Abstract
Background
The vacuoles, E1-enzyme, X linked, autoinflammatory and somatic (VEXAS) syndrome is an adult-onset autoinflammatory disease (AID) due to postzygotic UBA1 variants.
Objectives
To investigate the presence of VEXAS syndrome among patients with adult-onset undiagnosed AID. Additional studies evaluated the mosaicism distribution and the circulating cytokines.
Methods
Gene analyses were performed by both Sanger and amplicon-based deep sequencing. Patients’ data were collected from their medical charts. Cytokines were quantified by Luminex.
Results
Genetic analyses of enrolled patients (n=42) identified 30 patients carrying UBA1 pathogenic variants, with frequencies compatible for postzygotic variants. All patients were male individuals who presented with a late-onset disease (mean 67.5 years; median 67.0 years) characterised by cutaneous lesions (90%), fever (66.7%), pulmonary manifestations (66.7%) and arthritis (53.3%). Macrocytic anaemia and increased erythrocyte sedimentation rate and ferritin were the most relevant analytical abnormalities. Glucocorticoids ameliorated the inflammatory manifestations, but most patients became glucocorticoid-dependent. Positive responses were obtained when targeting the haematopoietic component of the disease with either decitabine or allogeneic haematopoietic stem cell transplantation. Additional analyses detected the UBA1 variants in both haematopoietic and non-haematopoietic tissues. Finally, analysis of circulating cytokines did not identify inflammatory mediators of the disease.
Conclusion
Thirty patients with adult-onset AID were definitively diagnosed with VEXAS syndrome through genetic analyses. Despite minor interindividual differences, their main characteristics were in concordance with previous reports. We detected for the first time the UBA1 mosaicism in non-haematopoietic tissue, which questions the previous concept of myeloid-restricted mosaicism and may have conceptual consequences for the disease mechanisms.
Keywords: polymorphism, genetic; immune system diseases; inflammation
WHAT IS ALREADY KNOWN ON THIS TOPIC
The vacuoles, E1 enzyme, X linked, autoinflammatory and somatic (VEXAS) syndrome is a late-onset autoinflammatory disease characterised by inflammatory manifestations and/or haematological perturbations, with poor or null response to different anti-inflammatory drugs.
This syndrome has been associated with myeloid-restricted mosaicism at UBA1 gene.
Recent investigations suggest that VEXAS syndrome may be one of the most frequent genetically determined autoinflammatory diseases since pathogenic UBA1 variants have been detected in up to 1/4269 males over 50 years.
WHAT THIS STUDY ADDS
We evaluated a cohort of patients with late-onset, undiagnosed autoinflammatory disease for the presence of UBA1 variants, identifying 30 patients with mosaicism at this gene that enable us to diagnose them as suffering from VEXAS syndrome.
The main features of genetically positive patients are in line with previous reports.
The response of patients treated with either hypomethylating drugs or allogeneic haematopoietic stem cell transplantation was evaluated, with the results showing a clear phenotype improvement.
Analysis of distribution of mosaicism revealed for the first time the presence of postzygotic UBA1 variants in non-haematopoietic tissue.
These data question the previous idea of myeloid-restricted mosaicism in VEXAS, and may have conceptual consequences for the disease mechanisms that are deeply discussed.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
On the basis of the known clinical spectrum of the VEXAS syndrome and considering that gene analyses represent the unique way to establish its diagnosis, in this study we propose to relax the criteria to perform the genetic test to identify as many patients as possible and as early as possible.
The genetic evidence described here suggests that the mutational event causing VEXAS probably occurs during embryonic development.
Consequently, VEXAS syndrome should be also considered in the differential diagnosis of young adults, or even paediatric, patients with otherwise undiagnosed haematological and/or inflammatory diseases.
The patients’ responses to treatments targeting the haematopoietic component of the disease were associated with a marked decrease or disappearance of mutant UBA1 allele.
These preliminary data suggest that monitoring the mutant UBA1 allele might be a potential future biomarker of the outcome of administered treatments.
Introduction
Autoinflammatory diseases (AIDs) are genetically determined conditions characterised by recurrent sterile inflammation mainly mediated by cells of innate immunity.1 Similar to most human inherited diseases, it has been admitted that most AIDs start during infancy or early childhood. This categoric statement is currently under revision as recent evidences support for a continuously growing group of patients with late-onset, but otherwise typical AIDs. In most of these patients, the disease is a consequence of postzygotic variants leading to gene mosaicism. This mechanism was well established in certain AIDs such as cryopyrin-associated periodic syndromes (CAPS), but it has been considered as minor due to the small number of reported patients.2–6 A notable earthquake in the field occurred when the vacuoles, E1 enzyme, X linked, autoinflammatory and somatic (VEXAS) syndrome was described.7 This condition has been described as a consequence of postzygotic UBA1 variants,7–10 and typically starts during late adulthood with inflammatory manifestations (fever, neutrophilic dermatosis, relapsing polychondritis, pulmonary manifestations, arthritis) and/or haematological perturbations (anaemia, macrocytosis, bone marrow vacuolisation, thrombocytopenia).7 11 In contrast to the rare AIDs due to gene mosaicism, VEXAS syndrome has been genetically confirmed in hundreds of patients around the world.12 13 Recent genetic investigations suggest that the pathogenic UBA1 variants may be present in up to 1/4269 male over 50 years,10 strongly suggesting that VEXAS syndrome may be the most frequent genetically determined AID described to date among human populations.
In this study, we investigated a cohort of unrelated adult patients with clinical similarities to those described in VEXAS syndrome. UBA1 analysis led to the genetic confirmation of this syndrome in 30 patients. These analyses identified four different postzygotic pathogenic variants at UBA1 gene, with different mutant allele frequency (MAF). Additional studies performed to evaluate the mosaicism distribution revealed the presence of the postzygotic UBA1 variants in non-haematopoietic tissue (nails), which was previously unreported and questions the original idea that VEXAS syndrome was due to a myeloid-restricted mosaicism at UBA1. These evidences led us also to hypothesise that the mutational event leading to the disease-causing UBA1 variants might occur early in life, probably during embryogenesis, rather than during adulthood, and we discuss the potential clinical and conceptual consequences that these novel insights have for this disease.
Methods
Patients
Among those patients with genetically undiagnosed AID received in our centre (Department of Immunology, Hospital Clínic, Barcelona, Spain), we selected a group according the following inclusion criteria: (i) onset of the disease during adulthood (≥18 years of age); (ii) recurrent episodes of sterile inflammation spanning a period >6 months of duration; (iii) presence of relapsing polychondritis and/or neutrophilic dermatosis proven by skin biopsies and/or venous thromboembolism and/or recurrent arthritis and/or pulmonary manifestations; (iv) plasma levels of C reactive protein >0.5 mg/dL and/or erythrocyte sedimentation rate (ESR) >10 mm/hour during active disease; (v) negative or non-confirmatory genotypes in genetic studies using a targeted gene panel for AIDs (online supplemental table S1); (vi) exclusion of infectious causes for the disease; (vii) absence or low titre (≤1/160) of circulating autoantibodies in repetitive tests and (viii) presence of haematological alterations (decreased haemoglobin level (<120 g/L) and/or increased mean corpuscular volume (MCV; >100 fL) and/or leucopenia (<4×109/L) and/or thrombocytopenia (<140×109/L)). Sex was not considered an inclusion criteria, and consequently, both male and female patients were potentially enrolled. Patients’ data were collected at the time of clinical diagnosis and/or at the time of genetic testing.
ard-2023-224460supp006.pdf (264.4KB, pdf)
Molecular genetics
Blood samples were collected in EDTA tubes by venous punction, and patients’ nail clippings were collected after their exhaustive cleaning to prevent contamination by potential scratching of skin lesions. Genomic DNA samples were prepared from whole peripheral blood, from isolated leucocyte subpopulations or from tissues using the QIAmp DNA Blood Mini Kit or the QIAamp DNA Investigator Kit (QIAGEN, Germany) according to the manufacturer’s instructions. UBA1 genotyping was performed by both the Sanger method of DNA sequencing and the amplicon-based deep sequencing (ADS) in all enrolled patients. For Sanger sequencing, all exons of UBA1 (RefSeq NM_003334.4) were PCR-amplified using in-house designed primers (online supplemental table S2), purified with Illustra ExoStar 1-Step kit (GE Healthcare), bidirectionally fluorescence sequenced using ABI BigDye Terminator V.3.1 Cycle Sequencing Kit (Applied Biosystems) and run on an automated ABI 3730XL DNA analyzer (Applied Biosystems). Sequence reads were analysed using the SeqPilot software (JSI Medical Systems, Germany), and detected variants were classified according to the previously published recommendations.14
Additional studies of mosaicism were performed using the ADS method as previously described.15 Briefly, amplicons covering the exons and intronic boundaries of UBA1 were generated by in-house-designed PCR. Deep sequencing was performed on a S5XL platform (Ion Torrent, Life Technologies, USA), with coverage >1000×. Reads were mapped against the GRCh37 (hg19) using the BWA, and variants were subsequently analysed using the Integrative Genomics Viewer. The MAF was calculated as the proportion of variant reads from the total reads and expressed as a percentage.
Cell isolation
Fresh whole blood samples were collected in EDTA tubes. Peripheral blood mononuclear cells (PBMCs) were isolated via Ficoll-Paque Premium (Cytiva, Freiburg, Germany) following the manufacturer’s instructions. For cell sorting, whole blood or PBMCs were incubated at 4°C with Fc-blocking reagent (Miltenyi Biotech) and stained for 30 min with the following monoclonal antibodies: anti-CD45 AF700, anti-CD14 APC-Cy7 and anti-CD19 PE-Cy7 (all three from Biolegend), anti-CD15 FITC and anti-CD3 AF67 (from Southern Biotech), anti-CD16 AF647 and anti-CD34 PE (both from BD Biosciences Pharmingen). After staining, erythrocytes from whole blood were lysed with the Red Blood Cell Lysis Solution (Miltenyi Biotech). CD45+ CD15+ CD16+ neutrophils from whole blood and CD3+ T cells, CD19+ B cells, CD14+ monocytes and CD34+ cells from PBMCs were FACS sorted in the BD FACSAriaII cell sorter (Beckton Dickinson, California, USA) after excluding dead cells by 4′,6-diamidino-2-phenylindole (DAPI) staining.
Cytokines profile
The serum concentrations of brain-derived neurotrophic factor, epidermal growth factor, eotaxin, fibroblast growth factor-2, granulocyte macrophage colony-stimulating factor, growth-related oncogene-α, hepatocyte growth factor, interferon (IFN)-α, IFN-γ, interleukin (IL)-1α, IL-1β, IL-1Ra, IL-2, IL-4, IL-5, IL-6, IL-7, IL-8, IL-9, IL-10, IL-12p70, IL-13, IL-15, IL-17A, IL-18, IL-21, IL-22, IL-23, IL-27, IL-31, inducible protein-10, leukaemia inhibitory factor, monocyte chemoattractant protein-1, macrophage-inflammatory protein (MIP)1-α, MIP1-β, nerve growth factor-β, platelet-derived growth factor-BB, placental growth factor 1, RANTES, stem cell factor, stromal cell-derived factor-α, tumour necrosis factor (TNF)-α, TNF-β, vascular endothelial growth factor (VEGF)-A and VEGF-D cytokines were measured using a custom bead-based multiplex Luminex immunoassay (eBioscience).
Statistical analysis
Statistics were calculated using Prism software (GraphPad). Analyses between two groups were performed with a two-tailed unpaired t-test. Differences were considered statistically significant at a p value of <0.01.
Results
Identification of patients with UBA1 variants
Forty-two unrelated patients fulfilling the inclusion criteria were enrolled in this study. UBA1 genotyping using Sanger sequencing detected 30 patients with postzygotic UBA1 variants (table 1 and online supplemental figure S1). These analyses detected four different single nucleotide variants at UBA1, all of which have been reported as causing VEXAS syndrome.7 8 According to the recommendations for gene variant classification of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology, the four detected UBA1 variants were classified as pathogenic (online supplemental table S3), thus supporting the definitive diagnosis of VEXAS syndrome in the 30 genetically positive patients. Genetic studies performed in eight healthy daughters from six UBA1-positive patients (P6, P8, P26, P29, P32, P38) tested all negative (online supplemental table S4).
Table 1.
Results of UBA1 genotyping in enrolled patients
| Pt | Sample | Sanger sequencing | ADS | ||
| Nucleotide UBA1
variant* |
Amino acid exchange* |
Mutant allele fraction—mean (SD)† |
Coverage—mean (SD)† | ||
| 1 | Peripheral blood | c.121A>G | p.Met41Val | 18.6% (1.0) | 1098× (55) |
| 2 | Peripheral blood | – | – | 0% | 1262× (50) |
| 3 | Peripheral blood | c.122T>C | p.Met41Thr | 76.3% (1.1) | 7486× (893) |
| 4 | Bone marrow aspirate | c.122T>C | p.Met41Thr | 40.5% (0.6) | 4519× (517) |
| 5 | Peripheral blood | – | – | 0% | 8335× (203) |
| 6 | Peripheral blood | c.122T>C | p.Met41Thr | 86.3% (0.3) | 6606× (2523) |
| 7 | Peripheral blood | – | – | 0% | 1538× (25) |
| 8 | Peripheral blood | c.121A>C | p.Met41Leu | 82.0% (0.8) | 1453× (207) |
| 9 | Peripheral blood | c.122T>C | p.Met41Thr | 60.7% (1.0) | 4621× (525) |
| 10 | Bone marrow aspirate | c.118-1G>C | Splice acceptor site variant | 71.8% (1.0) | 1876× (1588) |
| 11 | Peripheral blood | – | – | 0% | 1289× (38) |
| 12 | Peripheral blood | c.118-1G>C | Splice acceptor site variant | n.p. | n.p. |
| 13 | Peripheral blood | – | – | 0% | 6759× (268) |
| 14 | Peripheral blood | c.122T>C | p.Met41Thr | n.p. | n.p. |
| 15 | Peripheral blood | c.121A>C | p.Met41Leu | n.p. | n.p. |
| 16 | Peripheral blood | c.121A>G | p.Met41Val | 14.6% (0.7) | 5369× (2214) |
| 17 | Bone marrow aspirate | c.122T>C | p.Met41Thr | 76.2% (0.4) | 14 889× (769) |
| 18 | Peripheral blood | – | – | 0% | 1142× (36) |
| 19 | Peripheral blood | – | – | 0% | 1390× (104) |
| 20 | Peripheral blood | c.121A>G | p.Met41Val | 76.9% (0.5) | 5761× (82) |
| 21 | Peripheral blood | c.121A>C | p.Met41Leu | 68.5% (0.5) | 8036× (729) |
| 22 | Peripheral blood | c.121A>G | p.Met41Val | 55.6% (2.5) | 1043× (45) |
| 23 | Peripheral blood | – | – | 0% | 5443× (566) |
| 24 | Peripheral blood | c.121A>C | p.Met41Leu | 76.8% (2.9) | 5749× (1028) |
| 25 | Peripheral blood | c.122T>C | p.Met41Thr | 67.4% (0.3) | 4939× (578) |
| 26 | Bone marrow aspirate | c.122T>C | p.Met41Thr | 57.5% (0.3) | 5229× (2196) |
| 27 | Peripheral blood | – | – | 0% | 1254× (34) |
| 28 | Peripheral blood | – | – | 0% | 1383× (91) |
| 29 | Peripheral blood | c.122T>C | p.Met41Thr | 85.0% (0.2) | 2284× (78) |
| 30 | Peripheral blood | c.122T>C | p.Met41Thr | 64.7% (0.7) | 10 175× (1163) |
| 31 | Peripheral blood | – | – | 0% | 2465× (506) |
| 32 | Peripheral blood | c.121A>G | p.Met41Val | 45.3% (1.5) | 4425× (1421) |
| 33 | Peripheral blood | c.121A>C | p.Met41Leu | 60.0% (2.7) | 3039× (748) |
| 34 | Bone marrow aspirate | c.121A>C | p.Met41Leu | 76.1% (1.0) | 9619× (356) |
| 35 | Peripheral blood | c.122T>C | p.Met41Thr | n.p. | n.p. |
| 36 | Peripheral blood | c.122T>C | p.Met41Thr | 50.2% (1.6) | 1786× (415) |
| 37 | Peripheral blood | – | – | 0% | 1152× (19)19 |
| 38 | Bone marrow aspirate | c.121A>C | p.Met41Leu | 36.9% (1.4) | 5112× (1346) |
| 39 | Bone marrow aspirate | c.122T>C | p.Met41Thr | 75.5% (1.0) | 3180× (263) |
| 40 | Peripheral blood | c.121A>G | p.Met41Val | 48.1% (0.1) | 41 309× (2409) |
| 41 | Peripheral blood | c.122T>C | p.Met41Thr | 48.0% (1.0) | 13 789× (1676) |
| 42 | Peripheral blood | c.121A>C | p.Met41Leu | 54.0% (0) | 15 725× (1227) |
*RefSeq: NM_003334.4.
†Mean of data collected from three independent experiments.
ADS, amplicon-based deep sequencing; n.p., not performed; Pt, patient.
ard-2023-224460supp001.pdf (1.4MB, pdf)
Clinical features and analytical abnormalities in patients with VEXAS syndrome
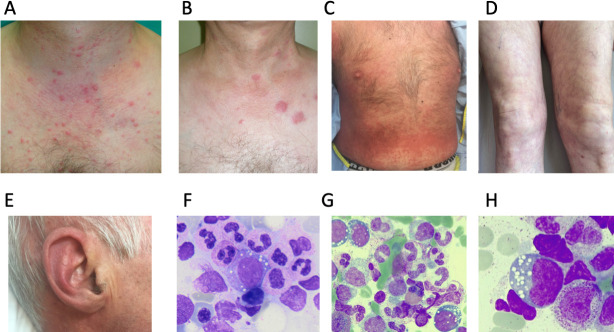
Among those patients with confirmed VEXAS syndrome, the mean and median age at disease onset was 67.5 and 67.0 years, respectively (range 52–86 years). At the time of writing the manuscript, the mortality rate was 40.0% (12 deceased). The mean time of disease evolution was slightly shorter in alive patients compared with that of deceased patients or until allogeneic haematopoietic stem cell transplantation (allo-HSCT) (4.0 vs 5.3 years, respectively; table 2 and online supplemental table S5). All participants displayed recurrent and simultaneously occurring inflammatory manifestations that are shown in detail in table 2 and online supplemental table S6. In this series, the cutaneous inflammatory manifestations were the most frequently detected (90%), and highly variable among patients. The most frequently detected skin lesion was the erythematous papules or nodules secondary to neutrophilic dermal infiltration (figure 1A–B). Other less frequent manifestations included urticaria-like skin rash (figure 1C) or livedo racemosa (figure 1D), and recurrent chondritis at ear and/or nose (figure 1E).
Table 2.
Demographics, clinical and analytical features of genetically confirmed patients with VEXAS syndrome allo-HSCT, allogeneic haematopoietic stem cell transplantation; MAS, macrophage activation syndrome; Hb, haemoglobin; MCV, mean corpuscular volume; WBC, white blood cells count; ESR, erythrocyte sedimentation rate; NSAID, non-steroid anti-inflammatory drugs; IVIG, intravenous immunoglobulins; TNF, tumour necrosis factor; IL-6, interleukin 6; IL-1, interleukin 1.
| General features | ||
| Gender (male/female)—n | 30/0 | |
| Alive/Deceased—n (%) | 18/12 (60.0%/40.0%) | |
| Mean age at disease onset (range) | 67.5 years (52–86) | |
| Mean age of alive patients (range) | 72.9 years (56–88) | |
| Mean age at death/allo-HSCT (range) | 71 years (55–79) | |
| Mean time of disease evolution of alive patients (range) | 4.0 years (0.5–8) | |
| Mean time of disease until death/allo-HSCT (range) | 5.3 years (1–11) | |
| Clinical manifestations | ||
| Recurrent skin manifestations—n (%) | 27/30 (90.0%) | |
| Erythematous papules/plaques/nodules—n (%) | 20/30 (66.6%) | |
| Purpuric papules/plaques—n (%) | 14/30 (46.7%) | |
| Panniculitis—n (%) | 9/30 (30.0%) | |
| Livedo racemosa—n (%) | 7/30 (23.3%) | |
| Recurrent fever—n (%) | 20/30 (66.7%) | |
| Pulmonary manifestations—n (%) | 20/30 (66.7%) | |
| Ocular manifestations—n (%) | 17/30 (56.7%) | |
| Palpebral oedema—n (%) | 8/30 (26.7%) | |
| Conjunctivitis—n (%) | 6/30 (20.0%) | |
| Uveitis—n (%) | 5/30 (16.7%) | |
| Arthritis—n (%) | 16/30 (53.3%) | |
| Recurrent chondritis (ear/nose)—n (%) | 16/30 (53.3%) | |
| Chondritis (non-ear/non-nose)—n (%) | 3/30 (10.0%) | |
| Venous thromboembolism—n (%) | 12/30 (40.0%) | |
| Arterial thrombosis—n (%) | 4/30 (13.3%) | |
| MAS-like episodes—n (%) | 2/30 (6.7%) | |
| Analytical parameters | Reference range | |
| Hb (mean; range) (g/L) | 99.2 (59–161) | 130–170 |
| Hb <120 g/L (%; n/total) | 86.9% (730/840) | |
| MCV (mean; range) (fL) | 104.5 (85–133) | 80–100 |
| MCV >100 fL (%; n/total) | 72.4% (608/840) | |
| Haematocrit <36% (%; n/total) | 83.8% (704/840) | |
| Platelets (mean; range) (109/L) | 150.5 (4–753) | 140–400 |
| Thrombocytopenia (platelet count <140.109/L) (%; n/total) | 48.2% (405/840) | |
| WBC (mean; range) (109/L) | 5.9 (0.2–33.3) | 4.0–11.0 |
| Leucopenia (WBC <4.109/L) (%; n/total) | 32.9% (271/823) | |
| Neutropenia (neutrophil count <2.5.109/L) (%; n/total) | 40.6% (333/821) | |
| Lymphopenia (lymphocyte count <0.9.109/L) (%; n/total) | 39.1% (321/821) | |
| Ferritin (mean; range) (ng/mL) | 1163 (54–11 931) | 20–400 |
| Increased ferritin (>400 ng/mL) (%; n/total) | 77.1% (195/253) | |
| High increased ferritin (>1000 ng/mL) (%; n/total) | 44.7% (113/253) | |
| ESR (mean; range) (mm/hour) | 78.5 (2–141) | <10 |
| Increased ESR (>10 mm/hour) (%; n/total) | 95.1% (365/384) | |
| High increased ESR (>50 mm/hour) (%; n/total) | 73.2% (281/378) | |
| Bone marrow vacuoles (%; n/total) | 100% (23/23) | |
| Treatments/Outcome | ||
| Colchicine | 11/30—complete response 0% | partial 18.2% | negative 81.8% | |
| NSAIDs | 17/30—complete response 0% | partial 52.9% | negative 47.1% | |
| Glucocorticoids | 30/30—complete response 70.0% | partial 26.7% | negative 3.3% | |
| Methotrexate | 17/30—complete response 0% | partial 35.3% | negative 64.7% | |
| Mycophenolate mofetil | 6/30—complete response 0% | partial 16.7% | negative 83.3% | |
| Azathioprine | 9/30—complete response 0% | partial 22.2% | negative 77.8% | |
| Cyclophosphamide | 2/30—complete response 0% | partial 50% | negative 50% | |
| IVIGs | 4/30—complete response 0% | partial 25% | negative 75% | |
| Anti-TNF | 3/30—complete response 0% | partial 0% | negative 100% | |
| Anti-IL-6 | 8/30—complete response 12.5% | partial 25% | negative 62.5% | |
| Anti-IL-1 | 5/30—complete response 0% | partial 60% | negative 40% | |
| Anti-CD20 | 4/30—complete response 0% | partial 75% | negative 25% | |
| Jak inhibitors | 5/30—complete response 20% | partial 40% | negative 40% | |
| Genetics | ||
| c.118-1G>C—n (%) | 2 (6.7%) | |
| p.Met41Leu—n (%) | 8 (26.7%) | |
| p.Met41Val—n (%) | 6 (20.0%) | |
| p.Met41Thr—n (%) | 14 (46.7%) | |
| Mutant allele frequency—haematopoietic tissues (mean; range) (%) | 60.5 (14.6–86.3) | |
| Mutant allele frequency—nails (mean; range) (%) | 24.2 (2.7–73.7) | |
allo-HSCT, allogeneic haematopoietic stem cell transplantation; ESR, erythrocyte sedimentation rate; Hb, haemoglobin; IL-1, interleukin 1; IL-6, interleukin 6; IVIG, intravenous immunoglobulins; MAS, macrophage activation syndrome; MCV, mean corpuscular volume; NSAID, non-steroid anti-inflammatory drugs; TNF, tumour necrosis factor; VEXAS, vacuoles, E1-enzyme, X linked, autoinflammatory and somatic syndrome; WBC, white blood cells count.
Figure 1.
Cutaneous lesions and cytosolic vacuoles in patients (Pt) with vacuoles, E1-enzyme, X linked, autoinflammatory and somatic syndrome. (A) Multiple erythematous papules on the chest and neck (Pt 10). (B) Erythematous and edematous plaques, and a few papules on the chest and neck (Pt 10). (C) Urticaria-like lesions on the abdomen (Pt 22). (D) Livedo racemosa (Pt 22). (E) Ear chondritis (Pt 22). (F, G, H) Cytosolic vacuoles in myeloid precursor cells in bone marrow aspirate from Pt 3.
All patients presented with severe and persistent perturbations of analytical parameters, even when they were treated with anti-inflammatory drugs. Among biochemical tests, the most frequent alterations included a marked increase of ESR, with 73.2% of all collected determinations with values >50 mm/hour, and a tendency to hyperferritinemia (44.7% of all determinations with values >1000 ng/mL) (table 2, online supplemental table S7 and online supplemental figure S2). The most frequently detected haematological abnormalities included low or very low haemoglobin concentration (mean 99.2 g/L; range 59–161), macrocytosis (mean 104.5 fL; range 85–133) and thrombocytopenia (48.2% of all determinations with platelet count <140×109/L) (table 2, online supplemental table S8 and online supplemental figures S3 and S4). As a consequence of their haematological disturbances, 23 patients underwent analysis of bone marrow, which demonstrated in all cases the cytosolic vacuolisation of erythroid and myeloid participants (figure 1F, G, H). The main features of patients who tested negative in the UBA1 analysis are shown in the online supplemental table S9.
ard-2023-224460supp002.pdf (451.3KB, pdf)
ard-2023-224460supp003.pdf (508.3KB, pdf)
ard-2023-224460supp004.pdf (638.6KB, pdf)
UBA1 mosaicism analyses
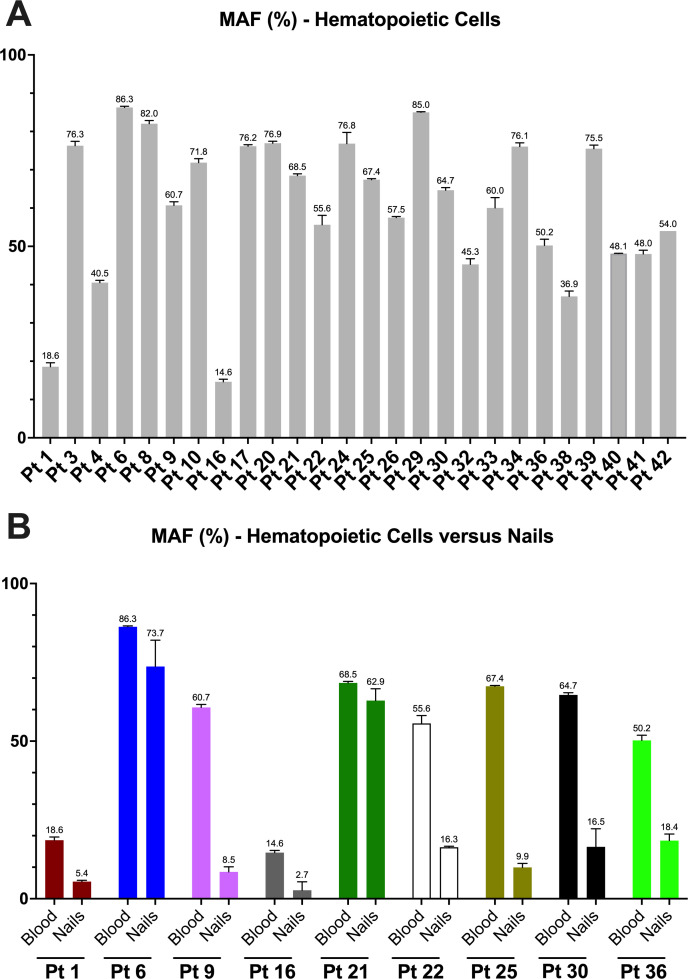
We further addressed the characterisation of the UBA1 mosaicism by quantifying the MAF using the ADS method. We first evaluated the MAF in haematopoietic tissues among patients. For this objective, DNA samples extracted from peripheral blood or from bone marrow aspirates from 26 patients were analysed, revealing a mean MAF in haematopoietic tissues of 60.5% (range 14.6%–86.3%; table 2 and figure 2A).
Figure 2.
Amplicon-based deep sequencing (ADS) of UBA1 gene in patients (Pt) with vacuoles, E1-enzyme, X linked, autoinflammatory and somatic syndrome. (A) ADS results obtained using DNA samples extracted from haematopoietic tissues. (B) Comparative results of ADS analyses using DNA samples extracted from haematopoietic tissues and non-haematopoietic tissues (nails). Columns represent the mean value of three independent experiments, and the error bars, the SD. MAF, mutant allele frequency.
In a second step, we evaluated the mosaicism distribution among the circulating leucocyte subpopulations. For this proposal, fresh peripheral blood samples from two patients (Pt 3 and Pt 9) were collected and the leucocyte subpopulations isolated by FACsorting. Sanger and ADS analyses performed on genomic DNA from each isolated subpopulation detected the UBA1 variant in CD34+ progenitor cells and in myeloid cells (neutrophils and monocytes), while only a trace of the variant (<1%) was found in lymphoid cells (T and B lymphocytes) (online supplemental table S10).
Finally, we evaluated the potential presence of the UBA1 mosaicism in non-haematopoietic tissue. For this proposal, we performed DNA extraction from nail samples, an ectodermic tissue that may be easily and repetitively collected with no risk of blood contamination. Nail samples were collected from nine patients with VEXAS syndrome, and ADS analyses revealed the presence of the respective UBA1 variants in these samples in all patients (mean MAF 24.2%; range 2.7%–73.7%), usually at lower percentages than that detected in haematopoietic samples (figure 2B and online supplemental table S11).
Outcome of medical treatments
During the course of their disease, the patients were treated with different anti-inflammatory drugs (median=5). Treatment with corticosteroids, often administered in doses ≥20 mg once daily, was the only therapeutic approach that improved the inflammatory manifestations, with mild or no improvement of haematological disturbances. Among those anti-inflammatory drugs administered to decrease the corticosteroids dose, partial or negative responses were often detected (table 2 and online supplemental table S12).
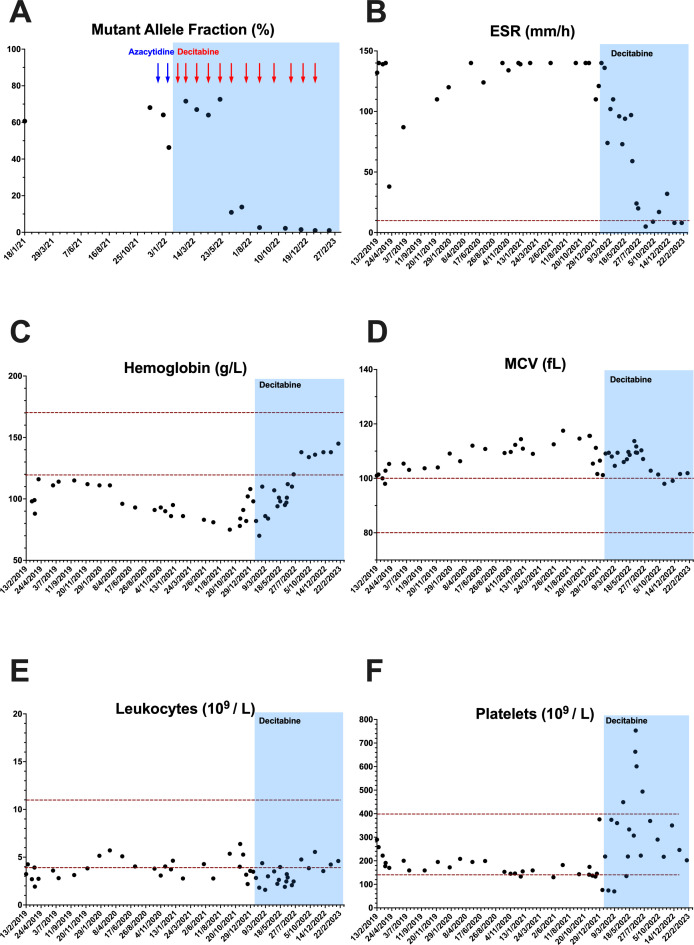
One patient of the cohort (Pt 9) is being treated with the hypomethylating drug decitabine (20 mg/m2 ×5 days; cycles of 28 days) in the last 17 months ago. During this period, the MAF in blood samples showed a progressive decrease after the sixth month of treatment until its near disappearance (MAF <2%) in the last 5 months (figure 3A). During the first two cycles, an episode of arthritis, which was well controlled with corticosteroids, appeared. However, from the fourth cycle onwards, the response was clearly positive, with no relapse of inflammatory manifestations, marked decrease of administered corticosteroids, suppression of transfusional support and a progressive improvement until normalisation of different haematological and biochemical parameters (figure 3B–F). Interestingly, a bone marrow aspiration (BMA) performed after the 11th cycle showed occasional cytosolic vacuoles in the promyelocytes and proerythroblasts in contrast with the numerous vacuoles observed in a BMA performed before starting the decitabine treatment (data not shown).
Figure 3.
Analytical results during treatment with the hypomethylating drug decitabine in patient 9. Evolution of mutant allele frequency of UBA1 gene (A), erythrocyte sedimentation rate (ESR) (B), haemoglobin concentration (C), mean corpuscular volume (MCV) (D) and leucocytes (E) and platelet (F) counts before and after treatment. Blue and red arrows indicate the different cycles of treatment with azacytidine and decitabine, respectively. The horizontal dotted red lines indicate the normal reference range for each parameter, and the blue rectangle indicates treatment with decitabine.
Phenotype and genotype rescue by allogeneic haematopoietic stem cell transplantation
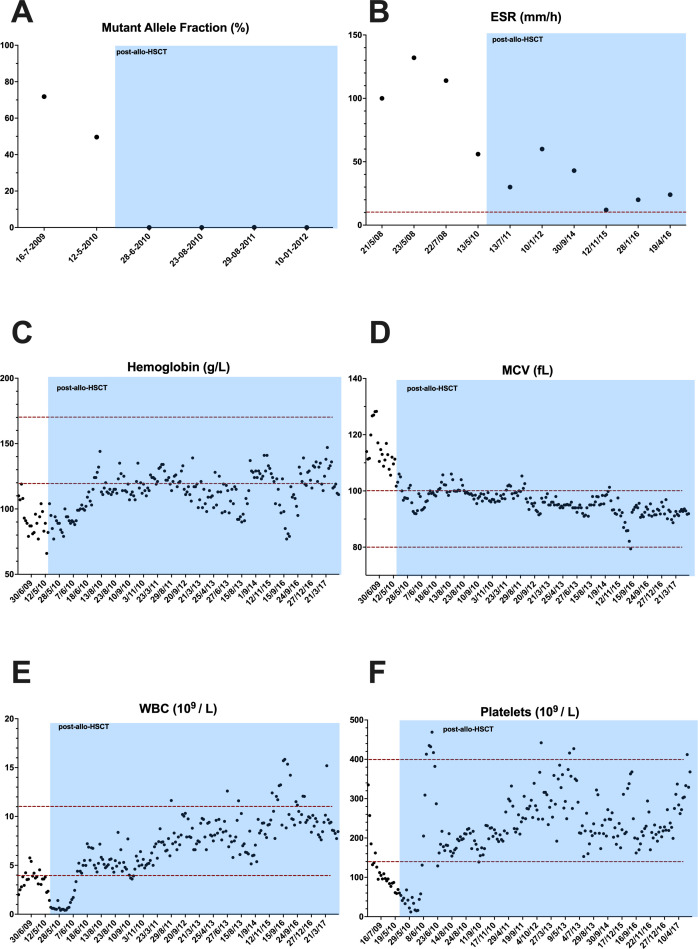
Among the patients with confirmed VEXAS, patient 10 underwent an human leukocyte antigen (HLA)-matched related allo-HSCT at the age of 55 years due to treatment-refractory haematological and inflammatory manifestations, years before VEXAS syndrome was described. After the procedure, all previous manifestations completely disappeared, but symptoms of a mild-to-moderate graft-versus-host disease were present during the following 7 years until the death of patient due to a solid neoplasia. We evaluated the outcome of allo-HSCT in this patient at different analytical levels. As DNA samples from haematopoietic cells collected before and after the transplant were available, we first investigated the evolution of UBA1 mosaicism along the time. ADS analyses detected the postzygotic UBA1 variant with a moderate-to-high MAF (49.2%–71.8%) before the transplantation, and its complete and permanent disappearance immediately after the procedure, remaining at undetectable level during the following years (figure 4A). The absence of UBA1 mosaicism in peripheral blood was associated with the disappearance of inflammatory manifestations as well as with the progressive normalisation of those haematological and biochemistry perturbations detected before the allo-HSCT (figure 4B–F). Altogether, these evidences support both a phenotype and a genotype rescue of the disease after allo-HSCT.
Figure 4.
Reversal of analytical abnormalities after HLA-matched related allogeneic haematopoietic stem cell transplantation (allo-HSCT) in patient 10. Evolution of mutant allele frequency of UBA1 gene (A), erythrocyte sedimentation rate (ESR) (B), haemoglobin concentration (C), mean corpuscular volume (MCV) (D) and leucocytes (E) and platelet (F) counts before and after allo-HSCT. The horizontal dotted red lines indicate the normal reference range for each parameter, and the blue rectangle indicates the post-allo-HSCT period. WBC, white blood cell.
Circulating cytokines
Finally, we quantified several circulating cytokines in serum samples collected from 14 patients to identify potential inflammatory mediators that may be involved in the disease pathogenesis. Unexpectedly, most of these cytokines were under the level of detection, or when detected, no significant statistical differences were observed with the values obtained in the control group (online supplemental figure S5).
ard-2023-224460supp005.pdf (415KB, pdf)
Discussion
Investigations here described allowed the definitive diagnosis of VEXAS syndrome to be made in 30 unrelated individuals previously classified as suffering from undiagnosed AID, representing one of the largest cohorts reported to date. The clinical features of these patients reflect interindividual differences, but the overall clinical picture is relatively homogenous and in concordance with the disease manifestations already described by several authors.7 11–13 Thus, all patients with VEXAS syndrome detected in our study were male in their 60s–80s, with a disease starting during late adulthood and mainly characterised by neutrophilic dermatosis, fever, pulmonary manifestations, arthritis, recurrent polychondritis, venous thromboembolism and a high mortality rate as a consequence of the evolution of the disease, infections or side events of administered treatments. Moreover, all patients also displayed haematological disturbances, with macrocytic anaemia, extremely high ESR values, moderate-to-high hyperferritinemia and a tendency to moderate-to-severe cytopenias requiring transfusional support as the most frequently detected. Despite the high number of patients with VEXAS we detected, we firmly believe that the disease has been underdiagnosed in our population on the basis of our inclusion criteria. At the beginning of this work, we established stringent inclusion criteria based on the main clinical and analytical manifestations reported at that time in the VEXAS syndrome.7 However, as novel studies have been published, it has become evident that there is a broader than expected clinical spectrum in the VEXAS syndrome, which ranges from the classical, highly inflammatory phenotype already reported in the first description of the disease and subsequently corroborated by different authors,8 9 12 13 until a recently described form in which all typical inflammatory manifestations are absent or under-represented, and the disease is mainly characterised by haematological perturbations and rheumatological manifestations (polymyalgia rheumatica, dermatomyositis, sarcoidosis, psoriasis).10 Those potential patients from our population with this ‘pauci-inflammatory’ phenotype of VEXAS syndrome did not fulfil our inclusion criteria and consequently, they were not enrolled in this work. This issue has to be addressed in the future to answer the questions of how many patients with the pauci-inflammatory phenotype could be identified, and whether there are clinical or therapeutic differences comparing them with the classical, highly inflammatory phenotype. On the basis of these reflections and considering that the UBA1 analyses represents a unique way to diagnose the VEXAS syndrome at present, we strongly suggest a relaxation of the criteria to perform this genetic test with the proposal to identify as many patients as possible and as early as possible. In this sense, candidates for UBA1 screening might include all those individuals with any sterile inflammatory manifestation associated with anaemia, highly increased ESR and/or ferritin level and a tendency of macrocytosis.
With regard to the response to treatments, the data we collected are also in line with previous reports.7 11–13 Thus, only high dose of corticosteroids ameliorated the inflammatory manifestations, with limited efficacy in the haematological perturbations, and often associated with serious side effects that prevented their long-lasting use. Among biological or synthetic drugs administered to decrease the corticosteroids dose, the responses were often negative or partial, and severe local side effects to anakinra were detected in some patients as has been previously described.7 In contrast, treatments targeting the haematological component of the disease seem to be more effective than the anti-inflammatory drugs. On one side, the unique patient of our series treated during a long period with the hypomethylating drug decitabine experienced a significant improvement of both inflammatory and haematological features, as has been also described in patients treated with the hypomethylating drug azacytidine.16 This clinical improvement was associated with a progressive and marked decrease of the MAF in peripheral blood. Despite this observation being based on a single patient, we propose that the monitorisation of MAF during administered treatments should be seriously considered as a potential biomarker in clinics to evaluate the patients’ responses. This particular issue should be addressed in future to confirm or to rule out its utility. On the other side, the results of allo-HSCT we collected are in line with previous reports, with a complete rescue of the clinical phenotype and analytical abnormalities associated with the disappearance of the mutant allele.17–19 Despite results of allo-HSCT being promising, the usual late-onset of the disease and the relevant comorbidities that most patients with VEXAS syndrome have prevent its use in several patients. In this sense, it is mandatory to obtain a diagnosis as early as possible through genetic analyses to be in a scenario to consider this type of curative approach with as many patients as possible.
From a genetic point of view, VEXAS syndrome is a consequence of UBA1 postzygotic variants that provoke the loss of the functional cytoplasmic UBA1b isoform. Most of the reported patients carried variants at the Met41 amino acid residue, which represents the main hot spot for the disease.7 10–13 The results of the UBA1 genotyping in our series are consistent with previously reported data similar to the similarities in patients’ clinical features and response to treatments mentioned earlier. Thus, in our series 93% patients carried pathogenic variants at the Met41 residue, with the remaining 7% carrying already known pathogenic variants at canonical splicing sites. Interesting and not previously reported data were obtained in the studies of distribution of mosaicism. On one side, the analyses of different isolated haematopoietic cell types showed that the UBA1 variant was present in all cell types with the exception of peripheral T cells and B cells, which are in line with previously published data.7 However, the most intriguing genetic data we obtained was the presence of the UBA1 variants in the DNA extracted from nail samples in nine patients. To reinforce these data, it would have been convenient to analyse nail samples from the entire cohort, but we were not able to collect them due to the death/allo-HSCT of some patients and to logistic issues in others alive patients. Moreover, it would have been also interesting to monitor the nail mosaicism during decitabine treatment, as was done in peripheral blood. However, the patients’ nail samples were not collected at different time points along the treatment. UBA1 variants had been previously detected in different non-haematopoietic tissue such as skin and muscle biopsies, but these results could have been false positive results as a consequence of blood contamination.20 21 The evidences we obtained in nails, a non-blood contaminated tissue collected in a manner that minimise potential contamination due to scratching of skin lesions, might indicate that both ectoderm and mesoderm, the germ layers that lead to nails and blood respectively, carry the UBA1 variants. Consequently, the mutational event at that gene would have occurred during an early phase of the embrionary development, and not during adulthood as has been classically thought. Consequently, our evidences do not concur with the previous idea that VEXAS syndrome is a consequence of a myeloid-restricted UBA1 mosaicism, which was mainly supported by the absence of UBA1 variants in patients’ fibroblasts.7 In this sense, it would have been of great interest to analyse isolated fibroblasts from fresh skin biopsies from patients of our cohort. However, the unique available skin biopsies were paraffined, a material from which we would have not been able to isolate the fibroblasts (the cell type of interest) from the myeloid cells (carrying the UBA1 variant) to evaluate the presence/absence of mosaicism at the UBA1 gene. This represents a limitation of the present study that should be addressed in future works. Despite this limitation, we realise that our conclusion of an early occurrence of the mutational event leading to VEXAS syndrome has important conceptual consequences for the disease pathogenesis, and both pro-argument and counterargument exist and should be carefully evaluated.
Among the favourable arguments for an early mutational event, we can indicate the following: (i) it has been previously reported that the UBA1 variants leading to VEXAS syndrome are present in all CD34+ cell types (haematopoietic stem cells, multipotent progenitors, megakaryocyte-erythroid progenitors, common myeloid progenitors, common lymphoid progenitors).7 This distribution in the CD34+ compartment is markedly different with that previously described in other AIDs due to myeloid-restricted mosaicism (eg, CAPS), where the unique type of CD34+ cells carrying the mosaicism were the common myeloid progenitors3; (ii) it has been recently reported that the haematological disturbances in the VEXAS syndrome may precede the inflammatory manifestations, which suggest the presence of the UBA1 variants in the haematopoietic compartment earlier than expected.10 This scenario may fit with a progressive, but undetermined course of the disease, which may be due to the expansion of a mutant clone through the well-known mechanism of clonal haematopoiesis of indeterminate potential.22 23 This mechanism typically appears with ageing, thus explaining the late onset of the disease in VEXAS syndrome, and does not evolve to a malignant condition as has been also reported in this syndrome.11 In contrast to these pro-arguments, there are also evidences against our mechanistic hypothesis of an early occurrence for the UBA1 mutational event associated with an expansion of the mutant clone, the following being the most relevant: (i) if our hypothesis is correct, we may have expected to identify individuals in large databases aged <40 years with these UBA1 variants at low MAF, before the expansion of the mutant clone occurred. We investigated this possibility by searching for postzygotic UBA1 variants with specific bioinformatic algorithms in the sequences of 1236 male individuals of the 1000 Genomes Project,24 and the results did not identify any individual with the UBA1 pathogenic variants. A limitation of these investigations was the total number of analysed individuals in this database, which may be solved in the future with specific investigations in larger repositories than 1000 Genomes Project (ie, gnomAD, UK Biobank) with appropriate bioinformatic pipelines to evaluate the presence of postzygotic UBA1 variants at low MAF among healthy adult or paediatric individuals; (ii) if our hypothesis is correct, it should be explained why no paediatric or young adults with VEXAS syndrome have been reported to date and (iii) if the mutational event occurred early as we propose, then there is the possibility that the postzygotic UBA1 variant was also present in the gonadal tissue. If this occurs, the individual has a moderate-to-high probability to transmit the mutant allele to his offspring, who will receive it as a germline variant. However, all available genetic evidence obtained in daughters and sons of affected patients, in previous studies and also in the present work, did not identify any individual with the UBA1 variant in germline status.
Finally, we tried to identify potential inflammatory mediators involved in the disease pathogenesis. We analysed serum samples from different patients collected during active disease (high ESR) and while patients were treated with steroids. The results of these investigations did not identify a clear mediator, a result that is in line with the absence of a positive response to the different administered treatments targeting inflammatory cytokines, but not in concordance with previous data.7 The differences with previous reports might be consequence of the disease activity at the time of blood collection, the years of disease evolution and most importantly the treatments that patients were receiving, because they often receive simultaneously different anti-inflammatory drugs that may modify the gene expression profile and consequently the pattern of circulating cytokines. Overall, these results raise the question about the type of mechanism involved in the (hyper)inflammation observed in the VEXAS syndrome. In this sense, the positive therapeutic responses observed when targeting the haematological component of the disease, and the previously mentioned poor responses to classical ant-inflammatory drugs seem to point towards inflammatory mechanisms other than the previously identified in the classical monogenic AID such as inflammasomopathies or type I interferonopathies.
In summary, we have identified a series of 30 patients with VEXAS syndrome by means of UBA1 analysis among adult patients with otherwise undiagnosed AID. Their clinical manifestations, results of laboratory tests and outcome of administered treatments are in line with previous reports. Genetically, all patients’ disease is a consequence of already described UBA1 pathogenic variants. Additional genetic investigations about mosaicism distribution support the early occurrence of the mutational event leading to mosaicism, in opposition to the previous concept of a myeloid-restricted mutational event occurring during adulthood. These novel evidences may have conceptual consequences for the mechanism causing the syndrome, and additional studies addressing this issue will be necessary.
Acknowledgments
The authors thank Dr Fernando Moldenhauer and Dr Javier Loscertales, both from Hospital de La Princesa (Madrid) for their collaboration in the search of patients, the patients and their families for their collaboration in this study, and William Sinclair for his assistance with English grammar review.
Footnotes
Handling editor: Josef S Smolen
Twitter: @gerardespinosa5
Correction notice: This article has been corrected since it published Online First. A second affiliation has been added for Dr Andrea Cerutti.
Contributors: Study conception and design: AM-V and JIA. Methodology—data collection: JMM, IR-P, GP, JF-M, LC-M, GE, JH-R, MD, JR-M, RS, SC, DC-A, RC, PF, JIE, SB, IF, FMM, MA, SS-D, AG-G, XF-N, CG-B, AG, NQ, MO-C, RGdlT, MCC, GE-F, AAA, EL, JRA-M. Methodology—samples collection: IR-P, GP, JF-M, LC-M, MD, SC, DC-A, RC, PF, JIE, IF, FMM, SS-D, AG-G, XF-N, CG-B, AG, MTF-F, MO-C, RGdlT, EL, MR, ML-G, PC, JRA-M, EV. Methodology—genetic studies: AM-V, EG-R, SPRA, VFRA, RLRA, NB, MS-M, FC, JY, JIA. Methodology—leucocyte subpopulation isolation: ST-V, GM, AC, OF. Formal analysis: all authors. Writing (original draft): JIA. Writing (revising, review and editing): all authors. All authors read and approved the final manuscript. JIA acts as guarantor for the study, had access to all data and controlled the decision to publish.
Funding: This work has been partially funded by RTI2018-096824-B-C21 (JIA) and RTI2018-096824-B-C22 (FC) grant from the Spanish Ministry of Science, Innovation and Universities co-funded by Agencia Estatal de Investigación/European Regional Development Fund una manera de hacer Europa. PID2021-125106OB-C31 (JIA) and PID2021-125106OB-C32 (FC) grants from the Ministerio de Ciencia e Innovación (MCIN) | Agencia Estatal de Investigación (AEI) | 10.13039/501100011033 | Fondo Europeo de Desarrollo Regional (FEDER), UE.PI19/01567 grant from Instituto de Salud Carlos III (ISCIII) co-funded by the European Union (AM-V).
Competing interests: None declared.
Patient and public involvement: Patients and/or the public were not involved in the design, or conduct, or reporting, or dissemination plans of this research.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
The data included in the present work are, by definition, deidentified, and are available upon reasonable request to, and after approval by, the study contributors and after a signed data access agreement.
Ethics statements
Patient consent for publication
Consent obtained directly from patient(s).
Ethics approval
The Ethical Review Board of Hospital Clínic, Spain approved the study (code HCB/2022/0855). All investigations were performed in accordance with the ethical standards of the 1964 Declaration of Helsinki and its later amendments. Written informed consent was obtained from patients or from patients' relatives at their respective medical centres.
References
- 1. Aksentijevich I, Schnappauf O. Molecular mechanisms of Phenotypic variability in Monogenic Autoinflammatory diseases. Nat Rev Rheumatol 2021;17:405–25. 10.1038/s41584-021-00614-1 [DOI] [PubMed] [Google Scholar]
- 2. Zhou Q, Aksentijevich I, Wood GM, et al. Cryopyrin-associated periodic syndrome caused by a myeloid-restricted somatic Nlrp3 Mutation. Arthritis Rheumatol 2015;67:2482–6. 10.1002/art.39190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mensa-Vilaro A, Teresa Bosque M, Magri G, et al. Brief report: late-onset Cryopyrin-associated periodic syndrome due to myeloid-restricted somatic Nlrp3 Mosaicism. Arthritis Rheumatol 2016;68:3035–41. 10.1002/art.39770 [DOI] [PubMed] [Google Scholar]
- 4. Rowczenio DM, Gomes SM, Aróstegui JI, et al. Late-onset Cryopyrin-associated periodic syndromes caused by somatic Nlrp3 Mosaicism-UK single center experience. Front Immunol 2017;8:1410. 10.3389/fimmu.2017.01410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kontzias A, Zarabi SK, Calabrese C, et al. Somatic Mosaicism in adult-onset TNF receptor-associated periodic syndrome (TRAPS). Mol Genet Genomic Med 2019;7:e791. 10.1002/mgg3.791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ionescu D, Peñín-Franch A, Mensa-Vilaró A, et al. First description of late-onset Autoinflammatory disease due to somatic Nlrc4 Mosaicism. Arthritis Rheumatol 2022;74:692–9. 10.1002/art.41999 [DOI] [PubMed] [Google Scholar]
- 7. Beck DB, Ferrada MA, Sikora KA, et al. Somatic mutations in Uba1 and severe adult-onset Autoinflammatory disease. N Engl J Med 2020;383:2628–38. 10.1056/NEJMoa2026834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Poulter JA, Collins JC, Cargo C, et al. Novel somatic mutations in Uba1 as a cause of VEXAS syndrome. Blood 2021;137:3676–81. 10.1182/blood.2020010286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Templé M, Duroyon E, Croizier C, et al. Atypical splice-site mutations causing VEXAS syndrome. Rheumatology (Oxford) 2021;60:e435–7. 10.1093/rheumatology/keab524 [DOI] [PubMed] [Google Scholar]
- 10. Beck DB, Bodian DL, Shah V, et al. Estimated prevalence and clinical manifestations of Uba1 variants associated with VEXAS syndrome in a clinical population. JAMA 2023;329:318–24. 10.1001/jama.2022.24836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Grayson PC, Patel BA, Young NS. VEXAS syndrome. Blood 2021;137:3591–4. 10.1182/blood.2021011455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Georgin-Lavialle S, Terrier B, Guedon AF, et al. Further characterization of clinical and laboratory features in VEXAS syndrome: large-scale analysis of a Multicentre case series of 116 French patients. Br J Dermatol 2022;186:564–74. 10.1111/bjd.20805 [DOI] [PubMed] [Google Scholar]
- 13. van der Made CI, Potjewijd J, Hoogstins A, et al. Adult-onset Autoinflammation caused by somatic mutations in Uba1: A Dutch case series of patients with VEXAS. J Allergy Clin Immunol 2022;149:432–9. 10.1016/j.jaci.2021.05.014 [DOI] [PubMed] [Google Scholar]
- 14. Richards S, Aziz N, Bale S, et al. ACMG laboratory quality assurance committee. standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American college of medical Genetics and Genomics and the Association for molecular pathology. Genet Med 2015;17:405–24. 10.1038/gim.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mensa-Vilaró A, Bravo García-Morato M, de la Calle-Martin O, et al. Unexpected relevant role of gene Mosaicism in patients with primary immunodeficiency diseases. J Allergy Clin Immunol 2019;143:359–68. 10.1016/j.jaci.2018.09.009 [DOI] [PubMed] [Google Scholar]
- 16. Comont T, Heiblig M, Rivière E, et al. Azacitidine for patients with Vacuoles, E1 enzyme, X-linked, Autoinflammatory, somatic syndrome (VEXAS) and myelodysplastic syndrome: data from the French VEXAS Registry. Br J Haematol 2022;196:969–74. 10.1111/bjh.17893 [DOI] [PubMed] [Google Scholar]
- 17. Loschi M, Roux C, Sudaka I, et al. Allogeneic stem cell transplantation as a curative therapeutic approach for VEXAS syndrome: a case report. Bone Marrow Transplant 2022;57:315–8. 10.1038/s41409-021-01544-y [DOI] [PubMed] [Google Scholar]
- 18. Al-Hakim A, Poulter JA, Mahmoud D, et al. Allogeneic haematopoietic stem cell transplantation for VEXAS syndrome: UK experience. Br J Haematol 2022;199:777–81. 10.1111/bjh.18488 [DOI] [PubMed] [Google Scholar]
- 19. Diarra A, Duployez N, Fournier E, et al. Successful allogeneic hematopoietic stem cell transplantation in patients with VEXAS syndrome: a 2-center experience. Blood Adv 2022;6:998–1003. 10.1182/bloodadvances.2021004749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zakine E, Schell B, Battistella M, et al. Uba1 variations in Neutrophilic Dermatosis skin lesions of patients with VEXAS syndrome. JAMA Dermatol 2021;157:1349–54. 10.1001/jamadermatol.2021.3344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cordts I, Hecker JS, Gauck D, et al. Successful treatment with Azacitidine in VEXAS syndrome with prominent Myofasciitis. Rheumatology (Oxford) 2022;61:e117–9. 10.1093/rheumatology/keab866 [DOI] [PubMed] [Google Scholar]
- 22. Steensma DP, Bejar R, Jaiswal S, et al. Clonal Hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood 2015;126:9–16. 10.1182/blood-2015-03-631747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Steensma DP. Clinical consequences of Clonal Hematopoiesis of indeterminate potential. Blood Adv 2018;2:3404–10. 10.1182/bloodadvances.2018020222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Auton A, Brooks LD, Durbin RM, et al. A global reference for human genetic variation. Nature 2015;526:68–74. 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
ard-2023-224460supp006.pdf (264.4KB, pdf)
ard-2023-224460supp001.pdf (1.4MB, pdf)
ard-2023-224460supp002.pdf (451.3KB, pdf)
ard-2023-224460supp003.pdf (508.3KB, pdf)
ard-2023-224460supp004.pdf (638.6KB, pdf)
ard-2023-224460supp005.pdf (415KB, pdf)
Data Availability Statement
The data included in the present work are, by definition, deidentified, and are available upon reasonable request to, and after approval by, the study contributors and after a signed data access agreement.