Visual Abstract
Abstract
The emergence and rapid development of single-cell technologies mark a paradigm shift in cancer research. Various technology implementations represent powerful tools to understand cellular heterogeneity, identify minor cell populations that were previously hard to detect and define, and make inferences about cell-to-cell interactions at single-cell resolution. Applied to lymphoma, recent advances in single-cell RNA sequencing have broadened opportunities to delineate previously underappreciated heterogeneity of malignant cell differentiation states and presumed cell of origin, and to describe the composition and cellular subsets in the ecosystem of the tumor microenvironment (TME). Clinical deployment of an expanding armamentarium of immunotherapy options that rely on targets and immune cell interactions in the TME emphasizes the requirement for a deeper understanding of immune biology in lymphoma. In particular, classic Hodgkin lymphoma (CHL) can serve as a study paradigm because of its unique TME, featuring infrequent tumor cells among numerous nonmalignant immune cells with significant interpatient and intrapatient variability. Synergistic to advances in single-cell sequencing, multiplexed imaging techniques have added a new dimension to describing cellular cross talk in various lymphoma entities. Here, we comprehensively review recent progress using novel single-cell technologies with an emphasis on the TME biology of CHL as an application field. The described technologies, which are applicable to peripheral blood, fresh tissues, and formalin-fixed samples, hold the promise to accelerate biomarker discovery for novel immunotherapeutic approaches and to serve as future assay platforms for biomarker-informed treatment selection, including immunotherapies.
Aoki and Steidl review the impact of single-cell technologies on the understanding of the heterogeneity of malignant cells and their microenvironment. Using classic Hodgkin lymphoma as a paradigm, the pair describe how single-cell sequencing and multiplexed imaging allows elucidation of the biology of a disease where a small number of tumor cells are embedded in a complex collection of nonmalignant cells in the tumor microenvironment.
Introduction
Recent technology development has significantly improved the comprehensiveness and accuracy of single-cell measurements in biology studies, and, in large part because of the progress in single-cell genomics and multiparametric imaging, single-cell studies are emerging as the new frontier and likely future standard in cancer research. A wide array of single-cell analytic tools offers broad-ranging molecular characterizations of cancer cells on the DNA, RNA, protein, and epigenetic levels. Single-cell measurements can overcome the major limitations of bulk characterizations of heterogenous cell populations, where measurements only represent averages and are usually dominated by only the most abundant cell populations in a given sample. Encouragingly, many studies using modern single-cell technologies successfully identified novel rare and/or previously hidden phenotypes of cancer cells and immune cells as part of the tumor microenvironment (TME).1, 2, 3, 4, 5, 6 In addition, from a historical perspective, single-cell analyses have played an important role in investigating the pathogenesis of rare malignant Hodgkin and Reed-Sternberg (HRS) cells of classic Hodgkin lymphoma (CHL). Of clinical importance, single-cell characterizations using patient samples have shown the potential to identify novel biomarkers for clinical decision-making in the field of cancer immunotherapy.6, 7, 8 In particular, lymphoid cancers have emerged as one of the most exciting application fields for single-cell profiling because of the importance of immune cell microenvironments for pathogenesis and as a therapeutic target for a wide variety of available immunotherapies.
Here, we will summarize technical aspects of modern single-cell technologies, including mass cytometry (CyTOF), single-cell RNA sequencing (scRNA-seq), and multiplexed imaging, and contextualize new insights from single-cell studies for lymphoma biology with an emphasis on CHL, TME cellular ecosystems, and potential implications for clinical treatment decisions.
Workflow of single-cell methodologies
scRNA-seq applications can serve to illustrate the multistep nature of a typical single-cell workflow encompassing sample processing, data preprocessing, and downstream computational analysis (Figure 1). Although initial single-cell technologies have faced significant challenges to obtain full transcripts from single cells at sufficient sequencing coverage, several robust technologies, such as Smart-seq and massively parallel RNA single-cell sequencing (MARS-Seq), have improved the overall quality of sequencing.9,10 For example, MARS-Seq increased technical stability and multiplex levels by employing 3 levels of barcording.9 Droplet-based approaches, including Drop-seq and inDrop, recently enabled interrogation of a high number of cells and are now widely used in various research fields.11, 12, 13
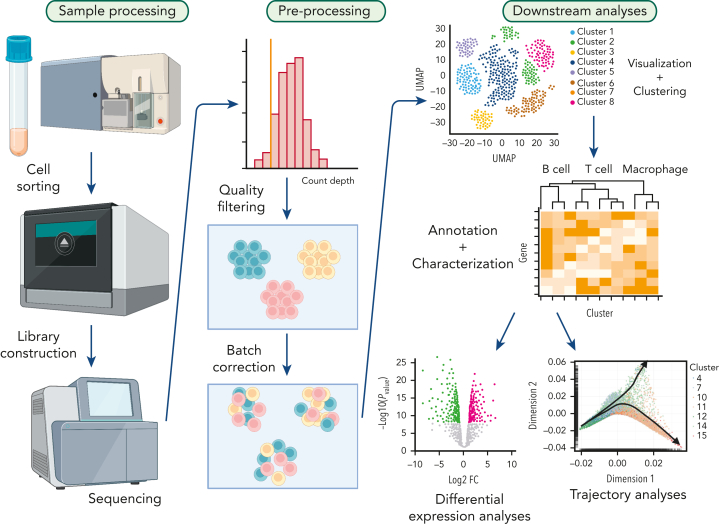
Figure 1.
Typical workflow for single-cell analysis. Target cell populations are purified from patient samples and constructed libraries from sorted cell populations are sequenced. Next, raw data is preprocessed to remove poor quality cells followed by normalization. Subsequently, batch correction is performed to remove technical variations due to multiple experiments. Finally, data are visualized by dimensionality reduction methods, and further downstream analyses are performed including differential expression analysis, cell-to-cell communication prediction and trajectory analysis.
Although high-dimensional data from single-cell technologies are extremely informative, there are many challenges for interpretation. For example, the limited sensitivity of mRNA detection in scRNA-seq data can lead to the presence of a large proportion of low (close to noise level) or zero values (measurement “drop-out”), representing a significant challenge. An elegant solution might be the target capture enrichment of single-cell genomic libraries to boost the signal of specific genes of interest.6 In addition, the complexity of single-cell data generated in the context of various physiological and disease conditions leads to inevitable technical variation between experimental batches, which has hampered the interpretability of results after data integration. To minimize these batch effects, a plethora of integration methods have been introduced and summarized in benchmarking studies.14,15 After quality control by preprocessing from raw outputs, unsupervised methods, such as phenograph clustering,16 are often used to discover phenotypic diversity in a sample, and cell types can be annotated by expression of canonical cell markers and other genes of interest. Subsequently, various types of downstream analyses can be performed to describe differential states and cell-to-cell interactions. However, a detailed description of analytical pipelines is out of scope for the present review, and we refer to excellent review articles that have summarized important methodologies and tools such as batch correction and trajectory analyses.14,17, 18, 19, 20
From an experimental point of view, there are 2 major types of samples to which we can apply single-cell technologies (Figure 2), both with various advantages and disadvantages. First, liquid biopsies (eg, blood) and disaggregated cell suspensions, that require prior dissociation of tumors, can be used for scRNA-seq and CyTOF as prominent application platforms. Second, single-cell characterizations can be performed in histologically intact tissues, allowing for single-cell measurements in a spatial context using, for example, mass spectrometry–based detection by laser abrasion (imaging mass cytometry [IMC]), multiplexed ion beam imaging,21 multiplexed imaging after DNA-based cell tagging, and high-plex in situ spatial multiomics analysis.22, 23, 24, 25, 26, 27, 28 Each of these multiplexed phenotyping approaches has their own strengths and limitations related to varying infrastructure requirements, antibody cost, the maximum number of protein targets, acquisition time during slide scanning, and data processing and analysis. For the latter, specific analysis requirements encompass solutions for image segmentation and cell annotations that are affected by limited resolution and low abundance of specific proteins because of a lack of effective signal amplification systems.21,25, 26, 27, 28, 29, 30, 31, 32 In particular, computational image segmentation, defining cellular boundaries, is challenging in all current methodologies.29,33 For instance, IMC analyses are limited to 1 μm imaging resolution, and although some analytic pipelines, such as ilastik and CellProfiler, help mitigate these problems,34,35 most of these algorithms do not fully overcome segmentation challenges, especially in high-density areas such as epithelial layers.29
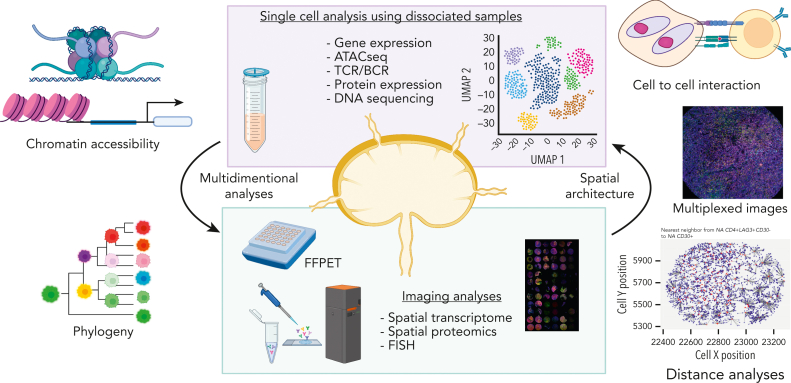
Figure 2.
Overview of single-cell and tissue architecture analyses. Two major specimen types are depicted to which single-cell technologies can be applied: (1) liquid biopsies (eg, blood) and disaggregated cell suspensions and (2) histologically intact tissues such as FFPET allowing for single-cell measurements in a spatial context. These 2 approaches work synergistically.
Considering the advantage of cell suspension–based analyses such as scRNA-seq, they can capture the transcriptome data of thousands of genes from a single cell, whereas cell suspension–based analyses do not take into account important information about the spatial architecture of the TME or phenotypically diverse malignant subclones.2,5,16,36,37 In contrast, multiplexed spatial assessment of immune cells in the TME has the potential to reveal a complex, spatially resolved TME ecosystem, whereas the dimensionality of data (ie, number of simultaneous measurements per cell) might be lower compared with dissociated cell-based methodology. Computational tools, such as spatial variance component analysis, can serve as a powerful tool to provide a quantitative assessment of the cell-to-cell interactions with spatially resolved molecular expression data.32 Another strength of many platforms with spatial resolution is the applicability to formalin-fixed paraffin-embedded tissue (FFPET) samples, which are routinely available from diagnostic pathology procedures in contrast to rarely available fresh frozen samples.27,28 Indeed, FFPET-based multiplexed imaging techniques have been used in the clinical trial setting for biomarker development.31 However, overall, the full potential of single-cell approaches in clinical studies and biomarker assay development is yet to be realized. Considering the unique advantages and limitations for each sample type and associated methodologies, synergistic study designs with the availability of both cell suspension and histologically intact tissues of the same study cases harbor the potential to overcome the above-described limitations (Figure 2).
Single-cell analyses in lymphoid cancers
The emergence of immunotherapy, including checkpoint inhibitors (CPIs) and chimeric antigen receptor therapy, as part of a new standard of care in lymphoma warrants a deeper understanding of cellular interactions between lymphoma cells and immune cells. Coinciding with this need, the number of studies using single-cell techniques in the lymphoma field are rapidly expanding4, 5, 6,36,38, 39, 40, 41, 42, 43, 44, 45 and the main published works are summarized in Table 1. Lymph node tissue as a primary site of lymphoma disease manifestation represents a tumor niche with a complex ecosystem of interacting immune cells, that changes during disease progression. The dynamics of TME changes can be categorized by the patterns of immune cell effacement, reeducation, and recruitment.46 These processes lead to significant interpatient and intrapatient heterogeneity of TME features in a given disease entity.
Table 1.
Summary of published literature related to single cell analyses in lymphoma
| Disease entity | Platforms | Main focus | Authors | Reference |
|---|---|---|---|---|
| DLBCL | scRNA-seq, cell-free DNA | Biomarkers of CAR T-cell therapy | Deng et al | 6 |
| DLBCL | scRNA-seq, bulk RNA-seq | Dissecting cellular heterogeneity | Steen et al | 44 |
| DLBCL, follicular lymphoma | scRNA-seq | Intratumor heterogeneity and personalized therapy | Roider et al | 43 |
| Follicular lymphoma | scRNA-seq | Subclones of cancer cells and Treg | Andor et al | 36 |
| Follicular lymphoma | scRNA-seq | Cancer cells and TFH cells | Haebe et al | 38 |
| Follicular lymphoma | scRNA-seq, MC-IF | Nonhematopoietic cells | Abe et al | 4 |
| Follicular lymphoma | scRNA-seq, MC-IF | Cytotoxic CD4+ cells and cancer cells with low MHC expression | Han et al | 45 |
| GCB cell | scRNA-seq, bulk RNA-seq | Cell of origin (follicular lymphoma) | Milpied et al | 40 |
| GCB cell | scRNA-seq, bulk RNA-seq | Cell of origin (DLBCL) | Holmes et al | 39 |
| Mantle cell lymphoma | scRNA-seq, bulk RNA-seq | Ibrutinib resistance | Zhang et al | 42 |
| DLBCL, CHL | IMC | Spatial classification | Colombo et al | 48 |
| CHL | scRNA-seq, IMC | MHC-II expression on HRS cells and LAG3+ Treg | Aoki et al | 5 |
| CHL | scRNA-seq, MC-IF | CXCL13+ helper T cells | Aoki et al | 41 |
| CHL | CyTOF, TCR | Biomarkers for PD-1 blockade | Cader et al | 79 |
| CHL | CyTOF | T-bet+ T cells | Cader et al | 76 |
| CHL | MC-IF | PDL1+ macrophages | Carey et al | 90 |
| CHL | MC-IF | CTLA4+CD4+ T cells | Patel et al | 91 |
| CHL | scRNA-seq, MC-IF | TOX2+CD4+ T cells | Veldman et al | 80 |
CAR, chimeric antigen receptor; DLBCL, diffuse large B-cell lymphoma.
Previous studies have identified variation in nonmalignant immune cell predominance and activation status to distinguish immunologically “hot” from “cold” TMEs,47,48 and a large body of biomarker literature exists that correlates TME composition with clinical outcome, such as tumor-associated macrophages, cytotoxic, and regulatory T cells (Treg).41,49, 50, 51, 52 Moreover, several studies have proposed microenvironment-driven classification systems in lymphoma.53,54 However, past studies are often limited by their reliance on identifying cell types using individual single markers, such as single stain immunohistochemistry (IHC), thus limiting insight into marker combinations and multidimensional phenotypes needed for more precise subsetting of cell populations and deriving functional state.
In contrast, scRNA-seq takes advantage of characterizing transcriptome states at the single-cell level, revealing previously unknown cellular diversity, including rare cell populations, in both cancer cells and immune cells in the TME. In particular, the emergence of novel drugs in the immunotherapy field warrants a more comprehensive understanding of TME biology in lymphoid malignancies because many drug targets are either expressed on immune cells in the TME or at the interface between malignant cells and neighboring cells. For example, CPIs are targeting “immune checkpoint molecules,” such as programmed death receptor 1 (PD-1), lymphocyte-activation gene 3 (LAG-3), and cytotoxic T lymphocyte antigen 4 (CTLA-4), which are negative regulators of T-cell activity.55 In fact, most single-cell studies in the lymphoma field have focused on studying TME interactions with the goal of developing biomarkers or uncovering novel treatment targets (Table 1). Furthermore, single-cell analyses have shed new light on the heterogeneity and developmental stages of germinal center B (GCB) cells and the dynamic states of GCB-derived B-cell lymphomas, with potential implications for future classification systems,39,40,44 and several studies have shown the potential of scRNA-seq to improve our understanding of treatment response and resistance through precise molecular profiling.42, 43, 44 Importantly, many studies are simultaneously using imaging techniques to describe the spatial context in which cellular populations, identified by single-cell sequencing, are situated4,5,41,48 (Figure 2).
Early single-cell studies in Hodgkin lymphoma (HL)
HL has a very distinctive TME that is unique among hematological malignancies. The malignant HRS cells of CHL, the main form of HL, typically only represent less than 1% of cells in tumor biopsies and are significantly outnumbered by nonmalignant cells, including lymphocytes, myeloid cells, and fibroblasts in the TME.56,57 Current pathogenesis models postulate that HRS cells “recruit and reeducate” their TME to create an immunosuppressive niche.58, 59, 60, 61 CHL can be further categorized into 4 subtypes mostly based on TME features: nodular sclerosis, mixed cellularity, lymphocyte-rich (LRCHL), and lymphocyte-depleted. PD-1 CPI, for example, nivolumab and pembrolizumab, targeting malignant cell to immune cell cross talk demonstrated high efficacy in CHL62, 63, 64, 65, 66 and are now standard-of-care treatments in relapsed/refractory CHL.
The most prominent technical obstacle for the study of HL is the scarcity of the malignant HRS cells, and single-cell sequencing was pivotal to demonstrate that HRS cells are derived from B cells in most of the cases. In 1994, Kuppers et al59 isolated single HRS cells from histological sections of 3 patients with HL by micromanipulation and analyzed individual HRS cells for immunoglobulin variable (V) gene rearrangement using polymerase chain reaction, establishing that HRS cells were indeed derived from a single malignant B-cell clone. Subsequently, Kanzler et al67 reported somatic mutations in the VH gene amplified from HRS cells together with nonfunctional Ig genes in most of the HL cases, suggesting GCB-cell derivation of HRS cells. In follow-up studies using similar approaches in paired samples from different biopsy sites, the persistence and spatial dissemination of a clonal HRS cell population were shown.68 Marafioti et al69 further confirmed these findings within a larger HL case series.
Moreover, key genetic features of HRS cells were identified through single-cell isolation of HRS cells,70, 71, 72, 73, 74, 75 thereby pointing to constitutive activation of the NF-κB pathway as one of the hallmarks of HL. First, several groups detected mutations in the NFKBIA gene from single HRS cells isolated from patients with HL,72, 73, 74, 75 and Schmitz et al71 further identified frequent somatic mutations in the TNFAIP3 (A20) gene from microdissected HRS cells, thus establishing these genes as tumor suppressors in the pathogenesis of HL. Intriguingly, TNFAIP3 mutations were observed at a significantly higher frequency in Epstein-Barr virus (EBV)–negative HL than in EBV-positive HL. This is consistent with the finding that somatic mutations in the NFKBIA gene were also predominantly identified in EBV-negative cases.72 This suggests complementary functions of inactivation of tumor suppressor genes of the NF-κB pathway and EBV infection, which also activates the NF-κB pathway through LMP1. Constitutive activation of the JAK/STAT pathway through SOCS1 mutations was also revealed by sequencing work using laser-microdissected HRS cells.70 Cumulatively, these studies described molecular similarities between HL and primary mediastinal large B-cell lymphoma70,71 and established the foundation for the current understanding of the molecular pathogenesis of HL.
In summary, these early studies have already shown the importance of genetic characterizations at the single-cell level, and, as described in the following sections, more recent studies using modern platforms have the transformational potential to describe the complexity of the cellular ecosystem in CHL at unprecedented spatial resolution and depth of simultaneous measurements.
Single-cell analyses using dissociated samples in CHL
As a high-dimensional single-cell analysis tool, CyTOF enables precise high-dimension protein detection. This single-cell suspension–based method takes advantage of metal-tagged antibodies to interrogate more than 50 protein markers simultaneously. In a first-of-its-kind study using a customized CyTOF panel for CHL, Cedar et al comprehensively analyzed the TME of 7 primary CHL samples.76 The study found a CHL-specific high abundance of Th1-polarized and terminally differentiated effector CD4+ T cells. Although Th1-polarized Treg cells generally did not express PD-1, most of the effector Th1-polarized CD4+ T cells in CHL showed high expression of PD-1, indicating an exhausted phenotype of this specific cell population. Moreover, the study confirmed an association between major histocompatibility complex class I (MHC-I) expression and B2M, a component of the MHC-I complex on HRS cells, with a possible association with latent EBV infection consistent with previous literature.77 These results indicate that, unlike in solid tumors, where PD-1 is mainly expressed on CD8+ T cells, PD-1–related cross talk might depend on MHC class II (MHC-II) restricted CD4+ T cells in CHL. Consistent with this hypothesis, the expression of MHC-II but not MHC-I on HRS cells was reported as a potential predictive biomarker for the clinical response to nivolumab treatment in CHL.78
The same group further expanded their CyTOF analyses to relapse patients with CHL treated with PD-1 blockade in the CheckMate 205 phase II clinical trial cohort (NCT02181738).79 Here, peripheral blood mononuclear cells were collected at multiple time points, including pretreatment (cycle 1 day 1) and at 2 time points during PD-1 blockade (cycle 2 day 1 and cycle 4 day 1), and compared with peripheral blood mononuclear cells obtained from patients with newly diagnosed CHL and healthy donors. CyTOF analyses revealed that the number of activated natural killer cells and a CD3−CD68+CD4+GrB+ cellular subset were associated with the response rate to PD-1 blockade. Simultaneously performed T-cell receptor (TCR) sequencing in the same patient cohort also revealed a correlation between TCR repertoire diversity and a favorable response to PD-1 blockade. These results motivate follow-up studies into the antigen specificity of T-cell clones and the dynamic assessment of clonal T-cell expansion during checkpoint blockade. Although several previous studies described that rosetting T cells in CHL are polyclonal,80, 81, 82 more comprehensive TCR analyses in specific T-cell subsets, possibly using single-cell TCR sequencing, are warranted.
As a complementary methodology, scRNA-seq provides an opportunity to describe the TME of CHL through the transcriptomic lineage assignment of individual cells. Aoki et al characterized the TME of 22 CHL samples and control cells from 5 reactive lymph nodes5 and described the phenotypic diversity of TME immune cell populations at a new level of resolution. As a major finding in this study, LAG3+ type-I regulatory T cells83 were identified as an expanded HL–specific cell population with immunosuppressive function in the TME. Because scRNA enables simultaneous measurement of typically more than 1000 genes per cell, this study also helped resolve coexpression patterns of phenotypic markers in T-cell subsets. In particular, a mutual exclusivity pattern between FOXP3 and LAG3 expression was revealed in CD4+ T cells, suggesting that the immunosuppressive population of LAG3+ cells might have been missed in previous single marker studies of immunomodulatory T cells using only FOXP3 for their identification.5 Additional context to the importance of LAG3+ CD4+ T cells was provided by trajectory analysis84,85 in which LAG3 positivity was identified as a late marker in the T-cell developmental trajectory, a finding that is consistent with a model of tumor cell–induced type-I regulatory T-cell phenotypes of terminally differentiated T cells.
scRNA-seq also helped to describe new contrast between histologically defined CHL subtypes. LRCHL is a rare, but highly instructive subtype, of CHL with a previously described high proportion of reactive B cells in the TME,86 relative to other CHL subtypes. Aoki et al again used scRNA-seq to compare LRCHL (n = 8) with other HL subtypes (n = 20), including mixed cellularity CHL and nodular sclerosis CHL.41 By analyzing the transcriptomes of more than 140 000 individual cells, an abundance of naïve B cells in the TME was confirmed, and a unique CD4+ helper T-cell subset with high expression of CXCL13 and PD-1 was found enriched in LRCHL compared with the other HL subtypes. In addition, a relative paucity of Treg cells indicated an overall very distinctive TME of LRCHL in the spectrum of CHL. Detailed analysis of expression patterns of helper T-cell subsets identified a unique CD4+PD-1+CXCL13+, but CXCR5-negative, T follicular helper (TFH)-like cell population, and bioinformatically inferred cell-to-cell interactions87 pointed to LRCHL-specific cross talk of CXCR5+ B cells with CXCL13+ T cells. Intriguingly, a similar CXCL13-producing TFH subset lacking CXCR5 expression was recently identified in the TME of breast cancer,88,89 indicating the pathogenic importance of the CXCR5/CXCL13 axis across cancer fields.
In summary, single-cell analysis of cell suspension samples provides new opportunities to identify precise marker combinations in cell populations and infer important cross talk between cancer cells and immune cells in the TME with unprecedented accuracy and measurement depth. These insights have led to significant refinement of CHL pathogenesis models (Figure 3). However, a number of limitations in published scRNA-seq studies need to be acknowledged, including the underrepresentation or complete lack of HRS cells in cell suspension samples and the absence of information identifying cells in a spatial context.5 In the following section, technology solutions in intact histology tissues are described that address these issues and synergize with cell-suspension approaches.
Figure 3.
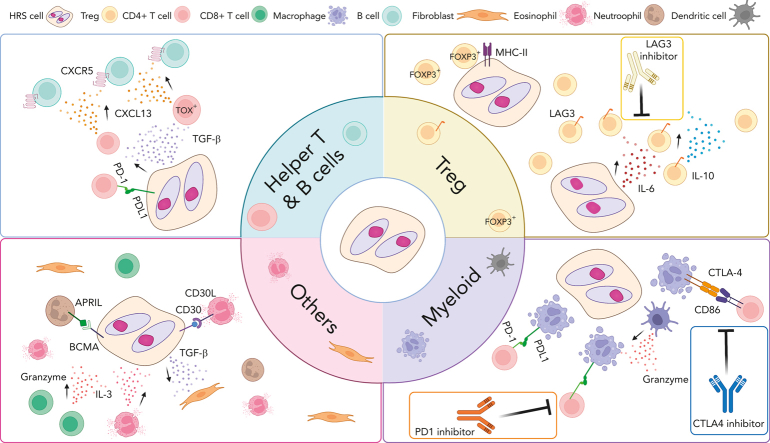
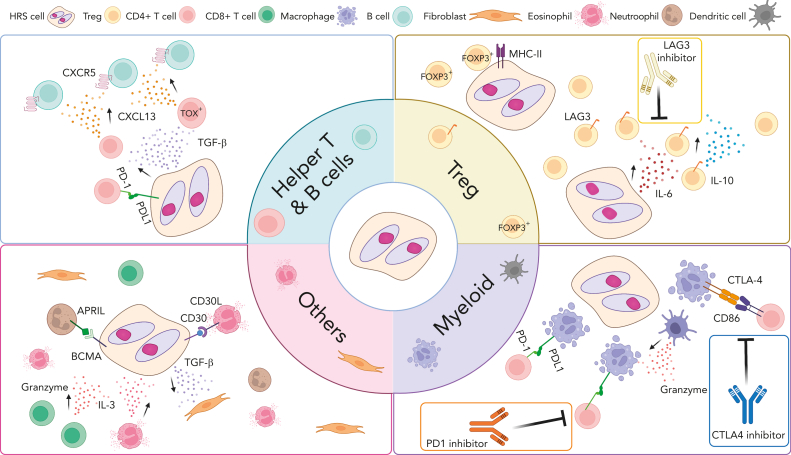
Overview of cross talk between tumor cells and noncancerous immune cells in the TME of CHL. Interaction between HRS cells and numerous nonmalignant immune cells in the TME are shown according to cell type. These interactions play an important role promoting acquired immune evasion and activation of pathways that support the growth and survival of HRS cells through (1) ligand and receptor interactions, such as PD-1/PD-L1, LAG-3/MHC-II and (2) cytokines and chemokine milieus (eg, CXCL13, TGF-β). Several cross talk mechanisms and related treatment targets are highlighted. CXCL13, chemokine C-X-C motif ligand 13; CXCR5, C-X-C chemokine receptor type 5; IL-3, interleukin 3; TGF- β, transforming growth factor beta.
Technologies using intact histology in CHL
Multicolor IHC (MC-IHC) and/or multicolor immunofluorescence (MC-IF) have been used to visualize specific cell populations using 4 to 7 markers in CHL studies.5,41,80,90,91 Carey et al gained important insights into tumor-associated macrophages that express PD-1 ligand (PD-L1) and described their spatial relationship with PD-1+ T cells.90 Intriguingly, they found that the abundance of PD-L1+ macrophages on average exceeded the number of PD-L1+ HRS cells that typically harbor copy number gains and amplifications of the PD-L1 locus on chromosome 9p24.1.90 In addition, their analysis confirmed the direct contact between PD-L1+ macrophages and PD-1+ T cells. These findings suggest that macrophages significantly contribute to PD-L1–mediated immune modulation in CHL. At the same time, their finding that PD-1+ CD4+ T cells were not more abundant than other CD4+ T cells in the direct vicinity of HRS cells leaves open questions about the dynamics of direct cellular interaction between HRS cells and PD1+ T cells. More recently, the same group further characterized the TME of CHL using MC-IF using different marker combinations.91 Patel et al91 found enrichment of CTLA-4+ CD4+ and CD8+ T cells in CHL tissues and their engagement with CTLA-4 ligand expressing CD86+ HRS cells and macrophages. Interestingly, the identified CTLA-4+ T-cell population mostly did not coexpress PD-1, and the CTLA-4-CD86 cellular interactions seemed to be distinct from the PD-1-PD-L1 interactions described previously.90 The identification of these potentially independent cross talk mechanisms suggest synergistic therapeutic efficacy of PD-1 and CTLA-4 targeting treatment in CHL92 and as shown in solid cancers.93,94
MC-IF has also been used in combination with scRNA-seq to validate coexpression patterns on the protein level and to delineate spatial relationships among target cell populations. Seven-marker MC-IF of FFPE cases on a tissue microarray, that matched cell suspension samples interrogated by scRNA-seq, confirmed the unique coexpression pattern of PD1+CXCL13+CD4+ TFH-like cells in LRCHL described in the previous section. Spatial analysis revealed that in 46% of cases, cells formed “rosettes” surrounding HRS cells and that CXCR5+ normal B cells were near CXCL13+ T cells by topological analysis. Of potential clinical importance, the abundance of PD-1+CXCL13+ CD4+ T cells in the TME was significantly associated with shorter progression-free survival in LRCHL (P = .032), whereas single IHC positivity of PD-1 and CXCL13 was not associated with outcome.
Rosetting CD4+ T cells that are in contact with the malignant HRS cells have been described as a unique feature of CHL. To understand the biology underlying this characteristic, Veldman et al80 effectively used MC-IF to further delineate the coexpression pattern and topology of a previously reported rosette-forming CD4+CD26− T-cell population.95 Here, the transcriptional factors, TOX and TOX2, which are involved in TFH cell development and T-cell exhaustion,96,97 were identified as key features of HRS cell–surrounding T cells. MC-IHC analyses further uncovered high expression of CXCL13 and PD-1 in this population, indicating an exhaustion phenotype of these T cells, and scRNA-seq data served as further validation of this unique phenotype. The translational significance of this cell population as a biomarker and treatment target needs to be investigated in future clinical trials.
Although conventional MC-IHC/IF is useful to investigate coexpression patterns and topography, the number of concurrent markers in antibody panels remains limited. However, recently introduced methodologies, such as IMC or antibody-based oligonucleotide tagging, enable simultaneous detection of more than 40 markers. Using a customized marker panel for CHL, immunosuppressive LAG3+ Tr1 cells were demonstrated to be predominantly in spatial contact with MHC-II–negative HRS cells, providing additional insight into the question which cell populations participate in rosette formation.5 In contrast, classic FOXP3+ Tregs were significantly enriched in the vicinity of MHC class-II–positive HRS cells. These data suggest that inducible Tr1 cells in the direct vicinity of malignant cells are only present if MHC-II, a ligand to LAG3, is not expressed on tumor cells, providing additional proof of concept that malignant cell phenotypes and TME architecture are linked in CHL. Moreover, functional validation experiments confirmed an inhibitory interaction between MHC-II–positive HRS and Tr1 cells in a coculture system with an MHC-II–negative CHL-derived L-428 cell line model.5
Future perspective
Single-cell analysis has already significantly added to the understanding of CHL biology (Figure 3) and might offer an opportunity to more effectively subdivide CHL based on TME biology for increased clinical utility. For instance, multiple rational TME-related treatment targets, including PD-1, CTLA-4, and LAG3, have now been identified,5,90,91 and refined classification might enable classification-driven treatment choices. However, at this time, the complex TME ecosystem of CHL has not been fully elucidated, and the most effective biomarker principles and assay platforms have not yet emerged from these studies.
An attractive pathogenesis model with a strong imprint on TME composition and function might be the predominance of certain cytokines and chemokines produced by HRS cells forming immunosuppressive niches.98 The most recent studies indeed focused on understanding the link between cytokine and chemokine expression and specific cellular subsets, with prominent examples being the importance of interleukin-6 for induction of LAG3+ Tregs and interleukin-10 and transforming growth factor β for CXCL13+ helper T-cell subsets.5,41 These patterns might contrast with inflammatory milieus characterized by high interferon gamma expression by various cell types.
Single-cell genomics and multiplexed imaging analyses provide new opportunities to characterize the TME, including architectural features, giving rise to the exciting new concept of developing “spatial biomarkers” in lymphoma. Cost effective and standardized implementation of biomarker assays gives strong consideration to FFPE-based solutions, as demonstrated in gene-expression–based predictive biomarker assays from FFPET in HL,99, 100, 101 and at the moment, this requirement points to limited marker designs currently achievable by multiparametric IHC. Considering the cost of single-cell methodologies, digital cytometry based on deconvolution of bulk sequencing data might be complementary to single-cell sequencing in clinical practice and trials for certain research questions.44,102
Moving forward, exciting new single-cell technologies are under development that hold the promise of deepening our understanding of lymphoma single-cell biology. Using DNA-barcoded antibodies, multiomics assays, such as RNA expression and protein sequencing and cellular indexing of transcriptomes and epitopes by sequencing, now enable simultaneous measurements of proteins and RNAs in single cells.25,103,104 Novel platforms also enable simultaneous interrogation of chromatin accessibility (ATAC) and gene expression, thus layering on insight into gene regulation at single-cell resolution.105 In addition, single-cell DNA sequencing is currently deployed to describe tumor heterogeneity and derive cellular phylogenies to infer temporal pathogenesis models. Spatial imaging technologies with single-cell resolution have also been introduced in several formats with more than 100-plex measurements.106 Furthermore, with the expansion of applicability to single nuclei from FFPET,107 unlocking multimodality single-cell analyses from clinical trial samples could provide a paradigm shift, enabling the discovery of predictors of immunotherapy response and treatment resistance where immunotherapy is showing curative potential for a subset of patients.108 In addition, obtaining multiple timepoint samples before and after treatment would be an ambitious but feasible approach for the discovery of novel dynamic biomarkers, as recently suggested in HL and other lymphomas.31,99
In conclusion, single-cell technologies have set new standards for the study of lymphoma biology with major new insights into cellular heterogeneity and TME interactions as exemplified in CHL. Although ongoing technology development will further increase the resolution and comprehensiveness of information, parallel biomarker translation studies and associated clinical utility studies are imperative.
Conflict-of-interest disclosure: C.S. received honoraria from Seattle Genetics, AbbVie, and Bayer, and holds research funding from Epizyme and Trillium Therapeutics Inc. T.A. declares no competing financial interests.
Acknowledgments
The figures were created with BioRender.com.
C.S. was supported by a Michael Smith Foundation for Health Research Career Investigator award and the British Columbia Cancer Foundation. T.A. was supported by a fellowship from the Canadian Institutes of Health Research and the Lymphoma Research Foundation (LRF). T.A. is the recipient of an LRF Lymphoma Scientific Research Mentoring Program Scholar award.
Authorship
Contribution: T.A. and C.S. reviewed the literature and wrote the paper.
References
- 1.Elyada E, Bolisetty M, Laise P, et al. Cross-species single-cell analysis of pancreatic ductal adenocarcinoma reveals antigen-presenting cancer-associated fibroblasts. Cancer Discov. 2019;9(8):1102–1123. doi: 10.1158/2159-8290.CD-19-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cohen M, Giladi A, Gorki AD, et al. Lung single-cell signaling interaction map reveals basophil role in macrophage imprinting. Cell. 2018;175(4):1031–1044.e1018. doi: 10.1016/j.cell.2018.09.009. [DOI] [PubMed] [Google Scholar]
- 3.Zheng C, Zheng L, Yoo JK, et al. Landscape of infiltrating T cells in liver cancer revealed by single-cell sequencing. Cell. 2017;169(7):1342–1356.e1316. doi: 10.1016/j.cell.2017.05.035. [DOI] [PubMed] [Google Scholar]
- 4.Abe Y, Sakata-Yanagimoto M, Fujisawa M, et al. A single-cell atlas of non-haematopoietic cells in human lymph nodes and lymphoma reveals a landscape of stromal remodelling. Nat Cell Biol. 2022;24(4):565–578. doi: 10.1038/s41556-022-00866-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aoki T, Chong LC, Takata K, et al. Single-cell transcriptome analysis reveals disease-defining T-cell subsets in the tumor microenvironment of classic Hodgkin lymphoma. Cancer Discov. 2020;10(3):406–421. doi: 10.1158/2159-8290.CD-19-0680. [DOI] [PubMed] [Google Scholar]
- 6.Deng Q, Han G, Puebla-Osorio N, et al. Characteristics of anti-CD19 CAR T cell infusion products associated with efficacy and toxicity in patients with large B cell lymphomas. Nat Med. 2020;26(12):1878–1887. doi: 10.1038/s41591-020-1061-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Au L, Hatipoglu E, Robert de Massy M, et al. Determinants of anti-PD-1 response and resistance in clear cell renal cell carcinoma. Cancer Cell. 2021;39(11):1497–1518.e1411. doi: 10.1016/j.ccell.2021.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krieg C, Nowicka M, Guglietta S, et al. High-dimensional single-cell analysis predicts response to anti-PD-1 immunotherapy. Nat Med. 2018;24(2):144–153. doi: 10.1038/nm.4466. [DOI] [PubMed] [Google Scholar]
- 9.Jaitin DA, Kenigsberg E, Keren-Shaul H, et al. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science. 2014;343(6172):776–779. doi: 10.1126/science.1247651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramskold D, Luo S, Wang YC, et al. Full-length mRNA-seq from single-cell levels of RNA and individual circulating tumor cells. Nat Biotechnol. 2012;30(8):777–782. doi: 10.1038/nbt.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng GX, Terry JM, Belgrader P, et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun. 2017;8:14049. doi: 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Macosko EZ, Basu A, Satija R, et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161(5):1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klein AM, Mazutis L, Akartuna I, et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 2015;161(5):1187–1201. doi: 10.1016/j.cell.2015.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tran HTN, Ang KS, Chevrier M, et al. A benchmark of batch-effect correction methods for single-cell RNA sequencing data. Genome Biol. 2020;21(1):12. doi: 10.1186/s13059-019-1850-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luecken MD, Buttner M, Chaichoompu K, et al. Benchmarking atlas-level data integration in single-cell genomics. Nat Methods. 2022;19(1):41–50. doi: 10.1038/s41592-021-01336-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levine JH, Simonds EF, Bendall SC, et al. Data-driven phenotypic dissection of AML reveals progenitor-like cells that correlate with prognosis. Cell. 2015;162(1):184–197. doi: 10.1016/j.cell.2015.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davis-Marcisak EF, Deshpande A, Stein-O'Brien GL, et al. From bench to bedside: single-cell analysis for cancer immunotherapy. Cancer Cell. 2021;39(8):1062–1080. doi: 10.1016/j.ccell.2021.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haghverdi L, Lun ATL, Morgan MD, Marioni JC. Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat Biotechnol. 2018;36(5):421–427. doi: 10.1038/nbt.4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trapnell C. Defining cell types and states with single-cell genomics. Genome Res. 2015;25(10):1491–1498. doi: 10.1101/gr.190595.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saelens W, Cannoodt R, Todorov H, Saeys Y. A comparison of single-cell trajectory inference methods. Nat Biotechnol. 2019;37(5):547–554. doi: 10.1038/s41587-019-0071-9. [DOI] [PubMed] [Google Scholar]
- 21.Angelo M, Bendall SC, Finck R, et al. Multiplexed ion beam imaging of human breast tumors. Nat Med. 2014;20(4):436–442. doi: 10.1038/nm.3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gouin KH, 3rd, Ing N, Plummer JT, et al. An N-Cadherin 2 expressing epithelial cell subpopulation predicts response to surgery, chemotherapy and immunotherapy in bladder cancer. Nat Commun. 2021;12(1):4906. doi: 10.1038/s41467-021-25103-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hwang WL, Jagadeesh KA, Guo JA, et al. Single-nucleus and spatial transcriptome profiling of pancreatic cancer identifies multicellular dynamics associated with neoadjuvant treatment. Nat Genet. 2022;54(8):1178–1191. doi: 10.1038/s41588-022-01134-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jackson HW, Fischer JR, Zanotelli VRT, et al. The single-cell pathology landscape of breast cancer. Nature. 2020;578(7796):615–620. doi: 10.1038/s41586-019-1876-x. [DOI] [PubMed] [Google Scholar]
- 25.Stoeckius M, Hafemeister C, Stephenson W, et al. Simultaneous epitope and transcriptome measurement in single cells. Nat Methods. 2017;14(9):865–868. doi: 10.1038/nmeth.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stahl PL, Salmen F, Vickovic S, et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science. 2016;353(6294):78–82. doi: 10.1126/science.aaf2403. [DOI] [PubMed] [Google Scholar]
- 27.Schurch CM, Bhate SS, Barlow GL, et al. Coordinated cellular neighborhoods orchestrate antitumoral immunity at the colorectal cancer invasive front. Cell. 2020;182(5):1341–1359. doi: 10.1016/j.cell.2020.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Black S, Phillips D, Hickey JW, et al. CODEX multiplexed tissue imaging with DNA-conjugated antibodies. Nat Protoc. 2021;16(8):3802–3835. doi: 10.1038/s41596-021-00556-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Z. Cell segmentation for image cytometry: advances, insufficiencies, and challenges. Cytometry A. 2019;95(7):708–711. doi: 10.1002/cyto.a.23686. [DOI] [PubMed] [Google Scholar]
- 30.Qian X, Harris KD, Hauling T, et al. Probabilistic cell typing enables fine mapping of closely related cell types in situ. Nat Methods. 2020;17(1):101–106. doi: 10.1038/s41592-019-0631-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Phillips D, Matusiak M, Gutierrez BR, et al. Immune cell topography predicts response to PD-1 blockade in cutaneous T cell lymphoma. Nat Commun. 2021;12(1):6726. doi: 10.1038/s41467-021-26974-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arnol D, Schapiro D, Bodenmiller B, Saez-Rodriguez J, Stegle O. Modeling cell-cell interactions from spatial molecular data with spatial variance component analysis. Cell Rep. 2019;29(1):202–211.e206. doi: 10.1016/j.celrep.2019.08.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Petukhov V, Xu RJ, Soldatov RA, et al. Cell segmentation in imaging-based spatial transcriptomics. Nat Biotechnol. 2022;40(3):345–354. doi: 10.1038/s41587-021-01044-w. [DOI] [PubMed] [Google Scholar]
- 34.Carpenter AE, Jones TR, Lamprecht MR, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7(10):R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sommer C, Straehle C, Kothe U, Hamprecht FA. 2011. Ilastik: interactive learning and segmentation toolkit. Paper presented at: 2011 8th IEEE International Symposium on Biomedical Imaging: From Nano to Macro; 30 March-2 April. Chicago, IL. [Google Scholar]
- 36.Andor N, Simonds EF, Czerwinski DK, et al. Single-cell RNA-Seq of follicular lymphoma reveals malignant B-cell types and coexpression of T-cell immune checkpoints. Blood. 2019;133(10):1119–1129. doi: 10.1182/blood-2018-08-862292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Azizi E, Carr AJ, Plitas G, et al. Single-cell map of diverse immune phenotypes in the breast tumor microenvironment. Cell. 2018;174(5):1293–1308.e1236. doi: 10.1016/j.cell.2018.05.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haebe S, Shree T, Sathe A, et al. Single-cell analysis can define distinct evolution of tumor sites in follicular lymphoma. Blood. 2021;137(21):2869–2880. doi: 10.1182/blood.2020009855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Holmes AB, Corinaldesi C, Shen Q, et al. Single-cell analysis of germinal-center B cells informs on lymphoma cell of origin and outcome. J Exp Med. 2020;217(10) doi: 10.1084/jem.20200483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Milpied P, Cervera-Marzal I, Mollichella ML, et al. Human germinal center transcriptional programs are de-synchronized in B cell lymphoma. Nat Immunol. 2018;19(9):1013–1024. doi: 10.1038/s41590-018-0181-4. [DOI] [PubMed] [Google Scholar]
- 41.Aoki T, Chong LC, Takata K, et al. Single-cell profiling reveals the importance of CXCL13/CXCR5 axis biology in lymphocyte-rich classic Hodgkin lymphoma. Proc Natl Acad Sci U S A. 2021;118(41) doi: 10.1073/pnas.2105822118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang S, Jiang VC, Han G, et al. Longitudinal single-cell profiling reveals molecular heterogeneity and tumor-immune evolution in refractory mantle cell lymphoma. Nat Commun. 2021;12(1):2877. doi: 10.1038/s41467-021-22872-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roider T, Seufert J, Uvarovskii A, et al. Dissecting intratumour heterogeneity of nodal B-cell lymphomas at the transcriptional, genetic and drug-response levels. Nat Cell Biol. 2020;22(7):896–906. doi: 10.1038/s41556-020-0532-x. [DOI] [PubMed] [Google Scholar]
- 44.Steen CB, Luca BA, Esfahani MS, et al. The landscape of tumor cell states and ecosystems in diffuse large B cell lymphoma. Cancer Cell. 2021;39(10):1422–1437.e1410. doi: 10.1016/j.ccell.2021.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Han G, Deng Q, Marques-Piubelli ML, et al. Follicular lymphoma microenvironment characteristics associated with tumor cell mutations and MHC class II expression. Blood Cancer Discov. 2022;3(5):428–443. doi: 10.1158/2643-3230.BCD-21-0075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scott DW, Gascoyne RD. The tumour microenvironment in B cell lymphomas. Nat Rev Cancer. 2014;14(8):517–534. doi: 10.1038/nrc3774. [DOI] [PubMed] [Google Scholar]
- 47.Ennishi D, Takata K, Beguelin W, et al. Molecular and genetic characterization of MHC deficiency identifies EZH2 as therapeutic target for enhancing immune recognition. Cancer Discov. 2019;9(4):546–563. doi: 10.1158/2159-8290.CD-18-1090. [DOI] [PubMed] [Google Scholar]
- 48.Colombo A, Hav M, Singh M, et al. Single-cell spatial analysis of tumor immune architecture in diffuse large B-cell lymphoma. Blood Adv. 2022;6(16):4675–4690. doi: 10.1182/bloodadvances.2022007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Steidl C, Lee T, Shah SP, et al. Tumor-associated macrophages and survival in classic Hodgkin's lymphoma. N Engl J Med. 2010;362(10):875–885. doi: 10.1056/NEJMoa0905680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Riihijarvi S, Fiskvik I, Taskinen M, et al. Prognostic influence of macrophages in patients with diffuse large B-cell lymphoma: a correlative study from a Nordic phase II trial. Haematologica. 2015;100(2):238–245. doi: 10.3324/haematol.2014.113472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tzankov A, Meier C, Hirschmann P, Went P, Pileri SA, Dirnhofer S. Correlation of high numbers of intratumoral FOXP3+ regulatory T cells with improved survival in germinal center-like diffuse large B-cell lymphoma, follicular lymphoma and classical Hodgkin's lymphoma. Haematologica. 2008;93(2):193–200. doi: 10.3324/haematol.11702. [DOI] [PubMed] [Google Scholar]
- 52.Autio M, Leivonen SK, Bruck O, et al. Immune cell constitution in the tumor microenvironment predicts the outcome in diffuse large B-cell lymphoma. Haematologica. 2021;106(3):718–729. doi: 10.3324/haematol.2019.243626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kotlov N, Bagaev A, Revuelta MV, et al. Clinical and biological subtypes of B-cell lymphoma revealed by microenvironmental signatures. Cancer Discov. 2021;11(6):1468–1489. doi: 10.1158/2159-8290.CD-20-0839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lenz G, Wright G, Dave SS, et al. Stromal gene signatures in large-B-cell lymphomas. N Engl J Med. 2008;359(22):2313–2323. doi: 10.1056/NEJMoa0802885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Waldman AD, Fritz JM, Lenardo MJ. A guide to cancer immunotherapy: from T cell basic science to clinical practice. Nat Rev Immunol. 2020;20(11):651–668. doi: 10.1038/s41577-020-0306-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alaggio R, Amador C, Anagnostopoulos I, et al. The 5th edition of the World Health Organization classification of haematolymphoid tumours: lymphoid neoplasms. Leukemia. 2022;36(7):1720–1748. doi: 10.1038/s41375-022-01620-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Campo E, Jaffe ES, Cook JR, et al. The international consensus classification of mature lymphoid neoplasms: a report from the Clinical Advisory Committee. Blood. 2022;140(11):1229–1253. doi: 10.1182/blood.2022015851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kuppers R. The biology of Hodgkin's lymphoma. Nat Rev Cancer. 2009;9(1):15–27. doi: 10.1038/nrc2542. [DOI] [PubMed] [Google Scholar]
- 59.Kuppers R, Rajewsky K, Zhao M, et al. Hodgkin disease: Hodgkin and Reed-Sternberg cells picked from histological sections show clonal immunoglobulin gene rearrangements and appear to be derived from B cells at various stages of development. Proc Natl Acad Sci U S A. 1994;91(23):10962–10966. doi: 10.1073/pnas.91.23.10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mottok A, Steidl C. Biology of classical Hodgkin lymphoma: implications for prognosis and novel therapies. Blood. 2018;131(15):1654–1665. doi: 10.1182/blood-2017-09-772632. [DOI] [PubMed] [Google Scholar]
- 61.Steidl C, Connors JM, Gascoyne RD. Molecular pathogenesis of Hodgkin's lymphoma: increasing evidence of the importance of the microenvironment. J Clin Oncol. 2011;29(14):1812–1826. doi: 10.1200/JCO.2010.32.8401. [DOI] [PubMed] [Google Scholar]
- 62.Moskowitz CH, Zinzani PL, Fanale MA, et al. Pembrolizumab in relapsed/refractory classical Hodgkin lymphoma: primary end point analysis of the phase 2 Keynote-087 Study. Blood. 2016;128(22):1107. [Google Scholar]
- 63.Timmerman JM, Engert A, Younes A, et al. Checkmate 205 update with minimum 12-month follow up: a phase 2 study of nivolumab in patients with relapsed/refractory classical Hodgkin lymphoma. Blood. 2016;128(22):1110. [Google Scholar]
- 64.Younes A, Santoro A, Shipp M, et al. Nivolumab for classical Hodgkin's lymphoma after failure of both autologous stem-cell transplantation and brentuximab vedotin: a multicentre, multicohort, single-arm phase 2 trial. Lancet Oncol. 2016;17(9):1283–1294. doi: 10.1016/S1470-2045(16)30167-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen R, Zinzani PL, Fanale MA, et al. Phase II study of the efficacy and safety of pembrolizumab for relapsed/refractory classic Hodgkin lymphoma. J Clin Oncol. 2017;35(19):2125–2132. doi: 10.1200/JCO.2016.72.1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Armand P, Engert A, Younes A, et al. Nivolumab for relapsed/refractory classic Hodgkin lymphoma after failure of autologous hematopoietic cell transplantation: extended follow-up of the multicohort single-arm phase II CheckMate 205 Trial. J Clin Oncol. 2018;36(14):1428–1439. doi: 10.1200/JCO.2017.76.0793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kanzler H, Kuppers R, Hansmann ML, Rajewsky K. Hodgkin and Reed-Sternberg cells in Hodgkin's disease represent the outgrowth of a dominant tumor clone derived from (crippled) germinal center B cells. J Exp Med. 1996;184(4):1495–1505. doi: 10.1084/jem.184.4.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vockerodt M, Soares M, Kanzler H, et al. Detection of clonal Hodgkin and Reed-Sternberg cells with identical somatically mutated and rearranged VH genes in different biopsies in relapsed Hodgkin's disease. Blood. 1998;92(8):2899–2907. [PubMed] [Google Scholar]
- 69.Marafioti T, Hummel M, Foss HD, et al. Hodgkin and reed-sternberg cells represent an expansion of a single clone originating from a germinal center B-cell with functional immunoglobulin gene rearrangements but defective immunoglobulin transcription. Blood. 2000;95(4):1443–1450. [PubMed] [Google Scholar]
- 70.Weniger MA, Melzner I, Menz CK, et al. Mutations of the tumor suppressor gene SOCS-1 in classical Hodgkin lymphoma are frequent and associated with nuclear phospho-STAT5 accumulation. Oncogene. 2006;25(18):2679–2684. doi: 10.1038/sj.onc.1209151. [DOI] [PubMed] [Google Scholar]
- 71.Schmitz R, Hansmann ML, Bohle V, et al. TNFAIP3 (A20) is a tumor suppressor gene in Hodgkin lymphoma and primary mediastinal B cell lymphoma. J Exp Med. 2009;206(5):981–989. doi: 10.1084/jem.20090528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jungnickel B, Staratschek-Jox A, Brauninger A, et al. Clonal deleterious mutations in the IkappaBalpha gene in the malignant cells in Hodgkin's lymphoma. J Exp Med. 2000;191(2):395–402. doi: 10.1084/jem.191.2.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Emmerich F, Theurich S, Hummel M, et al. Inactivating I kappa B epsilon mutations in Hodgkin/Reed-Sternberg cells. J Pathol. 2003;201(3):413–420. doi: 10.1002/path.1454. [DOI] [PubMed] [Google Scholar]
- 74.Emmerich F, Meiser M, Hummel M, et al. Overexpression of I kappa B alpha without inhibition of NF-kappaB activity and mutations in the I kappa B alpha gene in Reed-Sternberg cells. Blood. 1999;94(9):3129–3134. [PubMed] [Google Scholar]
- 75.Cabannes E, Khan G, Aillet F, Jarrett RF, Hay RT. Mutations in the IkBa gene in Hodgkin's disease suggest a tumour suppressor role for IkappaBalpha. Oncogene. 1999;18(20):3063–3070. doi: 10.1038/sj.onc.1202893. [DOI] [PubMed] [Google Scholar]
- 76.Cader FZ, Schackmann RCJ, Hu X, et al. Mass cytometry of Hodgkin lymphoma reveals a CD4(+) regulatory T-cell-rich and exhausted T-effector microenvironment. Blood. 2018;132(8):825–836. doi: 10.1182/blood-2018-04-843714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Reichel J, Chadburn A, Rubinstein PG, et al. Flow sorting and exome sequencing reveal the oncogenome of primary Hodgkin and Reed-Sternberg cells. Blood. 2015;125(7):1061–1072. doi: 10.1182/blood-2014-11-610436. [DOI] [PubMed] [Google Scholar]
- 78.Roemer MGM, Redd RA, Cader FZ, et al. Major histocompatibility complex class II and programmed death ligand 1 expression predict outcome after programmed death 1 blockade in classic Hodgkin lymphoma. J Clin Oncol. 2018;36(10):942–950. doi: 10.1200/JCO.2017.77.3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cader FZ, Hu X, Goh WL, et al. A peripheral immune signature of responsiveness to PD-1 blockade in patients with classical Hodgkin lymphoma. Nat Med. 2020;26(9):1468–1479. doi: 10.1038/s41591-020-1006-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Veldman J, Rodrigues Placa J, Chong L, et al. CD4+ T cells in classical Hodgkin lymphoma express exhaustion associated transcription factors TOX and TOX2: characterizing CD4+ T cells in Hodgkin lymphoma. Oncoimmunology. 2022;11(1):2033433. doi: 10.1080/2162402X.2022.2033433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Roers A, Montesinos-Rongen M, Hansmann ML, Rajewsky K, Kuppers R. Amplification of TCRbeta gene rearrangements from micromanipulated single cells: T cells rosetting around Hodgkin and Reed-Sternberg cells in Hodgkin's disease are polyclonal. Eur J Immunol. 1998;28(8):2424–2431. doi: 10.1002/(SICI)1521-4141(199808)28:08<2424::AID-IMMU2424>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 82.Trumper L, Jung W, Daus H, Mechtersheimer G, von Bonin F, Pfreundschuh M. Assessment of clonality of rosetting T lymphocytes in Hodgkin's disease by single-cell polymerase chain reaction: detection of clonality in a polyclonal background in a case of lymphocyte predominance Hodgkin's disease. Ann Hematol. 2001;80(11):653–661. doi: 10.1007/s002770100370. [DOI] [PubMed] [Google Scholar]
- 83.Gagliani N, Magnani CF, Huber S, et al. Coexpression of CD49b and LAG-3 identifies human and mouse T regulatory type 1 cells. Nat Med. 2013;19(6):739–746. doi: 10.1038/nm.3179. [DOI] [PubMed] [Google Scholar]
- 84.Haghverdi L, Buettner F, Theis FJ. Diffusion maps for high-dimensional single-cell analysis of differentiation data. Bioinformatics. 2015;31(18):2989–2998. doi: 10.1093/bioinformatics/btv325. [DOI] [PubMed] [Google Scholar]
- 85.Coifman RR, Lafon S, Lee AB, et al. Geometric diffusions as a tool for harmonic analysis and structure definition of data: diffusion maps. Proc Natl Acad Sci U S A. 2005;102(21):7426–7431. doi: 10.1073/pnas.0500334102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nam-Cha SH, Montes-Moreno S, Salcedo MT, Sanjuan J, Garcia JF, Piris MA. Lymphocyte-rich classical Hodgkin's lymphoma: distinctive tumor and microenvironment markers. Mod Pathol. 2009;22(8):1006–1015. doi: 10.1038/modpathol.2009.54. [DOI] [PubMed] [Google Scholar]
- 87.Wang Y, Wang R, Zhang S, et al. iTALK: an R package to characterize and illustrate intercellular communication. bioRxiv. Preprint posted online 4 January 2019 doi: 10.1101/507871. [DOI] [Google Scholar]
- 88.Gu-Trantien C, Migliori E, Buisseret L, et al. CXCL13-producing TFH cells link immune suppression and adaptive memory in human breast cancer. JCI Insight. 2017;2(11) doi: 10.1172/jci.insight.91487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Noel G, Fontsa ML, Garaud S, et al. Functional Th1-oriented T follicular helper cells that infiltrate human breast cancer promote effective adaptive immunity. J Clin Invest. 2021;131(19) doi: 10.1172/JCI139905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Carey CD, Gusenleitner D, Lipschitz M, et al. Topological analysis reveals a PD-L1-associated microenvironmental niche for Reed-Sternberg cells in Hodgkin lymphoma. Blood. 2017;130(22):2420–2430. doi: 10.1182/blood-2017-03-770719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Patel SS, Weirather JL, Lipschitz M, et al. The microenvironmental niche in classic Hodgkin lymphoma is enriched for CTLA-4-positive T cells that are PD-1-negative. Blood. 2019;134(23):2059–2069. doi: 10.1182/blood.2019002206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Diefenbach CS, Hong F, Ambinder RF, et al. Ipilimumab, nivolumab, and brentuximab vedotin combination therapies in patients with relapsed or refractory Hodgkin lymphoma: phase 1 results of an open-label, multicentre, phase 1/2 trial. Lancet Haematol. 2020;7(9):e660–e670. doi: 10.1016/S2352-3026(20)30221-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wolchok JD, Chiarion-Sileni V, Gonzalez R, et al. Overall survival with combined nivolumab and ipilimumab in advanced melanoma. N Engl J Med. 2017;377(14):1345–1356. doi: 10.1056/NEJMoa1709684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wolchok JD, Hodi FS, Weber JS, et al. Development of ipilimumab: a novel immunotherapeutic approach for the treatment of advanced melanoma. Ann N Y Acad Sci. 2013;1291(1):1–13. doi: 10.1111/nyas.12180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ma Y, Visser L, Roelofsen H, et al. Proteomics analysis of Hodgkin lymphoma: identification of new players involved in the cross-talk between HRS cells and infiltrating lymphocytes. Blood. 2008;111(4):2339–2346. doi: 10.1182/blood-2007-09-112128. [DOI] [PubMed] [Google Scholar]
- 96.Alfei F, Kanev K, Hofmann M, et al. TOX reinforces the phenotype and longevity of exhausted T cells in chronic viral infection. Nature. 2019;571(7764):265–269. doi: 10.1038/s41586-019-1326-9. [DOI] [PubMed] [Google Scholar]
- 97.Scott AC, Dundar F, Zumbo P, et al. TOX is a critical regulator of tumour-specific T cell differentiation. Nature. 2019;571(7764):270–274. doi: 10.1038/s41586-019-1324-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Connors JM, Cozen W, Steidl C, et al. Hodgkin lymphoma. Nat Rev Dis Primers. 2020;6(1):61. doi: 10.1038/s41572-020-0189-6. [DOI] [PubMed] [Google Scholar]
- 99.Chan FC, Mottok A, Gerrie AS, et al. Prognostic model to predict post-autologous stem-cell transplantation outcomes in classical Hodgkin lymphoma. J Clin Oncol. 2017;35(32):3722–3733. doi: 10.1200/JCO.2017.72.7925. [DOI] [PubMed] [Google Scholar]
- 100.Johnston RL, Mottok A, Chan FC, et al. A gene expression-based model predicts outcome in children with intermediate-risk classical Hodgkin lymphoma. Blood. 2022;139(6):889–893. doi: 10.1182/blood.2021011941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Scott DW, Chan FC, Hong F, et al. Gene expression-based model using formalin-fixed paraffin-embedded biopsies predicts overall survival in advanced-stage classical Hodgkin lymphoma. J Clin Oncol. 2013;31(6):692–700. doi: 10.1200/JCO.2012.43.4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Newman AM, Steen CB, Liu CL, et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat Biotechnol. 2019;37(7):773–782. doi: 10.1038/s41587-019-0114-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Peterson VM, Zhang KX, Kumar N, et al. Multiplexed quantification of proteins and transcripts in single cells. Nat Biotechnol. 2017;35(10):936–939. doi: 10.1038/nbt.3973. [DOI] [PubMed] [Google Scholar]
- 104.Cadot S, Valle C, Tosolini M, et al. Longitudinal CITE-seq profiling of chronic lymphocytic leukemia during ibrutinib treatment: evolution of leukemic and immune cells at relapse. Biomark Res. 2020;8(1):72. doi: 10.1186/s40364-020-00253-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Stuart T, Srivastava A, Madad S, Lareau CA, Satija R. Single-cell chromatin state analysis with Signac. Nat Methods. 2021;18(11):1333–1341. doi: 10.1038/s41592-021-01282-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Williams CG, Lee HJ, Asatsuma T, Vento-Tormo R, Haque A. An introduction to spatial transcriptomics for biomedical research. Genome Med. 2022;14(1):68. doi: 10.1186/s13073-022-01075-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Vallejo AF, Harvey K, Wang T, et al. snPATHO-seq: unlocking the FFPE archives for single nucleus RNA profiling. bioRxiv. Preprint posted online 24 August 2022 doi: 10.1101/2022.08.23.505054. [DOI] [Google Scholar]
- 108.Manson G, Herbaux C, Schiano JM, et al. Can nivolumab alone cure patients with relapse or refractory Hodgkin lymphoma? A 5-year analysis of the French early access program (EPA) Br J Haematol. 2022;198(1):203–206. doi: 10.1111/bjh.18198. [DOI] [PubMed] [Google Scholar]