to the collection sites; and to the personnel of the Ukrainian Center of Diseases Control and Monitoring, Ukraine Ministry of Health and laboratories in Volyn province for their contributions. This manuscript was prepared whilst KE and BPB held a National Research Council (NRC) Research Associateship Awards at the Walter Reed Biosystematics Unit, through the Walter Reed Army Institute of Research, Silver Spring, MD. NC is a fellow of the Army Educational Outreach Program (AEOP). The authors are grateful to David Pecor of WRBU for assistance with preparation of the graphical content. Preliminary findings of the study has been presented at the Entomology Society of America general meeting—Entomology 2023, November 5-8, National Harbor, MD.
Author contributions
Conceptualization: K.E., Y.M.L., D.D.R.W. Formal analysis: K.E., B.P.B., S.P.N., L.C.Q., N.G.C. Investigation: D.D.R.W., N.V., G.K., T.C., L.S., J.K.B. Resources: M.P.N., D.D.R.W. Supervision: C.L.T., Y.M.L., D.D.R.W. Project administration: M.P.N., C.L.T., Y.M.L., D.D.R.W. Funding acquisition: C.L.T., Y.M.L., D.D.R.W. Writing—original draft: K.E. Writing—review and editing: K.E., B.P.B., Y.M.L.
Data availability
Consensus DNA sequences, specimen images, and edited chromatograms are available on BOLD under project “SHTRP and SHTRB”. All data generated in this study are available in the NCBI Biosample and Sequence Read Archive under the BioProject PRJNA878809 (SRR21504630-SRR21504591) and GenBank with the accession numbers OP432043–OP432053.
Competing interests
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The funders of the research had no influence on results presented. The opinions or assertions contained herein are the private views of the authors, and are not to be construed as official, or as reflecting true views of the Department of the Army, Navy or the Department of Defense. Material contained within this publication has been reviewed by the Walter Reed Army Institute of Research. There is no objection to its presentation and / or publication.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-023-46879-2.
References
- 1.Ogden NH, Mechai S, Margos G. Changing geographic ranges of ticks and tick-borne pathogens: drivers, mechanisms and consequences for pathogen diversity. Front. Cell. Infect. Microbiol. 2013;3:1. doi: 10.3389/fcimb.2013.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vayssier-Taussat M, et al. How a multidisciplinary ‘One Health’ approach can combat the tick-borne pathogen threat in Europe. Future Microbiol. 2015;10:809–818. doi: 10.2217/fmb.15.15. [DOI] [PubMed] [Google Scholar]
- 3.Rochlin I, Toledo A. Emerging tick-borne pathogens of public health importance: A mini-review. J. Med. Microbiol. 2020;69:781–791. doi: 10.1099/jmm.0.001206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mansfield KL, Jizhou L, Phipps LP, Johnson N. Emerging tick-borne viruses in the twenty-first century. Front. Cell. Infect. Microbiol. 2017;7:1. doi: 10.3389/fcimb.2017.00298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ergunay K. Revisiting new tick-associated viruses: what comes next? Future. Virol. 2020;15:1. doi: 10.2217/fvl-2019-0149. [DOI] [Google Scholar]
- 6.Jia N, et al. Emergence of human infection with Jingmen tick virus in China: A retrospective study. EBioMedicine. 2019;43:317–324. doi: 10.1016/j.ebiom.2019.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang ZD, et al. A new segmented virus associated with human febrile illness in China. N. Engl. J. Med. 2019;380:2116–2125. doi: 10.1056/NEJMoa1805068. [DOI] [PubMed] [Google Scholar]
- 8.Liu X, et al. A tentative Tamdy orthonairovirus related to febrile illness in northwestern China. Clin. Infect. Dis. 2020;70:2155–2160. doi: 10.1093/cid/ciz602. [DOI] [PubMed] [Google Scholar]
- 9.Dong Z, et al. Human Tacheng tick virus 2 infection, China, 2019. Emerg. Infect. Dis. 2021;27:594–598. doi: 10.3201/eid2702.191486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma J, et al. Identification of a new orthonairovirus associated with human febrile illness in China. Nat. Med. 2021;27:434–439. doi: 10.1038/s41591-020-01228-y. [DOI] [PubMed] [Google Scholar]
- 11.Kartashov MY, et al. Novel Flavi-like virus in ixodid ticks and patients in Russia. Ticks. Tick. Borne. Dis. 2023;14:1. doi: 10.1016/j.ttbdis.2022.102101. [DOI] [PubMed] [Google Scholar]
- 12.Brinkmann A, Nitsche A, Kohl C. Viral metagenomics on blood-feeding arthropods as a tool for human disease surveillance. Int. J. Mol. Sci. 2016;17:1. doi: 10.3390/ijms17101743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ergunay K, et al. Impact of nanopore-based metagenome sequencing on tick-borne virus detection. Front. Microbiol. 2023;14:1. doi: 10.3389/fmicb.2023.1177651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pettersson JH, et al. Characterizing the virome of Ixodes ricinus ticks from northern Europe. Sci. Rep. 2017;7:1. doi: 10.1038/s41598-017-11439-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuhn JH, et al. 2022 taxonomic update of phylum Negarnaviricota (Riboviria: Orthornavirae), including the large orders Bunyavirales and Mononegavirales. Arch. Virol. 2022;67:2857–2906. doi: 10.1007/s00705-022-05546-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li, C. X. et al. Unprecedented genomic diversity of RNA viruses in arthropods reveals the ancestry of negative-sense RNA viruses. Elife. 4. 10.7554/eLife.05378 (2015). [DOI] [PMC free article] [PubMed]
- 17.Qin T, Shi M, Zhang M, Liu Z, Feng H, Sun Y. Diversity of RNA viruses of three dominant tick species in North China. Front. Vet. Sci. 2023;9:1. doi: 10.3389/fvets.2022.1057977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bratuleanu BE, et al. The virome of Rhipicephalus, Dermacentor and Haemaphysalis ticks from Eastern Romania includes novel viruses with potential relevance for public health. Transbound. Emerg. Dis. 2022;69:1387–1403. doi: 10.1111/tbed.14105. [DOI] [PubMed] [Google Scholar]
- 19.Brinkmann A, et al. A metagenomic survey identifies Tamdy orthonairovirus as well as divergent phlebo-, rhabdo-, chu- and flavi-like viruses in Anatolia. Turkey. Ticks. Tick. Borne. Dis. 2018;9:1173–1183. doi: 10.1016/j.ttbdis.2018.04.017. [DOI] [PubMed] [Google Scholar]
- 20.Dinçer E, et al. Several tick-borne pathogenic viruses in circulation in Anatolia. Turkey. Vector. Borne. Zoonotic. Dis. 2022;22:148–158. doi: 10.1089/vbz.2021.0082. [DOI] [PubMed] [Google Scholar]
- 21.Sameroff S, et al. Virome of Ixodes ricinus, Dermacentor reticulatus, and Haemaphysalis concinna ticks from Croatia. Viruses. 2022;14:1. doi: 10.3390/v14050929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, Jiang L, Yang Y, Xie S, Yuan W, Wang Y. A tick bite patient with fever and meningitis co-infected with Rickettsia raoultii and Tacheng tick virus 1: A case report. BMC. Infect. Dis. 2021;21:1. doi: 10.1186/s12879-021-06877-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ji N, et al. Tacheng tick virus 1 and songling virus infection in Great Gerbils (Rhombomys opimus) in Northwestern China. J. Wildl. Dis. 2023;59:138–142. doi: 10.7589/JWD-D-21-00137. [DOI] [PubMed] [Google Scholar]
- 24.Dincer E, et al. Molecular Detection of Tacheng Tick Virus-1 (TcTV-1) and Jingmen Tick Virus in ticks collected from wildlife and livestock in Turkey: First indication of TcTV-1 beyond China. Vector. Borne. Zoonotic. Dis. 2023 doi: 10.1089/vbz.2023.0029. [DOI] [PubMed] [Google Scholar]
- 25.Colmant, A. M. G., Charrel, R. N. & Coutard, B. Jingmenviruses: Ubiquitous, understudied, segmented flavi-like viruses. Front. Microbiol. 13. 10.3389/fmicb.2022.997058 (2022). [DOI] [PMC free article] [PubMed]
- 26.Emmerich P, et al. Viral metagenomics, genetic and evolutionary characteristics of Crimean-Congo hemorrhagic fever orthonairovirus in humans. Kosovo. Infect. Genet. Evol. 2018;65:6–11. doi: 10.1016/j.meegid.2018.07.010. [DOI] [PubMed] [Google Scholar]
- 27.Stanojević, M. et al. Depicting the RNA virome of hematophagous arthropods from Belgrade, Serbia. Viruses.12. 10.3390/v12090975 (2022). [DOI] [PMC free article] [PubMed]
- 28.Dinçer, E., et al. Survey and Characterization of Jingmen tick virus variants. Viruses. 11. 10.3390/v11111071 (2019). [DOI] [PMC free article] [PubMed]
- 29.Morozkin, E. S., et al. Integrated Jingmenvirus polymerase gene in Ixodes ricinus genome. Viruses. 14. 10.3390/v14091908 (2022). [DOI] [PMC free article] [PubMed]
- 30.Estrada-Peña, A., Mihalca, A. & Petney, T. (eds.) Ticks of Europe and North Africa: A Guide to Species Identification. 10.1007/978-3-319-63760-0 (2018).
- 31.Polsomboon Nelson, S. et al. Ticks (Acari: Ixodidae) and associated pathogens collected from domestic animals and vegetation in Stann Creek District, Southeastern Belize, Central America. J. Med. Entomol. 59, 1749–1755 (2022). [DOI] [PMC free article] [PubMed]
- 32.Polsomboon S, et al. Molecular detection and identification of Rickettsia species in ticks (Acari: Ixodidae) collected from Belize Central America. J. Med. Entomol. 2017;54:1718–1726. doi: 10.1093/jme/tjx141. [DOI] [PubMed] [Google Scholar]
- 33.Yu ZM, et al. Identification and characterization of Jingmen tick virus in rodents from Xinjiang China. Infect. Genet. Evol. 2020;84:1. doi: 10.1016/j.meegid.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wick RR, Judd LM, Holt KE. Performance of neural network basecalling tools for Oxford Nanopore sequencing. Genome. Biol. 2019;20:1. doi: 10.1186/s13059-019-1727-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De Coster W, D’Hert S, Schultz DT, Cruts M, Van Broeckhoven C. NanoPack: Visualizing and processing long-read sequencing data. Bioinformatics. 2018;34:2666–2669. doi: 10.1093/bioinformatics/bty149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Danecek P, et al. Twelve years of SAMtools and BCFtools. Gigascience. 2021;10:1. doi: 10.1093/gigascience/giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34:3094–3100. doi: 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buchfink B, Reuter K, Drost HG. Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat. Methods. 2021;18:366–368. doi: 10.1038/s41592-021-01101-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huson DH, et al. MEGAN Community Edition—interactive exploration and analysis of large-scale microbiome sequencing data. PLOS Comput. Biol. 2016;12:1. doi: 10.1371/journal.pcbi.1004957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 41.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic. Acids. Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martin, D.P., Murrell, B., Golden, M., Khoosal, A. & Muhire, B. RDP4: Detection and analysis of recombination patterns in virus genomes. Virus. Evol. 1, 1. 10.1093/ve/vev003 (2015). [DOI] [PMC free article] [PubMed]
- 43.Bateman A, et al. The Pfam protein families database. Nucleic. Acids. Res. 2002;30:276–280. doi: 10.1093/nar/30.1.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marchler-Bauer A, et al. CDD: NCBI's conserved domain database. Nucleic. Acids. Res. 2015;43:222–226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Minh BQ, et al. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020;37:1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2018;35:518–522. doi: 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Consensus DNA sequences, specimen images, and edited chromatograms are available on BOLD under project “SHTRP and SHTRB”. All data generated in this study are available in the NCBI Biosample and Sequence Read Archive under the BioProject PRJNA878809 (SRR21504630-SRR21504591) and GenBank with the accession numbers OP432043–OP432053.