Abstract
Fluid intelligence encompasses a wide range of abilities such as working memory, problem-solving, and relational reasoning. In the human brain, these abilities are associated with the Multiple Demand Network, traditionally thought to involve combined activity of specific regions predominantly in the prefrontal and parietal cortices. However, the structural basis of the interactions between areas in the Multiple Demand Network, as well as their evolutionary basis among primates, remains largely unexplored. Here, we exploit diffusion MRI to elucidate the major white matter pathways connecting areas of the human core and extended Multiple Demand Network. We then investigate whether similar pathways can be identified in the putative homologous areas of the Multiple Demand Network in the macaque monkey. Finally, we contrast human and monkey networks using a recently proposed approach to compare different species’ brains within a common organizational space. Our results indicate that the core Multiple Demand Network relies mostly on dorsal longitudinal connections and, although present in the macaque, these connections are more pronounced in the human brain. The extended Multiple Demand Network relies on distinct pathways and communicates with the core Multiple Demand Network through connections that also appear enhanced in the human compared with the macaque.
Keywords: white matter, connectivity fingerprint, fluid intelligence, comparative neuroscience, anatomy
Over the last two decades, functional neuroimaging has identified a set of cortical regions that form part of the brain’s response to many different types of cognitive challenge (Duncan 2010). This system, termed the Multiple Demand Network (MDN), consists mostly of specific lateral prefrontal, medial frontal and intraparietal regions, in addition to some temporal areas (Assem et al. 2020a), that are active when cognitive demands are high. The network’s similarity to patterns of activation correlating with performance on intelligence and working memory tests (Bishop et al. 2008) led to direct investigations that showed that the system is crucial for supporting fluid intelligence (Woolgar et al. 2018). It has been proposed that MDN regions lie at the heart of cognitive integration, selecting diverse components of cognitive operations across multiple brain systems and binding them together into appropriate roles and relations (Miller and Cohen 2001; Cole and Schneider 2007; Duncan 2010; Fusi et al. 2016).
Although the functional organization of the MDN is well characterized (Assem et al. 2020a, 2022), its structural connectivity is not. This is surprising given that the MDN consists of clusters of regions located in separate parts of the cortex, suggesting their communication relies on long-range fiber connections. Integrity of large white matter tracts has also been shown to correlate with general fluid intelligence abilities (Haász et al. 2013). Moreover, damage to connections between distant networks nodes, rather than damage to the gray matter itself, is now seen as a core contributor to many neurological syndromes (Thiebaut de Schotten et al. 2020). Indeed, white matter tracts hypothesized to underlie the MDN show a correlation of various indices of structural connectivity with a measure of fluid intelligence (Chen et al. 2020). These results indicate that identifying the structural connectivity underpinning the MDN would help further understand both its function and its variation across individuals.
Investigating structural connectivity also opens ways to compare the MDN across species. Many of the areas that form the MDN are regions that evolved or expanded in primates, such as posterior parietal cortex (Goldring and Krubitzer 2017) and parts of prefrontal cortex (Wise 2008; Passingham 2021). We should therefore expect that a homolog of MDN is present in other primate species. Indeed, electrophysiological studies in the macaque have shown promising results (Kadohisa et al. 2020). Although some early work focused on specifying the areas that form a putative macaque MDN (Mitchell et al. 2016), new methods allowing a quantitative comparison based on white matter tracts (Mars et al. 2018a) have the potential to make a more complete assessment of the similarity in organization of areas that belong to the MDN across species. Moreover, recently, a more fine-grained delineation of the MDN was presented by Assem et al. (2020a), who divided the network into a main set of 10 “core” regions and a set of 17 “extended” regions that likely provide support for the functioning of their core counterparts. It is therefore also useful to revisit previous between-species results in light of this new MDN characterization.
Here, we address both issues. We start by mapping the white matter tracts connecting regions of the MDN in healthy humans, using both the “core” and “extended” sets of regions assigned to the MDN in high detail in recent work (Assem et al. 2020a). We then repeat the analysis in the macaque monkey for regions previously identified as forming a putative MDN (Mitchell et al. 2016). We then apply a comparative mapping approach to compare the networks’ regions across species in terms of their structural connectivity profiles. Together, these results will allow us to both identify the structural basis of the MDN and identify which parts of the network are conserved across the human and macaque monkey.
Materials and methods
Human and macaque diffusion data
Human diffusion and structural brain data consist of eight healthy adult participants, selected at random (3 females; age range: 22–35 yr; mean = 29.6) from the S1200 subjects release of the Human Connectome Project (HCP) database (WU-Minn Consortium, funding provided by the NIH Neuroscience Blueprint Institutes and Centres (Van Essen et al. 2013)). We note that the sample of 8 human HCP brains was chosen to provide a sample size comparable to that of the macaques (see below). Although the sample size is smaller than in some studies on human connectivity, the protocols we used to identify white matter connections were designed to be robust across sample sizes; previous work showed that our group size shows 93% correlation in tract layout across subjects (Warrington et al. 2020, Supplementary Fig. 2).
The data consisted of isotropic 1.25-mm voxels acquired in a multi-shell sequence with b-values 1,000, 2,000, and 3,000 over 90 diffusion-weighting directions, as well as 8 non-diffusion-weighted (b-value = 0) volumes, in each shell in both left-right and right-left phase encoding directions. Data were acquired on a customized 3T Siemens Skyra scanner using a single-shot 2D spin-echo multiband EPI acquisition protocol. In addition, T1- and T2-weighted images were acquired with 0.7-mm isotropic voxels and a TR of 2.4 and 3.2 s, respectively. All data were preprocessed using the HCP’s minimal preprocessing pipeline, described in detail elsewhere (Glasser et al. 2013; Sotiropoulos et al. 2013). In brief, the data from the different modalities were registered to one another and to MNI standard space; the T1- and T2-weighted data were used to create cortical surfaces. Finally, the diffusion MRI data were preprocessed to create posterior distributions of fiber orientations for probabilistic tractography using FSL’s bedpostX (Behrens et al. 2007; Jbabdi et al. 2012).
Macaque data consist of post-mortem diffusion MRI data from eight (Macaca mulatta) brains (two females, age range at death 11–15 years old). Data were acquired and preprocessed as detailed in previous communications (Roumazeilles et al. 2022; Bryant et al. 2021). Formalin-fixed brains were rehydrated in a PBS solution one week prior to scanning and placed in fomblin or fluorinert for the scanning procedure. The diffusion-weighted MRI data were acquired from the whole brain using a 7T preclinical MRI scanner (Varian, Oxford UK) using a 2D diffusion-weighted spin-echo multi-slice protocol with single line readout (DW-SEMS; TR = 10 s; TE = 26 ms; Matrix size = 128 × 128 with a sufficient number of slices to cover the brain; 0.6 mm isotropic voxels). A total of 16 non-diffusion-weighted (b = 0 s/mm2) and 128 diffusion-weighted (b = 4,000 s/mm2) volumes were acquired with diffusion encoding directions evenly distributed over the whole sphere (single shell protocol).
Macaque data were preprocessed using the protocol implemented in the module “phoenix” of the MR Comparative Anatomy Toolbox (Mr Cat; www.neuroecologylab.org). We first converted the datasets to an NIFTI format, then built an image based on the volumes acquired without a diffusion gradient as well as a binary mask of this image. Using tools from FSL (www.fmrib.ox.ac.uk/fsl), we then fitted a diffusion tensor model using FSL’s dtifit. Following the preprocessing, bedpostX (Behrens et al. 2007) was used to fit a crossing fiber model to the data, allowing for three fiber orientations.
Identification of canonical MDNs
MDN maps for the human were derived in previous work by Assem et al. (2020a). The maps were created by conjunction of functional activation from fluid intelligence related tasks [working memory (hard vs easy), math vs story, relational reasoning (hard vs easy)]. These functional maps were then parcellated for each subject using an individual-specific multi-modally parcellated surface (MSMAll, Robinson et al. 2014, 2018; Glasser et al. 2016), whereby beta values across vertices were averaged resulting in one value per cortical area and per subject. The conjunction revealed a set of 27 parcels overall with 10 core MDN parieto-frontal regions surrounded by 17 extended MDN regions. The core MDN regions showed significantly higher correlation in functional activity among themselves, when compared with the extended regions and even more so when their functional connectivity was compared the with the rest of the brain (Assem et al. 2020a). The putative macaque MDN was derived in Mitchell et al. (2016), where a previously established human MDN map (Fedorenko et al. 2013) was warped onto a macaque atlas space (F99). The resulting warp was iteratively optimized, taking into consideration established borders of anatomical cortical regions in the macaque brain as well as the putative regions’ connectivity correlation strength with each other and compared with the rest of the brain, using data from resting state fMRI in 35 rhesus macaques under anesthesia. The identified MDN clusters most strongly overlapped with a set of 24 cortical regions. For our fingerprint connectivity analysis of these regions, we used the composite LV-FOA-PHT parcellation of macaque cortical surface created by Van Essen et al. (2012), based on landmark-constrained surface-based registration of serveral atlases.
Structural connectivity fingerprints
Structural connectivity fingerprints
Connectivity of each MDN area, on the right hemisphere of the brain, was assessed using connectivity fingerprints, which are here defined as surface descriptions of connections of each part of the cortical surface with the main white matter fiber bundles. The connectivity fingerprint is constructed as follows: first, each of the major fiber bundles is reconstructed in each brain using a series of standardized tractography recipes in standard space. These recipes have been validated previously (Mars et al. 2018a; Warrington et al. 2020) and are defined in such a way as to create homologous tracts across the human and macaque monkey brain. The reconstructed bundles were arcuate fascicle (AF); anterior and superior thalamic radiations (ATR, STR); peri-genual (CBP), dorsal (CBD), and temporal (CBT) subdivisions of the cingulum bundle; corticospinal tract (CST); frontal aslant (FA); forceps major (FMI) and minor (FMA); fornix (FX); inferior (ILF) and middle longitudinal fascicle (MDLF); first (SLF1), second (SLF2), and third (SLF3) branches of the superior longitudinal fascicle (SLF); middle cerebellar peduncle (MCP); optic (OR) and acoustic radiations (AR), uncinate fascicle (UF); vertical occipital fascicle (VOF).
All recipes were identical for the human and the macaque and similar to previous specification (Warrington et al. 2020), except for macaque SLF2 and SLF3. The dorsal longitudinal tracts are very difficult to reconstruct due to the presence of many crossing fibers in their territory (cf. Mars et al., 2019; Behrens et al. 2007). We therefore had to adjust our recipes and so instead of tracking SLF2 and SLF3 in a single recipe consisting of a central seed and anterior and posterior waypoint, we used a recipe consisting of a central seed and anterior target to reconstruct the SLFs for the frontal cortex only. Without this adjusment, these tracts would fail to reach anteriorly in the frontal lobe in a way that is analogous to the known course of the tracts (Schmahmann and Pandya 2006).
Tract reconstruction recipes were initially transformed from standard to subject native space where probabilistic tractography was performed. This resulted in tractograms, which were normalized according to the total number of valid streamlines delineated by the tractography algorithm and binarized (Warrington et al. 2020). Finally, we created a group averaged tractograms of each white matter tract per species.
Following creation of the tractograms, for each human and macaque subject, we performed tractography from each vertex of the cortical midthickness surface toward each brain voxel, to create a cortical area × brain connectivity matrix. We then created an average cortex × brain connectivity matrix across all subjects per species. Finally, the tractograms and this matrix were multiplied to create a tract × cortex matrix, the connectivity blueprint for both species (Mars et al. 2018a). From this connectivity blueprint, the connectivity profile of each region of the MDN was created by averaging the appropriate rows of the matrix. Each region’s connectivity profile describes the likelihood of it being reached by each of the major white matter fiber bundles. We display the connectivity profile or ‘connectivity fingerprint’ of a region as a polar plot, in which stronger connectivity is indicated by a line further away from the origin. The polar plot is normalized to the strongest connectivity of that particular fingerprint. Hence, the polar plot indicates relative strengths of tracts’ connectivity for a given region, but connectivity values of a particular tract cannot be compared across different regions.
The above was implemented using FSL’s XTRACT and XTRACT_BLUEPRINT features (Warrington et al. 2020) and customized Matlab functions from our in-house MR Comparative Anatomy Toolbox (Mr Cat). All tractography was run in individual space, following warping from the standard MNI, for human, and F99, for macaque, space masks using FNIRT, using the standard settings of PROBTRACKX2. Standard space surfaces consisted of the Freesurfer average 32 k surface used in the HCP (Glasser et al. 2013; MSMAll) for the human and the F99 surface (Van Essen et al. 2012) for the macaque.
To compare the connectivity fingerprints of the macaque and human, we used Kullback–Leibler (KL) divergence—a measure used to compare probability distributions. In this case, we use this measure to compare how the connectivity of an area with the white matter tracts identified above compares to the connectivity of other areas of the same species and, importantly, of another species (Mars et al. 2018a). One can then represent brain areas of the two species within a common connectivity space by submitting the matrix of KL values to a spectral reordering algorithm, whereby regions that have similar connectivity are clustered closer together in 2D space and those that are different are further away from each other (Higham et al. 2007; Mars et al. 2018a; Warrington et al. 2022). Furthermore, one can identify which areas in one species are most similar to any given area in the other species. In this instance, we estimate the top 2% closest areas in the macaque brain to each region belonging to the human MDN ROI.
Results
Connectivity profile of core human MD network regions
We first examined how the white matter tracts, common across human and macaque brains, drive the connectivity profiles of each of the 10 core MD network regions as delineated by Assem et al. (2020a) (Fig. 1A).
Fig. 1.
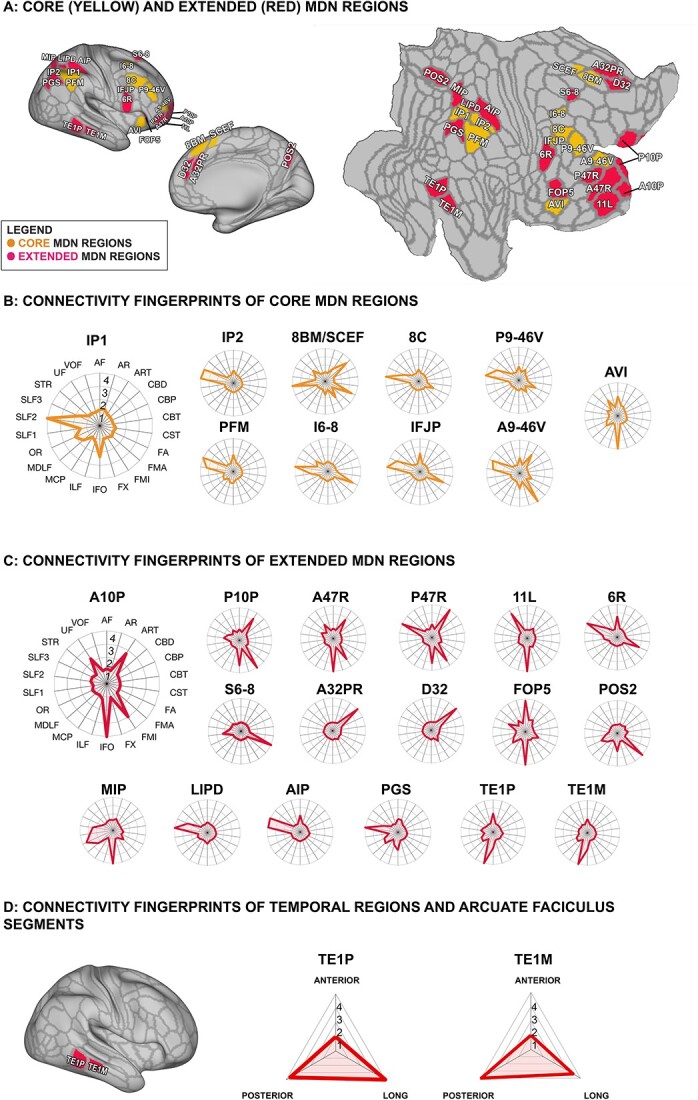
Connectivity profiles of each region in the MDN as defined by Assem et al. (2020a). (A) Core and extended MDN regions represented on an inflated and flat surface, right hemisphere. (B) Connectivity fingerprints of core MDN regions, normalized to show the relative degree to which each white matter tract contributes to the respective region. (C) Normalized connectivity fingerprints of extended MDN regions. (D) Connectivity fingerprints of temporal regions TEP1 and TEM1 as they relate to the three segments of the arcuate fasciculus. Abbreviations: AF (arcuate fasciculus); AR (acoustic radiation), ART (anterior radiations: thalamic); CBD (cingulate bundle dorsal); CBP (cingulate bundle peri-genual); CBT (cingulate bundle temporal); CST (corticospinal tract); FA (frontal aslant); FMA (forceps major); FMI (forceps minor); FX (fornix); IFO (inferior fronto-occipital fasciculus); ILF (inferior longitudinal fasciculus); MCP (middle cerebellar peduncle); MDLF (middle longitudinal fasciculus); OR (optic radiation); SLF1, SLF2, SLF3 (superior longitudinal fasciculus branches 1, 2, and 3); STR (superior thalamic radiation); UF (uncinate fasciculus); VOF (ventral occipital fasciculus).
The majority of areas in the core MD network form a set of lateral parietal-frontal regions. They are heavily interconnected by the two most lateral branches of the SLF (Schmahmann and Pandya 2006; Makris et al. 2005) (Fig. 1B). The origin points of SLF2, the second branch, are the anterior intraparietal sulcus and angular gyrus, and its terminations in the frontal lobe are near the superior and middle frontal gyri (Thiebaut de Schotten et al. 2011). This is reflected in the predominance of SLF2 in the connectivity profile of anterior intraparietal region IP1 and superior and middle frontal regions I6-8, 8C, P9-46V. Similarly, the third branch of the SLF, SLF3, originates from the parietal lobe but anteriorly to SLF2, near the supramarginal gyrus, and terminates near the inferior frontal gyrus (Thiebaut de Schotten et al. 2011). Our connectivity profiles reflect this with prevalence of SLF3 over the connectivity of inferior parietal region PFM and anterior inferior frontal region A9-46V. Intraparietal region IP2 seems to be influenced by both the second and third branches of the SLF. The AF, which contains both frontal-parietal connections and connections with the temporal lobe (Catani et al. 2005), also reaches parts of the lateral core MDN regions, although to a much lesser extent than the SLF2 and SLF3.
Within the frontal lobe, the frontal aslant, a tract important for dorsal-medial frontal communication, was present in all connectivity profiles, especially those located more posteriorly in the frontal lobe, likely aiding communication across the frontal MD regions. Region IFJP and dorsal region I6-8’s connectivity was most strongly influenced by this tract, consistent with its proposed anatomical location (Thiebaut de Schotten et al. 2012).
In addition to lateral fronto-parietal regions, Assem et al. (2020a) found that there was activation across all three fluid intelligence tasks in the middle segment of cingulate area SCEF and more dorsally in area 8BM, leading them to combine the anterior portion of SCEF and area 8BM into a single label, term 8BM/SCEF. We examined the connectivity profile of 8BM/SCEF accordingly. This area was reached by the most medial branch of the SLF, SLF1, but also by the dorsal segment of the cingulum bundle.
Finally, Assem et al. (2020a) delineated region AVI as a core insular component of the MD network. This region did not appear to have a strong connectivity with the SLFs. Rather, this region is in the vicinity of the course of the inferior fronto-occipital (IFO) fasciculus, the arcuate fasciculus, and the uncinate fasciculus.
Connectivity profile of extended human MDN regions
The extended MDN adds 17 additional cortical areas to the set of 10 core regions. Naturally, we observed that regions which clustered together in close proximity had a similar white matter profile, albeit not identical. Frontoparietal connections through the SLF fiber system again were a predominant feature for a number of these regions. Unlike the core MDN, however, in the extended set of regions dorsal longitudinal tracts were not always the only dominant tracts driving the connectivity fingerprints (Fig. 1C).
In the frontal cortex, extended MDN regions are mostly located in the ventral and polar aspects of lateral frontal cortex. Although the connectivity profile of the polar regions (A10P, P10P, A47R, P47R) bears some similarity to the connectivity profile of core area A9-46V, connections to fiber systems other than the SLF are more prevalent. Interhemispheric connections through the forceps minor (FMI) of the corpus callosum and thalamic connections through the anterior thalamic radiation (ART) are present in these regions’ fingerprints. Of note are the connections of some of the ventral and polar regions, namely A10P, A47R, and 11L, with the temporal lobe, though both the uncinate and inferior fronto-occipital (IFO) fascicles.
In parietal cortex, some of the extended regions, such as LIPD, AIP, occipito-parietal PGS, and, to a lesser extent, MIP, follow the pattern of predominant connectivity with the superior longitudinal fiber system. Lateral parietal extended MDN regions MIP and PGS show connectivity profiles influenced by occipitotemporal fibers systems, such as the inferior longitudinal fascicle (ILF) and IFO fascicles.
The ILF and IFO were also prominent in the connectivity profile of the two extended MD regions in the lateral temporal cortex, TE1M and TE1P. In addition, these regions, as well as some of the lateral parietal regions such as AIP and core region PFM, are reached by the AF. In the human brain, the AF consists of a number of subdivisions, namely a frontal-parietal “anterior” segment, a fronto-temporal “long” segment, and a parietal-temporal “posterior” segment (Catani et al. 2005). These segments were recently delineated in a format compatible with our connectivity fingerprint by Sierpowska et al. (2022), who suggested that the parietal–temporal connection is much stronger in the human and in the closely related chimpanzee. We therefore performed a follow-up analysis to establish which segments of the AF were most likely to connect to the temporal extended MDN regions. As can be seen in Fig. 1(D), the temporal MD regions are reached by both the long and posterior segments, connecting them to the lateral frontal and inferior parietal cortex, respectively.
Connectivity profile of putative macaque MD regions
We examined the connectivity profiles of a set of cortical regions that have been previously suggested to underlie the macaque MD network (Mitchell et al. 2016). Mitchell and colleagues identified 8 clusters that partially overlapped 24 regions from the LV_FOA_PHT atlas, spanning mainly frontal, parietal, and partially temporal cortices, by warping a human MD network map, derived from fMRI studies (Fedorenko et al. 2013), onto a standard macaque cortical surface based on multiple anatomical landmarks, then optimizing based on resting state interconnectivity. Here, we examine the structural connectivity profile of those regions.
In the regions overlapping frontal clusters of the putative macaque MDN (Fig. 2; 46V, 46P, 6VAM, 6VAL, 45, 12, 4C, 6DS), it is noticeable that the SLF is rarely the most dominant tract, unlike in the human. This is evident in regions within the lateral frontal clusters, such as 6VAM, 6VAL, 45, 4C, and 6DS, whose connectivity profile most strongly implicated the FA, whereas areas 46V and 46P had dominant connectivity via the FMI and the ART. Importantly, although the second and third branch of the SLF were prominent in the connectivity profiles of these regions, they overall made up a less substantial part of it.
Fig. 2.
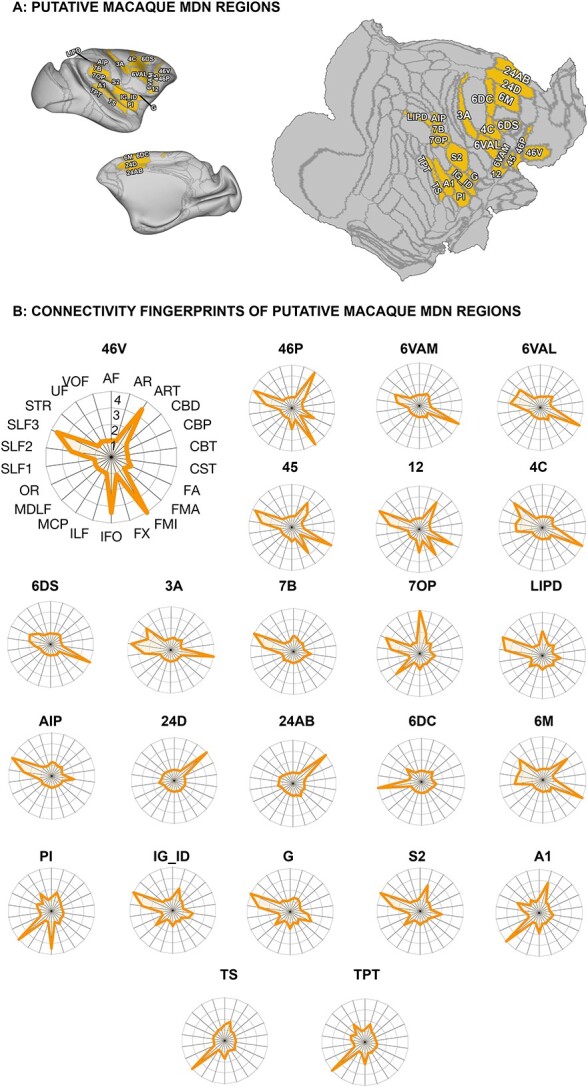
Connectivity profiles of regions overlapping frontal macaque MDN clusters as defined by Mitchell et al. (2016). (A) Anatomical regions overlapping putative macaque MDN clusters represented on an inflated and flat surface, right hemisphere. (B) Connectivity profiles, normalized to show the degree to which each white matter tract contributes to the connectivity of each anatomical regions overlapping frontal MDN clusters with strongest contribution (4), strong contribution (3), medium contribution (2), low contribution (1), no contribution (0). Abbreviations: AF (arcuate fasciculus); AR (acoustic radiation), ART (anterior radiations: thalamic); CBD (cingulate bundle dorsal); CBP (cingulate bundle peri-genual); CBT (cingulate bundle temporal); CST (corticospinal tract); FA (frontal aslant); FMA (forceps major); FMI (forceps minor); FX (fornix); IFO (inferior fronto-occipital fasciculus); ILF (inferior longitudinal fasciculus); MCP (middle cerebellar peduncle); MDLF (middle longitudinal fasciculus); OR (optic radiation); SLF1, SLF2, SLF3 (superior longitudinal fasciculus branches 1, 2, and 3); STR (superior thalamic radiation); UF (uncinate fasciculus); VOF (ventral occipital fasciculus).
In the macaque parietal lobe, the prominence of the SLF system is much more evident, with SLF2 and 3 most strongly represented in the connectivity profile of 3A, 7B, 7OP, LIPD, and AIP. In addition, we saw strong connectivity of the AF and dorsal part of the middle longitudinal fascicle in 7OP and to a lesser extent LIPD.
On the medial wall, Mitchell and colleagues identified the dorsal region 6M and the more ventrally located 24AB and 24D. Consistent with dorsally located human areas 8BM/SCEF, identified by Assem et al. (2020a) as part of the core MDN, of these regions 6M was most strongly connected via SLF1. The more ventral 24AB and 24D showed mostly connectivity to the dorsal part of the cingulum bundle. This connectivity pattern is similar in human extended MDN regions D32 and A32PR, although these are located more anteriorly at the edge of the projection zones of the dorsal and peri-genual parts of the cingulum bundle. Mitchell et al. (2016) additionally identified region 6DC that crosses over from dorso-lateral frontal cortex into anterior cingulate cortex. In our analysis this region displayed a distinct connectivity profile dominated by the first subdivision of the SLF. In the macaque, SLF1 projections reach frontal premotor and supplementary areas, which encompass area 6DC, and connect them to areas in dorso-lateral parietal cortex and precuneus (Schmahmann and Pandya 2006).
The study by Mitchell and colleagues also identified a cluster in the lateral fissure of the macaque that did not match human MD regions. This overlapped S2, and the primary auditory region A1, with spread into TS and TPT. Areas A1, TS and, TPT’s profiles mainly relied on MDLF, whereas region S2 was reached by the third division of SLF and, to a lesser extent, ART, MDLF, as well as the cortico-spinal tract.
In summary, the regions identified by Mitchell and colleagues as forming the putative MDN in the macaque brain show a pattern of white matter connectivity that is reminiscent of the human core MDN, with the superior longitudinal fiber system underlying most of the connections. It should be noted, however, that those connections are less dominant in the macaque frontal cortex than they are in the human.
Human MDN and putative macaque brain regions within a common connectivity space
The results above show that human core MDN regions are characterized predominantly by connectivity to the dorsal superior longitudinal fiber system. The same is true for putative macaque MDN regions, although in the frontal cortex the longitudinal tracts are supplemented by additional connections. The extended human MDN regions are served by different fiber systems that are not reminiscent of the putative macaque MDN regions identified by Mitchell and colleagues. To formalize these intuitions, we next created a connectivity embedding, in which we visualize human and macaque brain organization within the same space (Mars et al. 2018b). This is possible because we have defined regions in both species’ brains in terms of their connectivity to homologous white matter tracts (Mars et al. 2018a; Warrington et al. 2022).
First, we applied a data reduction to the human network, by clustering all areas of the core and extended MDN based on their connectivity fingerprints using kmeans clustering (Fig. 4). This revealed a clear organizational pattern in which regions belonging to the same networks in terms of their white matter connectivity were grouped together. This approach identified a fronto-parietal cluster (core MDN regions PFM, IP2, IFJP, 8C, P9-46V, I6-8, and extended 6R and LIPD), a dorso lateral prefrontal cluster (core region A9-46V and extended regions P47R and P10P), a dorso-medial frontal cluster (core MDN regions 8BM/SCEF and extended MDN region S6-8, D32, and A32PR), a ventral frontal cluster (extended MDN regions A47R, 11L, A10P, and FOP5 and core region AVI), a posterior parietal cluster (extended regions POS2, MIP, PGS and core IP1), and a temporal cluster (extended regions TE1 and TE2). The clustering provides a decent separation in clusters consisting of mostly core regions (the fronto-parietal cluster), hybrid clusters (dorsomedial frontal and dorsolateral prefrontal clusters), and predominantly extended regions (ventral frontal, posterior parietal, and temporal clusters).
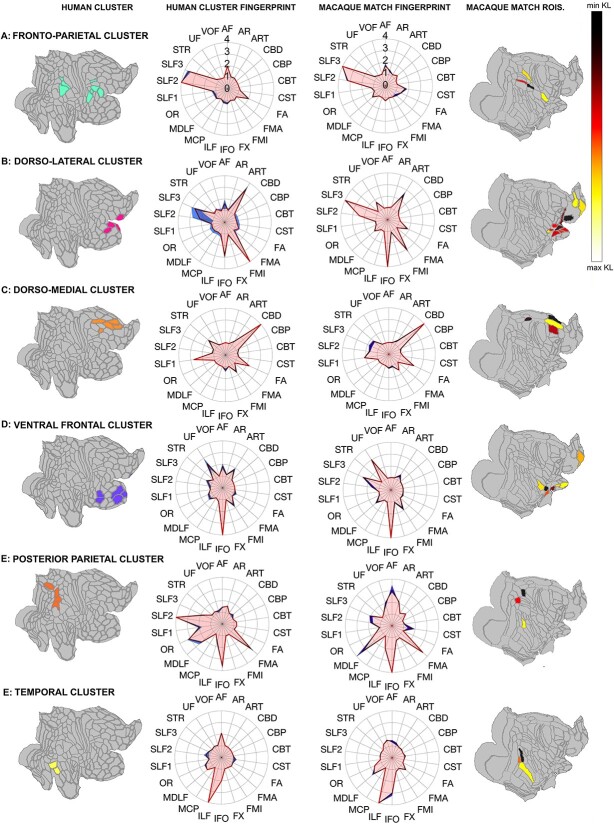
Fig. 4.
Direct match between human MDN clusters and top matching macaque regions. Left column depicts all human clusters, displayed on a flat surface; center-left column shows human cluster fingerprint and standard error; center-right column: Fingerprints of top 2% closest matched macaque regions and standard error; right column depicts top 2% closest matched macaque regions, displayed on a flat surface, with lowest KL values in darker colors and higher KL values in lighter colors. (A) Fronto-parietal cluster and top matching regions. (B) Dorso-lateral cluster and matching regions. (C) Dorso-medial cluster and matching regions. (D) Ventral frontal cluster and top matching regions. (E) Posterior parietal cluster and top matching regions. (F) Temporal cluster and top matching regions. Polar plots display normalized connectivity fingerprints. Abbreviations: AF (arcuate fasciculus); AR (acoustic radiation), ART (anterior radiations: thalamic); CBD (cingulate bundle dorsal); CBP (cingulate bundle peri-genual); CBT (cingulate bundle temporal); CST (corticospinal tract); FA (frontal aslant); FMA (forceps major); FMI (forceps minor); FX (fornix); IFO (inferior fronto-occipital fasciculus); ILF (inferior longitudinal fasciculus); MCP (middle cerebellar peduncle); MDLF (middle longitudinal fasciculus); OR (optic radiation); SLF1, SLF2, SLF3 (superior longitudinal fasciculus branches 1, 2, and 3); STR (superior thalamic radiation); UF (uncinate fasciculus); VOF (ventral occipital fasciculus).
We extracted the average connectivity profile of each of the human clusters and then calculated the KL divergence between the connectivity fingerprints of all human clusters and all macaque cortical regions. We then performed a spectral reordering to visualize all human clusters and macaque regions within the same two-dimensional connectivity space (Fig. 3) (Higham et al. 2007). This showed that the human fronto-parietal cluster occupies a similar position in connectivity space as most macaque lateral parietal areas, similar to our earlier observation that their connectivity is mostly driven by the SLF system.
Fig. 3.
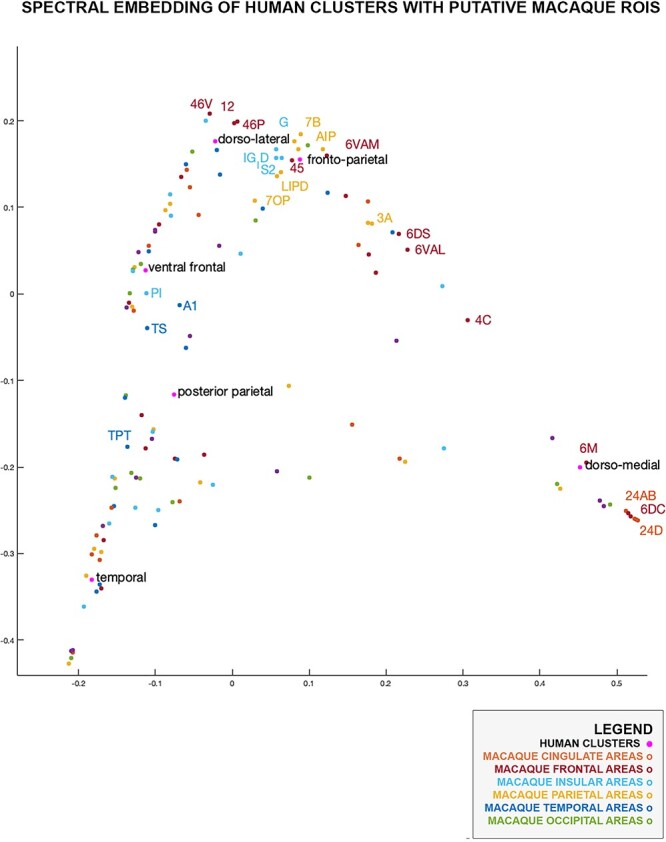
Human MDN and all macaque brain regions in a 2D common connectivity space. Labeled macaque regions are those listed by Mitchell et al. (2016) as partially overlapping with putative macaque MDN clusters.
Near the fronto-parietal cluster, the human dorso-lateral cluster congregates closely to macaque areas 46V, 46P, and 12, as predicted by Mitchell and colleagues, but also to regions along the anterior insula and frontal opercular area.
The human clusters containing mostly extended MDN regions did not cluster in the vicinity of macaque parietal or lateral frontal regions. The human medial frontal cluster assembled with areas on the macaque medial wall, including area 6M, 6DC, 24D, as identified by Mitchell and colleagues. The ventral frontal and posterior parietal clusters were situated closer to insular area PI and temporal areas TS and TPT. Interestingly, the human temporal cluster did not overlap with macaque MDN regions delineated by Mitchell and colleagues.
Direct matching of human MDN and macaque brain regions
The human-to-macaque transformation of MDN regions used by Mitchell et al. (2016) was based on a surface-based registration according to 23 landmark homologies as proposed by Van Essen and Dierker (2007), subsequently optimized based on their functional connectivity. The initial surface-based registration has also been used to project functional maps (Mantini et al. 2013) and well as cortical terminations of white matter tracts (Eichert et al. 2019) between species. An alternative approach is to use the connectivity profiles of the regions themselves. Just as the common definition of the white matter tracts across species allows the common connectivity space projection from the previous section, we can use them to assign to each human MDN cluster the most similar regions in the macaque brain (Mars et al. 2018a). Therefore, in our final analyses, we calculated KL divergence between human MDN kmeans clusters, identified in the previous section, and all macaque regions. We then identified the top 2% closest matching macaque regions for each human MDN cluster (Fig. 4), according to their KL values (Table 1).
Table 1.
Summary of 2% closest matched macaque regions per human MDN cluster and KL value.
| Human MDN cluster name | Top 2% matched macaque cortical regions and KL value | Average KL |
|---|---|---|
| Fronto-parietal | PRCO (1.5922), 7B (1.3767), LIPD (1.4903), 5V (1.5699) | 1.5073 |
| Dorso-lateral | 46V (0.7369), 46P (1.0120), 12 (1.3212), PROM (1.2438), GU (1.7542), 12 M (1.4696), 12O (1.2256), 12R (1.2734), 12L (0.8300), 10 M (1.8293), 10O (1.7281) | 1.3131 |
| Dorso-medial | 24AB (2.7409), 24D (3.2052), 6M (2.9178), PECG (2.9637) | 2.9069 |
| Ventral frontal | PI (2.0798), IPRO (1.3723), PIR (1.9234), 11L (2.1576), 13L (2.0629), IAI (1.5602), 10O (2.0178) | 1.8820 |
| Posterior parietal | MIP (2.5687), LOP (2.7042), FST (2.8217) | 2.6982 |
| Temporal | MT (1.1808), TEA_M (1.2870), V4TA (1.2584) | 1.2421 |
The fronto-parietal cluster (Fig. 4A) was defined by SLF2 and SLF3 connectivity and matched with a set of parietal regions and precentral opercular frontal area PrCO. One interesting difference was the proportion of SLF influence between the human and macaque cluster: in the human SLF2 had the highest number of streamlines that reached cortical surface in the MDN, however in the macaque this was SLF3.
The dorsolateral cluster (Fig. 4B), involving the anterior most frontal areas of the MDN, had the largest number of matches in the macaque brain. The fingerprints were strongly defined by SLF2 and SLF3, in addition to interhemispheric tract FMI, occipito-frontal IFO, and the ARTs. Among this cluster’s matches were area 46V, 46P, and 12, which were also identified by Mitchell et al. (2016). In addition, we identify extended area 12 with 12R, 12O, 12L, areas 10O and 10M as well as area PROM. Surprisingly, our matching also picked up gustatory area GU. Mitchell et al. (2016) previously identified a neighboring area, primary gustatory cortex G, as part of their putative macaque MDN map.
The dorso-medial frontal cluster, which was mainly driven by the dorsal cingulum bundle, matched with previously identified anterior cingulate areas 24AB, 24D, and 6M as well as posterior cingulate PECG. Posterior cingulate area PECG was identified presumably because the connectivity profile of this region is also mostly driven by the dorsal part of the cingulum bundle.
The ventral frontal cluster, involving anterior frontal areas A47R, A10P, 11L, and insular FOP and AVI, was mainly driven by IFO and uncinate fasciculus, both tracts connecting ventral prefrontal and temporal regions. Its closest macaque matches were mainly found in part of the orbitofrontal cortex, including parts of areas 11L, 13, polar area 10O, and insular regions PI, IPRO, PIR.
The human posterior parietal cluster, consisting of extended MDN regions, matched with only three macaque regions: macaque MIP, LOP, and temporal area FST. In the human connectivity profiles, SLF2 was much more present than in the macaque matches, whose connectivity profile was mainly defined by AF, MDLF, and IFO.
Our temporal cluster matched quite well with macaque areas MT, TEA_M, and V4TA; however, one important difference between the two species is the much stronger presence of AF in the human temporal cluster. As shown above, in the human temporal areas, AF connections were mainly subserved by the posterior and long AF segment allowing for communication with frontal and parietal cortices.
The above analyses were thresholded to match the human clusters with the top 2% of macaques regions. Changing this threshold does not dramatically alter the picture (Supplementary Figs. 1–3). In general, the additional regions shown at more lenient thresholds belong to the same anatomical networks as describe above.
Discussion
In this work, we have identified the white matter connections that underlie the communication of the areas of the MDN in the neocortex. Using an established set of protocols to reconstruct the major white matter fibers of the human brain, we established a connectivity fingerprint, describing each part of the neocortex in terms of its connectivity with the major white matter fiber pathways. This allowed us to establish the connectivity profile of each area of the MDN. Such a connectivity profile is important because it provides crucial information on the input an area receives and the influence it can exert on the rest of the brain (Passingham et al. 2002), providing a structural basis for its place in the larger cortical organization (Mars et al. 2018b). Similar reconstruction of the white matter pathways of the macaque monkey brain also allowed us to compare the human MDN regions directly to candidate MDN regions in the macaque brain, allowing a quantitative assessment of the potential similarity of this network across species.
We established that the lateral fronto-parietal areas that constitute the majority of the “core MDN” are connected through the second and third branches of the superior longitudinal fiber (SLF) system. The more medial frontal areas are served by the cingulum bundle and the first branch of the SLF. The superior longitudinal fiber system and its subdivision into three distinct branches is well established from macaque tracer data (Petrides and Pandya 1984; Schmahmann and Pandya 2006) and has been demonstrated repeatedly in the human using in-vivo tractography (Makris et al. 2005; Thiebaut de Schotten et al. 2011). Although there is some evidence of lateralization of the SLF in humans (Thiebaut de Schotten et al. 2011), given the lack of observed lateralization in the macaque and the averaging of results in the previous anatomical study on the MDN (Assem et al. 2020a), this study focused only on the non-language-dominant right hemisphere.
Genovesio et al. (2014) interpreted the parietal-frontal network in terms of cortical adaptations to the unique foraging niche of primates. Although some parietal cortex can be detected in other mammals (Goldring and Krubitzer 2017), the parietal-frontal system is much more developed in primates, most likely due to their adaptations to an arboreal niche (Wise et al. 2008; Murray et al. 2017). Genovesio and colleagues suggested parietal cortex’s visuomotor functions allow it to provide relational metrics such as order, duration, and distance to frontal areas involved in the computational of behavioral goals as well as a more complex interaction with the environment. Within the human brain, these systems might have expanded and their evolutionary early function co-opted for more general information processing.
In this context, it is interesting that the frontal connectivity of the dorsal longitudinal SLF is stronger in the human than in the macaque, as indicated by the SLF’s relative dominance of the connectivity fingerprints. Whether human prefrontal cortex is larger than expected for an ape brain of its size is a matter of debate (Barton and Venditti 2013; Donahue et al. 2018), but there is no doubt that in absolute numbers human prefrontal cortex is larger than that of non-human primates and that the ratio of prefrontal size to size of parietal and temporal inputs of prefrontal cortex is larger in the human than its closest animal relative (Passingham and Smaers 2014). It has also been suggested that prefrontal white matter is more extensive in the human brain (Donahue et al. 2018), which dovetails with our observation that the SLFs are more prominent in the human brain. It has been claimed that both the lateral frontal pole and inferior parietal cortex have expanded in the human (Van Essen and Dierker 2007), with the frontal pole containing an additional subdivision compared to the macaque (Neubert et al. 2014). Thus, although the overall organization of frontal-parietal connectivity is the same across species, differences in degree and arealization are present.
As noted in our methods, we choose to employ a slightly different strategy for reconstructing SFL2 and SLF3 in the macaque compared with the human brain. Tracing studies (Schmahmann and Pandya 2006) have clearly shown that, in the macaque brain, these two tracts extend from posterior parietal areas anteriorly to frontal regions 46, 9/46, and 6D for SLF2 as well as 6V, 44, and 9/46V for SLF3. Reconstruction of these tracts with probabilistic tractography can be difficult due to influence from crossing fibers, especially in the small macaque brain. Indeed, even in the much larger chimpanzee brain, these tracts are already difficult to reconstruct (Mars et al. 2019). We therefore choose to modify our recipes in a way that allowed us to access as much of the anterior fibers of the SLFs in the macaque brain and to avoid the effects of crossing fibers as much as possible. With this modification, we were able to obtain a more anatomically plausible reconstruction of macaque SLF2 and SLF3.
Regions belonging to the extended MDN do not always follow the pattern of the core regions. Indeed, their connectivity is partly driven by different fiber systems in many cases. For instance, regions in the more anterior parts of the frontal cortex, although still reached by the SLF to some extent, are also connected with other functional systems through the occipito-temporal-frontal fiber system often designated as the IFOF, IFO, or extreme capsule fiber complex (Forkel et al. 2014; Mars et al. 2016). A recent analysis by Nozais et al. (2021) linking white matter networks and functional task activations is consistent with our results. They found that activations by a working memory task, as often used to study the core MDN, are associated with the SLFs and FA, as is the case for our core MDN. Fibers connecting temporal cortex with areas in parietal cortex were associated with semantic task activations. This both fits with our interpretation of separate systems for the extended MDN and the observation that these systems are modified to some extent in the human brain.
Of particular note are the regions in the temporal lobe, TE1p and TE1m, which form part of the ventral visual stream subserved by the ILF (Latini et al. 2017; Roumazeilles et al. 2020). Assem and colleagues (2020) surmised that extended MDN regions are recruited in some cases to provide the core MDN with information required to solve the task at hand. This might be a clear case in which areas of the ventral visual stream interact with areas associated with dorsal stream and associated parietal regions. One potential avenue for this is a branch of the AF connecting inferior parietal cortex and temporal cortex (Catani et al. 2005). Such interactions are often hypothesized, but not often systematically studied. Interestingly, the temporal–parietal part of the AF is thought to be more pronounced in humans as compared with other primates (Sierpowska et al. 2022). Indeed, arcuate connectivity in the human brain is thought to reflect invasion of this tract into novel cortical territory compared with other primates, rather than simple cortical expansion (Eichert et al. 2020). This suggests that integration of these parts of the core and extended MDN might be more pronounced in the human. At present, however, this remains a hypothesis awaiting testing.
As such, this work shows that the anatomical organization of the core MDN is qualitatively similar in the human and the macaque monkey brain, but that the frontal connectivity to the dorsal longitudinal fiber system is more developed in the human brain, based on the dominance of these tracts in the connectivity fingerprints of the human in contrast to the macaque brain. The core and extended MDN rely on different fiber systems in the human brain, consistent with the suggestion that they process different types of information, with the extended MDN potentially feeding information into the core system based on task requirements (Assem et al. 2020a). The extended MDN regions have connectivity profiles similar to some found in the macaque brain, although modifications since the last common ancestor are evident as well.
The present study is purely anatomical. We take a known functional network and investigate its anatomical connections and their homologs across species. As such, we are reluctant to make strong inferences regarding the function of the macaque regions we identified in the context of the MDN. However, two issues deserve discussion. First, although we observe that most human core MDN regions are connected to dorsal fiber systems, some core regions were connected to more ventral fiber systems. This shows a dissociation between anatomical and functional networks. A second issue is whether the human MDN and its anatomical match in the macaque serve the same set of functions. Although fMRI research in humans indicates that the human MDN responds strongly to a variety of cognitive challenges, it is not certain which aspects of cognition they respond to or how exactly the accompanying computation is performed, although some potential mechanisms have been proposed (Duncan 2010). Furthermore, our matching regions in the macaque brain are involved in a variety of different functions, such as touch perception (parietal area 5), ecological actions (intraparietal areas LIPD), visuomotor transformations (area AIP), and tool use (intraparietal sulcus). The clarification of the functional roles of the macaque regions within the MDN context, and subsequent neuronal studies of their computational mechanisms, are at present the subject of future research.
On a more general level, this work shows how the technique of connectivity fingerprint matching can be used to compare functional networks between species through a common anatomical reference frame (Mars et al. 2021). Previous comparisons of brain networks across species are often performed only informally, without any quantitative assessment of whether results translate well across species. Such assessments often take the form of “same” or “different” statements about the similarity of regions across species, failing to acknowledge that homologous regions are likely to have undergone change in some aspects of their organization, be it connectivity or otherwise, as the last common ancestor of the species under comparison. The approach employed here allowed us to formally assess how different parts of the human MDN fit within the organization of the macaque monkey brain while highlighting the unique aspects of this network in the human, especially with respect to the strength of frontal longitudinal connections.
Supplementary Material
Acknowledgments
We thank Joanna Sierpowska for sharing the tractography recipes of the separate branch of the AF in humans.
Contributor Information
Katrin Karadachka, Donders Institute for Brain, Cognition and Behaviour, Faculty of Social Sciences, Radboud University Nijmegen, 6525HR Nijmegen, The Netherlands.
Moataz Assem, MRC Cognition and Brain Sciences Unit, University of Cambridge, Cambridge CB2 7EF, United Kingdom.
Daniel J Mitchell, MRC Cognition and Brain Sciences Unit, University of Cambridge, Cambridge CB2 7EF, United Kingdom.
John Duncan, MRC Cognition and Brain Sciences Unit, University of Cambridge, Cambridge CB2 7EF, United Kingdom.
W Pieter Medendorp, Donders Institute for Brain, Cognition and Behaviour, Faculty of Social Sciences, Radboud University Nijmegen, 6525HR Nijmegen, The Netherlands.
Rogier B Mars, Donders Institute for Brain, Cognition and Behaviour, Faculty of Social Sciences, Radboud University Nijmegen, 6525HR Nijmegen, The Netherlands; Wellcome Centre for Integrative Neuroimaging Nuffield Department of Clinical Neurosciences, John Radcliffe Hospital, University of Oxford, Headington, Oxford OX3 9DU, United Kingdom.
CRediT author statement
Katrin Karadachka (Formal analysis, Investigation, Methodology, Project administration, Visualization, Writing—original draft, Writing—review and editing), Moataz Assem (Conceptualization, Data curation, Methodology, Validation, Writing—review and editing), Daniel Mitchell (Conceptualization, Data curation, Methodology, Validation, Writing—review and editing), John Duncan (Conceptualization, Data curation, Investigation, Methodology, Validation, Writing—review and editing), W. Pieter Medendorp (Funding acquisition, Project administration, Resources, Supervision, Validation, Writing—review and editing), Rogier B. Mars (Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Software, Supervision, Validation, Writing—review and editing).
Funding
The work of K.K. was supported by a Donders Centre for Cognition (Ph.D. grant to R.B.M. and W.P.M.),. The work of R.B.M. was supported by EPA Cephalosporin Fund and Biotechnology and Biological Science Research Council (BBRSC) UK (BB/N019814/1 to R.B.M.) grants. The Wellcome Centre for Integrative Neuroimaging is supported by core funding from the Wellcome Trust (201319/Z/16/Z). M.A., D.J.M., and J.D. were supported by Medical Research Council Intramural Program MC_UU_00030/7.
Conflict of interest statement: None declared.
Data availability
All data and scripts are available in the following repository: Karadachka, K.V., Mars, R.B. (2023): Structural connectivity of the multiple demand network in humans and comparison to the macaque brain. Version 1. Radboud University. (dataset). https://doi.org/10.34973/xzd1-zs16.
References
- Assem M, Glasser MF, Van Essen DC, Duncan J. A domain-general cognitive core defined in multimodally parcellated human cortex. Cereb Cortex. 2020a:30(8):4361–4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assem M, Blank IA, Mineroff Z, Ademoglu A, Fedorenko E. Activity in the fronto-parietal multiple-demand network is robustly associated with individual differences in working memory and fluid intelligence. Cortex. 2020b:131:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assem M, Shashidhara S, Glasser MF, Duncan J. Precise topology of adjacent domain-general and sensory-biased regions in the human brain. Cereb Cortex. 2022:32(12):2521–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton RA, Venditti C. Human frontal lobes are not relatively large. Proc Natl Acad Sci U S A. 2013:110(22):9001–9006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens TEJ, Johansen Berg H, Jbabdi S, Rushworth MFS, Woolrich MW. Probabilistic diffusion tractography with multiple fibre orientations: what can we gain? NeuroImage. 2007:32(1):144–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop SJ, Fossella J, Croucher CJ, Duncan J. COMT val158 met genotype affects recruitment of neural mechanisms supporting fluid intelligence. Cereb Cortex. 2008:18(9):2132–2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant KL, Ardesch DJ, Roumazeilles L, Scholtens LH, Khrapitchev AA, Tendler BC, Wu W, Miller KL, Sallet J, van den Heuvel MP, et al. Diffusion MRI data, sulcal anatomy, and tractography for eight species from the Primate Brain Bank. Brain Struct Funct. 2021:226(8):2497–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catani M, Ffytche DH. The rises and falls of disconnection syndromes. Brain. 2005:128(10):2224–2239. [DOI] [PubMed] [Google Scholar]
- Catani M, Jones DK, Ffytche DH. Perisylvian language networks of the human brain. Ann Neurol. 2005:57(1):8–16. [DOI] [PubMed] [Google Scholar]
- Chen PY, Chen CL, Hsu YC, Tseng WYI. Fluid intelligence is associated with cortical volume and white matter tract integrity within multiple-demand system across adult lifespan. NeuroImage. 2020:212:116576. [DOI] [PubMed] [Google Scholar]
- Cole MW, Schneider W. The cognitive control network: integrated cortical regions with dissociable functions. NeuroImage. 2007:37(1):343–360. [DOI] [PubMed] [Google Scholar]
- Donahue CJ, Glasser MF, Preuss TM, Rilling JK, Van Essen DC. Quantitative assessment of prefrontal cortex in humans relative to nonhuman primates. Proc Natl Acad Sci U S A. 2018:115(22):E5183–E5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan J. The multiple-demand (MD) system of the primate brain: mental programs for intelligent behaviour. Trends Cogn. 2010:14(4):172–179. [DOI] [PubMed] [Google Scholar]
- Eichert N, Verhagen L, Folloni D, Jbabdi S, Khrapitchev AA, Sibson NR, Mantini D, Sallet J, Mars RB. What is special about the human arcuate fasciculus? Lateralization, projections, and expansion. Cortex. 2019:118:107–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichert N, Robinson EC, Bryant KL, Jbabdi S, Jenkinson M, Li L, Krug K, Watkins KE, Mars RB. Cross-species cortical alignment identifies different types of anatomical reorganization in the primate temporal lobe. Elife. 2020:9:e53232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorenko E, Duncan J, Kanwisher N. Broad domain generality in focal regions of frontal and parietal cortex. Proc Natl Acad Sci U S A. 2013:110(41):16616–16621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forkel SJ, Thiebaut de Schotten M, Kawadler JM, Dell’Acqua F, Danek A, Catani M. The anatomy of fronto-occipital connections from early blunt dissections to contemporary tractography. Cortex. 2014:56:73–84. [DOI] [PubMed] [Google Scholar]
- Fusi S, Miller EK, Rigotti M. Why neurons mix: high dimensionality for higher cognition. Curr Opin Neurol. 2016:37:66–74. [DOI] [PubMed] [Google Scholar]
- Genovesio A, Wise SP, Passingham RE. Prefrontal–parietal function: from foraging to foresight. Trends Cogn. 2014:18(2):72–81. [DOI] [PubMed] [Google Scholar]
- Glasser MF, Sotiropoulos SN, Wilson JA, Coalson TS, Fischl B, Andersson JL, Xu J, Jbabdi S, Webster M, Polimeni JR, et al. The minimal preprocessing pipelines for the Human Connectome Project. NeuroImage. 2013:80:105–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasser MF, Coalson TS, Robinson EC, Hacker CD, Harwell J, Yacoub E, Ugurbil K, Andersson J, Beckmann CF, Jenkinson M, et al. A multi-modal parcellation of human cerebral cortex. Nature. 2016:536:171–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldring AB, Krubitzer LA. Evolution of parietal cortex in mammals: from manipulation to tool use. In: Kaas JH, editors. Evolution of nervous systems. Elsevier; 2017. pp. 259–286 [Google Scholar]
- Haász J, Westlye ET, Fjær S, Espeseth T, Lundervold A, Lundervold AJ. General fluid-type intelligence is related to indices of white matter structure in middle-aged and old adults. NeuroImage. 2013:83:372–383. [DOI] [PubMed] [Google Scholar]
- Higham DJ, Kalna G, Kibble M. Spectral clustering and its use in bioinformatics. J Comput Appl Math. 2007:204:25–37. [Google Scholar]
- Jbabdi S, Sotiropoulos SN, Savio AM, Graña M, Beherens TEJ. Model-based analysis of multishell diffusion MR data for tractography: how to get over fitting problems. Magn Reson Med. 2012:68(6):1846–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadohisa M, Watanabe K, Kusunoki M, Buckley M, Duncan J. Focused representation of successive task episodes in frontal and parietal cortex. Cereb Cortex. 2020:30(3):1779–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latini F, Mårtensson J, Larsson EM, Fredrikson M, Åhs F, Hjortberg M, Aldskogius H, Ryttlefors M. Segmentation of the inferior longitudinal fasciculus in the human brain: a white matter dissection and diffusion tensor tractography study. Brain Res. 2017:1675:102–115. [DOI] [PubMed] [Google Scholar]
- Makris N, Kennedy DN, McInerney S, Sorensen AG, Wang R, Caviness VS Jr, Pandya DN. Segmentation of subcomponents within the superior longitudinal fascicle in humans: a quantitative, in vivo DT MRI study. Cereb Cortex. 2005:15(6):854–869. [DOI] [PubMed] [Google Scholar]
- Makris N, Kaiser J, Haselgrove C, Seidman LJ, Biederman J, Boriel D, Valera EM, Papadimitriou GM, Fischl B, Caviness VS, et al. Human cerebral cortex: a system for the integration of volume- and surface-based representations. NeuroImage. 2006:33(1):139–153. [DOI] [PubMed] [Google Scholar]
- Mantini D, Corbetta M, Romani GL, Orban GA, Vanduffel W. Evolutionarily novel functional networks in the human brain? J Neurosci. 2013:33(8):3259–3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mars RB, Foxley S, Verhagen L, Jbabdi S, Sallet J, Noonan MP, Neubert F-X, Andersson JL, Croxson PL, Dunbar RIM, et al. The extreme capsule fiber complex in humans and macaque monkeys: a comparative diffusion MRI tractography study. Brain Struct Funct. 2016:221(8):4059–4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mars RB, Sotiropoulos SN, Passingham RE, Sallet J, Verhagen L, Khrapitchev AA, Sibson N, Jbabdi S. Whole brain comparative anatomy using connectivity blueprints. elife. 2018a:7:e35237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mars RB, Passingham RE, Jbabdi S. Connectivity fingerprints: from areal descriptions to abstract spaces. Trends Cogn. 2018b:22(11):1026–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mars RB, O'Muircheartaigh J, Folloni D, Li L, Glasser MF, Jbabdi S, Bryant KL. Concurrent analysis of white matter bundles and grey matter networks in the chimpanzee. Brain Struct Funct. 2019:224:1021–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mars RB, Jbabdi S, Rushworth MFS. A common space approach to comparative neuroscience. Annu Rev Neurosci. 2021:44:69–86. [DOI] [PubMed] [Google Scholar]
- Miller EK, Cohen JD. An integrative theory of prefrontal cortex function. Annu Rev Neurosci. 2001:24:167–202. [DOI] [PubMed] [Google Scholar]
- Mitchell DJ, Bell AH, Buckley MJ, Mitchell AS, Sallet J, Duncan J. A putative multiple-demand system in the macaque brain. J Neurosci. 2016:36(33):8574–8585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray EA, Wise SP, Graham KS. The evolution of memory systems: ancestors, anatomy, and adaptations. Oxford (UK): OUP Oxford; 2017 [Google Scholar]
- Neubert F-X, Mars RB, Thomas AG, Sallet J, Rushworth MFS. Comparison of human ventral frontal cortex areas for cognitive control and language with areas in monkey frontal cortex. Neuron. 2014:81(3):700–713. [DOI] [PubMed] [Google Scholar]
- Nozais V, Forkel SJ, Foulon C, Petit L, Thiebaut de Schotten M. Functionnectome as a framework to analyse the contribution of brain circuits to fMRI. Commun Biol. 2021:4:1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passingham RE. Understanding the prefrontal cortex. Oxford (UK): Oxford Scholarship Online; 2021 [Google Scholar]
- Passingham RE, Smaers JB. Is the prefrontal cortex especially enlarged in the human brain allometric relations and remapping factors. Brain Behav. 2014:84(2):156–166. [DOI] [PubMed] [Google Scholar]
- Passingham RE, Stephan KE, Kötter R. The anatomical basis of functional localization in the cortex. Nat Rev Neurosci. 2002:3(8):606–616. [DOI] [PubMed] [Google Scholar]
- Paxinos G, Huang X-F, Toga A. The rhesus monkey brain in stereotaxic coordinates. Faculty of Health and Behavioural Sciences Papers (Archive). San Diego, CA: Academic Press; 2000 [Google Scholar]
- Petrides M, Pandya DN. Projections to the frontal cortex from the posterior parietal region in the rhesus monkey. J Comp Neurol. 1984:228(1):105–116. [DOI] [PubMed] [Google Scholar]
- Robinson EC, Jbabdi S, Glasser MF, Andersson J, Burgess GC, Harms MP, Smith SM, Van Essen DC, Jenkinson M. MSM: a new flexible framework for multimodal surface matching. Neuroimage. 2014:100:414–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson EC, Garcia K, Glasser MF, Chen Z, Coalson TS, Markopoulos A, Bozek J, Wright R, Schuh A, Webster M, et al. Multimodal surface matching with higher-order smoothness constraints. Neuroimage. 2018:167:453–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roumazeilles L, Eichert N, Bryant KL, Folloni D, Sallet J, Vijayakumar S, Foxley S, Tendler BC, Jbabdi S, Reveley C, et al. Longitudinal connections and the organization of the temporal cortex in macaques, great apes, and humans. PLoS Biol. 2020:18(7):e3000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roumazeilles L, Lange FJ, Benn RA, Andersson JLR, Bertelsen MF, Manger PR, Flach E, Khrapitchev AA, Bryant KL, Sallet J, et al. Cortical morphology and white matter tractography of three phylogenetically distant primates: evidence for a simian elaboration. Cereb Cortex. 2022:32(8):1608–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmahmann J, Pandya D. Fiber pathways of the brain. Oxford (UK): Oxford University Press; 2006 [Google Scholar]
- Sierpowska J, Bryant KL, Janssen N, Blazquez Freches G, Römkens M, Mangnus M, Mars RB, Piai V. Comparing human and chimpanzee temporal lobe neuroanatomy reveals modifications to human language hubs beyond the frontotemporal arcuate fasciculus. Proc Natl Acad Sci U S A. 2022:119(28):e2118295119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotiropoulos SN, Jbabdi S, Xu J, Andersson JL, Moeller S, Auerbach EJ, Glasser MF, Hernandez M, Sapiro G, Jenkinson M, et al. Advances in diffusion MRI acquisition and processing in the Human Connectome Project. NeuroImage. 2013:80:125–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiebaut de Schotten M, Dell’Acqua F, Forkel SJ, Simmons A, Vergani F, Murphy DGM, Catani M. A lateralized brain network for visuospatial attention. Nat Neurosci. 2011:4(10):10. [DOI] [PubMed] [Google Scholar]
- Thiebaut de Schotten M, Dell’Acqua F, Valabregue R, Catani M. Monkey to human comparative anatomy of the frontal lobe association tracts. Cortex. 2012:48(1):82–92. [DOI] [PubMed] [Google Scholar]
- Thiebaut de Schotten M, Foulon C, Nachev P. Brain disconnections link structural connectivity with function and behaviour. Nat Commun. 2020:11(1):5094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen DC, Dierker DL. Surface-based and probabilistic atlases of primate cerebral cortex. Neuron. 2007:56(2):209–225. [DOI] [PubMed] [Google Scholar]
- Van Essen DC, Glasser MF, Dierker DL, Harwell J. Cortical parcellations of the macaque monkey analyzed on surface-based atlases. Cereb Cortex. 2012:22(10):2227–2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen DC, Smith SM, Barch DM, Behrens TEJ, Yacoub E, Ugurbil K. The WU-Minn Human Connectome Project: an overview. NeuroImage. 2013:80:62–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrington S, Bryant KL, Khrapitchev AA, Sallet J, Charquero-Ballester M, Douaud G, Jbabdi S, Mars RB, Sotiropoulos SN. XTRACT - standardised protocols for automated tractography in the human and macaque brain. NeuroImage. 2020:217:116923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrington S, Thompson E, Bastiani M, Dubois J, Baxter L, Slater R, Jbabdi S, Mars RB, Sotiropoulos SN. Concurrent mapping of brain ontogeny and phylogeny within a common space: standardized tractography and applications. Sci Adv. 2022:8(42):eabq2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise SP. Forward frontal fields: phylogeny and fundamental function. Trends Cogn Sci. 2008:32(12):599–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise SP. Forward frontal fields: phylogeny and fundamental function. Trends Neurosci. 31:599–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolgar A, Duncan J, Manes F, Fedorenko E. Fluid intelligence is supported by the multiple-demand system not the language system. Nat Hum Behav. 2018:2:200–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data and scripts are available in the following repository: Karadachka, K.V., Mars, R.B. (2023): Structural connectivity of the multiple demand network in humans and comparison to the macaque brain. Version 1. Radboud University. (dataset). https://doi.org/10.34973/xzd1-zs16.