Abstract
A Pseudomonas putida strain, strain GB-1, oxidizes Mn2+ to Mn oxide in the early stationary growth phase. It also secretes a siderophore (identified as pyoverdine) when it is subjected to iron limitation. After transposon (Tn5) mutagenesis several classes of mutants with differences in Mn2+ oxidation and/or secretion of the Mn2+-oxidizing activity were identified. Preliminary analysis of the Tn5 insertion site in one of the nonoxidizing mutants suggested that a multicopper oxidase-related enzyme is involved in Mn2+ oxidation. The insertion site in another mutant was preliminarily identified as a gene involved in the general protein secretion pathway. Two mutants defective in Mn2+-oxidizing activity also secreted porphyrins into the medium and appeared to be derepressed for pyoverdine production. These strains were chosen for detailed analysis. Both mutants were shown to contain Tn5 insertions in the ccmF gene, which is part of the cytochrome c maturation operon. They were cytochrome oxidase negative and did not contain c-type cytochromes. Complementation with part of the ccm operon isolated from the wild type restored the phenotype of the parent strain. These results indicate that a functional ccm operon is required for Mn2+ oxidation in P. putida GB-1. A possible relationship between porphyrin secretion resulting from the ccm mutation and stimulation of pyoverdine production is discussed.
In a number of studies during the last three decades it has been shown that various microbial species are able to stimulate the oxidation of Mn2+ through direct catalysis. These organisms produce proteinaceous macromolecules which catalyze the oxidation reaction. Manganese oxidations by a soil Arthrobacter species (24), Oceanospirillum and Vibrio strains (2, 3), Pseudomonas putida MnB1 (22, 30), Leptothrix discophora SS-1 (1, 11), and marine Bacillus strain SG-1 (23) are examples in which enzymes are most likely involved in the process. P. putida MnB1 produces a soluble protein which catalytically oxidizes Mn2+ in cell extracts (22). Manganese-oxidizing proteins from L. discophora SS-1 (1, 11) and from the spore coats of Bacillus strain SG-1 (43) have been identified on polyacrylamide gels. The oxidizing proteins have not been quantitatively purified or analyzed so far. In Bacillus strain SG-1, an operon containing seven genes appears to be involved in Mn2+ oxidation (46). One of these genes encodes a 137-kDa protein related to the family of multicopper oxidases (47). In a previous study we reported the isolation of a structural gene and its promoter postulated to be involved in Mn2+ oxidation in L. discophora (19). The encoded protein also contains the copper-binding signatures of multicopper oxidases. The oxidase-related proteins may represent Mn2+-oxidizing enzymes (44), but evidence supporting this hypothesis is still lacking.
In this paper we describe a genetic analysis of Mn2+ oxidation in a freshwater Pseudomonas strain, strain GB-1. In a previous study (32) this strain was preliminarily identified as a Pseudomonas fluorescens strain, but more recent data (see Materials and Methods) indicate that it should be identified as a P. putida strain. When supplied with Mn2+ ions, the cells deposit manganese oxide around the outer membrane in the early stationary growth phase (32). They form brown colonies on Mn2+-containing agar. Experiments performed with cell extracts indicated that Mn2+ oxidation is catalyzed by a protein. The Mn2+-oxidizing factor was partially purified, and electrophoresis on an acrylamide gradient gel revealed oxidizing proteins with apparent molecular weights of ca. 250,000 and 180,000 (32). An additional oxidizing factor with a lower molecular weight (ca. 130,000) was identified in another study by using different isolation and electrophoretic procedures (16). It has been suggested that the Mn2+-oxidizing protein isolated is part of a larger complex which disintegrates into smaller fragments that retain activity (32). The protein is supposed to be located in the outer membrane of the bacteria. It has not been chemically characterized, and nothing is known about its cellular function or about the possible involvement of other cellular components, such as electron carriers, in Mn2+ oxidation.
We used transposon mutagenesis to identify genes relevant to the Mn2+-oxidizing process in P. putida GB-1. One of these genes appeared to be part of the cytochrome c maturation operon. Transposon insertion in this gene not only abolished Mn2+ oxidation but also led to secretion of siderophores and porphyrins.
An accompanying report on the involvement of the cytochrome c maturation operon in Mn2+ oxidation in P. putida MnB1 (14) supports our findings.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Pseudomonas strain GB-1 was kindly provided by K. H. Nealson (Jet Propulsion Laboratory, Pasadena, Calif.). Preliminary identification of this strain as a P. fluorescens strain (32) was based on sequence homology data for a short stretch of 16S ribosomal DNA. However, P. fluorescens strains are able to liquefy gelatin (13) and secrete lipase and protease (21), activities which appeared not to be displayed by strain GB-1. More extensive 16S ribosomal DNA sequence analysis identified GB-1 as a P. putida strain (42a). Consequently, the strain is renamed P. putida GB-1.
P. putida GB-1 was routinely grown at room temperature in LD medium as described previously for L. discophora SS-1 (11). This medium contains relatively low amounts of nutrients (0.05% [wt/vol] Casamino Acids, 0.05% [wt/vol] yeast extract, 25 mM glucose) and 10 μM Fe(III), added as Fe(III) EDTA or FeCl3. P. putida GB-1 can utilize both forms of Fe(III). In some experiments, cells were grown on CAA medium (15), which contains 0.5% (w/v) Casamino Acids, 6.6 mM KH2PO4, 1 mM MgSO4, and 30 μM FeCl3. Escherichia coli strains were cultured in Luria-Bertani (LB) medium (38) at 37°C.
Solid media were prepared by adding 1.8% (wt/vol) agar (Gibco BRL) prior to autoclaving. Solid LD medium contained 100 μM MnCl2.
Generation of Tn5 mutants of P. putida GB-1.
The selection markers used for transformation of P. putida GB-1 were resistance to tetracycline (25 μg/ml) and resistance to kanamycin (50 μg/ml). Like most pseudomonads, P. putida GB-1 is resistant to ampicillin; growth is observed at ampicillin concentrations up to 1 mg/ml. Growth is inhibited by streptomycin (100 μg/ml). On streptomycin-containing media, streptomycin-resistant (Smr) colonies were spontaneously generated at a frequency of about 5 × 10−7. One of these colonies (strain GB-1-002) was subcultured and used in experiments in which Smr was an extra phenotypic marker.
Cultures of P. putida GB-1-002 were transformed by electroporation by using standard procedures. Transposon mutants were generated by electroporation with plasmid pBR322::Tn5, which was obtained by insertion of Tn5 (29) at a random site in pBR322 (10). This construct still conferred tetracycline resistance to E. coli. As pBR322 does not replicate in P. putida GB-1-002, the transposants were Tcs and Kmr.
As kanamycin, as well as tetracycline and streptomycin, appeared to inhibit Mn2+ oxidation in cultures (and lysates) of actively oxidizing cells, the Kmr transposants had to be screened for a nonoxidizing phenotype by replica plating them on media without antibiotics. Nonoxidizing strains were checked for retention of kanamycin resistance. The nonoxidizing mutants were designated GB-1-003 through GB-1-009.
Preparation of cell lysates.
Cell lysates were obtained by ultrasonication of a 50-fold-concentrated early-stationary-phase culture in 10 mM HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid) buffer (pH 7.5) at 0°C for 5 min at 20-s intervals (Vibracell; Sonics & Materials Inc.) at maximum amplitude. For a quantitative assay of Mn2+ oxidation the original volume was restored by suspending the lysate in HEPES buffer. For electrophoretic analysis the lysates were centrifuged at 15,000 × g for 10 min, and the supernatants were mixed with an equal volume of 2× Laemmli sample buffer. The samples were not heat denatured in order to allow detection of Mn2+-oxidizing activity.
For spectrophotometric cytochrome analysis, lysates were prepared by passing 50-fold-concentrated cultures in 10 mM Tris buffer (pH 7.0) twice through a French pressure cell at 120 MPa. The lysates were clarified by centrifugation at 15,000 × g for 40 min.
Analytical assays.
Quantitative determination of Mn2+-oxidizing activity with the Leucoberbelin blue assay and analysis of the activity on polyacrylamide gels have been described previously (11, 32).
The cytochrome c oxidase activity of whole cells was determined by streaking colonies from LD medium plates onto filter paper soaked in a 1% tetramethyl-p-phenylenediamine solution (12). Differential spectra for reduced (with sodium dithionite) minus oxidized (with potassium ferricyanide) cytochromes in cell lysates were recorded with a Shimadzu model UV 2101PC double-beam spectrophotometer.
Secretion of siderophores by cells was detected on solid medium [LD medium without Fe(III), pH 7.0] supplemented with the ternary complex chrome azurol S-Fe(III)-hexamethylammonium bromide (CAS) (40). Extraction of Fe(III) from this complex resulted in a color change from blue-green to orange. Siderophores were identified by optical spectroscopy with the Shimadzu spectrophotometer and by fluorescence spectroscopy with a model Spex 1681 fluorometer.
Secreted porphyrins (coproporphyrin and protoporphyrin) were differentially extracted from ether extracts of culture media by using HCl solutions of increasing molarity as described by Tait (42) and were analyzed by optical spectroscopy.
Uptake of Fe by cells was measured in 250-ml cultures in LD medium. Samples (50 ml) were centrifuged, and the supernatants were acidified with HNO3. The cell pellets were washed once with 25 mM HCl, resuspended in 10 ml of 0.5 M HNO3, and heated at 120°C for 20 min. Particulate material was removed by filtration with glass fiber filters (type GFC; Schleicher and Schuell). The Fe contents of culture supernatants, HCl washes, and cell extracts were determined with a Perkin-Elmer model 3100 atomic absorption spectrometer.
DNA isolation.
Genomic DNA was isolated from P. putida GB-1-002 and was purified as described previously for L. discophora and Sphaerotilus natans genomic DNA (17).
Cloning of Tn5-containing fragments.
Genomic DNA of Tn5 mutants were digested with EcoRI and BamHI. Tn5 contains a unique BamHI restriction site upstream of the kanamycin resistance gene and no EcoRI site. The EcoRI-BamHI fragments of GB-1-004 through GB-1-007 and the EcoRI fragments of GB-1-003 and GB-1-008 were ligated into pUC19, transformed into E. coli DH5α, and selected for kanamycin resistance. The resulting plasmids were designated pPLH4EB through pPLH7EB and pPLH3E and pPLH8E, respectively. Plasmid pPLH9EB represented the cloned EcoRI-BamHI fragment from the 18-kb Tn5-containing fragment of GB-1-009.
Construction of a genomic library in the cosmid vector pLAFR3.
Genomic DNA of P. putida GB-1-002 was partially digested with Sau3A. Fragments that were 25 to 30 kb long, which were isolated from sucrose gradients (38), were ligated into BamHI-digested pLAFR3 arms, packaged in vitro as described by Staskewicz et al. (41), and transduced into E. coli DH5α. Transductants carrying a vector with an insert were selected on LB agar containing tetracycline (25 μg/ml) and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside; 50 μg/ml). A genomic library was constructed with 1,920 of the 7 × 104 transductants obtained. The total length of the inserts (average insert length, 27.5 kb) in the library corresponds to about 10 times the length of the genome, which was estimated to be 4 to 5 Mb.
Southern blotting and colony screening.
Southern blotting of genomic DNA digests of Tn5 mutants was performed by using standard methods and digoxigenin (DIG)-labelled pBR322::Tn5 as a probe.
The library was screened by using standard protocols (9). Cells were transferred to a nylon membrane covering solid LB medium containing tetracycline (25 μg/ml). Colonies were grown overnight, the cells were lysed, and the DNA were hybridized to DIG-labelled probes (see below). The DIG labels were visualized by using chemiluminescent or chromogenic detection (9).
Complementation.
Cosmid clones were mobilized from E. coli to nonoxidizing Tn5 mutants by triparental mating by using the helper strain E. coli HB101 harboring plasmid pRK2013 (25). Mixtures (100 μl) containing 108 donor cells, 108 recipient cells, and 108 helper cells were allowed to grow overnight on solid LB medium at 28°C. The cells were suspended in LD medium, plated onto solid LD medium containing tetracycline (25 μg/ml), kanamycin (50 μg/ml), streptomycin (100 μg/ml), and ampicillin (50 μg/ml), and incubated overnight at room temperature. Resistant colonies were screened for Mn2+ oxidation by replica plating on solid LD medium without antibiotics (see above). Complemented mutants were found to retain their antibiotic resistance characteristics during growth on antibiotic-free media. One of the complementing plasmids (pPLH34) was used for further analysis.
DNA sequencing and analysis.
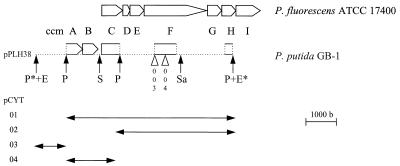
A 6.7-kb EcoRI fragment from pPLH34 was subcloned in pLAFR3 in two orientations, resulting in pPLH37 and pPLH38. Deletions of pPLH38 were obtained by partial digestion with PstI. The resulting cosmids were designated pCYT01 through pCYT04 (Fig. 1). The PstI fragments of pCYT02 and pCYT04 were cloned in pUC19 and (partially) sequenced by using the dideoxynucleotide chain termination method (39).
FIG. 1.
Partial sequence analysis of the cytochrome c maturation operon of P. putida GB-1 compared with that of P. fluorescens ATCC 17400 (27). The (partial) genes are designated ccmA through ccmI, as recently proposed by Page et al. (33). Restriction sites are indicated by arrows (E, EcoRI; P, PstI; S, SmaI; Sa, SalI). Sites marked by an asterisk are part of the multiple cloning region of the complementing cosmid pPLH38. The sites of Tn5 insertions in GB-1-003 and GB-1-004 are indicated by open triangles. Plasmids pCYT01 through pCYT04 were obtained by PstI deletion of pPLH38. The accession numbers for sequences are as follows: ccmA through ccmC, U85716; ccmF, U85717; and ccmH, U85718. b, bases.
The sites of Tn5 insertions in GB-1-004 through GB-1-007 and GB-1-009 were determined by performing a sequence analysis of pPLH4EB through pPLH7EB and pPLH9EB with a Tn5-specific primer (5′-CCGTTCAGGACGCTACTTGT-3′). This primer could not be used to analyze pPLH3E and pPLH8E. The insertion in GB-1-003 was localized by analyzing the EcoRI insert of pPLH3E with a primer (5′-AAGGCCCAGGCGACGATG-3′) based on the partial pCYT02 sequence.
Computer-assisted sequence analyses and homology searches were performed by using programs of the University of Wisconsin Genetics Computer Group (version 8.1).
RESULTS
Generation of nonoxidizing mutants.
Transposon mutagenesis of strain GB-1-002 with pBR322::Tn5 yielded 1,000 to 2,000 Kmr colonies per μg of plasmid DNA. Screening of the transposants for Mn2+-oxidizing activity resulted in seven white colonies out of a total of 11,000 Kmr clones. The white colonies were subcultured and designated GB-1-003 through GB-1-009. Their growth rates in the logarithmic phase did not differ significantly from the growth rate of the parent strain, but the lag phases of some of the mutants (GB-1-003, GB-1-004, and GB-1-007) were longer than the lag phase of GB-1-002.
A nonoxidizing phenotype may result from inactivation of the Mn2+-oxidizing process (the oxidizing factor or an ancillary component) and also from inhibited secretion of the factor. Previously, we described a spontaneous white mutant of L. discophora SS-1 which was unable to secrete the Mn2+-oxidizing activity (18). To screen for this type of mutant, suspensions of disrupted cells were assayed for Mn2+ oxidation. In three lysates (GB-1-006, GB-1-008, and GB-1-009) the activity was at least partially recovered; the activities of GB-1-008 and GB-1-009 approached the activity of the parent cells (Table 1). Lysates of GB-1-003, GB-1-004, and GB-1-005 did not oxidize Mn2+ at all, and the GB-1-007 lysate had only a low level of activity (1% of the activity of the parent strain). The absolute values for Mn2+ oxidation rates varied among different batches depending on the growth conditions (32). The relative activities in one experiment as presented in Table 1 were reproducible within a 5% range.
TABLE 1.
Mn2+-oxidizing activities of P. putida GB-1 and Tn5 mutants
At least two Mn2+-oxidizing factors were previously identified in lysates of P. putida GB-1 by electrophoretic analysis (16). These factors were also detected in lysates of GB-1-002 and in lysates of GB-1-006, GB-1-008, and GB-1-009 but not in lysates of the other mutant strains (Table 1). These data suggest that Mn2+ oxidation in P. putida GB-1 is (at least partially) mediated by these factors, since they could not be detected in strains in which the oxidizing activity was (almost) completely abolished.
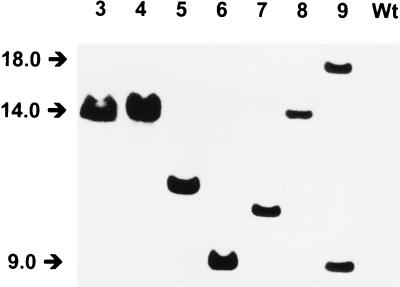
The genomic DNA of the Tn5 mutants were completely digested with EcoRI, which has no sites in Tn5, and the digests were analyzed on Southern blots by using pBR322::Tn5 as a probe (Fig. 2). Strains GB-1-003 through GB-1-008 each contained a single hybridizing fragment, which varied in length from 9.0 to 14.5 kb. Strain GB-1-009 contained two hybridizing fragments (9.0 and 18.0 kb), apparently resulting from a double Tn5 insertion. The Tn5-containing EcoRI fragments of GB-1-003 and GB-1-004 were of equal length (ca. 14 kb), and the hybridization patterns of EcoRI-BamHI double digests (Tn5 contains a unique BamHI restriction site) were similar (data not shown). The latter two mutants were obtained from independent transposition experiments. In GB-1-003 and GB-1-004 the transposon probably inserted in a narrow DNA region. The probe of plasmid pBR322, which was used as a control, gave no hybridization signal.
FIG. 2.
Southern blot of EcoRI digests of genomic DNA of P. putida GB-1 transposon mutants with pPBR322::Tn5 as a probe. Lanes 3 through 9, GB-1-003 through GB-1-009, respectively; lane Wt, GB-1-002. The positions of molecular weight markers are indicated by arrows.
Preliminary identification of inserted genes.
The sites of Tn5 insertion in five of the seven mutants were preliminarily identified by performing a partial sequence analysis of the corresponding EcoRI-BamHI fragments with a Tn5-specific primer. Analysis of the fragments of strains GB-1-005 and GB-1-006 did not reveal significant similarities with protein sequences obtained from databases. One of the insertions in strain GB-1-009 appeared to be located in an xcpT homolog. The insert in pPLH9EB contained a 280-bp open reading frame with 60% similarity (45% identity) (on an amino acid basis) to xcpT of Pseudomonas aeruginosa (4). GB-1-007 contained the Tn5 insertion in a region with two stretches encoding copper binding signatures of multicopper oxidases (HPIHLHGM and HCHVIDHMETG [conserved residues are in boldface type]) (44). Strain GB-1-004 appeared to be mutated in a gene homologous to ccmF of various microbial species (see below). Studies to conclusively analyze the mutations in GB-1-005 through GB-1-009 are under way, and the results will be reported elsewhere. Here we focused on the mutations in strains GB-1-003 and GB-1-004, which were postulated to be inserted in the same DNA region.
Complementation of the mutations in strains GB-1-003 and GB-1-004.
The GB-1-002 library was screened with the DIG-labelled probes of pPLH3E and pPLH4EB. Ten positive clones were isolated, and all hybridized with both inserts. The corresponding pLAFR constructs, pPLH25 through pPLH34, were mobilized to the nonoxidizing mutants. Eight of these cosmids restored the Mn2+-oxidizing phenotype in GB-1-003, as well as in GB-1-004. None of the other mutants was complemented by pPLH25 through pPLH34.
Restriction analysis showed that six of the complementing cosmids contained the same 6.7-kb EcoRI fragment. This fragment was isolated from pPLH34 and cloned into pLAFR in two orientations, which yielded pPLH37 and pPLH38. Only pPLH38 (Fig. 1) conferred the Mn2+-oxidizing phenotype to GB-1-003 and GB-1-004, indicating that expression was initiated from the cosmid vector. By PstI deletion of pPLH38, subclones pCYT01 through pCYT04 were obtained; only two of these subclones, pCYT01 and pCYT02, were capable of complementation (Fig. 1).
Analysis of the complementing insert.
Sequence analysis of the pCYT02 insert resulted in identification of two partial open reading frames, one of which exhibited high degrees of homology to the ccmF genes of various microbial strains, including P. fluorescens strains and P. putida MnB1 (Table 2). The ccmF gene was found to carry the Tn5 insertion in GB-1-004 (Fig. 1) (see above). This gene is part of the cytochrome c maturation operon (33, 45). In this study we used the nomenclature for the ccm genes (ccmA through ccmI) proposed by Page et al. (33). The partial CcmF protein of P. putida GB-1 contained the heme binding and/or ligation motif, which is conserved in all of the CcmF proteins analyzed so far (45). Analogous to the organization of the cytochrome c maturation operon of, for instance, P. fluorescens ATCC 17400 (27), a partial open reading frame was found downstream of the ccmF gene, and a preliminary sequence analysis of this partial open reading frame revealed 65% homology (on a nucleotide basis) to ccmH. The sequence determined was too short to allow detection of the motif LRCXXC, which is conserved in CcmH proteins. Sequence analysis of pCYT04 resulted in identification of ccmA, ccmB, and ccmC homologs upstream of the ccmF gene (Fig. 1 and Table 2). The CcmA protein contained two conserved nucleotide binding sites (Walker motifs) (48), and CcmB and CcmC both contained six putative membrane-spanning helices (data not shown) (45). The region between helix III and helix IV of CcmC contained a conserved tryptophan-rich motif thought to be involved in heme binding (45). The intergenomic region between ccmB and ccmC did not contain obvious transcription termination or promoter sequences.
TABLE 2.
Comparison of Ccm proteins
| Protein | Organism | Former designation | % Identity with P. putida GB-1 proteina | Conserved motif(s) | Reference |
|---|---|---|---|---|---|
| CcmA | P. putida GB-1b | GPNGSGKTSLLR LWILDE | This study | ||
| P. putida MnB1b | CycV | 85 | ...GSGKTSLLR | 14 | |
| E. coli | 52 | GSNGAGKTTLLR LWILDE | 8 | ||
| B. japonicum | CycV | 39 | GRNGSGKTSLLR IWLLDE | 36 | |
| R. capsulatus | HelA | 39 | GPNGIGKTTLLR VWVLDE | 5 | |
| CcmB | P. putida GB-1 | This study | |||
| E. coli | 52 | 8 | |||
| B. japonicum | CycW | 41 | 36 | ||
| R. capsulatus | HelB | 50 | 5 | ||
| CcmC | P. putida GB-1b | WGSWWVWDARLT | This study | ||
| P. fluorescens | CytA | 86 | WGSWWVWDARLT | 27 | |
| E. coli | 59 | WGTWWVWDARLT | 8 | ||
| B. japonicum | CycZ | 46 | WGTYWEWDARLT | 36 | |
| R. capsulatus | HelC | 43 | WGTWWEWDPRLT | 5 | |
| CcmF | P. putida GB-1b | WAYYELGWGGWWFWDPVEN | This study | ||
| P. putida MnB1b | CykK | 94 | 14 | ||
| P. fluorescens | CytD | 96 | WAYYELGWGGWWFWDPVEN | 27 | |
| E. coli | 80 | WAYYELGWGGWWFWDPVEN | 8 | ||
| B. japonicum | CykK | 74 | WAYYELGWGGWWFWDPVEN | 37 | |
| R. capsulatus | Ccl1 | 71 | WAYYELGWGGFWFWDPVEN | 5 |
Based on amino acid sequences.
Partial sequence.
As determined by restriction analysis of pPLH3E, the Tn5 insertion in GB-1-003 was preliminarily localized ca. 300 bp upstream of the GB-1-004 insertion (Fig. 1). The exact site of Tn5 insertion in pPLH3E could be resolved by primer walking by using a primer based on the 5′ sequence of the partial ccmF gene. In GB-1-003, the transposon appeared to be located in ccmF also.
Determination of c-type cytochromes.
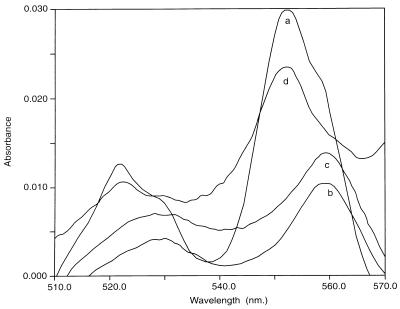
As the nonoxidizing mutants GB-1-003 and GB-1-004 appeared to be mutated in a gene involved in cytochrome c maturation, we compared their cytochrome oxidase activities (12) with those of the parent strain and the other mutants. Both GB-1-003 and GB-1-004 were cytochrome oxidase negative, whereas all other strains were cytochrome oxidase positive. Differential spectroscopy showed that the c-type cytochromes present in the parent strain (Fig. 3, line a) and in mutants GB-1-005 through GB-1-009 (data not shown) were absent in GB-1-003 and GB-1-004 (Fig. 3, lines b and c). Complemented strains GB-1-003 and GB-1-004 had oxidase activity and contained c-type cytochromes (Fig. 3, line d).
FIG. 3.
Differential spectra of reduced-minus-oxidized cytochromes in cell lysates of P. putida GB-1-002 (line a), GB-1-003 (line b), GB-1-004 (line c), and GB-1-003 complemented with pPLH38 (line d).
Other pleiotropic effects of the ccmF mutations.
A number of recent studies have described mutations in the ccm operons of various microorganisms which resulted in defective siderophore synthesis or secretion (27, 34, 50) and/or accumulation or secretion of heme precursors (6, 28, 50). We investigated whether P. putida mutants GB-1-003 and GB-1-004 displayed similar defects.
Strains GB-1-003 and GB-1-004 did not appear to be defective in siderophore secretion. On LD agar plates containing CAS the colonies produced bright halos, in contrast to the parent strain or any of the other mutants, which secreted only minor amounts of siderophore (Fig. 4). On LD medium without added Fe(III), GB-1-003 and GB-1-004 secreted green fluorescent pigments in the stationary growth phase, whereas the other strains hardly produced these substances. LD medium is not well-defined, since it contains yeast extract and a variety of trace elements. Moreover, all of the strains had low maximum densities (optical density at 660 nm, 0.3) due to the low nutrient concentration in LD medium. Apparently, none of the strains except both ccmF mutants experienced Fe limitation under the conditions used.
FIG. 4.
Secretion of siderophores by P. putida GB-1-003 and GB-1-004 compared to secretion of siderophores by GB-1-002 and GB-1-005 during growth on LD medium containing CAS. Extraction of iron from the ternary CAS complex resulted in a change in color from dark blue to bright orange.
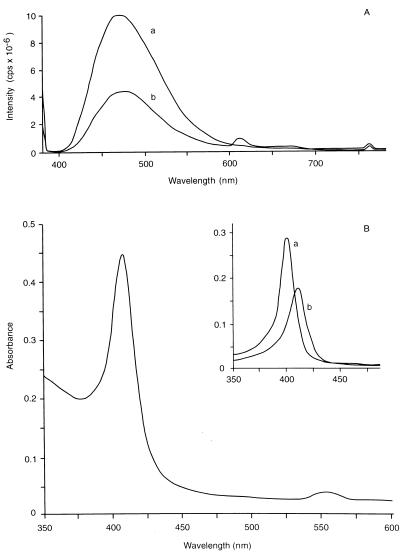
For identification and comparison of secreted siderophores, the cells were grown on CAA medium from which Fe(III) was omitted. All of the strains reached an optical density at 660 nm of 1.2 on this medium and secreted a green fluorescent pigment, which could be identified as pyoverdine on the basis of its spectroscopic properties. The absorption spectrum had a broad maximum at 409 nm, which shifted to lower values (ca. 400, 380, and 360 nm) upon acidification, as reported previously for pyoverdine of P. fluorescens (data not shown) (35). Addition of Fe(III) resulted in a loss of fluorescence, and a shoulder at 450 nm appeared in the absorption spectrum. Fluorescence spectroscopy revealed an excitation maximum at 410 nm and a broad emission maximum around 475 nm (Fig. 5A). These data are consistent with physicochemical parameters previously reported for pyoverdine (31). We concluded that the parent strain and all of the mutant strains could secrete pyoverdine under iron-limiting conditions. Strains GB-1-003 and GB-1-004 secreted green fluorescent siderophores under conditions which repressed secretion in the other strains. In complemented GB-1-003 and GB-1-004 cultures siderophore secretion was also repressed under these conditions.
FIG. 5.
Spectroscopic analysis of siderophores and porphyrins secreted by ccmF mutants of P. putida GB-1. (A) Fluorescence emission spectra of spent media (CAA medium without Fe) from GB-1-002 (line a) and GB-1-003 (line b) cultures, with excitation at 380 nm. (B) Optical spectrum of GB-1-003 medium (CAA medium with Fe) acidified with HCl, with an absorption maximum intermediate between that of coproporphyrin (401 nm) and that of protoporphyrin (410 nm). (Inset) Spectra of 0.15 M HCl (line a) and 1.5 M HCl (line b) extracts obtained by acid extraction of ether-dissolved porphyrins, representing coproporphyrin and protoporphyrin, respectively (42).
To investigate whether the deviant siderophore metabolism of GB-1-003 and GB-1-004 was due to defective Fe(III) uptake, we monitored the Fe concentrations in cells and culture media during growth on LD medium. Both mutant strains, as well as the parent strain, accumulated ca. 90% of the Fe from the medium within 3 days. The final cellular Fe contents in the three strains were the same, accounting for 60% of the total Fe added as far as could be determined by the extraction procedure used. The Fe was taken up and was not precipitated on or bound to the cell surface, since only 5 to 10% of the accumulated Fe could be removed by rinsing the cells with 25 mM HCl. Apparently, Fe uptake was not affected in the mutant strains.
The emission spectrum of the GB-1-003 culture supernatant shown in Fig. 5A revealed a small additional peak at 610 nm, which was absent in the spectrum of the parent strain. In contrast to all other strains, GB-1-003 and GB-1-004 also secreted a purple pigment in various amounts, depending on the growth phase and the culture media employed. Especially on CAA medium supplemented with Fe(III), the culture supernatants were intensely purple. The pigment was identified as a mixture of coproporphyrin and protoporphyrin by differential extraction of the culture media (42) and optical spectroscopy (Fig. 5B). The secreted porphyrins were not capable of extracting Fe(III) from the ternary CAS complex used to screen for siderophore secretion. Complemented GB-1-003 and GB-1-004 did not secrete porphyrins into the medium.
DISCUSSION
Mutations in the ccmF gene of the cytochrome c maturation (ccm) operon of the Mn2+-oxidizing organism P. putida GB-1 abolished cytochrome c production and Mn2+ oxidation. The organization of the GB-1 ccm operon, as far as it was analyzed, was very similar to the organization of the ccm operons of P. fluorescens strains (27, 49) and P. putida MnB1 (14). Homologs of the ccm genes (ccmA through ccmI) have been identified in a variety of bacterial species, and their characteristics and postulated functions have been reviewed recently (33, 45). The CcmA, CcmB, and CcmC proteins are supposed to form an ABC transporter, which might be involved in the translocation of heme through the plasma membrane. CcmE, CcmF, CcmH, and CcmI are postulated to be subunits of a cytochrome c heme lyase involved in the covalent attachment of heme to apocytochrome c (49). However, it has recently been proposed that CcmF functions as a transport protein, possibly, but not necessarily, for heme (33, 34). CcmG is thought to function in reduction of the apocytochrome c heme binding site, possibly by interacting with CcmH. Both of these proteins contain the sequence CXXC, which is typical of the thioredoxin-protein disulfide isomerase family.
A genomic P. putida GB-1 DNA fragment containing, inter alia, the complete ccmF gene and the 5′ end of the ccmH gene was found to restore cytochrome c production and Mn2+ oxidation in the ccmF mutants. Although the partial sequence did not allow localization of ccmG on this fragment, ccmG should be located between ccmF and ccmH, as it is in the P. fluorescens operon. Assuming that Tn5 insertion in ccmF abolishes expression of the downstream genes of the ccm operon, we cannot decide whether the observed mutant phenotype resulted from the Tn5 insertion in ccmF itself or from a polar effect on ccmG (or possibly ccmH) expression. Transposon insertions in the ccmA, ccmE, and ccmF genes of P. putida MnB1 (14) also resulted in defective cytochrome c synthesis with a concomitant loss of Mn2+-oxidizing activity. All of these data indicate that Mn2+ oxidation depends on an operational ccm operon that involves either a functional end product (c-type cytochromes) or a product of one of the genes in this cluster.
The first possibility mentioned above implies that cytochrome c is involved in electron transfer from Mn2+ to oxygen, possibly as an intermediate in an electron transport chain involving a terminal cytochrome c oxidase. Such an involvement may explain the inhibition of Mn2+ oxidation by NaN3 (32). Alternatively, c-type cytochromes may be part of an Mn2+-oxidizing complex. Such a complex may include an outer membrane, Cu-dependent oxidase. This suggestion is based on the results of preliminary analyses of some of the other P. putida GB-1 mutants. One of the secretion mutants (GB-1-009) appeared to be mutated in xcpT, which is supposed to encode a subunit of the outer membrane translocation pore involved in protein secretion (7). This supports localization of the Mn2+-oxidizing factor in the outer membrane, as previously proposed (32). The results of a partial sequence analysis of mutant GB-1-007, which revealed the presence of Cu binding sites in the gene product, suggest that a protein related to multicopper oxidases is required for Mn2+ oxidation in P. putida GB-1. As enzymes with similar Cu binding sites were identified in Leptothrix (19) and Bacillus (47) species, involvement of this type of oxidase in Mn2+ oxidation seems to be a general feature of Mn2+-oxidizing bacteria.
The second possibility is illustrated by the results of a report of pleiotropic effects of ccm mutations in P. fluorescens 09906 (49). Transposon insertions in most of the genes of the ccm cluster disrupted cytochrome c oxidase activity and copper resistance in this species. However, mutations localized in the 3′ end of ccmI (formerly cycH) resulted in copper sensitivity but not in a loss of oxidase activity, suggesting that not cytochrome c but one of the ccm products is required for copper resistance. It was suggested that one or both of the thioredoxin-related proteins (CcmG and CcmH) are involved in copper metabolism and that their CXXC domains may interact with copper ions. A dual role for periplasmic thioredoxin-like proteins in cytochrome c maturation and copper metabolism has also been reported in E. coli (20, 26). If the copper oxidase-related protein of P. putida GB-1 were part of the Mn2+-oxidizing complex of this species, disturbance of the copper metabolism via (polar) mutations in the ccm cluster might result in inactivation of the oxidizing process.
Other pleiotropic effects of ccm mutations involve metabolism of iron. In P. fluorescens ATCC 17400, transposon insertion in ccmC (cytA) resulted in strongly decreased pyoverdine production in addition to defective cytochrome c assembly (27). It has been suggested that CcmC has a function in the transport of pyoverdine in addition to heme translocation. Mutants of Rhizobium leguminosarum affected in ccmF (cycK) failed to make or export siderophores and also accumulated protoporphyrin, the immediate heme precursor, in the periplasm (50). Yeoman et al. (50) speculated that cells accumulating heme (precursors) at this site might sense an iron concentration higher than the true concentration, resulting in repression of siderophore biosynthesis. The ccmF mutants of P. putida GB-1 were able to synthesize and export pyoverdine when they were grown in iron-deficient CAA medium. On LD medium without added Fe(III) and LD medium containing CAS (both of which contain yeast extract, which supplies traces of available iron), the mutant strains did secrete siderophores, in contrast to the parent strain. Although we cannot completely exclude the possibility that a green fluorescent siderophore other than pyoverdine was produced under these circumstances, we propose that the ccmF mutants of P. putida GB-1 secreted pyoverdine under conditions which repressed production of the siderophore in the parent strain. These mutants secreted heme precursors into the medium. Secretion of overproduced heme precursors was also observed in ccmF (ccl1) and ccmABC (helABC) mutants of Rhodobacter capsulatus (6, 28). We suggest that secretion of porphyrins, possibly complexing iron, acted as a signal of iron deficiency, which resulted in activation of the pyoverdine biosynthetic machinery. This may be supported by our preliminary observation that mutant cells grown on CAA medium not only secreted large amounts of porphyrins but also seemed to secrete the Fe which was taken up initially into the medium at a later growth stage (data not shown). This explanation implies that there is a correlation between the amounts of pyoverdine and porphyrins produced, which will be the subject of future experiments. It is also possible that one of the ccm products of P. putida GB-1 not only functions in cytochrome c maturation but plays an additional role in the regulation of pyoverdine production. A role for one of the ccm gene products in siderophore production has also been proposed for P. fluorescens (27) (see above) and Paracoccus denitrificans (34).
ACKNOWLEDGMENTS
We thank Theo Goosen for supplying pBR322::Tn5 and Nora Goosen and Erik Vijgenboom for fruitful discussions during the course of this work. Jos van Brussel is gratefully acknowledged for technical assistance with atomic absorption measurements. We are grateful to Ron Caspi, Bradley Tebo, and Margo Haygood for giving us access to their data prior to publication.
REFERENCES
- 1.Adams L F, Ghiorse W C. Characterization of extracellular Mn2+-oxidizing activity and isolation of an Mn2+-oxidizing protein from Leptothrix discophora SS-1. J Bacteriol. 1987;169:1279–1285. doi: 10.1128/jb.169.3.1279-1285.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arcuri E J, Ehrlich H L. Cytochrome involvement in Mn(II) oxidation by two marine bacteria. Appl Environ Microbiol. 1979;37:916–923. doi: 10.1128/aem.37.5.916-923.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arcuri E J, Ehrlich H L. Electron transfer coupled to Mn(II) oxidation in two deep-sea Pacific Ocean isolates. In: Trudinger P A, Walter M R, Ralph B J, editors. Biogeochemistry of ancient and modern environments. Berlin, Germany: Springer-Verlag; 1980. pp. 339–344. [Google Scholar]
- 4.Bally M, Filloux A, Akrim M, Ball G, Lazdunski A, Tommassen J. Protein secretion of Pseudomonas aeruginosa: characterization of seven xcp genes and processing of secretory apparatus components by prepilin peptidase. Mol Microbiol. 1992;6:1121–1131. doi: 10.1111/j.1365-2958.1992.tb01550.x. [DOI] [PubMed] [Google Scholar]
- 5.Beckman D L, Trawick D R, Kranz R G. Bacterial cytochromes c biogenesis. Genes Dev. 1992;6:268–283. doi: 10.1101/gad.6.2.268. [DOI] [PubMed] [Google Scholar]
- 6.Biel S, Biel A J. Isolation of a Rhodobacter capsulatus mutant that lacks c-type cytochromes and excretes porphyrins. J Bacteriol. 1990;172:1321–1326. doi: 10.1128/jb.172.3.1321-1326.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bitter W, Koster M, Latijnhouwers M, de Cock H, Tommassen J. Formation of oligomeric rings by XcpQ and PilD, which are involved in protein transport across the outer membrane of Pseudomonas aeruginosa. Mol Microbiol. 1998;27:209–219. doi: 10.1046/j.1365-2958.1998.00677.x. [DOI] [PubMed] [Google Scholar]
- 8.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davus N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 9.Boehringer Mannheim GmbH. DIG nonradioactive DNA labelling and detection applications manual. Mannheim, Germany: Boehringer Mannheim GmbH; 1989. [Google Scholar]
- 10.Bolivar F, Rodriguez R L, Greene P J, Betlach M C, Heyneker H L, Boyer H W, Crosa J H, Falkow S. Construction and characterization of new cloning vehicles. II. A multiple cloning system. Gene. 1977;2:95. [PubMed] [Google Scholar]
- 11.Boogerd F C, de Vrind J P M. Manganese oxidation by Leptothrix discophora. J Bacteriol. 1987;169:489–494. doi: 10.1128/jb.169.2.489-494.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bott M, Bolliger M, Hennecke H. Genetic analysis of the cytochrome c-aa3 branch of the Bradyrhizobium japonicum respiratory chain. Mol Microbiol. 1990;21:777–785. doi: 10.1111/j.1365-2958.1990.tb00576.x. [DOI] [PubMed] [Google Scholar]
- 13.Breed R S, Murray E G D, Hitchens A P, editors. Bergey’s manual of determinative bacteriology. 6th ed. Baltimore, Md: The Williams and Wilkins Company; 1948. p. 97. [Google Scholar]
- 14.Caspi R, Tebo B M, Haygood M G. c-Type cytochromes and manganese oxidation in Pseudomonas putida MnB1. Appl Environ Microbiol. 1998;64:3549–3555. doi: 10.1128/aem.64.10.3549-3555.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cornelis P, Anjaiah V, Koedam N, Delfosse P, Jacques P, Thonart P, Neirinckx L. Stability, frequency and multiplicity of transposon insertions in the pyoverdine region in the chromosome of different fluorescent pseudomonads. J Gen Microbiol. 1992;138:1337–1343. doi: 10.1099/00221287-138-7-1337. [DOI] [PubMed] [Google Scholar]
- 16.Corstjens P L A M. Bacterial oxidation of iron and manganese—a molecular biological approach. Ph. D. thesis. Leiden, The Netherlands: Leiden University; 1993. [Google Scholar]
- 17.Corstjens P L A M, Muyzer G. Phylogenetic analysis of the metal-oxidizing bacteria Leptothrix discophora and Sphaerotilus natans using 16S rDNA sequencing data. Syst Appl Microbiol. 1993;16:219–223. [Google Scholar]
- 18.Corstjens P L A M, de Vrind J P M, Westbroek P, de Vrind-de Jong E W. Enzymatic iron oxidation by Leptothrix discophora: identification of an iron-oxidizing protein. Appl Environ Microbiol. 1992;58:450–454. doi: 10.1128/aem.58.2.450-454.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Corstjens P L A M, de Vrind J P M, Goosen T, de Vrind-de Jong E W. Identification and molecular analysis of the Leptothrix discophora SS-1 mofA gene, a gene putatively encoding a manganese-oxidizing protein with copper domains. Geomicrobiol J. 1997;14:91–108. [Google Scholar]
- 20.Crooke H, Cole J. The biogenesis of c-type cytochromes in Escherichia coli requires a membrane-bound protein, DipZ, with a protein disulphide isomerase-like domain. Mol Microbiol. 1995;15:1139–1150. doi: 10.1111/j.1365-2958.1995.tb02287.x. [DOI] [PubMed] [Google Scholar]
- 21.de Groot A, Filloux A, Tommassen J. Conservation of xcp genes, involved in the two-step protein secretion process, in different Pseudomonas species and other Gram-negative bacteria. Mol Gen Genet. 1991;229:278–284. doi: 10.1007/BF00272167. [DOI] [PubMed] [Google Scholar]
- 22.DePalma S R. Manganese oxidation by Pseudomonas putida. Ph. D. thesis. Cambridge, Mass: Harvard University; 1993. [Google Scholar]
- 23.de Vrind J P M, de Vrind-de Jong E W, de Voogt J-W H, Westbroek P, Boogerd F C, Rosson R A. Manganese oxidation by spores and spore coats of a marine Bacillus species. Appl Environ Microbiol. 1986;52:1096–1100. doi: 10.1128/aem.52.5.1096-1100.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ehrlich H L. Bacteriology of manganese nodules. II. Manganese oxidation by a cell-free extract from a manganese nodule bacterium. Appl Microbiol. 1968;16:197–202. doi: 10.1128/am.16.2.197-202.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Figurski D H, Helinski D R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fong S-T, Camakaris J, Lee B T O. Molecular genetics of a chromosomal locus involved in copper tolerance in Escherichia coli K-12. Mol Microbiol. 1995;15:1127–1137. doi: 10.1111/j.1365-2958.1995.tb02286.x. [DOI] [PubMed] [Google Scholar]
- 27.Gaballa A, Koedam N, Cornelis P. A cytochrome c biogenesis gene involved in pyoverdine production in Pseudomonas fluorescens ATCC 17400. Mol Microbiol. 1996;21:777–785. doi: 10.1046/j.1365-2958.1996.391399.x. [DOI] [PubMed] [Google Scholar]
- 28.Goldman B S, Beckman D L, Bali A, Monika E M, Gabbert K S, Kranz R G. Molecular and immunological analysis of an ABC transporter complex required for cytochrome c biogenesis. J Mol Biol. 1997;268:724–738. doi: 10.1006/jmbi.1997.0992. [DOI] [PubMed] [Google Scholar]
- 29.Jorgensen R A, Rothstein S J, Reznikoff W S. A restriction cleavage map of Tn5 and location of a region encoding neomycin resistance. Mol Gen Genet. 1979;177:65–72. doi: 10.1007/BF00267254. [DOI] [PubMed] [Google Scholar]
- 30.Jung W K, Schweisfurth R. Manganese oxidation by an intracellular protein of a Pseudomonas species. Z Allg Mikrobiol. 1979;19:107–115. [PubMed] [Google Scholar]
- 31.Marugg J D, van Spanje M, Hoekstra W P M, Schippers B, Weisbeek P J. Isolation and analysis of genes involved in siderophore biosynthesis in plant-growth-stimulating Pseudomonas putida WCS358. J Bacteriol. 1985;164:563–570. doi: 10.1128/jb.164.2.563-570.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okazaki M, Sugita T, Shimizu M, Ohode Y, Iwamoto K, de Vrind-de Jong E W, de Vrind J P M, Corstjens P L A M. Partial purification and characterization of manganese-oxidizing factors of Pseudomonas fluorescens GB-1. Appl Environ Microbiol. 1997;63:4793–4799. doi: 10.1128/aem.63.12.4793-4799.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Page M D, Sambongi Y, Ferguson S J. Contrasting routes of c-type cytochrome assembly in mitochondria, chloroplasts and bacteria. Trends Biochem Sci. 1998;23:103–108. doi: 10.1016/s0968-0004(98)01173-6. [DOI] [PubMed] [Google Scholar]
- 34.Pearce D A, Page M D, Norris H A C, Tomlinson E J, Ferguson S J. Identification of the contiguous Paracoccus denitrificans ccmF and ccmH genes: disruption of ccmF, encoding a putative transporter, results in formation of an unstable apocytochrome c and deficiency in siderophore production. Microbiology. 1998;144:467–477. doi: 10.1099/00221287-144-2-467. [DOI] [PubMed] [Google Scholar]
- 35.Philson S B, Llinás M. Siderochromes from Pseudomonas fluorescens. I. Isolation and characterization. J Biol Chem. 1982;257:8081–8085. [PubMed] [Google Scholar]
- 36.Ramseier T M, Winteler H V, Hennecke H. Discovery and sequence analysis of bacterial genes involved in the biogenesis of c-type cytochromes. J Biol Chem. 1991;266:7793–7803. [PubMed] [Google Scholar]
- 37.Ritz D, Bott M, Hennecke H. Formation of several bacterial c-type cytochromes requires a novel membrane-anchored protein that faces the periplasm. Mol Microbiol. 1993;9:729–740. doi: 10.1111/j.1365-2958.1993.tb01733.x. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 39.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schwyn B, Neilands J B. Universal chemical assay for the detection and determination of siderophores. Anal Biochem. 1986;160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- 41.Staskawicz B, Dahlbeck D, Keen N, Napoli C. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J Bacteriol. 1987;169:5789–5794. doi: 10.1128/jb.169.12.5789-5794.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tait G H. Coproporphyrinogenase activities in extracts of Rhodopseudomonas spheroides and Chromatium strain D. Biochem J. 1972;128:1159–1169. doi: 10.1042/bj1281159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42a.Tebo, B. M. Personal communication.
- 43.Tebo B M, Mandernack K, Rosson R A. Abstracts of the 88th Annual Meeting of the American Society for Microbiology 1988. Washington, D.C: American Society for Microbiology; 1988. Manganese oxidation by a spore coat or exosporium protein from spores of a manganese(II) oxidizing marine Bacillus, abstr. no. I-121. [Google Scholar]
- 44.Tebo B M, Ghiorse W C, van Waasbergen L G, Siering P L, Caspi R. Bacterially-mediated mineral formation: insights into manganese(II) oxidation from molecular genetic and biochemical studies. In: Banfield J F, Nealson K H, editors. Geomicrobiology: interactions between microbes and minerals. Washington, D.C: Mineralogical Society of America; 1997. pp. 225–266. [Google Scholar]
- 45.Thöny-Meyer L. Biogenesis of respiratory cytochromes in bacteria. Microbiol Mol Biol Rev. 1997;61:337–376. doi: 10.1128/mmbr.61.3.337-376.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Waasbergen L G, Hoch J A, Tebo B M. Genetic analysis of the marine manganese-oxidizing Bacillus sp. strain SG-1: protoplast transformation, Tn917 mutagenesis, and identification of chromosomal loci involved in manganese oxidation. J Bacteriol. 1993;175:7594–7603. doi: 10.1128/jb.175.23.7594-7603.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Waasbergen L G, Hildebrand M, Tebo B M. Identification and characterization of a gene cluster involved in manganese oxidation by spores of the marine Bacillus sp. strain SG-1. J Bacteriol. 1996;178:3517–3530. doi: 10.1128/jb.178.12.3517-3530.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walker J E, Saraste M, Runswick M J, Gay N J. Distantly related sequences in the a and b subunits of ATP synthase, myosin kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang C-H, Azad H R, Cooksey D A. A chromosomal locus required for copper resistance, competitive fitness, and cytochrome c biogenesis in Pseudomonas fluorescens. Proc Natl Acad Sci USA. 1996;93:7315–7320. doi: 10.1073/pnas.93.14.7315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yeoman K H, Delgado M-J, Wexler M, Allan Downie J, Johnson A W B. High affinity iron acquisition in Rhizobium leguminosarum requires the cycHJKL operon and the feuPQ gene products, which belong to the family of two-component transcriptional regulators. Microbiology. 1997;143:127–134. doi: 10.1099/00221287-143-1-127. [DOI] [PubMed] [Google Scholar]