Abstract
Melanoma brain metastases (MBM) are clinically challenging to treat and exhibit variable responses to immune checkpoint therapies. Prior research suggests that MBM exhibit poor tumor immune responses and are enriched in oxidative phosphorylation. Here, we report results from a multi-omic analysis of a large, real-world melanoma cohort. MBM exhibited lower interferon-gamma (IFNγ) scores and T cell-inflamed scores compared to primary cutaneous melanoma (PCM) or extracranial metastases (ECM), which was independent of tumor mutational burden. Among MBM, there were fewer computationally inferred immune cell infiltrates, which correlated with lower TNF and IL12B mRNA levels. Ingenuity pathway analysis (IPA) revealed suppression of inflammatory responses and dendritic cell maturation pathways. MBM also demonstrated a higher frequency of pathogenic PTEN mutations and angiogenic signaling. Oxidative phosphorylation (OXPHOS) was enriched in MBM and negatively correlated with NK cell and B cell-associated transcriptomic signatures. Modulating metabolic or angiogenic pathways in MBM may improve responses to immunotherapy in this difficult-to-treat patient subset.
Subject terms: Melanoma, Cancer genomics, Cancer microenvironment
Introduction
The advent of novel systemic therapies, including immune checkpoint blockade, has raised the 5-year survival for patients with metastatic melanoma from 15% in 2004, up to 30% in the period between 2011 and 20171. However, approximately half of all patients with advanced melanoma will develop brain metastases, leading to death in up to 70% of these patients2,3. The management of melanoma brain metastases (MBM) is clinically challenging due to a number of factors, including the heterogeneity of presentation, the presence of neurological symptoms, patterns of intracranial relapse, and the complexity of multi-disciplinary care spanning a combination of surgery, radiotherapy, and systemic therapies2. The use of immune checkpoint blockade has demonstrated efficacy in treating some patients with MBM. Indeed, the phase 2 CheckMate 204 trial evaluated the efficacy of combining PD-1 and CTLA-4 inhibition in 94 patients with asymptomatic, non-steroid dependent, untreated MBM and reported an intracranial response rate of 55%, with a partial response rate of 30% and complete response rate of 26%4; these response rates are similar to that reported in CheckMate 067, where PD-1 and CTLA-4 combination therapy induced a response rate of 57.6% among patients with extracranial metastases (ECM)5. Meanwhile, single-agent PD-1 immunotherapy appears to be less effective in treating MBM, with a response rate (complete or partial) of 26% compared to ECM, where there is a roughly 40% objective response rate6,7. While these response rates may not be directly comparable due to differences in prior therapies received among cohorts, there remains much room for improvement in MBM responses to immune checkpoint blockade, which may be aided by a better understanding of the biological differences between MBM and ECM or primary cutaneous melanoma (PCM).
Melanoma is characterized by driver mutations in BRAF (47–59%), NRAS (22–28%), and NF1 (14–17%); these mutations have been described in MBM as well, with similar frequencies8–13. In addition, PTEN alterations (loss-of-function mutations or deletions) are present in 7.3% of primary melanomas and 15.2% of metastatic melanomas and lead to constitutive phosphoinositide 3-kinase (PI3K) pathway activation14. The loss of PTEN tumor suppressor activity is important in the early stages of metastasis and is associated with a shorter time to MBM but not ECM15–17. Interestingly, higher tumor infiltrating lymphocyte (TIL) density was observed in MBM compared to brain metastases from other cancers18, in accordance with the highly immunogenic nature of melanoma that results in greater response rates to immunotherapy19. In studies investigating the tumor immune microenvironment of MBM, CD8 + T cells were observed in 90.7% of tumors20, and tumors with higher ImmuneScores (a gene expression score representative of greater immune cell infiltrate), were associated with improved overall survival21. Despite the contribution of the PI3K pathway to the initiation of MBM and the correlation of PTEN loss with reduced TILs in PCM and local metastases22, PTEN protein loss was not found to be related to ImmuneScores in MBM21, suggesting other mechanisms at play that remain to be elucidated. Comparison of MBM with patient-matched ECM has revealed that despite a strong presence of immune infiltrate, there are nonetheless significantly lower ImmuneScores and T cell infiltrate in MBM than in ECM21. Fischer and colleagues also reported enrichment of oxidative phosphorylation (OXPHOS) and increased immunosuppression in MBM compared to ECM21, suggesting that altered metabolism could potentially shape the tumor microenvironment (TME) in MBM. Ultimately, a greater understanding of the TME in MBM is required to optimize therapies for these patients.
In the current study, we sought to compare the transcriptomic and immunologic landscape of MBM relative to PCM and ECM in a large, multi-institutional clinical melanoma cohort, with the goal to consolidate and further clarify recent reports on the immunosuppressive features of MBM. This analysis of a real-world MBM cohort integrating diverse molecular features identified through comprehensive genomic and transcriptomic testing provides advantages over smaller cohorts or clinical trial cohorts that exclude certain subsets of patients. Investigation of the molecular profile and TME unique to MBM can provide further insight into possible treatment vulnerabilities and avenues for future clinical trials.
Results
Patient population
In total, 1,314 melanoma samples were available, comprising 94 MBM, 350 PCM, and 870 ECM samples. 48.9% of patients were younger than 67 years old and 61.7% were male, consistent with the established greater incidence of cutaneous melanoma in men than women23. ECM comprised tumors from lymph node, lung, liver, and other diverse sites of metastatic disease (Supplementary Table 1). In all samples, DNA was assessed using a 592 gene panel or by whole exome sequencing (WES) and RNA was assessed by whole transcriptome sequencing (WTS) at Caris Life Sciences (Phoenix, AZ).
Characterization of the immunogenic TME in MBM
To determine whether patients with MBM would likely benefit from immunotherapy, we assessed the immunogenicity of MBM at the transcriptomic level by calculating the interferon-gamma (IFNγ)24 score and the T cell-inflamed signature score for each tumor sample in our melanoma cohort25. These signatures correlate clinically with response to anti-PD1 therapy independent of tumor mutational burden (TMB)24 and immunogenic antigen density25. These approaches identified lower IFNγ signature scores (Fig. 1a) and fewer tumors with T cell-inflamed signatures (Fig. 1b) in MBM with respect to PCM and ECM (p < 0.05 for IFNγ score and p < 0.0001 for percentage of tumors with T cell-inflamed signature). To explore the possibility of lower baseline IFNG gene expression in normal brain versus other anatomic sites, we analyzed the GENT2 database of normal and cancer gene expression26. Interestingly, IFNG gene expression was significantly lower at baseline in normal skin than brain, liver, or lung (Supplementary Fig. 1a). While this data does not consider potential differences in IFNγ scores or IFNγ protein levels, it supports the conclusion that lower IFNγ scores in brain are not simply a reflection of lower baseline levels in normal brain tissue relative to other sites.
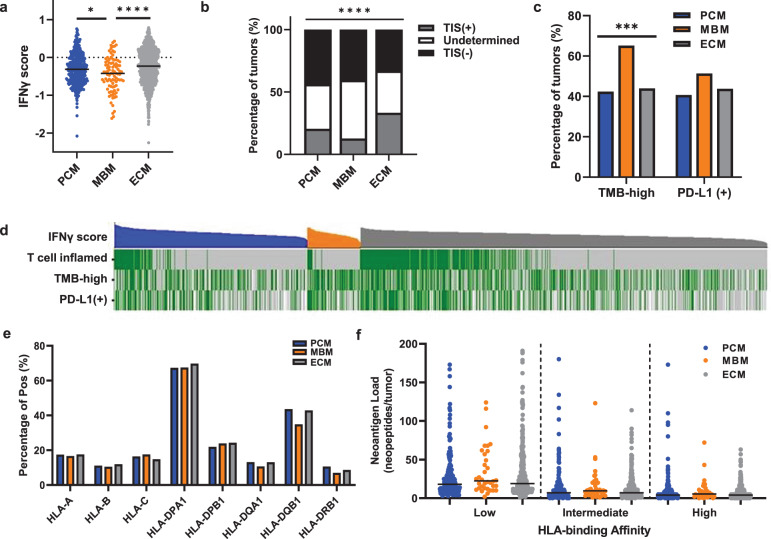
Fig. 1. MBM with comparable HLA homozygosity and neoantigen load are less immunogenic than PCM or ECM.
a IFNγ scores for PCM (blue, n = 350), MBM (orange, n = 94) and ECM (gray, n = 870). Black lines indicate median IFNγ score values. Kruskal–Wallis test with Benjamini-Hochberg correction; ****, corrected p < 0.0001; *, corrected p < 0.05. b PCM, MBM, and ECM were evaluated and classified by T cell-inflamed signature. The percentages of tumors with T cell-inflamed signatures (TIS(+); gray), undetermined (white), and lacking a T cell-inflamed signature (TIS(-); black). Chi-square test; ****p < 0.0001. c The percentage of PCM (n = 276), MBM (n = 72), and ECM (n = 681) that are TMB-high and PD-L1(+). Chi-square test; ***, p < 0.0005. d Oncoplot including IFNγ scores (top, highest to lowest within PCM, blue; MBM, orange; ECM, gray), T cell-inflamed signature (green, T cell-inflamed tumors; gray, others), TMB (high, green; low, gray; white, data not available) and PD-L1 IHC staining (positive, green; negative, gray; white, data not available). e Percentage of tumors that are homozygous at HLA-I (HLA-A, HLA-B, and HLA-C) and HLA-II (HLA-DPA1, HLA-DPB1, HLA-DQB1, and HLA-DRB1) loci in PCM, MBM, and ECM. Chi-square test, not significant (p > 0.05). f Assessment of neoantigen load with low (left), intermediate (middle), and high (right) HLA-binding affinity for PCM (blue), MBM (orange), and ECM (gray). Kruskal–Wallis test; not significant (p > 0.05).
Despite these lower IFNγ and T cell-inflamed signature scores, MBM had a higher TMB relative to PCM and ECM (p < 0.0001; Fig. 1c), which supports our previous study reporting increased TMB in MBM compared to primary tumors13. However, no significant differences were observed in PD-L1 positivity by IHC (Fig. 1c). Oncoplot further showed that tumors with T cell-inflamed signatures clustered together with higher IFNγ scores (Fig. 1d) and significantly correlated in PCM, MBM, and ECM (Supplementary Fig. 1b–d), which has been demonstrated in melanoma and other tumor types. However, neither of these transcriptomic signatures were associated with TMB, indicating that other molecular features may affect immune responses beyond TMB.
We next sought to evaluate the distribution of homozygosity (which can represent parental homozygosity or loss of heterozygosity [LOH]) for human leukocyte antigen (HLA)-I and HLA-II and neoantigen load, which is associated with TMB status in various cancer types27,28. Though TMB and total neoantigen load were positively correlated (Pearson r = 0.96, p < 0.0001; Supplementary Fig. 2), we failed to detect a significant difference between PCM, MBM, or ECM in the percentage of HLA homozygosity at any major loci (Fig. 1e). Likewise, there was no difference in neoantigen load among PCM, MBM, or ECM with different MHC binding affinities (Fig. 1f). This observation agrees with a previous study showing that immunogenic antigen density is not associated with a T cell-inflamed TME in melanoma25. Together, we conclude that MBM exhibit a less immunogenic microenvironment identified by both IFNγ score and T cell-inflamed signature, which are independent of the distribution of HLA subtypes, neoantigen load, or TMB.
Computationally inferred immune cell infiltrates among MBM
The immune cell populations for PCM, MBM, and ECM were inferred using quanTIseq, a high-throughput computational pipeline specifically designed for RNA-seq data that quantifies the absolute fractions of ten different intratumoral immune cell types29. Transcriptomic signatures indicative of B cells, M2 macrophages, natural killer (NK) cells, T regulatory cells (Tregs) and myeloid dendritic cells (mDC) were significantly reduced in MBM compared to PCM (B cells, p < 0.0001; M2, p < 0.0001; NK cells, p < 0.005; Tregs, p < 0.0001; mDCs, p < 0.05) or ECM (B cells, p < 0.0001; M2, p < 0.05; Tregs, p < 0.005; Fig. 2a). Interestingly, M1 macrophages were significantly higher among MBM relative to PCM (p < 0.05).
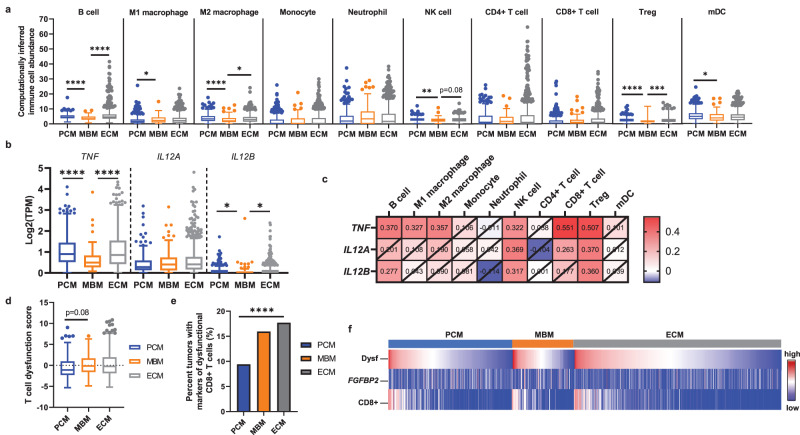
Fig. 2. MBM have significantly fewer computationally inferred immune cell infiltrates and more T cell dysfunction.
a Box plots showing computationally inferred immune cell abundance in PCM, MBM, and ECM using quanTIseq analysis. The data is displayed using the Tukey method for box and whiskers, with the center line indicating the median. Kruskal-Wallis test with Benjamini-Hochberg correction; *, corrected p < 0.05; **, corrected p < 0.005; ***, corrected p < 0.0005; ****, corrected p < 0.0001. b The mRNA levels of TNF, IL12A, and IL12B were compared between PCM, MBM, and ECM tumor samples. The data is displayed using the Tukey method for box and whiskers, with the center line indicating the median. Kruskal–Wallis test with Benjamini-Hochberg correction; **, corrected p < 0.005; ****, corrected p < 0.0001 c Heat map of Spearman rank correlation coefficients between TNF, IL12A, and IL12B mRNA and immune cell infiltrates calculated from bulk transcriptomic data in MBM. The numbers in each box show the correlation coefficient, where crossed boxes indicate non-significant correlation values (p > 0.05). d Box plots of T cell exhaustion scores in PCM, MBM, and ECM. The data is displayed using the Tukey method for box and whiskers. Kruskal–Wallis test with Benjamini–Hochberg correction. e Percentage of PCM, MBM, and ECM with CD8 + T cells that have high dysfunction scores (composite z-score > 1.0) but low FGFBP2 mRNA levels (less than the median for the whole cohort). χ2 test; ****p < 0.0001. f Oncoplot with T cell dysfunction score (top, highest to lowest within PCM, blue; MBM, orange; ECM, gray), FGFBP2 mRNA levels (middle), and CD8 + T cell abundance (bottom).
To better understand cytokine shifts contributing to the altered TME in MBM, we analyzed the gene expression of several critical cytokines between PCM, MBM, and ECM. Interestingly, TNF and IL12B gene expression in MBM was significantly lower compared to PCM (TNF, p < 0.0001; IL12B, p < 0.005) and ECM (TNF, p < 0.0001; IL12B, p < 0.005; Fig. 2b), and significantly correlated with computationally inferred B cell, NK cell, and Treg immune cell fractions in MBM (p < 0.05; Fig. 2c). Although we observed no difference in CD8+ immune cell fraction measured by quanTIseq between PCM, MBM, and ECM, TNF also correlated with CD8 + T cells in MBM tumors (Fig. 2c). Together, these results suggest that immune cell infiltration is overall reduced in MBM especially compared to PCM, and associated reductions in cytokine levels in MBM may contribute to the more immune excluded phenotype.
T cell dysfunction and STING scores among MBM
Within the heterogeneous TME, dysfunctional CD8 + T cells may be observed, as characterized by increased expression of inhibitory checkpoint molecules such as TIM3, LAG3, and PD-1, as well as the loss of classical CD8 + T cell effector functions including proliferative capacity and cytotoxicity30–32. Therefore, we were interested in further defining the state of the CD8 + T cells infiltrated in MBM, even though the transcriptomic data suggested that the total abundance of CD8 + T cells was comparable between PCM, MBM, and ECM (Fig. 2a). We evaluated the T cell dysfunction scores—defined by the sum of z-scores of HAVCR2 (TIM3), LAG3, and PDCD1 (PD-1) mRNA levels—as well as mRNA levels for the cytotoxic T cell marker FGFBP230. T cell dysfunction scores trended toward higher in MBM relative to PCM (p = 0.08; Fig. 2d), and among tumors with non-zero CD8 + T cell infiltration, the percentage of tumors with transcriptomic signatures indicating non-functional CD8 + T cells was significantly lower among MBM (p < 0.0001; non-functional CD8 + T cells were defined by high dysfunction scores [composite z-score >1.0] but low FGFBP2 mRNA levels [less than median TPM for cohort]; Fig. 2e). These results contribute toward a picture of MBM as possessing a more immune-suppressed TME, particularly relative to PCM. Tumors with transcriptomes associated with high CD8 + T cell infiltration also exhibited greater dysfunction scores regardless of anatomic site (i.e., PCM, ECM, MBM; Fig. 2f).
T cell responses have also been shown to be affected by STING signaling33,34, with various predicted outcomes on immune response and metastasis35–37. A positive association was observed between composite STING scores (comprising mRNA levels of key regulators of the STING pathway [TMEM173, CGAS, CCL5, CXCL10 and IRF3]), computationally inferred CD8 + T cell infiltration, and T cell dysfunction scores among all melanoma tumors (p < 0.0001; Supplementary Fig. 3a–c) and among each anatomic site (p < 0.0001; Supplementary Fig. 3d). Despite this correlation, T cell dysfunction scores trended toward higher among MBM compared to PCM (Fig. 2d), while STING (TMEM173) mRNA levels and STING scores were significantly reduced in MBM compared to both PCM and ECM (p < 0.05; Supplementary Fig. 3e, f). These data suggest that further analysis of the effect of STING signaling on tumor immunity in MBM may be warranted.
PTEN mutation-associated angiogenic factors among MBM
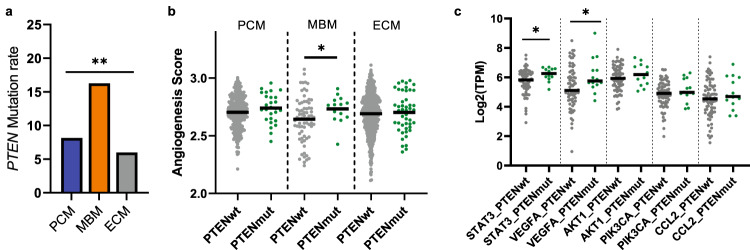
To determine the frequency of non-synonymous somatic mutations enriched in MBM that could possibly contribute to reduced immune responses or altered immune cell trafficking, DNA sequencing data was analyzed. BRAF, NRAS, and NF1 are the most commonly mutated genes in cutaneous melanomas38, while PTEN loss is associated with brain metastasis15; therefore, we examined mutations in these key genes among PCM, MBM, and ECM. Among MBM samples with available NGS data, there were 44/92 samples with pathogenic mutations affecting BRAF (47.8%), 20/91 with NRAS (22.0%), and 28/41 with NF1 (68.3%; Supplementary Data 2). When assessing for molecular or immunologic correlates by driver mutation status, TMB-high tumors were more prevalent in BRAF-mut MBM relative to BRAF-mut PCM, and neutrophils were enriched in NRAS-mut MBM relative to NRAS-mut PCM (Supplementary Fig. 4). Driver mutations notwithstanding, our data demonstrate significantly higher pathogenic PTEN mutation rates in MBM (16.3%) with respect to PCM (8.1%) and ECM (6.0%, p < 0.005; Fig. 3a and Supplementary Data 2). Previous studies found that PTEN-deficient tumors have higher levels of VEGFA and STAT3, which promote angiogenesis and impair the trafficking of immune cells into the TME22,39. In addition, inhibition of the JAK-STAT3 pathway in PTEN-null prostate cancers increased immune infiltration and promoted a more immunogenic TME40. Therefore, we sought to evaluate the correlation of pathogenic (loss-of-function) PTEN mutations with angiogenesis via single sample gene set enrichment analysis (ssGSEA) in PCM, MBM, and ECM. A significant enrichment of angiogenic signaling was observed in PTEN-mut compared to PTEN wild-type (wt) MBM (p < 0.05), but not in PCM or ECM (Fig. 3b). Moreover, simultaneous elevation of VEGFA (p < 0.05) and STAT3 (p < 0.05) mRNA levels was observed in PTEN-mut MBM (Fig. 3c), suggesting a relationship between PTEN-mut and angiogenic signaling uniquely in MBM, which may contribute to its altered TME.
Fig. 3. Association of angiogenic factors with PTEN mutations in MBM.
a The percentage of PCM (blue, n = 332), MBM (orange, n = 86), and ECM (gray, n = 801) with PTEN mutations. Chi-square test; **p < 0.005. b ssGSEA analysis for normalized enrichment scores (NES) of hallmark angiogenesis pathway comparing tumors with PTEN mutations (green) and without PTEN mutations (gray) within PCM, MBM, and ECM groups. Black line indicates median. Mann-Whitney test; *p < 0.05. c The mRNA levels of key regulatory genes (STAT3, VEGFA, AKT1, PIK3CA, and CCL2) in the angiogenesis pathway were cross compared between MBM tumors with PTEN mutations (green) and without PTEN mutations (gray). Black line indicates median. Mann-Whitney test; *p < 0.05.
Differentially expressed genes and associated pathways in MBM
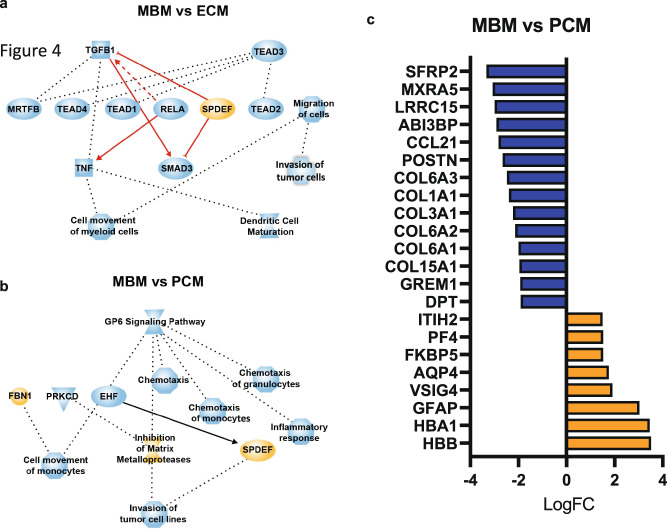
Transcriptomic profiles were assessed to determine if they would explain the differences in inferred immune cell abundance between MBM and PCM or ECM. Significantly differentially expressed genes (DEGs) were identified between MBM and ECM and between MBM and PCM (Supplementary Data 3), followed by the assessment of differentially regulated pathways via Ingenuity Pathway Analysis (IPA). Movement of myeloid cells and DC maturation pathways were less enriched in MBM compared to ECM, and chemotaxis and inflammatory responses were less enriched in MBM compared to PCM (Fig. 4a). In addition, signaling involving the collagen receptor platelet glycoprotein VI (GP6) was less enriched in MBM compared to PCM (Fig. 4b), which likely represents the differing milieu of the brain relative to collagen-rich skin.
Fig. 4. Differentially expressed genes in MBM compared to PCM and ECM.
Ingenuity pathway analysis (IPA) shows a significant enrichment of pathways in MBM compared to ECM (a) and PCM (b). Orange node, enrichment in MBM; blue node, enrichment in ECM or PCM. c Differentially expressed genes (Log2FC; adjusted p < 0.01) in MBM compared to PCM. Only genes that were differentially expressed in MBM compared to both PCM and ECM are shown. Orange bars indicate genes that were upregulated in MBM and blue bars indicate genes that were downregulated in MBM.
To better understand the unique transcriptomic features of MBM, we further examined DEGs between MBM and PCM/ECM (Supplementary Data 3). Hemoglobin subunit beta (HBB) and hemoglobin subunit alpha 1 (HBA1) mRNA levels were significantly upregulated in MBM (Fig. 4c), although their upregulation in MBM may reflect a hemorrhagic component of the tumor as opposed to a true biological difference. Furthermore, v-set and immunoglobulin domain containing 4 (VSIG4) and platelet factor 4 (PF4), which are negatively associated with T cell function and proliferation41–43, were overexpressed in MBM relative to PCM (Fig. 4c). Conversely, extracellular matrix-related genes, including several collagens, were downregulated in MBM relative to PCM, as expected due to anatomic site (Fig. 4c). Notably, CCL21, which stimulates chemotaxis of mDCs and helps leukocytes traverse the blood-brain barrier44, was downregulated among MBM compared to PCM, illustrating potential cytokine shifts supporting altered immune infiltration in MBM. While there was no correlation of mDC abundance measured by quanTIseq with CCL21 mRNA, there was a moderate positive correlation between CCL21 mRNA and CD8 + T cell abundance in MBM (Spearman r = 0.32, p = 0.0017; Supplementary Fig. 5a, b). These observations suggest a distinct environment in MBM that might impact on both the abundance and functionality of infiltrating immune cells.
Enrichment of oxidative phosphorylation pathway in MBM
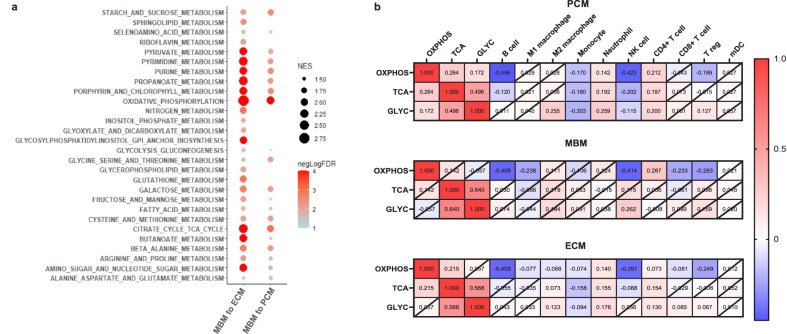
Fischer et al. and Biermann et al. reported significant immunosuppression and enrichment of oxidative phosphorylation (OXPHOS) in MBM compared to ECM21,45. To confirm these findings in our large melanoma cohort, we performed GSEA analysis for KEGG metabolic pathways. This analysis demonstrated that the most significant pathways enriched in MBM were OXPHOS, the TCA cycle, various amino acid metabolism, followed by glycolysis (Fig. 5a and Supplementary Data 4), which was particularly striking when comparing MBM to ECM. Interestingly, individual normalized enrichment scores (NES) for OXPHOS and the TCA cycle were not correlated in MBM and only weakly in PCM and ECM (Fig. 5b). In agreement with the lack of correlation between OXPHOS and the TCA cycle in MBM, we noted that OXPHOS, but not TCA cycle or glycolysis enrichment, significantly correlated with transcriptional signatures indicating a lower abundance of B cells (Spearman r = −0.41), M1 macrophages (Spearman r = −0.238), NK cells (Spearman r = −0.41), CD8 + T cells (Spearman r = −0.23), and Tregs (Spearman r = −0.26, p < 0.05 for all), demonstrating a potential negative impact of OXPHOS on these immune infiltrates in a TCA-independent manner (Fig. 5b).
Fig. 5. MBM show enrichment in oxidative phosphorylation, TCA cycle, and glycolysis gluconeogenesis pathways.
a GSEA analysis demonstrates a significant enrichment in of KEGG metabolic pathways in MBM versus PCM (left lane) or ECM (right lane). The normalized enrichment score (NES) is indicated by the size of the dot and the -log10 p value (cut off FDR < 0.05) is indicated by the color of the dot. b Heat maps of Spearman rank correlations between NES of OXPHOS, TCA cycle (TCA), and glycolysis (GLYC) determined from ssGSEA and computationally inferred immune cell abundance in PCM, MBM, and ECM. The numbers in each box show the correlation coefficient, where crossed boxes indicate non-significant correlation values (p > 0.05).
Discussion
The management of MBM remains an area of complexity and clinical challenge for healthcare providers treating patients with advanced melanoma. While the promise of immune checkpoint blockade in treating MBM has been an important breakthrough, there are still many unanswered questions, including how to select patients who will benefit from immunotherapy and how to overcome resistance. As such, an improved understanding of the unique biological features of MBM and the brain microenvironment is critical to identifying additional barriers and opportunities for therapy. In this study, we utilized a large cohort of 94 MBM, 350 PCM, and 870 ECM samples for which comprehensive genomic and transcriptomic profiling was readily available, in order to assess underlying biological differences in MBM compared to PCM and ECM. To our knowledge, this is one of the largest such studies to date.
Given the molecular heterogeneity of brain metastases, the role of putative biomarkers such as PD-L1 and TMB may have limited predictive value for MBM. Prior studies have shown that PD-L1 expression may vary between anatomic sites and does not clearly correlate with the presence of CD8 + T cells and other immune infiltrates13,20,46. Meanwhile, TMB is related to neoantigen load and correlates with improved outcomes in several cancer types, but the absolute relationship between TMB and immunogenicity or response to immunotherapy may also depend upon the tumor type or the specific genomic landscape and immune repertoire of each patient47. For instance, Biermann and colleagues demonstrated spatial heterogeneity of type-I IFN responses in MBM despite broad expression of antigen presentation genes45. The current study further corroborates that PD-L1, TMB status, or neoantigen load are not necessarily representative of functional immunogenicity, and may be poor biomarkers of immunotherapy response in MBM.
While a healthy blood-brain barrier normally restricts T cell extravasation into the brain, the high concentration of tumor-associated T cells in MBM illustrates a breakdown of this barrier18,48 and necessitates a thorough understanding of factors affecting the quantity and function of MBM immune infiltrate. Despite a robust T cell infiltrate in MBM, studies have found reduced ImmuneScores, mDCs, NK/T cells, B lineage cells, and neutrophils and increased exhausted T cells in matched and unmatched MBM cohorts relative to PCM or ECM21,45,49. Alvarez-Breckenridge et al. furthermore found that while absolute T cell infiltration did not predict response to immune checkpoint blockade in MBM, peripheral T cell clonal expansion was associated with improved responses, and intracranial clonally expanded T cells were more likely to be exhausted50. Our analysis is consistent with the above findings, identifying reduced immune cell recruitment, lower IFNγ and T cell-inflamed signature scores, and transcriptional signatures suggesting a trend toward increased dysfunctional CD8 + T cells in MBM. The decrease in computationally inferred B cells in MBM was the most pronounced and significant immune cell difference relative to both PCM and EMC, meriting further investigation as humoral immunity is increasingly recognized to play a significant role in promoting an inflammatory milieu and helping mediate responses to immune checkpoint blockade in melanoma51,52. These data are a timely addition to single-cell analyses of MBM showing the full range of intratumoral differentiation from naïve B cells to activated B cells to plasma cells, with increased plasma cell aggregates present in MBM compared to ECM45,49.
While multiple mechanisms may contribute to the distinct TME of MBM, our correlative analysis suggests that cytokine shifts, genomic differences in rates of pathogenic PTEN mutations, and altered metabolism may each contribute to these observations. Interestingly, Fischer and colleagues found no differences in PTEN protein loss that could explain the reduced immunogenicity they observed in MBM21. Notwithstanding, their group previously reported that loss of PTEN upregulated VEGF and CCL2 and resulted in lower responses to immunotherapy in a mouse model of melanoma and that VEGF and PTEN protein expression inversely correlated in clinical samples22. Together with our findings that pathogenic PTEN mutations correlate with angiogenic signaling in MBM, these results emphasize the potential role of unrestricted PI3K-AKT signaling in dampening immune responses. Therefore, we hypothesize that patients exhibiting loss of PTEN tumor suppressor activity may be more likely to benefit from a combined therapeutic approach involving anti-angiogenesis therapy with immunotherapy, as is currently being investigated in a phase 2 clinical trial enrolling untreated MBM patients (NCT03175432).
The most striking finding from GSEA was the enrichment of OXPHOS metabolic pathways in MBM, agreeing with major findings by Fischer et al. and Biermann et al. in smaller cohorts21,45. Interestingly, OXPHOS may be a mechanism of metastatic spread53 and resistance to MAPK inhibitors in melanoma54,55. IACS-010759, an OXPHOS inhibitor currently in early-phase trials, inhibited MBM formation and prolonged survival in a spontaneous MBM mouse model21. Our data suggest that the metabolic diversity enacted through OXPHOS and glycolysis fluctuation present in a large percentage of melanomas56 may also occur in MBM, which displayed significant enrichment of OXPHOS and the TCA cycle relative to PCM and ECM, and to a lesser degree glycolysis and gluconeogenesis relative to ECM tumors. This conclusion is further supported by findings by Biermann et al. indicating heterogeneity of OXPHOS metabolism in MBM at both a single cell level and spatially45. Beyond metabolic flexibility, the emerging field of immunometabolism suggests there may be a connection between metabolism and immune cell effector functions57, potentially explaining the negative correlation between OXPHOS and NK cell, M1 macrophage, B cell, CD8 + T cell, and Treg abundance determined from bulk transcriptomic data in MBM. In agreement with these findings, Li et al. determined that a subset of CD8 + T cells with high OXPHOS promotes immunotherapy resistance in melanoma58. Taken together, our findings highlight several possible immunosuppressive mechanisms in MBM that underscore the need for a deeper understanding of potential therapeutic vulnerabilities and biomarkers of immunotherapy response in MBM.
Considering that multiple immune inhibitory mechanisms may be simultaneously active within MBM, a number of clinical trials are investigating whether combinations of BRAF, MEK1/2, AKT/PI3K, CTLA-4, PD-1, and VEGF targeted therapy will be more successful, as opposed to monotherapy approaches2,59,60. Investigation of mechanisms contributing to dampened immune responses in MBM, as we present here, is essential to improve long-term responses to these therapies and expand the number of eligible patients. To this point, two phase I/II trials are enrolling patients to evaluate the combination of metformin (an inhibitor of OXPHOS) with vemurafenib (NCT01638676) or dabrafenib and trametinib (NCT02143050) in patients with metastatic melanoma with activating mutations in the BRAF gene. However, additional pharmacologic approaches for modulating OXPHOS and tumor immunogenicity in MBM should also be explored.
One major limitation of the present study is that this retrospective, correlative analysis does not allow the precise determination of mechanisms connecting cytokine signaling, PTEN-mut/angiogenesis, and OXPHOS to reduced immunogenicity in MBM; the bulk-profiling nature of the study also further limits our understanding of intra-lesional heterogeneity. Likewise, while we only detected a trend toward increased T cell dysfunction scores in MBM relative to PCM, examination of alternative transcriptional signatures representing T cell exhaustion or dysfunction could be informative. Future experimental studies can utilize the hypothesis-generating results from this large, “real-world” MBM cohort to decipher mechanisms involved in regulating the TME in MBM. An additional limitation is the lack of clinical annotation for this melanoma cohort, including stage and detailed treatment information (i.e., whether patients had received therapy prior to tissue collection), as systemic targeted therapy or immunotherapy is known to affect the tumor microenvironment of MBM49. Finally, the ECM and MBM cohorts were not strictly exclusive of non-cutaneous primary sites; however, the rarity of such tumors likely limits any potential effect on results due to our relatively large cohort size. Despite these limitations, the overall concordance of our results with the two prior studies by Fischer et al. and Biermann et al., each of which used smaller sample sizes and different analytic techniques21,45, substantially supports our conclusions regarding immunosuppression in MBM.
In summary, this retrospective study of 1314 melanoma samples contributes to existing data that TMB, PD-L1, and other traditional biomarkers are poor predictors of immunogenicity in MBM, which instead may engage multiple intersecting pathways for immune evasion, including cytokine signaling, angiogenesis, and OXPHOS metabolism. Future studies must define how targeting OXPHOS affects the tumor immune microenvironment mechanistically and determine the clinical benefit of targeting such pathways to improve immunotherapy responses in MBM.
Methods
Patient samples
A multi-institutional cohort of 1,314 patient samples, including PCM (n = 350), MBM (n = 94), and ECM (n = 870) from the Caris Life Sciences database were analyzed in a CLIA/CAP-certified laboratory (Caris Life Sciences, Phoenix, AZ). Analyses performed include next-generation sequencing (NGS), whole transcriptome sequencing (WTS), and immunohistochemistry (IHC) for molecular and genomic features including tumor mutational burden, tumor mutations, HLA homozygosity, neoantigen load, PD-L1 positivity, and mRNA levels. A breakdown of patient specimens, including the number of samples, tumor site, age, and sex, is shown in Supplementary Table 1. The data was not further analyzed according to age or sex.
Next-generation sequencing
NGS was performed on genomic DNA isolated from formalin-fixed paraffin-embedded (FFPE) tumor samples using the NextSeq or NovaSeq 6000 platforms (Illumina, Inc., San Diego, CA). Prior to molecular testing, tumor enrichment was achieved by harvesting targeted tissue using manual microdissection techniques. For NextSeq-sequenced tumors, a custom-designed SureSelect XT assay was used to enrich 592 whole-gene targets (Agilent Technologies, Santa Clara, CA). For NovaSeq whole exome-sequenced tumors, a hybrid pull-down panel of baits designed to enrich for more than 700 clinically relevant genes at high coverage and high read-depth was used, along with another panel designed to enrich for additional >20,000 genes at a lower depth. A 500 Mb SNP backbone panel (Agilent Technologies, Santa Clara, CA) was added to assist with gene amplification/deletion measurements and other analyses. All variants were detected with >99% confidence, with an average sequencing depth of coverage of > 500 and an analytic sensitivity of 5%. This test has a sensitivity to detect as low as approximately 10% population of cells containing a mutation in all exons from the high read-depth clinical genes and 99% of all exons in the 20 K whole exome regions. Genetic variants identified were interpreted by board-certified molecular geneticists and categorized as ‘pathogenic,’ ‘likely pathogenic,’ ‘variant of unknown significance,’ ‘likely benign,’ or ‘benign,’ according to the American College of Medical Genetics and Genomics (ACMG) standards. When assessing mutation frequencies of individual genes, ’pathogenic,’ and ‘likely pathogenic’ were counted as mutations while ‘benign’, ‘likely benign’ variants, and ‘variants of unknown significance’ were excluded. Variants detected were mapped to reference genome (hg19), and well-established bioinformatics tools such as BWA, SamTools, GATK, and snpFF were incorporated to perform variant calling functions; germline variants were filtered with various germline databases, including 1000 Genomes and dbSNP.
Tumor mutational burden
Tumor mutational burden (TMB) was measured (592 genes and 1.4 megabases [MB] sequenced per tumor) by counting all non-synonymous missense mutations found per tumor that had not been previously described as germline alterations according to dbSNP and 1KG databases. A cutoff of ≥10 mutations per MB (mt/MB) was used based on the KEYNOTE-158 pembrolizumab trial, which showed that patients with a TMB of ≥ 10 mt/MB across several tumor types had higher response rates than patients with a TMB of <10 mt/MB61.
Neoantigen load
HLA genotyping and immune epitope prediction of peptides was performed to quantify neoantigen load. HLA genotyping was performed using arcasHLA62. If a single HLA genotype was detected, the specimen was classified as “homozygous”, which can occur due to parental homozygosity or HLA loss of heterozygosity (LOH). MHC-I binding prediction was performed for all HLA allele/peptide combinations for each tumor using the NetMHCpan v4.0 method in the Immune Epitope Database (IEDB) online resource at http://tools.iedb.org/mhci. Peptide affinity for MHC-I molecules was categorized based on guidelines described on the IEDB website: high affinity [inhibitory concentration 50% (IC50) < 50 nmol/L], intermediate affinity (IC50 > 50 nmol/L and < 500 nmol/L), low affinity (IC50 > 500 and < 5000 nmol/L), and no affinity (IC50 > 5000 nmol/L).
Whole transcriptome sequencing
Tumor-specific RNA was extracted from microdissected FFPE specimens with at least 10% tumor content using a Qiagen RNeasy FFPE Kit. RNA quality and quantity were determined using the Agilent TapeStation. Biotinylated RNA baits were hybridized to the synthesized and purified cDNA targets, and the bait-target complexes were amplified in a post-capture PCR reaction. WTS was performed using the Illumina Novaseq 6500 to an average of 60 M reads. Raw data were demultiplexed by Illumina Dragen BioIT accelerator, trimmed, counted, PCR-duplicates removed, and aligned to the human reference genome hg19 by STAR aligner. Transcripts per million (TPM) were calculated using the Salmon expression pipeline. Differentially expressed genes (DEGs) were determined by calculating log2 fold-change between defined groups, and genes were considered significantly different when adjusted p < 0.05. Ingenuity Pathway Analysis (IPA) was used to determine differentially regulated pathways between PCM and MBM and between MBM and ECM. Significantly enriched pathways in MBM relative to PCM or ECM were determined using Gene Set Enrichment Analysis (GSEA) to calculate normalized enrichment score (NES) and significance level adjusted for multiple hypothesis testing63. Single sample GSEA (ssGSEA) was used to calculate individual enrichment scores for samples relative to specific gene sets (e.g., angiogenesis score).
Immune cell abundance and scoring
Immune cell abundance was calculated from WTS data using quanTIseq29. QuanTIseq is a computational pipeline that measures the abundance of ten immune cell populations from bulk transcriptomic data. The algorithm is highly validated against flow cytometry and immunohistochemistry for the deconvolution of absolute fractions of myeloid dendritic cells (DCs), T regulatory (Tregs), CD8+ and CD4 + T cells, natural killer (NK) cells, neutrophils, monocytes, M1 and M2 macrophages, and B cells. Signature scores were calculated using defined gene sets for interferon (IFN)24, T cell-inflammation25, exhausted T cells (HAVCR2, LAG3, PDCD1), and STING pathway (TMEM173, CGAS, CCL5, CXCL10, IRF3)64,65.
Immunohistochemistry
Immunohistochemistry (IHC) was performed on FFPE sections on glass slides. Automated staining techniques were utilized per the manufacturer’s instructions and were optimized and validated per CLIA/CAP and ISO requirements. The primary antibody used for PD-L1 was clone SP142 (Roche/Ventana Medical Systems Inc., Oro Valley, AZ, cat. #740-4859), which is provided by the manufacturer at the optimal dilution for use. A board-certified pathologist evaluated all IHC results independently, and the staining was regarded as positive if its intensity on the membrane of the tumor cells was ≥ 2+ (on a semiquantitative scale of 0–3: 0 for no staining, 1+ for weak staining, 2+ for moderate staining, or 3 + for strong staining) and the percentage of positively stained cells was ≥ 5%.
Statistics
Kruskal-Wallis test with Benjamini-Hochberg post-hoc correction or two-sided Mann-Whitney test were used to determine significant differences in continuous data between groups analyzed. Fisher exact or Chi-square tests were used to determine significant differences in the prevalence of binary variables between groups. Spearman rank or Pearson correlation were used for correlation analyses. All measurements were taken from distinct samples. Standard statistical methods in R (version 4) and GraphPad Prism (version 9) were employed to analyze the data. For all tests, p < 0.05 (or corrected p < 0.05, where applicable) was considered statistically significant.
Ethics statement
This study was conducted in accordance with guidelines set forth in the Declaration of Helsinki, Belmont report, and U.S. Common rule. Per 45 CFR 46.101(b)(4) for retrospective studies using de-identified biospecimens and clinical data, this study was considered IRB exempt by Western IRB and informed consent was not required.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Differentially expressed genes between MBM and ECM or PCM
Acknowledgements
This work was supported by NIH grant number P30 CA014089 (G.K.I.), R01CA255240-01A1 (F.I.) and Adelson Medical Research Foundation (D.S.B.H.).
Author contributions
G.K.I. was responsible for conceptualization, methodology, investigation, supervision, writing-original draft, and writing-review/editing. J.Y. and J.X. performed data curation and formal analysis. J.R.R. performed formal analysis and was responsible for writing-original draft/review/editing. F.I., F.C., G.Z., L.H., S.D., M.L.A., and K.M. contributed to investigation, supervision, and writing-review/editing. A.K.S.S., S.P., J.C.M., M.A.B., D.S.B.H., E.D., J.L., G.T.G., B.I., M.B.A., and A.M.V. contributed to conceptualization, methodology, investigation, supervision, and writing-review/editing. All authors read and approved the manuscript.
Data availability
The datasets generated and/or analyzed during the current study are available for replication and verification purposes from the corresponding author on reasonable request. The de-identified DNA and RNA sequencing data are owned by Caris Life Sciences and cannot be publicly shared due to the data usage agreement signed by Dr. Heinz-Josef Lenz at Keck School of Medicine of USC. Qualified researchers can apply for access to these data by contacting Joanne Xiu, PhD at jxiu@carisls.com, submitting a brief proposal, and signing a data usage agreement. The summarized sequencing data are available within the Supplementary Materials for this paper and at https://figshare.com/s/4f0dbb3aaf58d17f4615.
Competing interests
G.K.I.: Research funding (paid to institution): Regeneron, Array, Idera, Genentech, Replimune, Xencor, InstilBio, Pfizer, Checkmate Pharmaceuticals; Consulting or Advisory role: Bristol Myers Squibb, Regeneron, Sanofi, Replimune, Pfizer, Novartis. J.Y., J.X., J.R.R., and A.M.V.: employees of Caris Life Sciences. S.D.: Oncolens, Bayer, and BostonGene. A.K.S.S.: Research funding (paid to institution): Ascentage, Bristol Myers Squibb, Ideaya, Immunocore, Merck, Olatec Therapeutics, Regeneron, Replimune, Seagen; consultant or advisory role: Bristol Myers Squibb, Iovance, Regeneron, Replimune, Novartis, Pfizer. J.C.M.: Research funding (paid to institution): NovoCure (Inst), Genentech, Alpine Immune Sciences, Amgen, Trishula Therapeutics, BioEclipse Therapeutics, FujiFilm, ImmuneSensor, Simcha, Repertoire Immune Sciences, Nektar Therapeutics, Synthorx Inc, Istari Oncology, Ideaya Biosciences, Rubius, University of Arizona, Senwha, Storm Therapeutics, Werewolf Therapeutics, Fate Therapeutics, Y-Mab, Agenus; consultant or advisory role: BMS, Amunix, Thirona Bio, Adagene, Imaging Endpoints, Boxer Capitol, Oberland Capital, IQVIA, Caris Life Sciences, Genome Insight; speakers Bureau: Caris Life Sciences, Immunocore. S.J.P.: Advisory role: Regeneron. B.I.: Consulting fees/honoraria: Volastra Therapeutics Inc, Merck, AstraZeneca, Eisai and Janssen Pharmaceuticals; research funding (paid to institution): Agenus, Alkermes, Arcus Biosciences, Checkmate Pharmaceuticals, Compugen, Immunocore, and Synthekine.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41698-023-00471-z.
References
- 1.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J. Clin. 2022;72:7–33. doi: 10.3322/caac.21708. [DOI] [PubMed] [Google Scholar]
- 2.Gutzmer R, et al. Melanoma brain metastases - Interdisciplinary management recommendations 2020. Cancer Treat. Rev. 2020;89:102083. doi: 10.1016/j.ctrv.2020.102083. [DOI] [PubMed] [Google Scholar]
- 3.Janavicius M, Lachej N, Anglickiene G, Vincerzevskiene I, Brasiuniene B. Outcomes of treatment for melanoma brain metastases. J. Skin Cancer. 2020;2020:7520924. doi: 10.1155/2020/7520924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tawbi HA, et al. Combined nivolumab and ipilimumab in melanoma metastatic to the brain. N. Engl. J. Med. 2018;379:722–730. doi: 10.1056/NEJMoa1805453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Larkin J, et al. Combined nivolumab and ipilimumab or monotherapy in untreated melanoma. N. Engl. J. Med. 2015;373:23–34. doi: 10.1056/NEJMoa1504030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robert C, et al. Nivolumab in previously untreated melanoma without BRAF mutation. N. Engl. J. Med. 2015;372:320–330. doi: 10.1056/NEJMoa1412082. [DOI] [PubMed] [Google Scholar]
- 7.Kluger HM, et al. Long-term survival of patients with melanoma with active brain metastases treated with pembrolizumab on a phase II trial. J. Clin. Oncol. 2019;37:52–60. doi: 10.1200/JCO.18.00204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.The Cancer Genome Atlas Network. Genomic classification of cutaneous melanoma. Cell161, 1681–1696 (2015). [DOI] [PMC free article] [PubMed]
- 9.Rabbie R, et al. The mutational landscape of melanoma brain metastases presenting as the first visceral site of recurrence. Br. J. Cancer. 2021;124:156–160. doi: 10.1038/s41416-020-01090-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hayward NK, et al. Whole-genome landscapes of major melanoma subtypes. Nature. 2017;545:175–180. doi: 10.1038/nature22071. [DOI] [PubMed] [Google Scholar]
- 11.Curtin JA, et al. Distinct sets of genetic alterations in melanoma. N. Engl. J. Med. 2005;353:2135–2147. doi: 10.1056/NEJMoa050092. [DOI] [PubMed] [Google Scholar]
- 12.Sperduto PW, et al. The prognostic value of BRAF, C-KIT, and NRAS mutations in melanoma patients with brain metastases. Int. J. Radiat. Oncol. Biol. Phys. 2017;98:1069–1077. doi: 10.1016/j.ijrobp.2017.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.In GK, et al. Molecular profiling of melanoma brain metastases compared to primary cutaneous melanoma and to extracranial metastases. Oncotarget. 2020;11:3118–3128. doi: 10.18632/oncotarget.27686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aguissa-Touré A-H, Li G. Genetic alterations of PTEN in human melanoma. Cell. Mol. Life Sci. 2012;69:1475–1491. doi: 10.1007/s00018-011-0878-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bucheit AD, et al. Complete loss of PTEN protein expression correlates with shorter time to brain metastasis and survival in stage IIIB/C melanoma patients with BRAFV600 mutations. Clin. Cancer Res. 2014;20:5527–5536. doi: 10.1158/1078-0432.CCR-14-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tehranian C, et al. The PI3K/Akt/mTOR pathway as a preventive target in melanoma brain metastasis. Neuro Oncol. 2022;24:213–225. doi: 10.1093/neuonc/noab159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nguyen B, et al. Genomic characterization of metastatic patterns from prospective clinical sequencing of 25,000 patients. Cell. 2022;185:563–575.e511. doi: 10.1016/j.cell.2022.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berghoff AS, et al. Density of tumor-infiltrating lymphocytes correlates with extent of brain edema and overall survival time in patients with brain metastases. Oncoimmunology. 2016;5:e1057388. doi: 10.1080/2162402X.2015.1057388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mukherji B. Immunology of melanoma. Clin. Dermatol. 2013;31:156–165. doi: 10.1016/j.clindermatol.2012.08.017. [DOI] [PubMed] [Google Scholar]
- 20.Berghoff AS, et al. Tumour-infiltrating lymphocytes and expression of programmed death ligand 1 (PD-L1) in melanoma brain metastases. Histopathology. 2015;66:289–299. doi: 10.1111/his.12537. [DOI] [PubMed] [Google Scholar]
- 21.Fischer GM, et al. Molecular profiling reveals unique immune and metabolic features of melanoma brain metastases. Cancer Discov. 2019;9:628–645. doi: 10.1158/2159-8290.CD-18-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peng W, et al. Loss of PTEN promotes resistance to T cell-mediated immunotherapy. Cancer Discov. 2016;6:202–216. doi: 10.1158/2159-8290.CD-15-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bellenghi M, et al. Sex and gender disparities in melanoma. Cancers (Basel) 2020;12:1819. doi: 10.3390/cancers12071819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cristescu R, et al. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science. 2018;362:eaar3593. doi: 10.1126/science.aar3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spranger S, et al. Density of immunogenic antigens does not explain the presence or absence of the T-cell-inflamed tumor microenvironment in melanoma. Proc. Natl Acad. Sci. USA. 2016;113:E7759–e7768. doi: 10.1073/pnas.1609376113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park S-J, Yoon B-H, Kim S-K, Kim S-Y. GENT2: an updated gene expression database for normal and tumor tissues. BMC Med. Genomics. 2019;12:101. doi: 10.1186/s12920-019-0514-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mardis ER. Neoantigens and genome instability: impact on immunogenomic phenotypes and immunotherapy response. Genome Med. 2019;11:71. doi: 10.1186/s13073-019-0684-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shim JH, et al. HLA-corrected tumor mutation burden and homologous recombination deficiency for the prediction of response to PD-(L)1 blockade in advanced non-small-cell lung cancer patients. Ann. Oncol. 2020;31:902–911. doi: 10.1016/j.annonc.2020.04.004. [DOI] [PubMed] [Google Scholar]
- 29.Finotello F, et al. Molecular and pharmacological modulators of the tumor immune contexture revealed by deconvolution of RNA-seq data. Genome Med. 2019;11:34. doi: 10.1186/s13073-019-0638-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H, et al. Dysfunctional CD8 T cells form a proliferative, dynamically regulated compartment within human melanoma. Cell. 2019;176:775–789.e718. doi: 10.1016/j.cell.2018.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thommen DS, Schumacher TN. T cell dysfunction in cancer. Cancer Cell. 2018;33:547–562. doi: 10.1016/j.ccell.2018.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wherry EJ, Kurachi M. Molecular and cellular insights into T cell exhaustion. Nat. Rev. Immunol. 2015;15:486–499. doi: 10.1038/nri3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.An M, et al. Induction of necrotic cell death and activation of STING in the tumor microenvironment via cationic silica nanoparticles leading to enhanced antitumor immunity. Nanoscale. 2018;10:9311–9319. doi: 10.1039/C8NR01376D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Woo SR, et al. STING-dependent cytosolic DNA sensing mediates innate immune recognition of immunogenic tumors. Immunity. 2014;41:830–842. doi: 10.1016/j.immuni.2014.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li J, et al. Metastasis and immune evasion from extracellular cGAMP hydrolysis. Cancer Discov. 2021;11:1212–1227. doi: 10.1158/2159-8290.CD-20-0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bakhoum SF, et al. Chromosomal instability drives metastasis through a cytosolic DNA response. Nature. 2018;553:467–472. doi: 10.1038/nature25432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li S, et al. STING-induced regulatory B cells compromise NK function in cancer immunity. Nature. 2022;610:373–380. doi: 10.1038/s41586-022-05254-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Davis EJ, Johnson DB, Sosman JA, Chandra S. Melanoma: what do all the mutations mean? Cancer. 2018;124:3490–3499. doi: 10.1002/cncr.31345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.George S, et al. Loss of PTEN is associated with resistance to anti-PD-1 checkpoint blockade therapy in metastatic uterine leiomyosarcoma. Immunity. 2017;46:197–204. doi: 10.1016/j.immuni.2017.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Toso A, et al. Enhancing chemotherapy efficacy in Pten-deficient prostate tumors by activating the senescence-associated antitumor immunity. Cell Rep. 2014;9:75–89. doi: 10.1016/j.celrep.2014.08.044. [DOI] [PubMed] [Google Scholar]
- 41.Vogt L, et al. VSIG4, a B7 family-related protein, is a negative regulator of T cell activation. J. Clin. Investig. 2006;116:2817–2826. doi: 10.1172/JCI25673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu CY, et al. Platelet factor 4 differentially modulates CD4+CD25+ (regulatory) versus CD4+CD25- (nonregulatory) T cells. J. Immunol. 2005;174:2680–2686. doi: 10.4049/jimmunol.174.5.2680. [DOI] [PubMed] [Google Scholar]
- 43.Fleischer J, et al. Platelet factor 4 inhibits proliferation and cytokine release of activated human T cells. J. Immunol. 2002;169:770–777. doi: 10.4049/jimmunol.169.2.770. [DOI] [PubMed] [Google Scholar]
- 44.Nguyen T, et al. Insights into CCL21’s roles in immunosurveillance and immunotherapy for gliomas. J. Neuroimmunol. 2017;305:29–34. doi: 10.1016/j.jneuroim.2017.01.010. [DOI] [PubMed] [Google Scholar]
- 45.Biermann J, et al. Dissecting the treatment-naive ecosystem of human melanoma brain metastasis. Cell. 2022;185:2591–2608.e2530. doi: 10.1016/j.cell.2022.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weiss SA, et al. Melanoma brain metastases have lower T-cell content and microvessel density compared to matched extracranial metastases. J. Neurooncol. 2021;152:15–25. doi: 10.1007/s11060-020-03619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jardim DL, Goodman A, de Melo Gagliato D, Kurzrock R. The challenges of tumor mutational burden as an immunotherapy biomarker. Cancer Cell. 2021;39:154–173. doi: 10.1016/j.ccell.2020.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tawbi HA, Boutros C, Kok D, Robert C, McArthur G. New era in the management of melanoma brain metastases. Am. Soc. Clin. Oncol. Educ. Book. 2018;38:741–750. doi: 10.1200/EDBK_200819. [DOI] [PubMed] [Google Scholar]
- 49.Smalley I, et al. Single-cell characterization of the immune microenvironment of melanoma brain and leptomeningeal metastases. Clin. Cancer Res. 2021;27:4109–4125. doi: 10.1158/1078-0432.CCR-21-1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alvarez-Breckenridge C, et al. Microenvironmental landscape of human melanoma brain metastases in response to immune checkpoint inhibition. Cancer Immunol. Res. 2022;10:996–1012. doi: 10.1158/2326-6066.CIR-21-0870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Willsmore ZN, et al. B cells in patients with melanoma: implications for treatment with checkpoint inhibitor antibodies. Front. Immunol. 2020;11:622442. doi: 10.3389/fimmu.2020.622442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Griss J, et al. B cells sustain inflammation and predict response to immune checkpoint blockade in human melanoma. Nat. Commun. 2019;10:4186. doi: 10.1038/s41467-019-12160-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.LeBleu VS, et al. PGC-1α mediates mitochondrial biogenesis and oxidative phosphorylation in cancer cells to promote metastasis. Nat. Cell Biol. 2014;16:992–1003. doi: 10.1038/ncb3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang G, et al. Targeting mitochondrial biogenesis to overcome drug resistance to MAPK inhibitors. J. Clin. Investig. 2016;126:1834–1856. doi: 10.1172/JCI82661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gopal YV, et al. Inhibition of mTORC1/2 overcomes resistance to MAPK pathway inhibitors mediated by PGC1α and oxidative phosphorylation in melanoma. Cancer Res. 2014;74:7037–7047. doi: 10.1158/0008-5472.CAN-14-1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vazquez F, et al. PGC1α expression defines a subset of human melanoma tumors with increased mitochondrial capacity and resistance to oxidative stress. Cancer Cell. 2013;23:287–301. doi: 10.1016/j.ccr.2012.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gardiner CM. NK cell metabolism. J. Leukoc. Biol. 2019;105:1235–1242. doi: 10.1002/JLB.MR0718-260R. [DOI] [PubMed] [Google Scholar]
- 58.Li C, et al. A high OXPHOS CD8 T cell subset is predictive of immunotherapy resistance in melanoma patients. J. Exp. Med. 2022;219:e20202084. doi: 10.1084/jem.20202084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rulli E, Legramandi L, Salvati L, Mandala M. The impact of targeted therapies and immunotherapy in melanoma brain metastases: a systematic review and meta-analysis. Cancer. 2019;125:3776–3789. doi: 10.1002/cncr.32375. [DOI] [PubMed] [Google Scholar]
- 60.Salvati L, Mandalà M, Massi D. Melanoma brain metastases: review of histopathological features and immune-molecular aspects. Melanoma Manag. 2020;7:Mmt44. doi: 10.2217/mmt-2019-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Marabelle, A. et al. Association of tumor mutational burden with outcomes in patients with select advanced solid tumors treated with pembrolizumab in KEYNOTE-158. Ann. Oncol.30, v475–532 (2019).
- 62.Orenbuch R, et al. arcasHLA: high-resolution HLA typing from RNAseq. Bioinformatics. 2020;36:33–40. doi: 10.1093/bioinformatics/btz474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.An X, et al. An analysis of the expression and association with immune cell infiltration of the cGAS/STING pathway in pan-cancer. Mol. Ther.-Nucleic Acids. 2019;14:80–89. doi: 10.1016/j.omtn.2018.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Della Corte CM, et al. STING pathway expression identifies NSCLC with an immune-responsive phenotype. J. Thorac. Oncol. 2020;15:777–791. doi: 10.1016/j.jtho.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Differentially expressed genes between MBM and ECM or PCM
Data Availability Statement
The datasets generated and/or analyzed during the current study are available for replication and verification purposes from the corresponding author on reasonable request. The de-identified DNA and RNA sequencing data are owned by Caris Life Sciences and cannot be publicly shared due to the data usage agreement signed by Dr. Heinz-Josef Lenz at Keck School of Medicine of USC. Qualified researchers can apply for access to these data by contacting Joanne Xiu, PhD at jxiu@carisls.com, submitting a brief proposal, and signing a data usage agreement. The summarized sequencing data are available within the Supplementary Materials for this paper and at https://figshare.com/s/4f0dbb3aaf58d17f4615.