Abstract
The introduction of the in-vitro evolution method known as SELEX (Systematic Evolution of Ligands by Exponential enrichment) more than 30 years ago led to the conception of versatile synthetic receptors known as aptamers. Offering many benefits such as low cost, high stability and flexibility, aptamers have sparked innovation in molecular diagnostics, enabled advances in synthetic biology and have facilitated new therapeutic approaches. The SELEX method itself is inherently adaptable and offers near limitless possibilities in yielding functional nucleic acid ligands. This Primer serves to provide guidance on experimental design and highlight new growth areas for this impactful technology.
Introduction
The concept of single-stranded RNA or DNA oligonucleotides serving as molecular recognition agent akin to antibodies was unprecedented until the late 1980s. Then, the pioneering concept of Systematic Evolution of Ligands by EXponential enrichment (SELEX) led to the discovery of oligonucleotide-based molecular affinity agents, now known as aptamers, 1, 2 SELEX enabled isolation of high affinity-binding oligonucleotides from a large population of random sequences, expanding the known functions of DNA and RNA into unexpected roles. Aptamers offer specificity and affinity normally expected of antibodies but in small, chemically synthesized molecules free from cell-culture-derived contaminants. Additionally, aptamers can be easily synthesized and chemically modified, can be reversibly denatured and have tuneable affinity and kinetic properties. The in vitro nature of SELEX offers opportunities to control the conditions of the binding event to best suit a goal. The process follows the rules of combinatorial chemistry and directed evolution 3,4. Chemically, SELEX uses a heterogeneous library [G] of nucleic acids that folds into conformations unique to their sequence identity. SELEX starts with the premise that within this multitude of three-dimensional nanostructures, an oligonucleotide with exclusive affinity to nearly any target molecule exists and can be sequestered. The affinity between an aptamer and its target is often described using the equilibrium dissociation constant (), where lower values signify tighter binding:
where A is the aptamer, T is the target and AT is the aptamer-target complex. can also be described using the ratio of the on-rate and off-rate,
where is the rate of association and is the rate of disassociation.
Determination of the dissociation constant by either method reveals the optimal working conditions of an aptamer for detecting its cognate target. Most commonly, aptamers are reported with an apparent when working with concentration-based measurements, owing to the fact that a true is not affected by varying concentrations of the limiting reagent5. The aptamer concentration should be much less than the 6. However, in aptamer systems it is not always possible to control experimental parameters to sufficiently eliminate this variable6.
A typical SELEX process is shown in Figure 1. The main steps include incubation, partitioning [G] and amplification. The idea of molecular evolution stems from using a polymerase reaction to generate a screened library with potential winners, for example aptamers with strong binding affinity, to be utilized in the next round of SELEX. Multiple SELEX cycles generate a competition among active sequences, ideally enriching the highest affinity sequences in an evolved SELEX library. Success is measured by selection of aptamers with high affinity to their cognate target, high specificity, as well as other features, for example tolerance to nuclease digestion.
Figure 1. General outline of the SELEX process and aptamer identification.
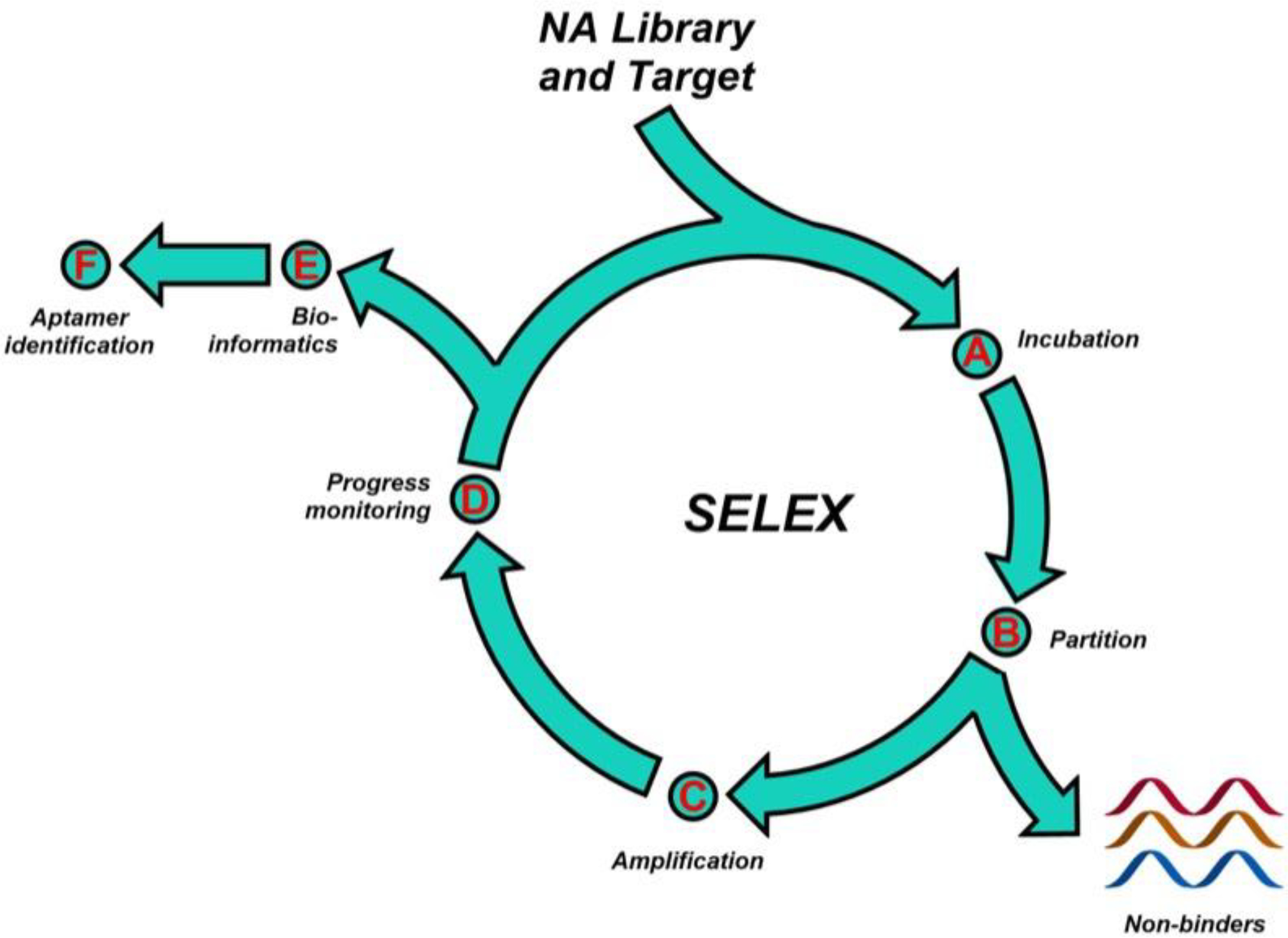
A SELEX library is chemically synthesized using a DNA synthesizer, and PCR optimization prior to SELEX ensures efficient amplification. The SELEX library is incubated with the target, and binders are separated from non-binders using an appropriate partitioning method. The SELEX library with potential binders is then amplified and utilized in the next round of SELEX. The evolved libraries are monitored for enrichment and sequenced to identify hit aptamer candidates.
The elegance of SELEX is in its simplicity – powerful molecular recognition agents can be generated at low cost, using basic biochemistry/molecular biology equipment (for example, thermocycler, gel electrophoresis unit) with techniques taught in most undergraduate science curricula, such as polymerase chain reaction (PCA) and sequencing. The accessible nature of SELEX has democratized molecular affinity and led to a surge of start-up companies and academic labs working on aptamer development and applications, after the initial patents around the process expired. Oligonucleotides with these unique properties are the exception, not the rule. While SELEX is extremely proficient at finding these exceedingly rare molecules, careful consideration of the conditions and design of the experiment could help avoid potential pitfalls and greatly improve the likelihood of success. This provides guidance on experimental design and data analysis, while providing a snapshot of nascent opportunities for aptamer applications. While the recommendations described here could equally be applied to the generation of other functional nucleic acids, for example DNAzymes and riboswitches, this Primer focuses on nucleic acid aptamer selection, characterization, and applications. Considerations to improve reproducibility in experiments, limitations and workarounds, as well as the future outlook of the field are also discussed.
Experimentation
Proper preparation greatly improves the likelihood of success in a SELEX experiment. Ultimately, a comprehensive SELEX plan will require considerations involving nine aspects of the selection experiment: library design; incubation conditions; partitioning method [G]; stringency [G]; amplification; generation of single stranded nucleic acids; monitoring enrichment [G]; sequencing and bioinformatics; characterization. The flow chart in Figure 2 offers a blueprint to guide the planning process for a successful selection, including examples of queries and how they may influence the SELEX experiment. Considering the nature of the target, the desired application for the aptamer and the medium within which the aptamer will encounter the target, balanced by an understanding of the resources available for the experiment, will help guide decisions about all aspects of the SELEX experiment. For example, the end-application of the aptamer should be carefully considered prior to starting a SELEX experiment. Before starting an experiment, the ideal affinity of the aptamer to be successful in a particular application should be considered. The desired affinity could, for example, stem from the required limit of detection of the aptamer-based assay, or the biological concentration of a key target of the aptamer’s therapeutic intervention. Understanding this value can help inform what partitioning method should be employed and what the stringency of the selections must be to achieve that desired affinity. Furthermore, it impacts choices on characterization methods that are used to assess the selected hits. For example, techniques to estimate binding constants are limited in the range of values they can assess by their own sensitivity,7 thus the choice of method used should be guided by the range of values expected and required. The following sections give more detail about the critical parameters in a typical SELEX process and give some examples of the typical data generated for both aptamer selection and characterization experiments.
Figure 2. Decision-making for SELEX design.
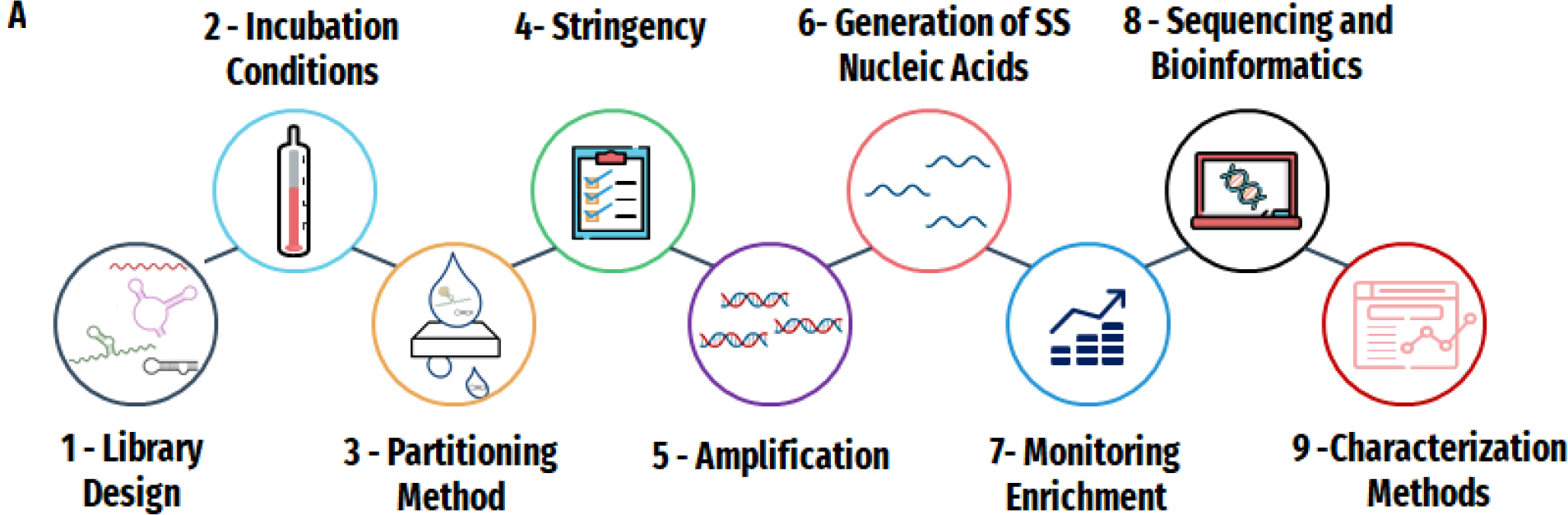
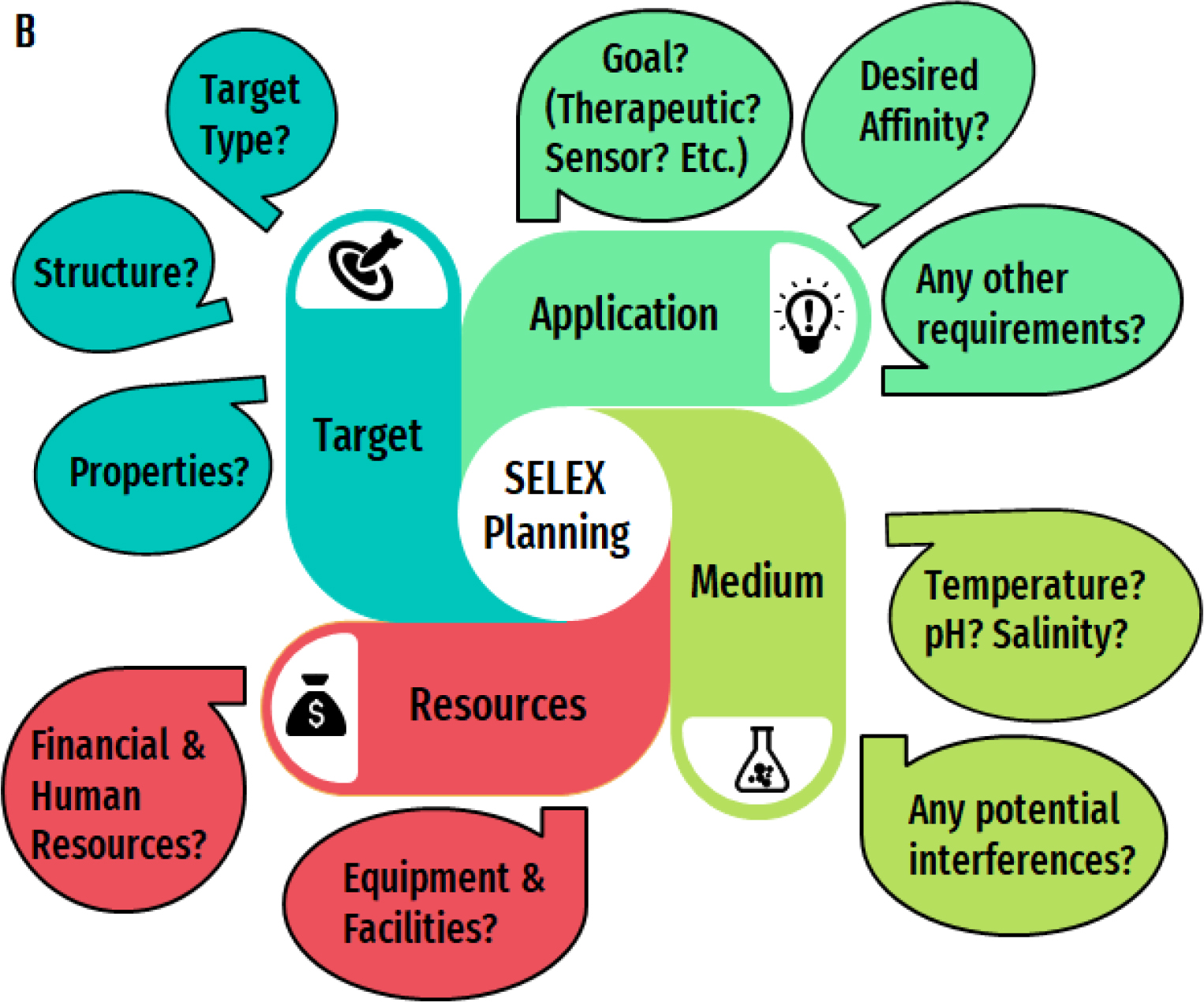
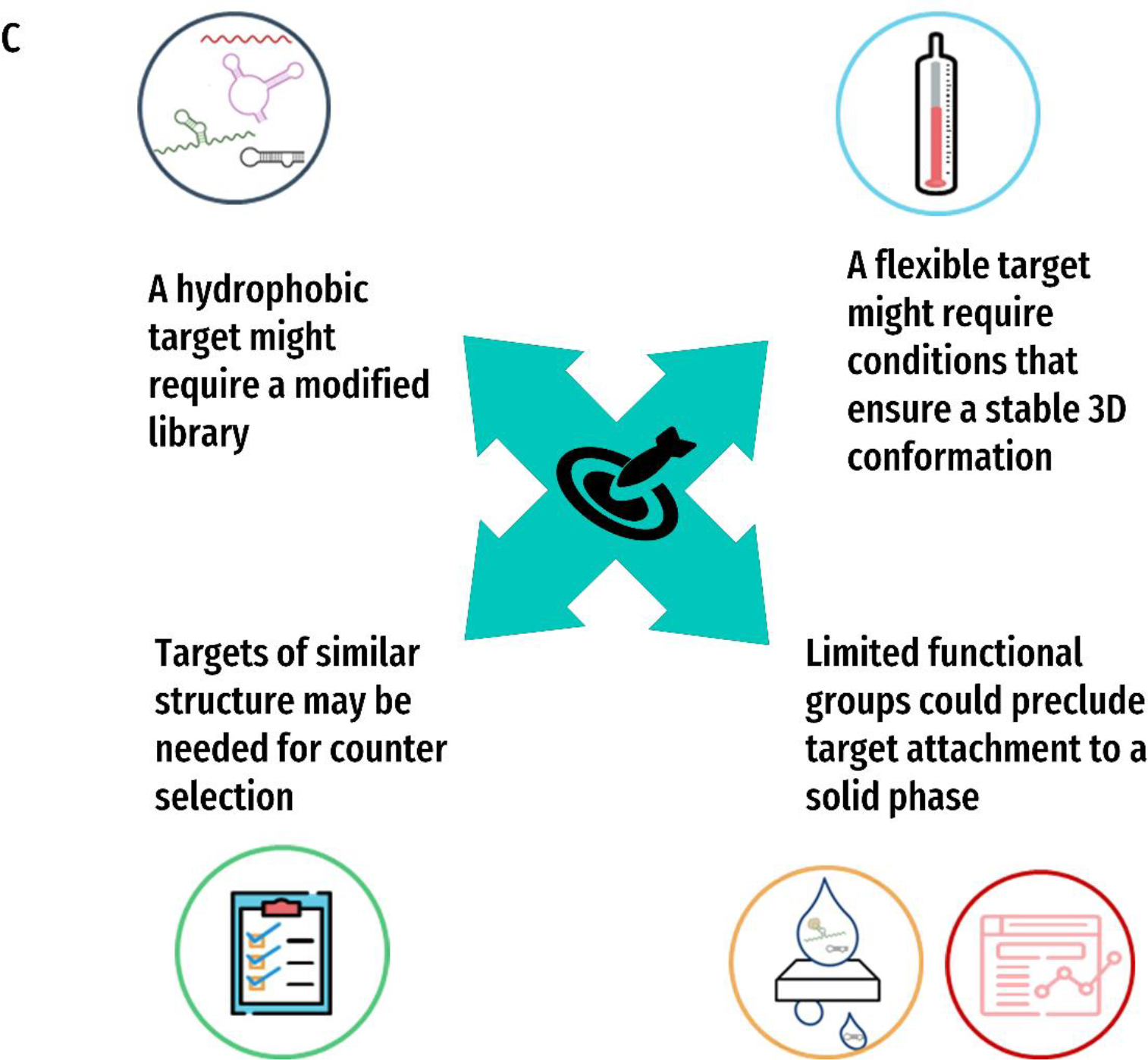
A) The nine main aspects of a selection experiment: 1) Library Design 2) Incubation Conditions 3) Partitioning Method 4) Stringency 5) Amplification 6) Generation of Single Stranded Nucleic acids 7) Monitoring Enrichment 8) Sequencing and Bioinformatics and 9) Characterization Methods. B) The four key factors that will inform those decisions: 1) the target of interest 2) the eventual application of the aptamer 3) the medium and 4) the resources available. C)-F) Examples of ways these queries may influence the different aspects of the SELEX experiment for each of the four key factors C) Target D) Application E) Medium and F) Resources
Library Design
SELEX starts with naïve nucleic acid library, from which the aptamer hits will be revealed. The SELEX library is typically designed with a central randomized region of a variable number of nucleotides flanked by two fixed primer regions at both ends. The fixed regions facilitate the amplification of the library using PCR. The base composition, as well as the length of the randomized region of a SELEX library governs the extent of diversity of the initial library. A typical SELEX library consists of A, G, T/U, C. Considering an ‘n’ nt randomized region, the number of unique sequences that could be generated is 4n. For example, a 40 nt randomized region can generate 440 unique sequences. Notably, the sequence space [G] that can be sampled in a SELEX experiment is governed by the practical limitations of DNA synthesis; any library with a random region of more than 25 random nucleotides would not be expected to contain every possible random sequence (see box 1). Nevertheless, strategies for increasing sequence space coverage exists, such as using error-prone PCR conditions8 to introduce point mutations. Additionally, a growing number of available modified bases have been utilized in SELEX libraries to generate aptamers with high nuclease stability and affinity. For example, to increase nuclease stability, efforts have been primarily focused on RNA SELEX, substituting pyrimidines with Xeno Nucleic Acid (XNA) bases9,10. Several XNA bases are currently available, including, 2’-F11, 2’O-Me12, 2’-NH213, LNA14, TNA15, FANA10, and HNA16 monomers. Additionally, while this approach is less common, the nuclease stability of a SELEX library can be enhanced by backbone modifications by using nucleic acids with phosphorothioate backbones17. Strategies to enhance diversity of a DNA-SELEX library and to produce protein-like binding have been accomplished by incorporating Unnatural Base Pairs (UBPs). These bases are decorated with hydrophobic or hydrophilic residues to enhance the chemical space of a SELEX library. For example, Ds: Px18, AEGIS bases19,20, and dnAM: d5SICS, dPTMO and dMTMO bases21, have been used to select high-affinity aptamers. Chemical diversity can also be expanded by synthesizing uridine analogs with substituted groups at the 5th position on the purine ring22. This strategy led to the generation of SOMAmers with high specificity and high affinity against a large array of soluble biomarker proteins22. Furthermore, libraries can be designed with reactive functional groups, such as an alkyne, followed by the introduction of functional arms decorated with azide groups added via bioorthogonal reactions, such as click chemistry, to select high-affinity aptamers with novel functional groups23,24. The synthesis of a typical, unbiased SELEX library should yield an equal ratio of A:G:T:C in the random region; to achieve an equal nucleotide distribution, the molar ratio of A:C:G:T phosphoramidites on the DNA synthesizer should be optimized to account for the different reactivity of these building blocks. For example, ratios of A:C:G:T have been varied from 1.30:1.25:1.45:1.00,25 or 1.50:1.25:1.15:1.0026. These ratios can be achieved using a hand-mix in which an appropriate number of moles of AGTC phosphoramidites are premixed and added to a single position in the DNA synthesizer or machine-mix by programing the synthesizer to deliver an appropriate volume of each base to the column during synthesis. Generating RNA or XNA libraries is done by synthesizing a DNA library and then transcribing it into RNA/XNA libraries.10 Since most UBPs are based on DNA bases, UBP-containing libraries are directly synthesized using a standard DNA synthesizer.
Design of fixed regions
Regardless of the composition of the random region, the fixed region typically contains natural RNA or DNA bases because of their compatibility with polymerases utilized to amplify SELEX libraries. The primer region normally contains 18–20 nts with a nucleic acid base composition optimized to achieve high PCR efficiency. Factors in primer design include optimal melting points, no primer-dimer formation and avoidance of secondary structure formation within the primer sequence. Several commercial sources are available for primer design, such as the Oligoanalyzer tool in IDTDNA, Bio-RAD, and Thermo Fisher. When the SELEX library is composed of RNA/XNA molecules, most XNA libraries are generated by primer-extension, though rarely an additional promoter sequence is included in the 5’-end for the T7 RNA polymerase reaction. A note of caution when designing multiple SELEX experiments that may be running concurrently: always design the libraries to have either different primer pairs or unique barcodes to avoid cross-contamination during parallel experiments. When multiple selections are on-going in the same space, it is very easy to cross contaminate selections, through shared instrumentation and reagents. Though good lab practices for handling nucleic acids and consistent decontamination (of pipettes, shared spaces, shared equipment) with ethanol and bleach can usually eliminate these issues, using unique libraries is an effective strategy to eliminate this problem entirely.
In addition to the fixed primer regions, the library may also contain fixed regions for other purposes. For example, a capture sequence may be included to facilitate binding of the library to a solid support such as capture SELEX, vide infra. A SELEX library can also be predesigned to select the desired aptamers with unique structural and functional properties. For example, structural components, such as stem-loop,27 and three-way junctions28, can be added within the randomized region to select aptamers containing predefined structures. A sequence can also be modified to facilitate the selection of aptamers that can bind to recalcitrant targets, for example, a library with predefined hairpin structures incorporating clustered glycans,24 was used to select aptamers for carbohydrate-binding proteins. Predesigned SELEX libraries can also be centered on parent natural nucleic acid ligands by partially randomizing or by adding a doping mixture of nucleic acid bases to a set position.29 This unconventional approach has been successful in generating second-generation aptamers with higher affinities and functionalities. Furthermore, a SELEX library can be designed for post-SELEX selection to add functional arms to already selected aptamers to enhance their affinity.29,30
Incubation conditions
The incubation of a SELEX library and the target can be achieved with both free in solution or by immobilizing either the target or the SELEX library on a solid support. This choice is usually based on the nature of the target and end-use of the aptamer. The conditions (pH, salinity, temperature) of this step should match those of the application as best as possible to ensure optimal fit for purpose.
During selection, it is necessary to present a target with consistent and uniform structure. Therefore, target preparation must be predominantly aimed at optimizing the conditions to facilitate generating a conformation like one in its natural environment. Several types of targets have been utilized in SELEX, and preparation steps are unique to the target.
Small molecules
Generally, when the target of interest is a small molecule, SELEX is more challenging.31 However, the target molecule can be immobilized on a solid support, such as a bead, using standard bioconjugation procedures.32 Here, it is essential to ensure that the structure of the target is not perturbed by immobilization and that the target functional groups used for immobilization are not critical for ensuring the specific binding of the aptamer. If the target molecules cannot be immobilized on a solid support, the SELEX library can be immobilized on a solid support.33 This strategy is known as capture SELEX, and it has proven to be effective in generating structure-switching aptamers against various targets. Here, the SELEX library is hybridized to a bead-bound short complementary strand, and upon binding, the sequences undergo a conformational switch allowing the sequences to bind to the target, followed by spontaneous dissociation of the complex from the beads that leads to elution.34 If the target or the SELEX library cannot be immobilized, then the binding can be done in a free solution, but here, the separation of bound sequences to the target is needed.
Proteins
In contrast, when the target is a protein, the SELEX experiment will have a higher expectation of success. However, the buffer conditions need to be optimized to ensure that protein folding is consistent and stable, enabling unperturbed presentation of the target’s binding moieties to the SELEX library.35 The protein target can be presented free in solution, allowing unrestricted binding of potential aptamers followed by partitioning. Proteins can also be immobilized on a solid support using several immobilization methods, such as Ni columns36, biotin-streptavidin37, magnetic beads38 or direct chemical attachment39,40. In general, care must be taken to avoid selecting aptamers toward a protein’s conformation that may not exist in its natural environment. If the protein is a membrane protein, a practical method is to use whole cells with endogenous expression levels of the target or whole cells engineered to express the protein of interest.40,41
Cells and tissues
A particular advantage of SELEX is that the target need not be known to carry out the selection. This feature is commonly exploited in cell-SELEX. The challenges and opportunities of whole-cell SELEX for bacteria, parasites and animal cells has been reviewed recently42. Generally, for cell selections, the selection matrix needs to be carefully considered to accommodate for potential challenges, for example nucleases or highly abundant non-targets that may deplete the library. As a specific example, when whole cells are used, the culture conditions need to be optimized to avoid generating aptamers with a high bias towards cell-surface proteins only expressed on cultured cells. One way to avoid this bias is to use primary cells known to express the desired target.43 Another consideration is the number of dead cells that could remove potential binders from the SELEX pool.44 Use of fluorescence-activated cell sorting (FACS) to remove dead cells from the population helps increase SELEX efficiency. In addition to whole cells, SELEX can be used to select aptamers against whole tissues. When the target is whole tissue, the selection can be performed either in vitro or in vivo using animal models.45,46 When using animal models, the SELEX library is injected systemically into the animal and allowed to circulate, subsequently removing a tissue of interest to extract bound sequences.45
Partitioning
Partitioning of winning sequences with high affinity is an essential part of the SELEX process. Partition efficiency47 can be defined as , where is the transmittance of binders and is the transmittance of non-binders after a partitioning step. Better partitioning efficiency leads to fewer SELEX cycles generating aptamers with desired affinity with low background sequences. Partition efficiencies vary widely by selection method, ranging from 10–100 for traditional methods33 such as magnetic bead separation, to 105-106 for more advanced methods such as microfluidic-SELEX or capillary electrophoresis SELEX.39,48,49
If the target is immobilized on a solid support,50 nucleic acids can be separated by simple washing, while bound aptamers can be eluted by applying heat51, chaotropic agents or using enzymatic approaches.52 Capture-SELEX employs a unique way of partitioning that involves a conformational switch facilitating the elution of an aptamer/small-molecule complex from the beads.33 The most common method of partitioning a protein/aptamer complex free in solution is the use of nitrocellulose filters.53,54 Free nucleic acids show minimal non-specific binding to nitrocellulose, while proteins are often tightly held. Thus, only oligonucleotides in a tight complex with the target protein will be expected to bind to the nitrocellulose, and weakly bound sequences are washed away. The retained nucleic acid sequences with target can be extracted using standard nucleic acid extraction methods. In addition, based on the changes of electrophoretic mobility, protein/nucleic acid complexes can be separated by gel electrophoresis or capillary electrophoresis.55,56 When whole cells are the target, the nucleic acid sequences bound to cells can be separated by simply heating or using a higher affinity competitor, such as a monoclonal antibody.57,58 In the case of tissue-SELEX, of the bound nucleic acid sequences can be eluted using standard nucleic acid extraction methods, such as homogenization and organic solvent extraction, or commercially available kits.59,60
Stringency
Stringency of the selection can be judiciously controlled in the planning phase. To complement the conditions of the positive selection [G] with the chosen target, counter and negative selections are frequently included. Counter-selections [G] involve incubating the selection library with a similar molecule to remove aptamers with cross reactivity, whereas negative selections [G] are done to remove sequences with affinity to the selection matrix (for example, magnetic beads). The target concentration is also a key parameter of a selection experiment and the optimal target concentration will depend on a variety of factors.61 In theory, the higher the target concentration, the greater the possibility of isolating aptamers with low to moderate affinity. In practice, however, low frequency but high affinity binders could be easily lost in early rounds if target concentration is too low. Gradually reducing the target concentration throughout the selection can be one approach to increasing stringency to ensure enrichment of true binders. Several studies have suggested that the optimal library to target ratio for SELEX experiments should range from 100:1–1000:162,63 Setting the final target concentration to be in the range of the desired , and tracking your optimal target concentrations with the changes in the bulk of the enriched pool, is a sound strategy.22,64 To minimize enriching aptamers against the matrix, such as the solid support or the nitrocellulose filters, the performance of a negative selection step, using the matrix, is necessary.65 When whole cells are utilized, a similar cell line that lacks the target receptor of interest to remove binders common to both cell types should be used for a counter-selection step.66 When the whole cell is engineered to express a high level of target protein, a native cell line that does not express the same target is used to remove amplicons in the library.67 Implementing a negative selection step has been proven to remove a significant number of nonspecific sequences from the pool. Other counter selection steps against potential cross-reacting species can also be very effective. When a specific application is desired, it is advisable to introduce conditions related to that application (pH, salinity, temperature, interferences) in the later rounds of SELEX to ensure the enrichment of robust, versatile aptamers.43
Amplification
Once the winning sequences are partitioned using an appropriate method, sequences are ready for PCR amplification. Since only a handful of truly functional aptamers to the target is present at any given early round of a SELEX pool, careful optimization of the amplification step is crucial for the successful selection of aptamers against the desired target.68,69 At the beginning of SELEX, the nucleic acid library is heterogeneous and may contain sequences that are difficult to amplify, with high GC content, tending toward the formation of exotic secondary structures.70 Therefore, at the beginning of SELEX (approximately rounds one to five), the probability of forming primer-dimers and overamplifying sequences with high PCR bias, also known as PCR parasites, is high. This could lead to a potential loss of aptamer sequences present in the pool. Therefore, it is essential to optimize PCR conditions to account for the concentration of primers and the number of cycles to minimize the overamplification of PCR parasites. Strategies to minimize PCR bias include limiting the amplification cycles, monitoring the amount of template, decreasing primer concentrations, and utilizing emulsion PCR.71 In particular, quantitative (q)PCR is particularly useful for quantifying target-bound ssDNA after each round and revealing the optimal (minimal) number of PCR cycles for amplifying the target-bound fractions for the next round of SELEX.72 Because PCR depends on the efficiency of recovering winning sequences at the partitioning step, no standard number of PCR cycles can be recommended. Therefore, when the copies of winning sequences are low during the initial rounds of selection, performing a two-step PCR reaction is advisable to avoid the potential loss of aptamer sequences.73 When UBP’s are used in the SELEX library, the PCR procedure needs to be optimized to ensure that the modified SELEX library is uniformly amplified.74,75,76,77 When an RNA library is used first, reverse transcription (RT)-PCR converts the RNA library into a DNA library. RT-PCR is followed by PCR to generate amplified double-stranded DNA pools subsequently converted into the evolved RNA library using transcription. When modified bases are used in a SELEX library, engineered polymerases are available mostly as plasmids from respective academic labs, which requires expression and the purification prior to beginning of SELEX.78 PCR product can be detected using either a polyacrylamide or agarose gel electrophoresis. Typically, the number of PCR cycles required to obtain a reasonable amount of an evolved SELEX pool is relatively high during the first rounds of SELEX, but then decreases and eventually plateaus. When using modified bases or UBPs, at special polymerases are sometimes required so experiments should be planned accordingly. 79
Generation of single-stranded DNA
When an RNA library is used, transcription of the amplified DNA library typically generates single-stranded RNA. The product of the amplified DNA library is double-stranded, which then needs to be converted to single-stranded aptamer sequences. One common method of generating a single-stranded SELEX pool is using a biotinylated reverse primer captured by streptavidin-coated magnetic or agarose beads, followed by alkaline denaturation.80 The released sense strand is collected, desalted, and quantified. There are some challenges with this method, including carry over contamination of the selection library with streptavidin, which can interfere with the selection (an example that illustrates the importance of counter-selections). Only a few other approaches, such as asymmetric PCR,81 Lambda exonuclease digestion23, and denaturing PAGE separation82,83, are commonly used, usually based on the availability of resources. Each of these methods have their own advantages and drawbacks.80 Asymmetric PCR can generate a much higher concentration of single-stranded DNA than other methods, but PCR or proofreading issues can introduce bias. Exonuclease digestion is efficient in the sense that it produces a relatively high yield compared to magnetic beads, but it is a multiple step process. Finally, denaturing PAGE methods are useful in physically separating DNA strands of different sizes, but they are also laborious to carry out.
Monitoring enrichment
When selecting aptamers from large libraries, it is critical to remember that you get what you select for. As such, it is essential to evaluate whether the selection is working to maximize the chance of identifying aptamer candidates exhibiting the desired functional properties following the months-long selection experiment. Enrichment can be monitored by measuring the amount of target-bound nucleic acid, by measuring the affinity of the oligonucleotide pool to the target or by assessing the decrease in the diversity of the selected library, after each round. This term is also used for individual aptamer sequences or clusters from high throughput sequencing data by comparing cycle-to-cycle abundance. Using the following equation84, relative sequence enrichment over each round can be compared and calculated:
First, for researchers employing typical, primer-based SELEX, confirmation of PCR amplification product by gel electrophoresis remains the method of choice. The PCR amplification product from the selection round should show a band matching the size as the original library (positive control), without any by-product formation and no band in the negative control (non-template control) lane. By-products or undesired amplicons may result in parasitic elongated or ladder-like products.68 Figure 3A shows an example of an ideal PCR amplification analyzed by gel electrophoresis.
Figure 3. Representative analysis of the in vitro selection process.
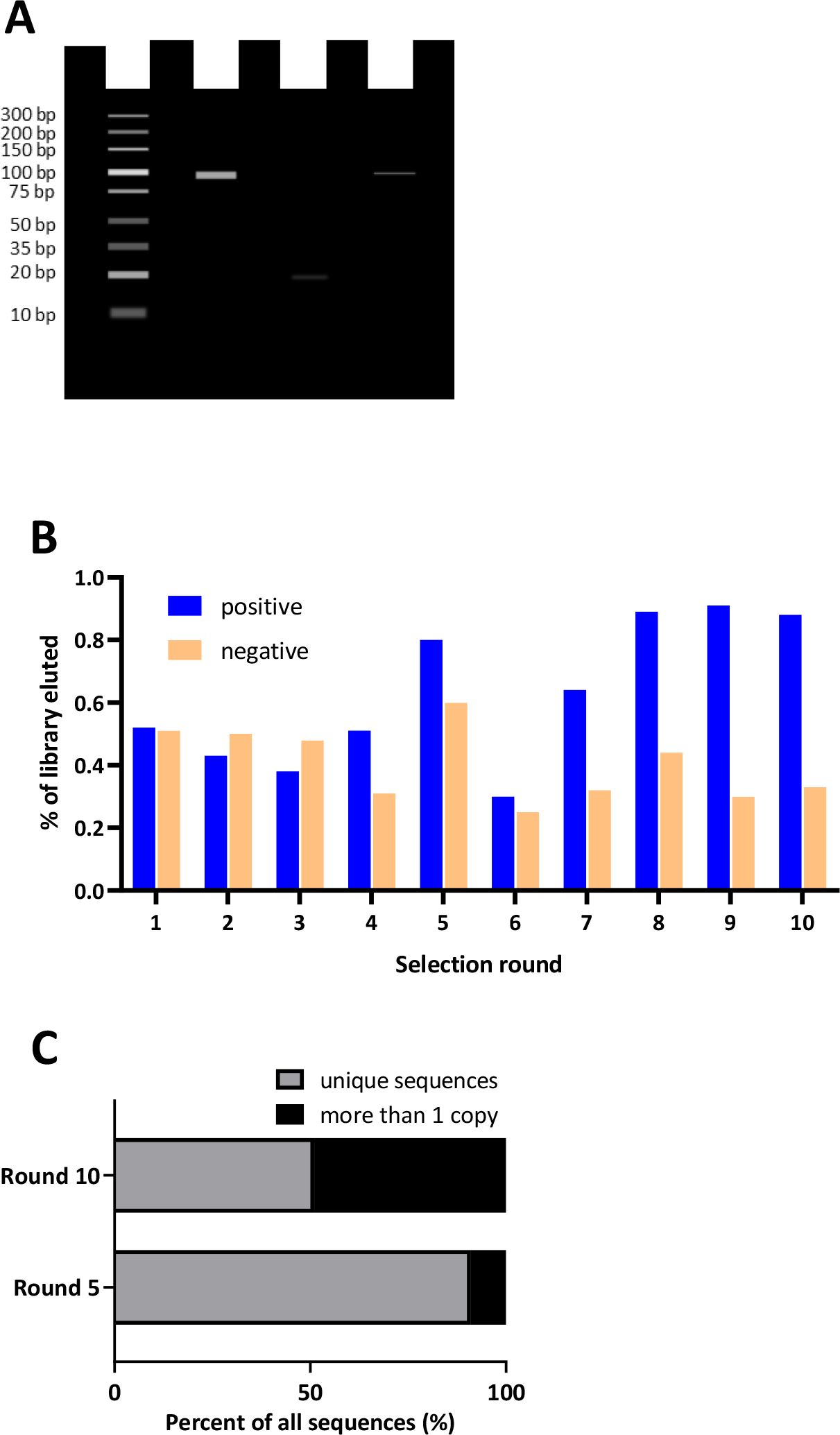
A. Ideal Polymerase Chain Reaction (PCR) amplification analyzed by gel electrophoresis. Lane 1 ladder, lane 2 positive control, lane 3 negative control, lane 4 amplification of the library following a round of selection. B. One method for monitoring the in vitro selection process is by comparing the quantity of library that interacts with the target compared to controls. Blue bars represent percent of library from each positive selection round (the target of interest); orange bars represent the library recovered from a parallel negative control selection round (absence of target). C. Landscape of sequence enrichment throughout rounds ofin vitro selection. In early rounds, the library is not enriched and therefore sequencing yields a diverse sequence pool with very few copies of the same sequence and are therefore mostly unique sequences. In later rounds, after enrichment has occurred, more sequences will emerge from the data that have numerous copies, thus resulting in fewer unique sequences.
Next, as the selection proceeds, it is critical to monitor the round-to-round selection progress by assessing both library diversity and relative affinity. The progress of SELEX should be assessed to understand the enrichment of winning aptamer sequences, to decide when to introduce more stringent conditions and to avoid amplification of nonspecific sequences in the SELEX pool. Several methods are available to assess SELEX progress.85–87 With the use of high-throughput sequencing (HTS – vide infra), the ability to deconvolute a SELEX library has become feasible. qPCR-based remelting curve analysis has also been used to track changes in sequence diversity as a surrogate for enrichment.71,88 However, because enriched sequences may not represent putative aptamers,89 it is ideal to also monitor and measure the relative affinity of the library to the target-of-interest.90 The method of assessment of affinity depends on the target and end-use of the aptamer. One simple indirect method to do this involves quantifying the amount of library that is recovered or eluted from the target each round compared to the amount of library recovered from a control experiment – such as a counter target or a negative control.91 As an alternative, in the later rounds, the entire library can be subjected to a head-to-head binding affinity experiment in bulk to determine the relative affinity of the enriched library to the target compared to the original library or a random sequence. For example, in a protein-SELEX experiment, affinity can be assessed by Surface Plasmon Resonance (SPR) analysis,92,93 biolayer interferometry94,95qPCR,96 enzyme-linked oligonucleotide assay,97 capillary electrophoresis,98 or flow cytometry43,99 and high-throughput sequencing43. The SELEX library can also be labeled with a fluorophore or 32P, while nitrocellulose filter binding100 or gel mobility shift assays101 can be utilized to assess the affinity of an evolved SELEX pool. When the target is a whole cell, the most common method of affinity analysis is flow cytometry73 While there is no definitive fold change or clear cut-off value to strive for (an enrichment value of 80% has been proposed as optimal in stopping SELEX43). Monitoring both the diversity and affinity throughout the entire selection process can provide important insight into the enrichment success, suggesting whether the stringency is suitable, provide encouraging support that high affinity and selective aptamer candidates are emerging in the library and help determine which sequences should be ultimately moved forward to the aptamer characterization phase. (Figure 3B shows one method of monitoring the enrichment process by comparing the quantity of library-target versus library-control interactions). Notably, high affinity aptamers that bind to small molecules have emerged from enriched libraries demonstrating as low as 3-fold increased bulk binding to the target compared to the negative control91, while a 10-fold or greater interaction is expected for aptamer selection against protein and cell targets compared to a negative control.22,96 On the other hand, sequencing results processed via the bioinformatics platforms may reveal as little as two102 or hundreds of potential aptamer families with fold enrichments varying between 1 (not at all) to 105 compared to the previous round (see Figure 3C for an example of library landscape comparisons used to show sequence enrichment throughout rounds of SELEX). As such, the more putative candidates that can be individually screened, the higher the probability of identifying a high-affinity aptamer.
Results
Sequencing and Bioinformatics
HTS is the most common method to sequence a SELEX library, allowing comprehensive coverage of all sequences in a SELEX library. Sequencing of selection libraries is typically achieved using HTS. The 454 Roche,103 was the first commercially available HTS platform, although it is no longer commonly used. More recent platforms include the Illumina platform,89 the SOLiD sequencer104 and the Ion Torrent sequencing platform105. Details of HTS chemistry are reviewed elsewhere.106,104 All sequencing platforms require converting the SELEX library to a double-stranded SELEX library with an adaptor sequence to facilitate sequencing. Such aptamers can be added to a SELEX library using commercial kits provided by vendors or by simply using the adapter sequences tethered to the primers utilized in SELEX. The Illumina sequencing platform is widely known for HTS in SELEX among other sequencing platforms.89 SELEX-derived libraries can be prepared for Illumina HTS by introducing a two-step PCR approach. First, PCR is performed to introduce overhang adapter sequences to an enriched library to amplify. This step is known as amplicon PCR. The amplicon PCR is purified to remove primers and primer-dimers, ready for the second PCR called index PCR. Indexing allows the pooling of several samples, providing a cost advantage when analyzing multiple samples. Upon completion of index PCR, the product is purified and then quantified or characterized by gel electrophoresis to verify the size. Quality control can be performed prior to sequencing. More recently, nanopore sensing of DNA has been described. Rather than rely on some variation of sequencing by synthesisDNA amplification, each individual DNA molecule is unraveled and passed through the nanopore, during which time a characteristic disruption in the current across the nanopore is converted into an electrical readout of the DNA sequence.107Though this method has a lot of potential it has not yet been widely incorporated into SELEX experiments...
Processing sequencing data requires bioinformatics approaches to analyze the information obtained from HTS. The processing of data depends on the sequencing method employed. However, in general, initial preprocessing is required for all sequencing data, including removing sequencing artifacts, removing constant regions, such as primer and adapter sequences, and performing filtration of sequences for expected length. Once the HTS data is preprocessed, readily available bioinformatics software platforms can be used to analyze sequencing data. Features of available bioinformatics platforms are outlined in Table 1.86,87,108–115 The most common approach of selecting aptamers involves looking at the fold enrichment of individual sequences as a function of the SELEX round. Levenshtein distance [G] is one of the most common strategies of clustering sequences, while the use of shared motifs, consensus sequences, and secondary structure can also be utilized to cluster and identify potential aptamer families. When modified libraries with UBPs are used, the SELEX libraries are first converted to natural libraries, followed by conversion of selected aptamers back to modified sequences using customized algorithms.116
Table 1:
Bioinformatic platforms utilized in analyzing SELEX libraries
| Software | Input file format | Operating System | Data Analysis | Clustering Method |
|---|---|---|---|---|
| Galaxy86,87 | FASTQ | Web Linux Mac OS Windows |
Preprocessing, enrichment ratio, read count, filtering and sorting data | Sequence-based Structure-based |
| FASTAptamer108 | FASTQ | Linux Mac OS Windows |
Preprocessing, enrichment ratio, read count, sequence clustering based on Levenshtein edit distance | Sequence-based |
| APTANI109 | FASTQ | Linux Mac OS |
Count enrichment, identifies binding motifs and target-specific aptamers | Structure-based |
| MEMESuite/ GLAM110 | FASTA | Web Linux Mac OS Windows |
Identifies sequence motifs | Sequence-based |
| MPBind111 | Plain text | Linux Mac OS |
Identifies sequence motifs and ranks potential binding of aptamers | Sequence-based |
| AptCompare112 | FASTQ | Linux Mac OS Windows |
Identifies sequence motifs and count enrichment | Sequence-based |
| MEMERIS113 | FASTA | Linux Mac OS |
Identifies sequence motifs and secondary structure analysis | Structure-based |
| RaptRanker114 | FASTQ FASTA | Linux Mac OS |
Preprocessing, count, enrichment, identifies motifs based on CapR algorithms | Sequence-based/Structure-based |
| AptaSUITE115 (AptaPLEX, AptaCluster, AptaTRACE, AptaMUT) | FASTQ FASTA | Linux Mac OS Windows |
Preprocessing, count enrichment, identifies binding and structural motifs | Sequence-based Structure-based |
Characterization methods
Once the lead aptamer sequences are deciphered from the sequencing data, there are a wide variety of methods that can be used to characterize structure, affinity and other key features of aptamers. Thorough validation of individual aptamer sequences and characterization of their metrics are essential for implementing functional aptamers into diverse applications.117 The most important aptamer property is its affinity for the target. Several other properties, such as specificity, structure, and stability may also be assessed, depending on the application.
Binding affinity
A plethora of assays are available to assess aptamer binding affinity, but no single technique can be considered generally applicable to all aptamer cases;31,117,118 therefore, multiple methods must be employed to fully validate aptamer binding affinity. The dissociation constant is used to describe the affinity, and represents the concentration in which 50% of the aptamer-target interaction dissociates to its separate constituents.119 Bioinformatics analysis suggests that both RNA and DNA aptamers produce high affinity binding. In contrast, the range of possible is dependent on the target type.35 As an example, a successful small molecule binding aptamer will display in the micromolar to high nanomolar range35, whereas aptamers to large targets such as proteins and cell surfaces can exhibit in the low nanomolar and picomolar range. Additional binding characteristics that are more rarely reported but can be very important for downstream applications include binding kinetics (on rate and off rate) and thermodynamics (change in enthalpy (ΔH) and entropy (ΔS) of binding), which can be measured using specialized methods such as surface plasmon resonance120 and isothermal calorimetry121, respectively.
Specificity/selectivity
Aptamer specificity is the second most important property of aptamers and thus must be carefully assessed. All the binding methods described to-date can be used to measure the or relative binding of the aptamers to a suite of targets that are similar in structure or function to the target-of-interest. See Figure 4A and 4B for examples of methods used to characterize aptamer binding and specificity.
Figure 4. Representative characterization techniques of aptamer candidates.
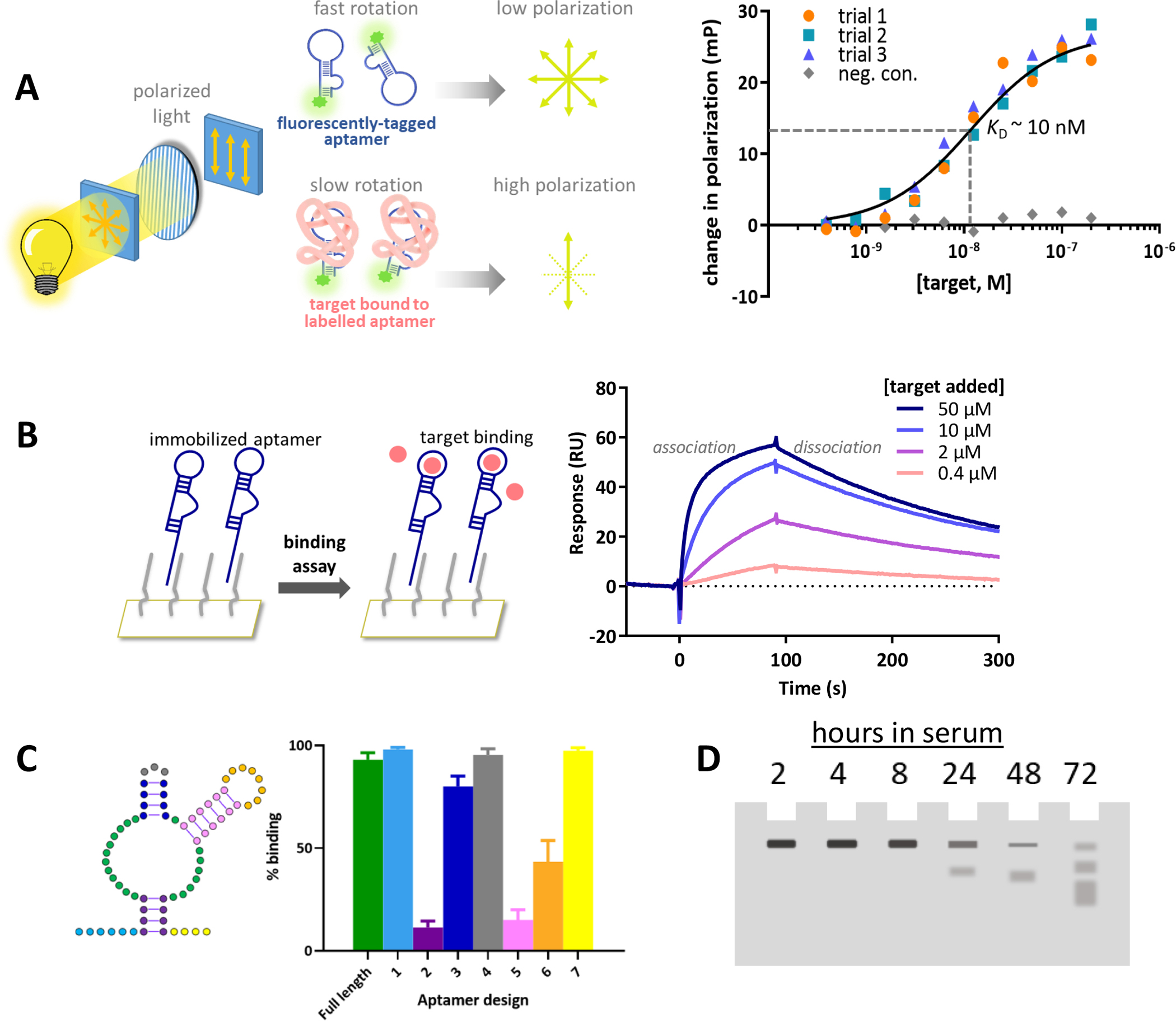
A) Aptamer binding measured using fluorescence polarization. The aptamer is modified with a fluorophore and is excited by polarized light. In solution, the aptamer rotates thus resulting in a depolarized emission of light. As increasing concentrations of the target are added and bind to the aptamer, the larger aptamer-target complex moves slower thus the emitted fluorescence remains more polarized. The change in polarization with increasing concentrations of target binding to the aptamer compared to a negative control sequence can be plotted as a binding isotherm to solve for the dissociation constant. B) Aptamer binding kinetics measured using a real-time surface plasmon resonance assay. The aptamer is immobilized to a surface and the target is flowed over a surface plasmon biosensor chip. The interaction at the surface is detected in real time resulting in on and off binding curves that can be fit to obtain kinetic data. C). Secondary structure prediction of an aptamer. By synthesizing truncated sequences, a simple binding assay can determine the importance of different regions of the full-length aptamer necessary for binding. Here, aptamer designs are synthesized with each colored region removed and then tested for relative binding compared to the full-length sequence. D). Aptamer stability and half-life measured using denaturing gel electrophoresis. A labelled aptamer can be incubated with serum or nucleases and loaded into a gel at various time points to determine the time required to fully degrade the aptamer of interest.
By comparing the binding affinity, it is common for aptamers to display up to 10,000 fold improved binding to the target of interest compared to similar compounds.122 Competition assays are also employed to demonstrate high affinity binding of the target-of-interest despite the presence of other interfering targets.22 In general, achieving 100% specificity [G] is unlikely, and thus a more reasonable goal is to achieving sufficient selectivity [G] needed for the particular application of interest. There are several selection strategies, post-selection methods, and application designs that can be implemented to help achieve maximal selectivity.7
Aptamer structure
Of the thousands of aptamers reported to-date, the length and sequence composition varies immensely.35 However, the functional aptamers described to date typically fold into only a few common structural motifs, with G-quadruplexes123 and stem-loops representing the majority. In fact, more complex aptamer structures, such as 5-way junctions, are very rare and not well-sampled in a typical SELEX library.124 Identifying the structural motifs or overall structure of aptamers is extremely useful in aptamer development. For example, knowledge of the structure can be used to create aptamer switches, to rationally modify the aptamer with chemical tags or other biomolecules, or to minimize the functional sequence to allow scalable synthesis at higher yields.
Several methods are available to probe aptamer structure in varying levels of depth and detail. As a starting point, almost all researchers will employ a variety of secondary structure prediction programs such as MFold and RNAstructure. However, most programs do not capture the entire global 3-D structure or interactions with the target molecules, and therefore require follow-up laboratory experimentation. The most common means for obtaining some structural information is by synthesizing a variety of truncated125 or mutated126 versions of a given aptamer sequence (Figure 4C) and performing a structural activity relationship study using a quick binding assay or by measuring the for each variant (see 127for example). Figure 4B shows a surface plasmon resonance binding assay set-up and the subsequent kinetic analysis from which a can be determined. Using this strategy, the initial aptamers emerging from a SELEX experiment, that are between 40 and 100 nucleotides in length, can be reduced to the minimal stable stem-loop or G-quadruplex scaffold representing as little as 15 nucleotides.35 Another frequent method involves circular dichroism (CD), which has long been employed to determine the signature of a given nucleic secondary structure or monitoring structural changes resulting from changes to the environment.128 As such, some aptamer structural insight can be gained using CD. In particular, CD is very powerful in identifying different G-quadruplex aptamers, where certain spectral features have been associated with different with quadruplex topologies.129 Finally, the gold standards for structural determination are NMR spectroscopy and X-ray crystallography and have been extensively reviewed.130 However these methods are labour intensive, and require specialized equipment, explaining why there are relatively few in vitro selected aptamer structures available via PDB.
Stability
A final parameter that is frequently characterized for therapeutic or in vivo applications is the stability of the aptamer. These studies are typically performed on chemically-modified aptamers given that RNA is highly unstable131 and even DNA aptamers are quickly degraded in vivo.132 It is relatively straight forward to assess aptamer stability: each candidate is incubated with different serum compositions or with specific nucleases, or in different environmental conditions with varying temperatures and pH for hours or days at a time. Then, the integrity of the full length sequence is examined via denaturing gel electrophoresis91 or by performing an affinity experiment to assess whether binding has been impacted.133 Figure 4D shows an example of aptamer stability characterization through denaturing gel electrophoresis analysis.
Applications
Aptamers can be antibody substitutes in most routine of applications in research and development 134. However, their distinct differences from antibodies also mean that they can be used in unique applications as well. An important advantage that aptamers have over antibodies is they are made of DNA, a predictable and programmable molecule. This can be exploited as aptamers themselves can be amplified, and they are compatible with multiple nanotechnologies that can amplify a molecular recognition event.135 Specifically, amplification strategies such as PCR, rolling circle amplification, strand-displacement amplification, and other methods can be used to directly amplify molecular recognition. Alternatively, aptamers can also be easily chemically modified, with an enzyme or nanoparticle for example, that can indirectly amplify molecular recognition. This section will delve into some of these examples in more detail, focusing mostly on cancer and drug delivery. It is not possible to review the entire breadth of aptamer applications herein. Generally, aptamers have found wide applications ranging from environmental monitoring136 to food safety137,138 to medicine,139,140 and many interesting reviews have been published on these topics.
Diagnostics
Aptamers have long been considered as replacements in applications typically the mainstay of monoclonal and polyclonal antibodies such as immunohistochemistry, immunophenotyping by flow cytometry and enzyme-linked immunosorbent assays (ELISA) 141. With the ease of adding functional reporter molecules on to the end of aptamers, such as fluorophores and biotin, aptamers have seen use in these applications 142–144.
There are numerous instances where complex staining protocols are required to determine the phenotype of specific cells, whether they are cells of the immune system or cancer cells. This can often be challenging with antibodies, due to the need for antibodies to be raised in different species or have a different isotype. The larger size of the antibody can also impede binding to cell surface receptors. Complex protocols have been generated that involve sequential antibody staining then stripping to accomplish this, even for fluorescent microscopy to ensure complete phenotyping 145. Three key unique properties of aptamers make this process less complex. Aptamers can be easily chemically synthesized, labelled with fluorophores during synthesis with limited or no loss of functionality (Figure 5A and 5B) and they are much smaller than antibodies, meaning more aptamers can attach to different receptors on the cell surface. This latter property could also lead to an increase in sensitivity 146. Aptamers can be easily adapted for functional cell sorting via magnetic beads or FACS. The benefits of using aptamers are that the aptamer can be easily removed from the cell surface, either through nuclease treatment or the use of an antidote sequence that is complementary to the aptamer, allowing normal receptor functionality for downstream applications, including cell transplants that are contraindicated in antibody-based separation strategies 147.
Figure 5:
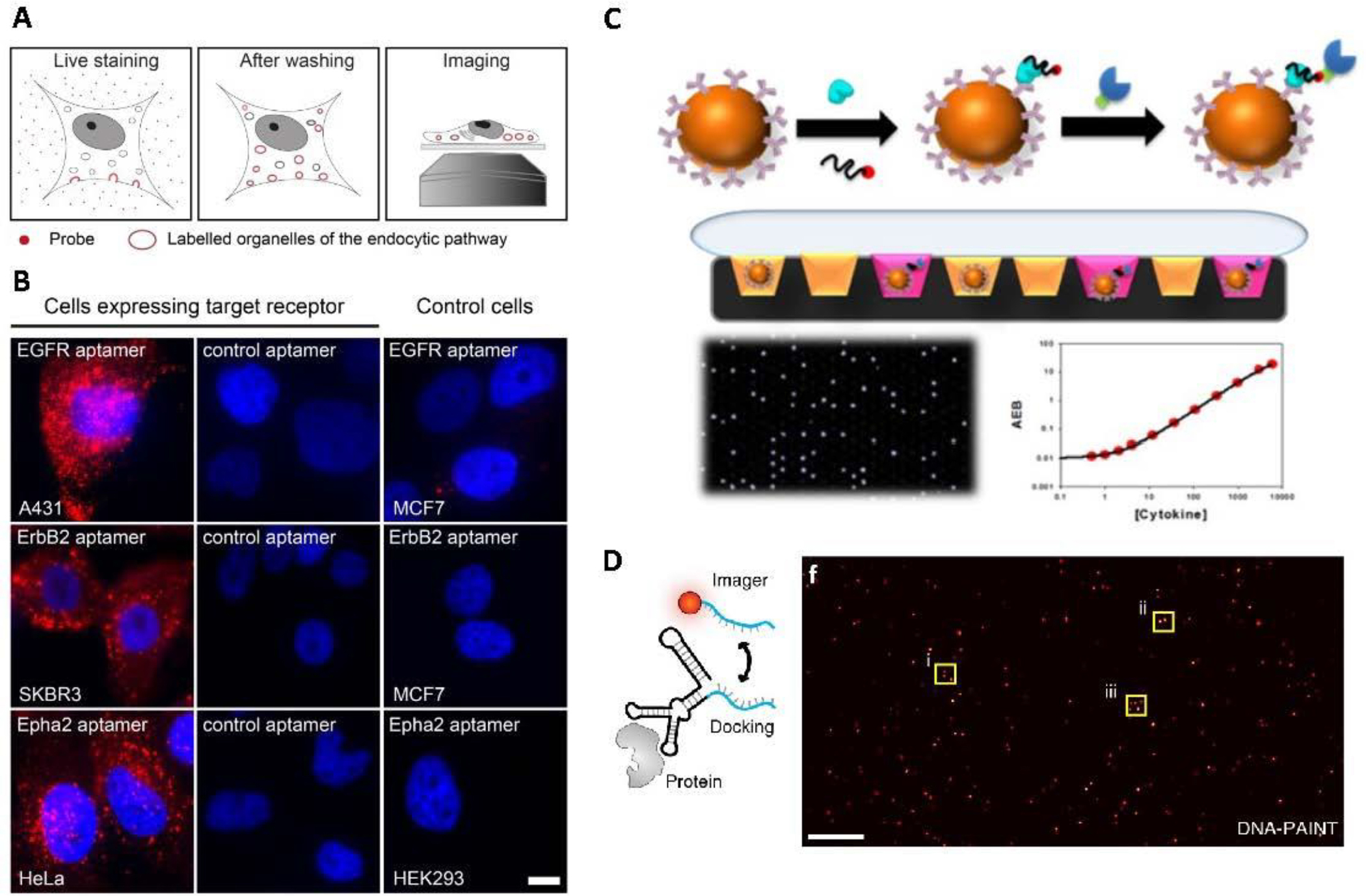
Aptamers are powerful tools for molecular imaging. A) Schematic of staining live cells with fluorescently labeled aptamer. B) Aptamers that bind to epidermal growth factor receptor (EGFR), and two other proteins were compared to control cells to demonstrate the ability of aptamers for molecular imaging.. C) Schematic illustration of the Simoa assay,. Briefly, capture antibodies are immobilized on microbeads, which are then incubated with the target protein and biotinylated SOMAmer. Finally a streptavidin modified beta–galactosidase enzyme is added. The beads are distributed in microwells, and the presence of the target is revealed in the presence ofresorufin β-D-galactopyranoside substrate. Digital detection (black panel with lit up wells) is then translated into a concentration based reading (calibration curve shown). D) Schematic illustration of EGFR labelling by SOMAmer. The SOMAmer binds to the target protein, and a fluorescently labelled imager strand binds to a docking region of the SOMAmer. The SOMAmer DNA-PAINT based method allows for sensitive fluorescent detection of target protein expressed in cells. Panel A and B were reproduced under Creative Commons Attribution License from Gomes de Castro et al., 2017146. Part C reproduced with permission from Wu et al., 2016150. Part D Reproduced with permission from Strauss et al., 2018.157
One area where aptamers could surpass expectations and overtake antibodies is in single molecule array assays (Simoa).148 These are adaptions of ELISA that offer increased sensitivity. The basis of the technique is that the molecule is captured by a paramagnetic bead labelled with a ligand that recognises the small molecule. An excess of beads is included to ensure that only one or no molecule binds per bead. A second biotinylated ligand that recognises the molecule is then added to the mix and they are then incubated with a fluorogenic substrate solution and the mixture is loaded into an array of microwells that can only hold one bead each. The enzymatic reaction proceeds, which is then detected with a CCD camera 149. Biotinylated slow off-rate aptamers (SOMAmers) to six cytokine targets 150 were recently tested using this approach (Figure 5C) and showed an ultralow detection limit and comparable sensitivity to the antibody pairs, while also minimising the issues associated with antibodies of cross-reactivity 151. It is important to note that it is rare to be able to mutate antibodies to increase both affinity and specificity, and that this can be a time consuming process 152. While mutating aptamers is not without these same problems, it can be a much easier and quicker process 153,154.
Another single molecule application studies the interaction in cellular events that are initiated through ligand-receptor interactions at the plasma membrane. For successful localisation to occur, the fluorophore must be as close to their targets as possible, which is achievable as aptamers are typically far smaller than antibodies. A study in 2012 compared aptamers with antibodies for this purpose and demonstrated superior performance of the aptamers. In addition, as aptamer size can be modified through truncation or coupling additional nucleotides, the authors found that a size of 15 kDa or lower led to superior image quality 155. This technique has also been used to visualise individual insulin receptors in the plasma membrane and study real-time dynamics 156, as well as imaging and tracking of individual epidermal growth factor receptor (EGFR) molecules in their native state in live cells, as shown in Figure 5D 157. The mutation of an aptamer for lower binding affinity and faster dissociation rate from EGFR to allow for no perturbation of endogenous activity and faster tracking without the need for photobleaching were also observed 158.
On the topic of small molecules, it is generally considered easier to develop aptamers to these rather than antibodies, owing to the inherent low immunogenicity of small molecules, resulting in an upsurge in reports on biosensors that utilise aptamers, (aptasensors). The challenge with small molecule detection is that their size often means that they only have one binding region, which has made it challenging to develop highly sensitive sensors. One development in the field of aptamers has been the split aptamer, pioneered in 2000 159. In this application, a complete aptamer is split into two or more aptamer fragments and denoted as Apt1 and Apt2. When the target is then incubated with them, Apt1 and Apt2 are drawn close to each other, thus activating a reporter molecule for optical aptasensors, or the refractory index for surface plasmon resonance or electrochemiluminescence aptasensors 160. The additional benefit of these aptasensors is the stability of the aptamers, meaning they can be used for in-field testing without the requirement for cold-chain.
An interesting development in the selection of aptamers was the ability to generate aptamers to fluorophores, leading to several new aptamers named after vegetables and fruits (See Figure 6). These RNA aptamers exhibit fluorescence when bound to small molecules and have allowed the imaging and tracking of RNA in living cells. The first of these to be developed was named Spinach and was selected to the green fluorescent protein derivative, 3,5-difluoro-4-hydroxybenzylidene imidazolinone (DFHBI) 161. Owing to some instabilities of the original aptamer, mutagenesis led to the development of Spinach 2 162 and truncation generated Baby Spinach 163. The Broccoli aptamer was generated to DFHBI, with a superior fluorescence when compared to Spinach 2 164. Other aptamers have also been developed that produce yellow, red, and orange fluorescence and a full list is provided in Table 2.
Figure 6:
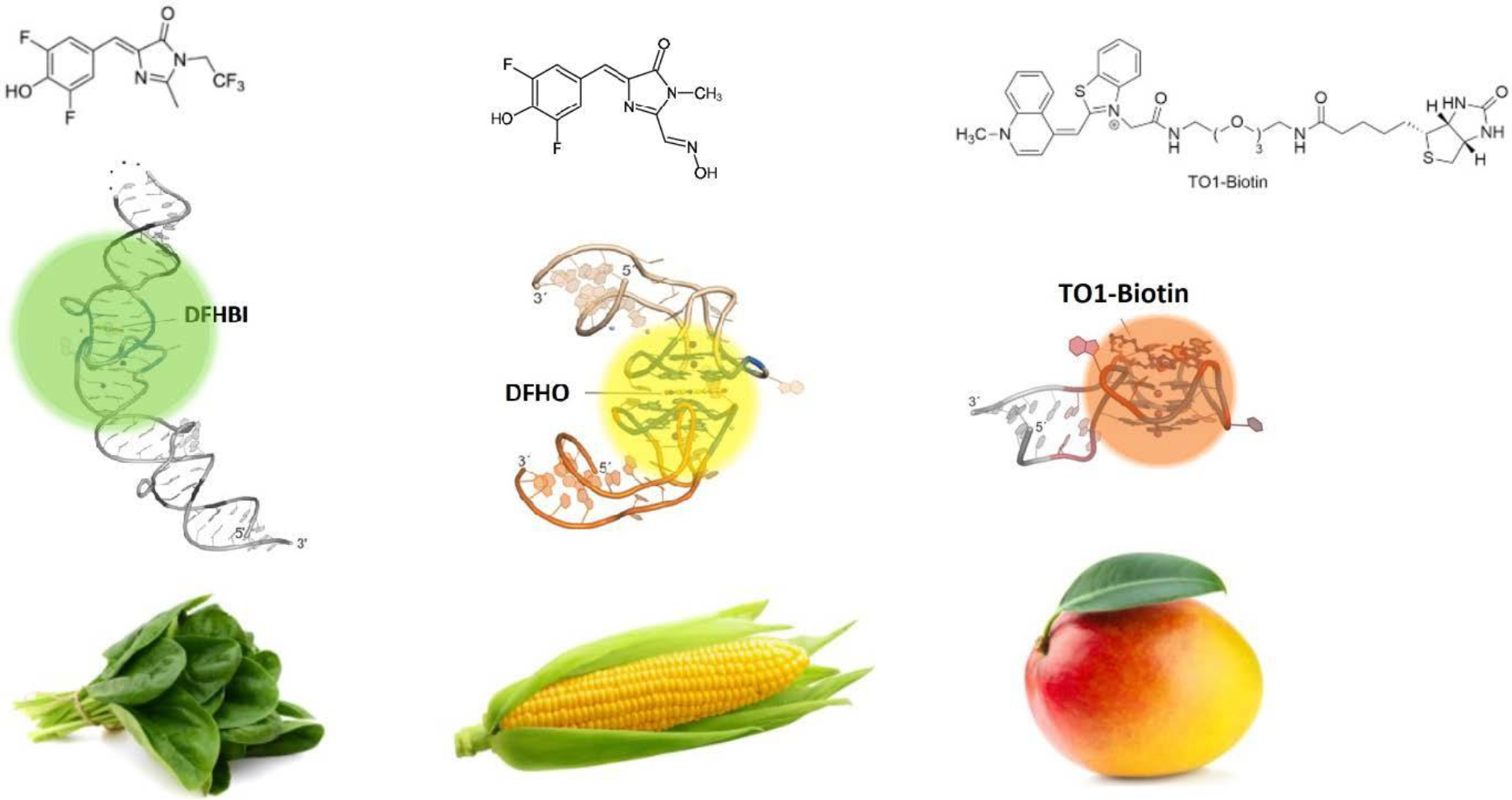
Aptamers that have been selected to bind to light up fluorophores When bound to a fluorogen (DFHBI, DFHO, and TO1-biotin), aptamers can substantially enhance their emitted fluorescence. The spinach (a), corn (b), and mango (c) aptamers are shown Adapted from Neubacher and Hennig (2018). 237
Table 2.
Aptamers that have been selected to bind to “light up” fluorophores, the resulting fluorescence color, and their targets
| Colour | Name | Target | Reference |
|---|---|---|---|
| Green | Broccoli | 3,5-difluoro-4-hydroxybenzylidene imidazolinone | 164 |
| Green | Spinach | 3,5-difluoro-4-hydroxybenzylidene imidazolinone | 161 |
| Green | Spinach 2 | 3,5-difluoro-4-hydroxybenzylidene imidazolinone | 224 |
| Green | Baby Spinach | 3,5-difluoro-4-hydroxybenzylidene imidazolinone | 163 |
| Green | Bunch of baby spinach | 3,5-difluoro-4-hydroxybenzylidene imidazolinone | 163 |
| Yellow | Corn | 3,5-difluoro-4-hydroxybenzylidene-imidazolinone-2-oxime | 225 |
| Red and Orange | Red Broccoli | 3,5-difluoro-4-hydroxybenzylidene-imidazolinone-2-oxime | 225 |
| Red and Orange | Orange Broccoli | 3,5-difluoro-4-hydroxybenzylidene-imidazolinone-2-oxime | 225 |
| Red and Orange | Mango | Thiazole orange | 226 |
Advancements in technology have also opened up an attractive and novel area of wearable biosensors 165 that are non-invasive and collect health information rapidly. While this is a relatively new concept, it would revolutionise our understanding of health. Issues with real time monitoring include, but are not limited to, drift and the need to perform several calibrations a day, no sample preparation, stability with high signal to noise ratios and biofouling. One of the first devices demonstrated that aptamers could be readily used in the microfluidic electrochemical detector for in vivo continuous monitoring (MEDIC) for the real-time monitoring of doxorubicin in human whole blood, as well as live animals drawing blood into the chip using an indwelling catheter 166. Further updates to the technology using a dual-frequency approach allowed for the continuous real-time monitoring of doxorubicin and cocaine for 8 hours 167. Further work has been performed to identify sensors capable of real-time monitoring of cytokines 168,169
Therapeutics
Aptamers have shown great potential as therapeutic agents in preclinical trials, with some aptamers entering clinical trials, either as the therapy itself, as a diagnostic, or as part of biomarker discovery. To date, however, only one aptamer has been approved for use by the US FDA. Macugen was approved for use for age-related macular degeneration in 2004 following successful clinical trials. According to the U.S. National Library of Medicine’s online database, there are a total of 54 registered studies involving aptamers. Consistent with Macugen, the studies that have progressed the furthest, into phase 3 or phase 4, are mostly for conditions relating to the eye (macular degeneration, macular edema, and diabetic retinopathy). Certainly, the next few years may see a boom in oligonucleotide therapeutics with the approval of the mRNA vaccines and the success of antisense oligonucleotides, however there are still several associated challenges to explore collectively for oligonucleotide therapeutics and individually for each technology.
There are certain advantages that make aptamers particularly suited for therapeutics. Typically, aptamers are suggested to be a much safer version of monoclonal antibodies owing to the lack of or very low immunogenicity. However, as more has become known about immune cell triggers, it has been discovered that aptamers can also be immunogenic if the right sequence is chosen, suggesting they can also be used for immunotherapy 170. Another major advantage is that aptamers are much smaller than antibodies, which means better penetrance away from blood vessels. This is especially important for the targeted delivery of chemotherapeutic agents for solid tumors 171. Finally, the tunable binding affinity of aptamers also offers advantages for receptors that are over-expressed on the target population but are expressed at a lower level on normal healthy cells. Table 3 lists some of the uses of aptamers as therapeutic agents with key references provided.
Table 3.
Therapeutic applications of aptamers. Further information can be found in these review articles184,227–229
| Modality | Example | Target (Aptamer) | Reference |
|---|---|---|---|
| Aptamer | Antagonist | Nucleolin (AS1411) | 230 |
| Aptamer | Blocking receptor for viral entry into cell | Hemagglutinin (HA12-16) | 231 |
| Aptamer-drug conjugates | Targeted delivery of chemotherapeutic agent (Doxorubicin) | Prostate specific membrane antigen (A10) | 232 |
| Aptamer-radioligand conjugates | Targeted delivery of radiotherapeutic agents for imaging | Tenascin-C (TTA1-99mTc) | 233 |
| Aptamer-RNAi | Targeted delivery of short interfering RNA | Prostate specific membrane antigen for knockdown of Lamin A (A9) | 234 |
| Aptamer-RNAi | Targeted delivery of microRNA | Tyrosine kinase Axl for knock-in of tumor suppressor gene (GL21.T-let7g) | 235 |
| Aptamer functionalised nanoparticles | Targeted delivery of liposomes | CEM-CCRF cells (Sgc8) | 183 |
| Aptamer functionalised nanoparticles | Targeted delivery of gold nanoparticles | Prostate specific membrane antigen (A9) | 236 |
Perhaps one of the most striking advantages of aptamers in therapeutics is the availability of a ready-made antidote (complementary sequence) 172 for the aptamer. Aptamer/antidote pairs have mostly been developed for the use in haematology as anti-coagulant agents173. The first aptamer/antidote pair to enter clinical trials was pegnivacogin (aptamer)/anivamersen (antidote). Success was seen in terms of efficacy and safety in Phase 1 and 2a trials. This was also observed in a Phase 2b clinical trial, where it was compared to unfractionated heparin. Unfortunately, a large Phase 3 clinical trial was halted in 2014 owing to a small number of allergic reactions, though these were likely caused by the polyethylene glycol modification rather than the aptamer itself 174. In fact, a similar allergic reaction was observed upon administration of COVID-19 vaccines containing polyethylene glycol.175 Regardless of the challenges associatied with the delivery vehicle, these studies indicate the uniqueness of aptamers and how easy the generation of antidotes can be.
Theranostics
The combination of diagnostic and therapeutic agents is another area where aptamers are demonstrating their utility.176 Molecular imaging is required to diagnose various diseases and can be used to assess treatment efficacy. This field has seen several technological developments where imaging has progressed from radioisotopes to magnetic particles to fluorescence-based assessment. While some of these modalities are currently restricted to animal models for classical imaging,177–179 they are being pioneered for intraoperative uses 180 Traditional radiolabelling of aptamers is typically a simpler process than for antibodies, given the temperature stability 181. Conjugation to nanoparticles can use the same chemical reactions for attachment or through click-chemistry, with companies such as Baseclick™ licensing their technology. Depending on the attachment to the aptamer, these can be used for molecular imaging of the tumour and treatment at the same time, which offers benefit to the patient. These attachments can include lutetium, a radioisotope that emits both beta and gamma particles or specific types of nanoparticles that have both diagnostic and therapeutic capabilities. There is also potential for aptamer chimeras; bivalent molecules that contain a therapeutic aptamer and a delivery aptamer in one, or therapeutic OR delivery aptamer conjugated to another type of functional nucleic acid.182,183Some of the aptamers and their mechanism of action are listed in Table 3, with a full review provided in this reference 184.
Not strictly fitting in the theranostics field but providing a combination of diagnostic and therapeutic effects is the use of a companion diagnostic test, a principle first drafted by the FDA in 2016. In terms of targeted therapeutics, it is important to demonstrate the presence of the receptor on the target cell population. Typically, this would involve a biopsy being taken from the patient for confirmation of the presence, using basic diagnostic techniques. In the case of antibodies, it is possible that one antibody may be used for diagnostics but a different one used for therapy as it may not be possible to functionalize the first antibody, or it may not show favorable kinetics for in vivo use. As the antibodies may bind to different epitopes of the receptor, a false positive result may be observed, resulting in treatment failure. As aptamers can be easily functionalized, the same one can be used for both applications 185,186.
Reproducibility and data deposition
Reproducibility
The interest in aptamer selections has grown, producing many synthetic ligands with antibody-like properties. However, scrutiny has been directed on certain aptamers owing to lack of reproducibility and with only one aptamer achieving clinical success.187,188 Several reports offer minimal publication guidelines189 or specific guidelines to assess aptamers selected against cell-surface proteins,190–192 There are two primary contributing factors in aptamer’s binding properties. Importantly, an aptamer’s structure is highly variable, and its binding and function depend on several factors.193 It is essential that the correct buffer systems are used to ensure a uniform functional fold of aptamers. Variations to buffer composition, such as the concentration of divalent metals, which is known to stabilize the secondary structures, can impact generating the uniform functional fold of the aptamer. Furthermore, the three-dimensional structure of the target governs the aptamer binding; maintenance of the structure of the target presented during SELEX is essential in reproducing the aptamers’ similar binding and functional properties. Variations of the structural stability of the target are often disregarded in many SELEX studies, even though the structures of targets can also change based on salt and pH.194,195 One way of minimizing these variabilities is to use multiple controls and conditions when assessing the aptamer binding followed by adequately reporting these conditions. To assess the background and propensity for non-specific nucleic acid binding to the target, use of a scrambled sequence of the aptamer is advisable. Moreover, binding should be assessed using multiple related samples such as isoforms of the protein used196 structurally similar small molecules117 or multiple cells lines that express the same protein to a different degree; cell lines that do not express the same protein must be used to ensure that the aptamer’s binding is specific.197 Finally, the specificity of an aptamer can be assessed by investigating the aptamer’s biochemical function.198 Competition assays using a secondary ligand against the same molecule or cross-competition experiments between aptamers from the same family can provide further clarity.57,199 Additionally, interlaboratory testing117 can be a powerful approach to confirming reproducibility. The field would benefit greatly from a database that is like the protein data bank. In this database, each aptamer could be given a unique identifier, like an accession number, that would allow all experimental data to be linked to the sequence. Further, the requirement of this sort of entry before publication would go a long way in improving the reproducibility of aptamer data.
Data deposition
Several aptamer data deposition platforms have been made available for data related to aptamer sequences and other parameters. These databases, demonstrated precursors to an aptamer data bank, have attempted to collect the data from multiple selections, and offer researchers varying degrees of access to the selection information. The aptamer database initiated by the Ellington lab consisted of comprehensive sequence information on aptamers and ribozymes identified using in vitro selection methods.200 PPAI, is a web-based server allowing users to predict aptamer and protein-aptamers interactions.201 RiboaptDB, includes comprehensive sequence information on aptamers and ribozymes generated using in vitro selection experiments.202 Biclusters allows users to identify similarities with aptamer sequences that have been reported in other studies.203 Aptamer base was developed to allow investigators to report and access detailed information on experimental conditions and SELEX outcomes.204 With the incorporation of HTS, currently, it is possible to deposit all sequences in SELEX libraries and the identified aptamer sequences in the Sequence Read Archive of the National library of medicine as reported in ref 43. Two significant challenges exist with the maintenance of these databases, the first being that there is no recognized centralized database, so information curation is up to a subset of researchers. The other significant challenge of many of the aptamer databases has been keeping them current, as several have gone dormant over time.
Limitations and optimizations
Since the establishment of the SELEX method, thousands of aptamers have been developed to a wide range of different targets, demonstrating the remarkable power of this approach. However, in most cases the SELEX process remains relatively blind to the researcher, yields low affinity aptamers to certain targets (such as small molecules), is low throughput and sometimes does not produce application-ready aptamers. Although some challenges are due to lack of resources, most limitations can be optimized by more judicious experimental planning.205 The past three decades have seen numerous modifications to the SELEX method with the goal of addressing some key limitations.
First, the SELEX process consists of so many different parameters and oftentimes limited predictive understanding of which parameters should be tuned for a given target or application of interest.35 As such, an entire selection experiment, using published protocols may not see enrichment nor yield desired aptamers.102 The way forward is to select aptamers with an end application in mind, and to combine methods that monitor the evolving affinity, diversity and enrichment of the library throughout the entire SELEX experiment.206,207 Additionally this information should be used to tune the stringency of selection, PCR amplification, number of rounds and counter selections. Largely this becomes a challenge of resources. If researchers could easily (and inexpensively) sequence every round, and monitor enrichment by real-time PCR, the selection could be adapted in real time to improve stringency and increase the probability of success. With next generation sequencing technologies becoming more accessible, quick and cheap sequencing may eliminate several barriers to successful selections.
Second, SELEX is low throughput. Most new aptamers described represent several years of effort, involving iterative trial-and-error experiments. In an effort to overcome the bottleneck of low-throughput experimentation, there has been remarkable progress involving the use of particle display,208,209 microfluidics,210,211 and automation212,213 that can be leveraged for multiplexed aptamer selection. For example, a new selection procedure called de novo rapid in vitro evolution of RNA biosensors (DRIVER) uses aptamer-coupled ribozyme libraries to discover aptamer sequences in high throughput. 214 Additionally, the success and efficiency of SELEX experiments is often limited by availability of bioinformatic and computational tools. Interdisciplinary research is bridging the gap between bench science and big data. An example in aptamer selection experiments was the switch from cloning and sanger sequencing, where a small subset of sequences (typically <100) from the initial and final rounds were examined, to high throughput sequencing methods where millions of sequences from each round were computationally processed for trends in enrichment, secondary structure and common motifs.
A third critical limitation of SELEX is that aptamers are often not immediately ready for their application of interest. In particular, the SELEX process usually involves identifying candidates in an ideal buffering environment and using highly purified targets. However, many applications require function at a variety of temperatures and pH, in the presence of organic solvents and in highly complex environments with molecular crowding and mechanical forces at play. Several aptamers function in complex matrices such as waste water, river water, or soil,91, 136, 215, 216, but most aptamers must be heavily adapted, truncated, and/or modified to function in these differing environments. A more recent trend is to perform the selection experiment using conditions that closely mimic the environment of the intended application. This approach is especially important for health related applications that are not biosensing in nature. If aptamers are going to work in a live cell or animal they should be selected in conditions that mimic these environments as closely as possible, and very likely will require post-selection modifications for increased biostability. Most aptamer applications fall into biosensing, likely because biosensing conditions more closely resemble a simple buffer system. A future goal could be to continue to innovate SELEX approaches to minimize post-SELEX modifications and adaptations to yield a functional end-product from environmental monitoring to oligonucleotide therapeutics.
The final challenge is that academic researchers are often concerned with demonstrating the innovation of aptamer technology leading to novel assay designs that are overly complex and pay little attention to real world applicability, thus negating the inherent benefits of aptamer technology. Aptamers can take advantage of their nature as oligonucleotides to conceive of new uses and assays designs that would not be possible with antibody counterparts. Moreover, aptamers will always be well-suited for applications that benefit from their low cost of production and their extended shelf-lives, for example to develop economical screening technology for testing that require extensive sampling. Aptamers have been compared to antibodies since their earliest days, yet conventional antibody-based applications are well-established and their supremacy in certain applications is uncontested. Finding ways to capitalize on the distinct advantages of aptamers and to find unique niches for their application are keys to their future success.
Outlook
The last thirty years have seen enormous progress in aptamer research. The priorities for aptamer selections and applications for the next 5–10 years should be to address the select limitations of aptamers, to focus on areas of strength and to push for adoption and commercialization of aptamer technology. Despite some limitations, there is enormous potential for aptamer selections and applications. Only recently have new aptamer publications reported in depth analysis of selection experiments, in part due to the quantity of data afforded by next generation sequencing. However, with numerous bioinformatics tools available, such as in silico molecular docking (for aptamer design)84 the application of machine learning to aptamers (to improve library design and sequencing data analysis),217,218,219 and other computational methods220–222 it is expected that understanding of the selection process will improve, further improving reproducibility and enabling the strengths of aptamer technology. Moreover, like all realms of life, machine learning, artificial intelligence, and other computational methods likely lie at the intersection of challenge and opportunity. Predictably, the time will come that machine learning and AI may be able to predict aptamers for specific targets, but the success of such endeavors will most likely be dependent on careful and thorough curation of aptamer and selection data in a centralized data bank.
While translation of the strengths of aptamer technology from the lab into the real world has been somewhat elusive to date, the future looks bright. The expiration of the original patents in the early 2010s sparked an increase in academic and commercial interest in aptamer applications and the last ten to fifteen years have been marked with exciting advances in aptamer-based technology. Aptamer-based commercial products have emerged including New England Biolab’s anti-Taq aptamers for Hot-Start PCR, and SOMAlogic’s Slow-Off rate Modified Aptamer (SOMAmer) technology for proteomics platforms, culminating with the recent listing of SOMAlogic on the NASDAQ in 2021. As illustrated by the extensive demonstration of aptamers in biosensing for environmental monitoring and food safety, the best practice is to keep things simple. A renewed emphasis on simple, portable and inexpensive screening assays could allow aptamers to profit from their strengths. Gleaning potential from the COVID-19 pandemic, there is clear need for rapid and robust solution and paper-based assays that can be shipped and stored at room temperature for extended periods of time. To reiterate an advantage aptamers have over antibodies to this end is that aptamers are much more chemically stable and compatible with devices that meet these criteria.223 Realizing the full promise of aptamer technology will hinge upon the rational design of effective SELEX experiments, a continued focus on aptamer reproducibility and the design of applications that take full advantage of all the assets that aptamers can offer.
Supplementary Material
Acknowledgements
The authors thank the editorial team and the reviewers for their valuable feedback in the preparation of this article. MCD and MM thank NSERC for Discovery Grant funding. MM thanks the CRC program for her Tier II Research Chair. PM acknowledges grant #R35 GM139336 from NIGMS.
Glossary
- Heterogeneous library
The pool of oligonucleotide sequences from which aptamers are selected, typically consisting of a random nucleotide core of between 20–60 nt, flanked by fixed regions used for library amplification. In a typical SELEX experiment, the library contains 1014-1015 random sequences (1–10 nmol)
- Partitioning
The process by which target binding oligonucleotide sequences are separated from non-binding sequences.
- Stringency
Selection pressure used during the SELEX experiment to increase the competition between strong and weak binders and ultimately increase aptamer suitability (affinity and specificity). Stringency can depend on several factors, including the partitioning method, target concentration, buffer composition, counter selection rounds
- Positive-selection
the incubation of the selection library with the desired
- Counter- selection
Incubation of the library with a similar molecule(s) allows for increased stringency as sequences that have cross reactivity to the similar molecule(s) is removed from the selection. Typically the counter-selection would occur after a negative selection and before a positive selection
- Negative-selection
It is important to remove sequences from the library that may bind to the selection matrix (such as magnetic beads, agarose beads, nitrocellulose paper). This is achieved by incubating the library with only the selection matrix prior to the positive selection
- Enrichment
A measure of the progress of a selection, library enrichment describes the degree of convergence of the library on sequences with the desired properties
- Selectivity
the ability of an aptamer to differentiate between components in a mixture
- Sequence Space
A library synthesis yield of 1 nmol of DNA would be composed of approximately 6 × 1014 sequences. A library with a random region of 24 nt long would have 424 different possibilities = 2.8 × 1014 sequences. If the random region is increased by 1 nt, the number of possibilities (425) = 1.1 × 1015. Thus, any SELEX library designed with a random region longer than about 25 nt will likely not contain every possible random sequence. Nevertheless, there may be reasons to consider a longer library length. There is some evidence to suggest that longer pools could be better for smaller targets.28 Furthermore, libraries built from longer oligos will have a greater diversity of possible 3D structures.
- Specificity
the ultimate selectivity, the ability of an aptamer to interact with only a single component in a mixture
- Levenshtein distance
an algorithm that measures the difference between two sequences. In sequence comparison, the number of substitutions, insertions or deletions will affect the value. A common example of where this is used is for the BLAST: Basic Local Alignment Search Tool maintained by the NIH
Footnotes
The authors declare no competing interests.
Peer review information
Nature Reviews XXX thanks [Referee#1 name], [Referee#2 name] and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Related links
Galaxy: https://usegalaxy.org/
FASTAptamer: https://burkelab.missouri.edu/fastaptamer.html
APTANI: http://aptani.unimore.it/downloads.php
MEMESuite: https://meme-suite.org/meme/doc/download.html
MPBind: https://morgridge.org/research/regenerative-biology/software-resources/mpbind/
AptCompare: https://bitbucket.org/shiehk/aptcompare/src/master/
MEMERIS: https://cs.stanford.edu/people/hillerm/Data/MEMERIS/
RaptRanker: https://github.com/hmdlab/RaptRanker
AptaSUITE: https://drivenbyentropy.github.io/
Supplementary information
Supplementary information is available for this paper at https://doi.org/10.1038/s415XX-XXX-XXXX-X
References:
- 1. Tuerk C & Gold L Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249, 505–510 (1990). a. One of the original three selection/aptamer papers, this work that describes in vitro selection of RNA aptamers that binds T4 DNA polymerase that coined the term ‘SELEX’, is required reading by anyone interested in the aptamer field.
- 2. Ellington AD & Szostak JW In vitro selection of RNA molecules that bind specific ligands. Nature 346, 818–822, doi: 10.1038/346818a0 (1990). a. One of the original three selection/aptamer papers, this work that describes in vitro selection of RNA aptamers that binds organic dyes and coins the term ‘aptamer’, is required reading by anyone interested in the aptamer field.
- 3.Mills DR, Peterson R & Spiegelman S An extracellular Darwinian experiment with a self-duplicating nucleic acid molecule. Proceedings of the National Academy of Sciences 58, 217–224 (1967). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Robertson DL & Joyce GF Selection in vitro of an RNA enzyme that specifically cleaves single-stranded DNA. Nature 344, 467–468, doi: 10.1038/344467a0 (1990). a. One of the original three selection/aptamer papers, this work that describes in vitro selection of an RNA enzyme that can cleave single-stranded DNA, is required reading by anyone interested in the functional nucleic acid field.
- 5.Jarmoskaite I, AlSadhan I, Vaidyanathan PP & Herschlag D How to measure and evaluate binding affinities. eLife 9, e57264, doi: 10.7554/eLife.57264 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jing M & Bowser MT Methods for measuring aptamer-protein equilibria: a review. Anal Chim Acta 686, 9–18, doi: 10.1016/j.aca.2010.10.032 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kalra P, Dhiman A, Cho WC, Bruno JG & Sharma TK Simple Methods and Rational Design for Enhancing Aptamer Sensitivity and Specificity. Front Mol Biosci 5, 41, doi: 10.3389/fmolb.2018.00041 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weill L Selection and evolution of NTP-specific aptamers. Nucleic Acids Research 32, 5045–5058, doi: 10.1093/nar/gkh835 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu H, Zhang S & Chaput JC Darwinian evolution of an alternative genetic system provides support for TNA as an RNA progenitor. Nat Chem 4, 183–187, doi: 10.1038/nchem.1241 (2012). [DOI] [PubMed] [Google Scholar]
- 10.Pinheiro VB et al. Synthetic genetic polymers capable of heredity and evolution. Science 336, 341–344, doi: 10.1126/science.1217622 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tahiri-Alaoui A et al. High affinity nucleic acid aptamers for streptavidin incorporated into bi-specific capture ligands. Nucleic Acids Res 30, e45, doi: 10.1093/nar/30.10.e45 (2002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burmeister PE et al. Direct in vitro selection of a 2’-O-methyl aptamer to VEGF. Chem Biol 12, 25–33, doi: 10.1016/j.chembiol.2004.10.017 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Jellinek D et al. Potent 2’-amino-2’-deoxypyrimidine RNA inhibitors of basic fibroblast growth factor. Biochemistry 34, 11363–11372, doi: 10.1021/bi00036a009 (1995). [DOI] [PubMed] [Google Scholar]
- 14.Barciszewski J, Medgaard M, Koch T, Kurreck J & Erdmann VA Locked nucleic acid aptamers. Methods Mol Biol 535, 165–186, doi: 10.1007/978-1-59745-557-2_10 (2009). [DOI] [PubMed] [Google Scholar]
- 15.Ichida JK et al. An in vitro selection system for TNA. J Am Chem Soc 127, 2802–2803, doi: 10.1021/ja045364w (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eremeeva E et al. Highly stable hexitol based XNA aptamers targeting the vascular endothelial growth factor. Nucleic Acids Res 47, 4927–4939, doi: 10.1093/nar/gkz252 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.King DJ et al. Combinatorial selection and binding of phosphorothioate aptamers targeting human NF-kappa B RelA(p65) and p50. Biochemistry 41, 9696–9706, doi: 10.1021/bi020220k (2002). [DOI] [PubMed] [Google Scholar]
- 18.Matsunaga K et al. Architecture of high-affinity unnatural-base DNA aptamers toward pharmaceutical applications. Sci Rep 5, 18478, doi: 10.1038/srep18478 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sefah K et al. In vitro selection with artificial expanded genetic information systems. Proc Natl Acad Sci U S A 111, 1449–1454, doi: 10.1073/pnas.1311778111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zumrut H et al. Ligand-Guided Selection with Artificially Expanded Genetic Information Systems against TCR-CD3epsilon. Biochemistry 59, 552–562, doi: 10.1021/acs.biochem.9b00919 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dhami K et al. Systematic exploration of a class of hydrophobic unnatural base pairs yields multiple new candidates for the expansion of the genetic alphabet. Nucleic Acids Res 42, 10235–10244, doi: 10.1093/nar/gku715 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gold L et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS One 5, e15004, doi: 10.1371/journal.pone.0015004 (2010). a. Using modified aptamers, called Slow Off-Rate Modified Aptamers, a proteomics platform able to detect over 800 proteins with median pM detection limit was demonstrated for the first time.
- 23.Pfeiffer F et al. Identification and characterization of nucleobase-modified aptamers by click-SELEX. Nat Protoc 13, 1153–1180, doi: 10.1038/nprot.2018.023 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Temme JS & Krauss IJ SELMA: Selection with Modified Aptamers. Curr Protoc Chem Biol 7, 73–92, doi: 10.1002/9780470559277.ch140233 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Unrau PJ & Bartel DP RNA-catalysed nucleotide synthesis. Nature 395, 260–263, doi: 10.1038/26193 (1998). [DOI] [PubMed] [Google Scholar]
- 26.Hall B et al. Design, Synthesis, and Amplification of DNA Pools for In Vitro Selection. Current Protocols in Molecular Biology 88, 24.22.21–24.22.27, doi: 10.1002/0471142727.mb2402s88 (2009). [DOI] [PubMed] [Google Scholar]
- 27.Davis JH & Szostak JW Isolation of high-affinity GTP aptamers from partially structured RNA libraries. Proc Natl Acad Sci U S A 99, 11616–11621, doi: 10.1073/pnas.182095699 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang KA, Pei R, Stefanovic D & Stojanovic MN Optimizing cross-reactivity with evolutionary search for sensors. J Am Chem Soc 134, 1642–1647, doi: 10.1021/ja2084256 (2012). [DOI] [PubMed] [Google Scholar]
- 29.Eaton BE et al. Post-SELEX combinatorial optimization of aptamers. Bioorg Med Chem 5, 1087–1096, doi: 10.1016/s0968-0896(97)00044-8 (1997). [DOI] [PubMed] [Google Scholar]
- 30.Futami K, Kimoto M, Lim YWS & Hirao I Genetic Alphabet Expansion Provides Versatile Specificities and Activities of Unnatural-Base DNA Aptamers Targeting Cancer Cells. Mol Ther Nucleic Acids 14, 158–170, doi: 10.1016/j.omtn.2018.11.011 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McKeague M & Derosa MC Challenges and opportunities for small molecule aptamer development. J Nucleic Acids 2012, 748913, doi: 10.1155/2012/748913 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huizenga DE & Szostak JW A DNA aptamer that binds adenosine and ATP. Biochemistry 34, 656–665, doi: 10.1021/bi00002a033 (1995). [DOI] [PubMed] [Google Scholar]
- 33.Stoltenburg R, Nikolaus N & Strehlitz B Capture-SELEX: Selection of DNA Aptamers for Aminoglycoside Antibiotics. J Anal Methods Chem 2012, 415697, doi: 10.1155/2012/415697 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nutiu R & Li Y In vitro selection of structure-switching signaling aptamers. Angew Chem Int Ed Engl 44, 1061–1065, doi: 10.1002/anie.200461848 (2005). a. The method reported in this work has been widely used to develop aptamer biosensors that are dependent on conformational changes of the aptamer in the presence of the target.
- 35. McKeague M et al. Analysis of In Vitro Aptamer Selection Parameters. J Mol Evol 81, 150–161, doi: 10.1007/s00239-015-9708-6 (2015). a. One challenge of aptamer work is that the apparent can change depending on the method used to assess aptamer-target binding; this paper gives examples of this challenge and also makes suggestions on best practises for affinity characterization.
- 36.Murphy MB, Fuller ST, Richardson PM & Doyle SA An improved method for the in vitro evolution of aptamers and applications in protein detection and purification. Nucleic Acids Res 31, e110, doi: 10.1093/nar/gng110 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mayer G & Hover T In vitro selection of ssDNA aptamers using biotinylated target proteins. Methods Mol Biol 535, 19–32, doi: 10.1007/978-1-59745-557-2_2 (2009). [DOI] [PubMed] [Google Scholar]
- 38. Stoltenburg R, Reinemann C & Strehlitz B FluMag-SELEX as an advantageous method for DNA aptamer selection. Anal Bioanal Chem 383, 83–91, doi: 10.1007/s00216-005-3388-9 (2005). a. Widely cited as a followed method, this paper reported two innovations on the original selection approach: using magnetic beads for immobilization of the target, and enrichment monitoring and recovery of single-stranded DNA by fluorescently labeled primer.
- 39.Lou X et al. Micromagnetic selection of aptamers in microfluidic channels. Proc Natl Acad Sci U S A 106, 2989–2994, doi: 10.1073/pnas.0813135106 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hicke BJ et al. Tenascin-C aptamers are generated using tumor cells and purified protein. J Biol Chem 276, 48644–48654, doi: 10.1074/jbc.M104651200 (2001). [DOI] [PubMed] [Google Scholar]
- 41.Morris KN, Jensen KB, Julin CM, Weil M & Gold L High affinity ligands from in vitro selection: complex targets. Proc Natl Acad Sci U S A 95, 2902–2907, doi: 10.1073/pnas.95.6.2902 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Duan Y, Zhang C, Wang Y & Chen G Research progress of whole-cell-SELEX selection and the application of cell-targeting aptamer. Molecular Biology Reports 49, 7979–7993, doi: 10.1007/s11033-022-07317-0 (2022). [DOI] [PubMed] [Google Scholar]
- 43.Zumrut HE et al. Integrating Ligand-Receptor Interactions and In Vitro Evolution for Streamlined Discovery of Artificial Nucleic Acid Ligands. Mol Ther Nucleic Acids 17, 150–163, doi: 10.1016/j.omtn.2019.05.015 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mayer G et al. Fluorescence-activated cell sorting for aptamer SELEX with cell mixtures. Nat Protoc 5, 1993–2004, doi: 10.1038/nprot.2010.163 (2010). [DOI] [PubMed] [Google Scholar]
- 45.Cheng C, Chen YH, Lennox KA, Behlke MA & Davidson BL In vivo SELEX for Identification of Brain-penetrating Aptamers. Mol Ther Nucleic Acids 2, e67, doi: 10.1038/mtna.2012.59 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li S et al. Identification of an aptamer targeting hnRNP A1 by tissue slide-based SELEX. J Pathol 218, 327–336, doi: 10.1002/path.2543 (2009). [DOI] [PubMed] [Google Scholar]
- 47.Le ATH et al. How to Develop and Prove High-Efficiency Selection of Ligands from Oligonucleotide Libraries: A Universal Framework for Aptamers and DNA-Encoded Small-Molecule Ligands. Analytical Chemistry 93, 5343–5354, doi: 10.1021/acs.analchem.1c00601 (2021). [DOI] [PubMed] [Google Scholar]
- 48.Ouellet E, Foley JH, Conway EM & Haynes C Hi-Fi SELEX: A high-fidelity digital-PCR based therapeutic aptamer discovery platform. Biotechnology and Bioengineering 112, 1506–1522, doi: 10.1002/bit.25581 (2015). [DOI] [PubMed] [Google Scholar]
- 49.Berezovski M et al. Nonequilibrium Capillary Electrophoresis of Equilibrium Mixtures: A Universal Tool for Development of Aptamers. Journal of the American Chemical Society 127, 3165–3171, doi: 10.1021/ja042394q (2005). [DOI] [PubMed] [Google Scholar]
- 50.Mondal B, Ramlal S, Lavu PS, Murali HS & Batra HV A combinatorial systematic evolution of ligands by exponential enrichment method for selection of aptamer against protein targets. Appl Microbiol Biotechnol 99, 9791–9803, doi: 10.1007/s00253-015-6858-9 (2015). [DOI] [PubMed] [Google Scholar]
- 51.Shangguan D et al. Aptamers evolved from live cells as effective molecular probes for cancer study. Proc Natl Acad Sci U S A 103, 11838–11843, doi: 10.1073/pnas.0602615103 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thiel WH et al. Cell-internalization SELEX: method for identifying cell-internalizing RNA aptamers for delivering siRNAs to target cells. Methods Mol Biol 1218, 187–199, doi: 10.1007/978-1-4939-1538-5_11 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wong I & Lohman TM A double-filter method for nitrocellulose-filter binding: application to protein-nucleic acid interactions. Proc Natl Acad Sci U S A 90, 5428–5432, doi: 10.1073/pnas.90.12.5428 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.White R et al. Generation of species cross-reactive aptamers using “toggle” SELEX. Mol Ther 4, 567–573, doi: 10.1006/mthe.2001.0495 (2001). [DOI] [PubMed] [Google Scholar]
- 55.Mendonsa SD & Bowser MT In vitro evolution of functional DNA using capillary electrophoresis. J Am Chem Soc 126, 20–21, doi: 10.1021/ja037832s (2004). [DOI] [PubMed] [Google Scholar]
- 56.Tsai RY & Reed RR Identification of DNA recognition sequences and protein interaction domains of the multiple-Zn-finger protein Roaz. Mol Cell Biol 18, 6447–6456, doi: 10.1128/MCB.18.11.6447 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zumrut HE, Ara MN, Fraile M, Maio G & Mallikaratchy P Ligand-Guided Selection of Target-Specific Aptamers: A Screening Technology for Identifying Specific Aptamers Against Cell-Surface Proteins. Nucleic Acid Ther 26, 190–198, doi: 10.1089/nat.2016.0611 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zumrut HE et al. Ligand-guided selection of aptamers against T-cell Receptor-cluster of differentiation 3 (TCR-CD3) expressed on Jurkat.E6 cells. Anal Biochem 512, 1–7, doi: 10.1016/j.ab.2016.08.007 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mi J et al. In vivo selection of tumor-targeting RNA motifs. Nat Chem Biol 6, 22–24, doi: 10.1038/nchembio.277 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Udofot O et al. Delivery of Cell-Specific Aptamers to the Arterial Wall with an Occlusion Perfusion Catheter. Mol Ther Nucleic Acids 16, 360–366, doi: 10.1016/j.omtn.2019.03.005 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Komarova N & Kuznetsov A Inside the Black Box: What Makes SELEX Better? Molecules 24, 3598, doi: 10.3390/molecules24193598 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Irvine D, Tuerk C & Gold L Selexion: Systematic evolution of ligands by exponential enrichment with integrated optimization by non-linear analysis. Journal of molecular biology 222, 739–761 (1991). [DOI] [PubMed] [Google Scholar]
- 63.Vant-Hull B, Payano-Baez A, Davis RH & Gold L The mathematics of SELEX against complex targets. Journal of molecular biology 278, 579–597 (1998). [DOI] [PubMed] [Google Scholar]
- 64.Aita T, Nishigaki K & Husimi Y Theoretical consideration of selective enrichment in in vitro selection: Optimal concentration of target molecules. Mathematical Biosciences 240, 201–211 (2012). [DOI] [PubMed] [Google Scholar]
- 65.Ray P & White RR Cell-SELEX Identifies a “Sticky” RNA Aptamer Sequence. J Nucleic Acids 2017, 4943072, doi: 10.1155/2017/4943072 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tang Z et al. Selection of aptamers for molecular recognition and characterization of cancer cells. Anal Chem 79, 4900–4907, doi: 10.1021/ac070189y (2007). [DOI] [PubMed] [Google Scholar]
- 67.Thiel KW et al. Delivery of chemo-sensitizing siRNAs to HER2+-breast cancer cells using RNA aptamers. Nucleic Acids Res 40, 6319–6337, doi: 10.1093/nar/gks294 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tolle F, Wilke J, Wengel J & Mayer G By-product formation in repetitive PCR amplification of DNA libraries during SELEX. PLoS One 9, e114693, doi: 10.1371/journal.pone.0114693 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tsuji S et al. Effective isolation of RNA aptamer through suppression of PCR bias. Biochem Biophys Res Commun 386, 223–226, doi: 10.1016/j.bbrc.2009.06.013 (2009). [DOI] [PubMed] [Google Scholar]
- 70.Levay A et al. Identifying high-affinity aptamer ligands with defined cross-reactivity using high-throughput guided systematic evolution of ligands by exponential enrichment. Nucleic Acids Res 43, e82, doi: 10.1093/nar/gkv534 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vanbrabant J, Leirs K, Vanschoenbeek K, Lammertyn J & Michiels L reMelting curve analysis as a tool for enrichment monitoring in the SELEX process. Analyst 139, 589–595 (2014). [DOI] [PubMed] [Google Scholar]
- 72.Takahashi M et al. High throughput sequencing analysis of RNA libraries reveals the influences of initial library and PCR methods on SELEX efficiency. Sci Rep 6, 33697, doi: 10.1038/srep33697 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sefah K, Shangguan D, Xiong X, O’Donoghue MB & Tan W Development of DNA aptamers using Cell-SELEX. Nat Protoc 5, 1169–1185, doi: 10.1038/nprot.2010.66 (2010). [DOI] [PubMed] [Google Scholar]
- 74.Yang Z, Chen F, Alvarado JB & Benner SA Amplification, mutation, and sequencing of a six-letter synthetic genetic system. J Am Chem Soc 133, 15105–15112, doi: 10.1021/ja204910n (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kimoto M, Kawai R, Mitsui T, Yokoyama S & Hirao I An unnatural base pair system for efficient PCR amplification and functionalization of DNA molecules. Nucleic Acids Res 37, e14, doi: 10.1093/nar/gkn956 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Musheev MU & Krylov SN Selection of aptamers by systematic evolution of ligands by exponential enrichment: addressing the polymerase chain reaction issue. Anal Chim Acta 564, 91–96, doi: 10.1016/j.aca.2005.09.069 (2006). [DOI] [PubMed] [Google Scholar]
- 77.Wang T, Chen C, Larcher LM, Barrero RA & Veedu RN Three decades of nucleic acid aptamer technologies: Lessons learned, progress and opportunities on aptamer development. Biotechnol Adv 37, 28–50, doi: 10.1016/j.biotechadv.2018.11.001 (2019). [DOI] [PubMed] [Google Scholar]
- 78.Liu Z, Chen T & Romesberg FE Evolved polymerases facilitate selection of fully 2’-OMe-modified aptamers. Chem Sci 8, 8179–8182, doi: 10.1039/c7sc03747c (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lapa SA, Chudinov AV & Timofeev EN The Toolbox for Modified Aptamers. Molecular Biotechnology 58, 79–92, doi: 10.1007/s12033-015-9907-9 (2016). [DOI] [PubMed] [Google Scholar]
- 80.Svobodova M, Pinto A, Nadal P & CK OS Comparison of different methods for generation of single-stranded DNA for SELEX processes. Anal Bioanal Chem 404, 835–842, doi: 10.1007/s00216-012-6183-4 (2012). [DOI] [PubMed] [Google Scholar]
- 81.Tolnai Z et al. A simple modification increases specificity and efficiency of asymmetric PCR. Anal Chim Acta 1047, 225–230, doi: 10.1016/j.aca.2018.10.017 (2019). [DOI] [PubMed] [Google Scholar]
- 82.Civit L, Fragoso A & O’Sullivan CK Evaluation of techniques for generation of single-stranded DNA for quantitative detection. Anal Biochem 431, 132–138, doi: 10.1016/j.ab.2012.09.003 (2012). [DOI] [PubMed] [Google Scholar]
- 83.Hwang C & Carothers JM Label-free selection of RNA aptamers for metabolic engineering. Methods 106, 37–41, doi: 10.1016/j.ymeth.2016.06.016 (2016). [DOI] [PubMed] [Google Scholar]
- 84.Bell DR et al. In silico design and validation of high-affinity RNA aptamers targeting epithelial cellular adhesion molecule dimers. Proc Natl Acad Sci U S A 117, 8486–8493, doi: 10.1073/pnas.1913242117 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mohammadinezhad R, Jalali SAH & Farahmand H Evaluation of different direct and indirect SELEX monitoring methods and implementation of melt-curve analysis for rapid discrimination of variant aptamer sequences. Anal Methods 12, 3823–3835, doi: 10.1039/d0ay00491j (2020). [DOI] [PubMed] [Google Scholar]
- 86.Thiel WH Galaxy Workflows for Web-based Bioinformatics Analysis of Aptamer High-throughput Sequencing Data. Mol Ther Nucleic Acids 5, e345, doi: 10.1038/mtna.2016.54 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Thiel WH & Giangrande PH Analyzing HT-SELEX data with the Galaxy Project tools--A web based bioinformatics platform for biomedical research. Methods 97, 3–10, doi: 10.1016/j.ymeth.2015.10.008 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kolm C et al. DNA aptamers against bacterial cells can be efficiently selected by a SELEX process using state-of-the art qPCR and ultra-deep sequencing. Scientific reports 10, 1–16 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Schutze T et al. Probing the SELEX process with next-generation sequencing. PLoS One 6, e29604, doi: 10.1371/journal.pone.0029604 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ruscito A et al. In Vitro Selection and Characterization of DNA Aptamers to a Small Molecule Target. Current protocols in chemical biology 9, 233–268, doi: 10.1002/cpch.28 (2017). [DOI] [PubMed] [Google Scholar]
- 91.Mastronardi E, Cyr K, Monreal CM & DeRosa MC Selection of DNA Aptamers for Root Exudate l-Serine Using Multiple Selection Strategies. J Agric Food Chem 69, 4294–4306, doi: 10.1021/acs.jafc.0c06796 (2021). [DOI] [PubMed] [Google Scholar]
- 92.Misono TS & Kumar PK Selection of RNA aptamers against human influenza virus hemagglutinin using surface plasmon resonance. Anal Biochem 342, 312–317, doi: 10.1016/j.ab.2005.04.013 (2005). [DOI] [PubMed] [Google Scholar]
- 93.Lamberti I et al. In vitro selection of RNA aptamers against CA125 tumor marker in ovarian cancer and its study by optical biosensing. Methods 97, 58–68, doi: 10.1016/j.ymeth.2015.10.022 (2016). [DOI] [PubMed] [Google Scholar]
- 94.Gao S et al. A novel biosensing platform for detection of glaucoma biomarker GDF15 via an integrated BLI-ELASA strategy. Biomaterials 294, 121997, doi: 10.1016/j.biomaterials.2023.121997 (2023). [DOI] [PubMed] [Google Scholar]
- 95.Poolsup S et al. Discovery of DNA aptamers targeting SARS-CoV-2 nucleocapsid protein and protein-binding epitopes for label-free COVID-19 diagnostics. Molecular Therapy -Nucleic Acids 31, 731–743, doi: 10.1016/j.omtn.2023.02.010 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Esposito CL et al. Identification of a novel RNA aptamer that selectively targets breast cancer exosomes. Mol Ther Nucleic Acids 23, 982–994, doi: 10.1016/j.omtn.2021.01.012 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bai C et al. Aptamer selection and application in multivalent binding-based electrical impedance detection of inactivated H1N1 virus. Biosens Bioelectron 110, 162–167, doi: 10.1016/j.bios.2018.03.047 (2018). [DOI] [PubMed] [Google Scholar]
- 98.Mosing RK, Mendonsa SD & Bowser MT Capillary electrophoresis-SELEX selection of aptamers with affinity for HIV-1 reverse transcriptase. Anal Chem 77, 6107–6112, doi: 10.1021/ac050836q (2005). [DOI] [PubMed] [Google Scholar]
- 99.Davis KA, Abrams B, Lin Y & Jayasena SD Use of a high affinity DNA ligand in flow cytometry. Nucleic Acids Res 24, 702–706, doi: 10.1093/nar/24.4.702 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fitzwater T & Polisky B A SELEX primer. Methods Enzymol 267, 275–301, doi: 10.1016/s0076-6879(96)67019-0 (1996). [DOI] [PubMed] [Google Scholar]
- 101.Jensen KB, Atkinson BL, Willis MC, Koch TH & Gold L Using in vitro selection to direct the covalent attachment of human immunodeficiency virus type 1 Rev protein to high-affinity RNA ligands. Proc Natl Acad Sci U S A 92, 12220–12224, doi: 10.1073/pnas.92.26.12220 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Oteng EK, Gu W & McKeague M High-efficiency enrichment enables identification of aptamers to circulating Plasmodium falciparum-infected erythrocytes. Sci Rep 10, 9706, doi: 10.1038/s41598-020-66537-1 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Stoltenburg R & Strehlitz B Refining the Results of a Classical SELEX Experiment by Expanding the Sequence Data Set of an Aptamer Pool Selected for Protein A. Int J Mol Sci 19, doi: 10.3390/ijms19020642 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ambardar S, Gupta R, Trakroo D, Lal R & Vakhlu J High Throughput Sequencing: An Overview of Sequencing Chemistry. Indian J Microbiol 56, 394–404, doi: 10.1007/s12088-016-0606-4 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Soldevilla MM et al. Identification of LAG3 high affinity aptamers by HT-SELEX and Conserved Motif Accumulation (CMA). PLoS One 12, e0185169, doi: 10.1371/journal.pone.0185169 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Kohlberger M & Gadermaier G SELEX: Critical factors and optimization strategies for successful aptamer selection. Biotechnology and Applied Biochemistry, doi: 10.1002/bab.2244 (2021). a. This comprehensive review offers helpful considerations and quality control measures for SELEX experiments.
- 107.MacKenzie M & Argyropoulos C An Introduction to Nanopore Sequencing: Past, Present, and Future Considerations. Micromachines 14, 459 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Alam KK, Chang JL & Burke DH FASTAptamer: A Bioinformatic Toolkit for High-throughput Sequence Analysis of Combinatorial Selections. Mol Ther Nucleic Acids 4, e230, doi: 10.1038/mtna.2015.4 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Caroli J, Taccioli C, De La Fuente A, Serafini P & Bicciato S APTANI: a computational tool to select aptamers through sequence-structure motif analysis of HT-SELEX data. Bioinformatics 32, 161–164, doi: 10.1093/bioinformatics/btv545 (2016). [DOI] [PubMed] [Google Scholar]
- 110.Bailey TL et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37, W202–208, doi: 10.1093/nar/gkp335 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Jiang P et al. MPBind: a Meta-motif-based statistical framework and pipeline to Predict Binding potential of SELEX-derived aptamers. Bioinformatics 30, 2665–2667, doi: 10.1093/bioinformatics/btu348 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Shieh KR et al. AptCompare: optimized de novo motif discovery of RNA aptamers via HTS-SELEX. Bioinformatics 36, 2905–2906, doi: 10.1093/bioinformatics/btaa054 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hiller M, Pudimat R, Busch A & Backofen R Using RNA secondary structures to guide sequence motif finding towards single-stranded regions. Nucleic Acids Res 34, e117, doi: 10.1093/nar/gkl544 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ishida R et al. RaptRanker: in silico RNA aptamer selection from HT-SELEX experiment based on local sequence and structure information. Nucleic Acids Res 48, e82, doi: 10.1093/nar/gkaa484 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Hoinka J, Backofen R & Przytycka TM AptaSUITE: A Full-Featured Bioinformatics Framework for the Comprehensive Analysis of Aptamers from HT-SELEX Experiments. Mol Ther Nucleic Acids 11, 515–517, doi: 10.1016/j.omtn.2018.04.006 (2018). a. The paper describes bioinformatics software that is freely available to aptamer researchers and has multiple functions including: processing sequencing data, clustering similar sequences, predicting secondary structure, etc.
- 116.Yang Z et al. Conversion strategy using an expanded genetic alphabet to assay nucleic acids. Anal Chem 85, 4705–4712, doi: 10.1021/ac400422r (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. McKeague M et al. Comprehensive analytical comparison of strategies used for small molecule aptamer evaluation. Anal Chem 87, 8608–8612, doi: 10.1021/acs.analchem.5b02102 (2015). a. One challenge of aptamer work is that the apparent KD can change depending on the method used to assess aptamer-target binding; this paper gives examples of this challenge and also makes suggestions on best practises for affinity characterization.
- 118.Bottari F et al. Do Aptamers Always Bind? The Need for a Multifaceted Analytical Approach When Demonstrating Binding Affinity between Aptamer and Low Molecular Weight Compounds. J Am Chem Soc 142, 19622–19630, doi: 10.1021/jacs.0c08691 (2020). [DOI] [PubMed] [Google Scholar]
- 119.Hirka S & McKeague M Quantification of aptamer-protein binding with fluorescence anisotropy. Aptamers 5, 1–6 (2021). [Google Scholar]
- 120.Chang AL, McKeague M, Liang JC & Smolke CD Kinetic and equilibrium binding characterization of aptamers to small molecules using a label-free, sensitive, and scalable platform. Analytical chemistry 86, 3273–3278, doi: 10.1021/ac5001527 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Slavkovic S et al. Thermodynamic analysis of cooperative ligand binding by the ATP-binding DNA aptamer indicates a population-shift binding mechanism. Sci Rep 10, 18944, doi: 10.1038/s41598-020-76002-8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Jenison RD, Gill SC, Pardi A & Polisky B High-resolution molecular discrimination by RNA. Science 263, 1425–1429, doi: 10.1126/science.7510417 (1994). [DOI] [PubMed] [Google Scholar]
- 123. Roxo C, Kotkowiak W & Pasternak A G-Quadruplex-Forming Aptamers-Characteristics, Applications, and Perspectives. Molecules 24, doi: 10.3390/molecules24203781 (2019). a. G-quadruplexes are a common secondary structure for DNA aptamers, this review summarizes several important aspects of working with G-quadruplexes.
- 124.Luo X et al. Computational approaches toward the design of pools for the in vitro selection of complex aptamers. Rna 16, 2252–2262, doi: 10.1261/rna.2102210 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Le TT, Chumphukam O & Cass AEG Determination of minimal sequence for binding of an aptamer. A comparison of truncation and hybridization inhibition methods. RSC Advances 4, 47227–47233, doi: 10.1039/C4RA08243E (2014). a. There are several practical advantages of understanding exactly what bases are implicated in target binding and which are important for secondary structure, this paper provides insights into investigating minimal sequences.
- 126.Kinghorn AB et al. Aptamer Affinity Maturation by Resampling and Microarray Selection. Analytical chemistry 88, 6981–6985, doi: 10.1021/acs.analchem.6b01635 (2016). [DOI] [PubMed] [Google Scholar]
- 127.Earnest KG et al. Development and characterization of a DNA aptamer for MLL-AF9 expressing acute myeloid leukemia cells using whole cell-SELEX. Sci Rep 11, 19174, doi: 10.1038/s41598-021-98676-4 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Bishop GR & Chaires JB Characterization of DNA structures by circular dichroism. Curr Protoc Nucleic Acid Chem Chapter 7, Unit 7 11, doi: 10.1002/0471142700.nc0711s11 (2003). [DOI] [PubMed] [Google Scholar]
- 129.Del Villar-Guerra R, Trent JO & Chaires JB G-Quadruplex Secondary Structure Obtained from Circular Dichroism Spectroscopy. Angew Chem Int Ed Engl 57, 7171–7175, doi: 10.1002/anie.201709184 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zhang N et al. Structural Biology for the Molecular Insight between Aptamers and Target Proteins. Int J Mol Sci 22, doi: 10.3390/ijms22084093 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kratschmer C & Levy M Effect of Chemical Modifications on Aptamer Stability in Serum. Nucleic Acid Ther 27, 335–344, doi: 10.1089/nat.2017.0680 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Griffin LC, Tidmarsh GF, Bock LC, Toole JJ & Leung LL In vivo anticoagulant properties of a novel nucleotide-based thrombin inhibitor and demonstration of regional anticoagulation in extracorporeal circuits. Blood 81, 3271–3276 (1993). [PubMed] [Google Scholar]
- 133.Rangel AE, Chen Z, Ayele TM & Heemstra JM In vitro selection of an XNA aptamer capable of small-molecule recognition. Nucleic Acids Res 46, 8057–8068, doi: 10.1093/nar/gky667 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bauer M, Strom M, Hammond SD & Shigdar S Anything You Can Do, I Can Do Better: Can Aptamers Replace Antibodies in Clinical Diagnostic Applications? Molecules 24, doi: 10.3390/molecules24234377 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Li F et al. Aptamers Facilitating Amplified Detection of Biomolecules. Analytical Chemistry 87, 274–292, doi: 10.1021/ac5037236 (2015). [DOI] [PubMed] [Google Scholar]
- 136. McConnell EM, Nguyen J & Li Y Aptamer-Based Biosensors for Environmental Monitoring. Frontiers in Chemistry 8, doi: 10.3389/fchem.2020.00434 (2020). a. A comprehensive review of aptamer applications for environmental monitoring sorted into examples of water, soil, and air monitoring.
- 137.Kalita JJ, Sharma P & Bora U Recent developments in application of nucleic acid aptamer in food safety. Food Control 145, 109406, doi: 10.1016/j.foodcont.2022.109406 (2023). [DOI] [Google Scholar]
- 138.Mohamad N et al. Future perspectives on aptamer for application in food authentication. Analytical Biochemistry 656, 114861, doi: 10.1016/j.ab.2022.114861 (2022). [DOI] [PubMed] [Google Scholar]
- 139.Hu Z et al. Aptamer based biosensor platforms for neurotransmitters analysis. TrAC Trends in Analytical Chemistry 162, 117021, doi: 10.1016/j.trac.2023.117021 (2023). [DOI] [Google Scholar]
- 140.Futane A, Narayanamurthy V, Jadhav P & Srinivasan A Aptamer-based rapid diagnosis for point-of-care application. Microfluidics and Nanofluidics 27, 15, doi: 10.1007/s10404-022-02622-3 (2023).( [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Waldmann TA Monoclonal Antibodies in Diagnosis and Therapy. Science 252, 1657–1662, doi: 10.1126/science.2047874 1991). [DOI] [PubMed] [Google Scholar]
- 142.Meyer M, Scheper T & Walter J-G Aptamers: versatile probes for flow cytometry. Appl. Microbiol. Biotechnol. 97, 7097–7109, doi: 10.1007/s00253-013-5070-z (2013). [DOI] [PubMed] [Google Scholar]
- 143.Bauer M, Macdonald J, Henri J, Duan W & Shigdar S The Application of Aptamers for Immunohistochemistry. Nucleic Acid Ther. 26, 120–126, doi: 10.1089/nat.2015.0569 (2016). [DOI] [PubMed] [Google Scholar]
- 144.Toh SY, Citartan M, Gopinath SCB & Tang T-H Aptamers as a replacement for antibodies in enzyme-linked immunosorbent assay. Biosens. Bioelectron. 64, 392–403, doi: 10.1016/j.bios.2014.09.026 (2015). [DOI] [PubMed] [Google Scholar]
- 145.Bolognesi MM et al. Multiplex Staining by Sequential Immunostaining and Antibody Removal on Routine Tissue Sections. J. Histochem. Cytochem. 65, 431–444, doi: 10.1369/0022155417719419 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Gomes de Castro MA, Höbartner C & Opazo F Aptamers provide superior stainings of cellular receptors studied under super-resolution microscopy. PLoS One 12, e0173050–e0173050, doi: 10.1371/journal.pone.0173050 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Gray BP, Requena MD, Nichols MD & Sullenger BA Aptamers as Reversible Sorting Ligands for Preparation of Cells in Their Native State. Cell Chem. Biol. 27, 232–244.e237, doi: 10.1016/j.chembiol.2019.12.004 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Mora J et al. Next Generation Ligand Binding Assays—Review of Emerging Technologies’ Capabilities to Enhance Throughput and Multiplexing. The AAPS Journal 16, 1175–1184, doi: 10.1208/s12248-014-9660-1 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wang X, Cohen L, Wang J & Walt DR Competitive Immunoassays for the Detection of Small Molecules Using Single Molecule Arrays. J Am Chem Soc 140, 18132–18139, doi: 10.1021/jacs.8b11185 (2018). [DOI] [PubMed] [Google Scholar]
- 150. Wu D, Katilius E, Olivas E, Dumont Milutinovic M & Walt DR Incorporation of Slow Off-Rate Modified Aptamers Reagents in Single Molecule Array Assays for Cytokine Detection with Ultrahigh Sensitivity. Anal. Chem. 88, 8385–8389, doi: 10.1021/acs.analchem.6b02451 (2016). a. Here SOMAmers are combined with the single molecule detection platform called Simoa, which affords excellent detection sensitivity via digital detection.
- 151.Wu D, Milutinovic MD & Walt DR Single molecule array (Simoa) assay with optimal antibody pairs for cytokine detection in human serum samples. Analyst 140, 6277–6282, doi: 10.1039/C5AN01238D (2015). [DOI] [PubMed] [Google Scholar]
- 152.Rabia LA, Desai AA, Jhajj HS & Tessier PM Understanding and overcoming trade-offs between antibody affinity, specificity, stability and solubility. Biochem Eng J 137, 365–374, doi: 10.1016/j.bej.2018.06.003 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Shigdar S et al. Aptamers as Theranostic Agents: Modifications, Serum Stability and Functionalisation. Sensors 13, 13624 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Elskens JP, Elskens JM & Madder A Chemical Modification of Aptamers for Increased Binding Affinity in Diagnostic Applications: Current Status and Future Prospects. Int J Mol Sci 21, doi: 10.3390/ijms21124522 (2020). a. This review offers a comprehensive account of aptamers for diagnostic applications, sorted by type of modification (modified base, sequence truncation, extended genetic alphabet, etc).
- 155.Opazo F et al. Aptamers as potential tools for super-resolution microscopy. Nat. Methods 9, 938–939, doi: 10.1038/nmeth.2179 (2012). [DOI] [PubMed] [Google Scholar]
- 156.Minhyeok C et al. Aptamer-based single-molecule imaging of insulin receptors in living cells. J. Biomed. Opt. 19, 1–7, doi: 10.1117/1.JBO.19.5.051204 (2013). [DOI] [PubMed] [Google Scholar]
- 157.Strauss S et al. Modified aptamers enable quantitative sub-10-nm cellular DNA-PAINT imaging. Nat Methods 15, 685–688, doi: 10.1038/s41592-018-0105-0 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Delcanale P et al. Aptamers with Tunable Affinity Enable Single-Molecule Tracking and Localization of Membrane Receptors on Living Cancer Cells. Angew Chem Int Ed Engl 59, 18546–18555, doi: 10.1002/anie.202004764 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Stojanovic MN, de Prada P & Landry DW Fluorescent Sensors Based on Aptamer Self-Assembly. J Am Chem Soc 122, 11547–11548, doi: 10.1021/ja0022223 (2000). [DOI] [PubMed] [Google Scholar]
- 160.Qi X, Yan X, Zhao Y, Li L & Wang S Highly sensitive and specific detection of small molecules using advanced aptasensors based on split aptamers: A review. TrAC - Trends Anal. Chem 133, 116069, doi: 10.1016/j.trac.2020.116069 (2020). [DOI] [Google Scholar]
- 161. Paige JS, Wu KY & Jaffrey SR RNA mimics of green fluorescent protein. Science 333, 642–646, doi: 10.1126/science.1207339 (2011). a. The first in a series of fruit and vegtable inspired RNA aptamers (in this case termed spinach) that light-up when bound to specific fluorogens is reported here.
- 162.Kellenberger CA, Wilson SC, Sales-Lee J & Hammond MC RNA-based fluorescent biosensors for live cell imaging of second messengers cyclic di-GMP and cyclic AMP-GMP. J Am Chem Soc 135, 4906–4909, doi: 10.1021/ja311960g (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Warner KD et al. Structural basis for activity of highly efficient RNA mimics of green fluorescent protein. Nat Struct Mol Biol 21, 658–663, doi: 10.1038/nsmb.2865 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Filonov GS, Moon JD, Svensen N & Jaffrey SR Broccoli: Rapid Selection of an RNA Mimic of Green Fluorescent Protein by Fluorescence-Based Selection and Directed Evolution. J Am Chem Soc 136, 16299–16308, doi: 10.1021/ja508478x (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Qiao L, Benzigar MR, Subramony JA, Lovell NH & Liu G Advances in Sweat Wearables: Sample Extraction, Real-Time Biosensing, and Flexible Platforms. ACS Appl. Mater. Interfaces 12, 34337–34361, doi: 10.1021/acsami.0c07614 (2020). [DOI] [PubMed] [Google Scholar]
- 166.Ferguson BS et al. Real-time, aptamer-based tracking of circulating therapeutic agents in living animals. Sci Transl Med 5, 213ra165–213ra165, doi: 10.1126/scitranslmed.3007095 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Li H, Dauphin-Ducharme P, Ortega G & Plaxco KW Calibration-Free Electrochemical Biosensors Supporting Accurate Molecular Measurements Directly in Undiluted Whole Blood. J Am Chem Soc 139, 11207–11213, doi: 10.1021/jacs.7b05412 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Wang Z et al. A Wearable and Deformable Graphene-Based Affinity Nanosensor for Monitoring of Cytokines in Biofluids. Nanomaterials 10, doi: 10.3390/nano10081503 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Hao Z et al. Measurement of cytokine biomarkers using an aptamer-based affinity graphene nanosensor on a flexible substrate toward wearable applications. Nanoscale 10, 21681–21688, doi: 10.1039/C8NR04315A (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Bruno JG Potential Inherent Stimulation of the Innate Immune System by Nucleic Acid Aptamers and Possible Corrective Approaches. Pharmaceuticals 11, doi: 10.3390/ph11030062 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Xiang D et al. Superior Performance of Aptamer in Tumor Penetration over Antibody: Implication of Aptamer-Based Theranostics in Solid Tumors. Theranostics 5, 1083–1097, doi: 10.7150/thno.11711 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Oney S et al. Development of universal antidotes to control aptamer activity. Nat. Med. 15, 1224–1228, doi: 10.1038/nm.1990 (2009). a. Aptamers have unique therapeutic potential as they have a complemetary antidote by nature, however this paper describes university antidote molecules that can be applied in a sequence-independent manner.
- 173.Chabata CV, Frederiksen JW, Sullenger BA & Gunaratne R Emerging applications of aptamers for anticoagulation and hemostasis. Curr. Opin. Hematol. 25 (2018). [DOI] [PubMed] [Google Scholar]
- 174.Lincoff AM et al. Effect of the REG1 anticoagulation system versus bivalirudin on outcomes after percutaneous coronary intervention (REGULATE-PCI): a randomised clinical trial. The Lancet 387, 349–356, doi: 10.1016/S0140-6736(15)00515-2 (2016). [DOI] [PubMed] [Google Scholar]
- 175.Greenhawt M et al. Diagnostic accuracy of vaccine and vaccine excipient testing in the setting of allergic reactions to COVID-19 vaccines: A systematic review and meta-analysis. Allergy 78, 71–83, doi: 10.1111/all.15571 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Torabi R, Ranjbar R, Halaji M & Heiat M Aptamers, the bivalent agents as probes and therapies for coronavirus infections: A systematic review. Molecular and Cellular Probes 53, 101636, doi: 10.1016/j.mcp.2020.101636 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Koudrina A et al. Fibrinogen aptamer functionalized gold-coated iron-oxide nanoparticles for targeted imaging of thrombi. Chemical Communications 58, 2870–2873, doi: 10.1039/D1CC03817F (2022). [DOI] [PubMed] [Google Scholar]
- 178.Koudrina A & DeRosa MC Advances in Medical Imaging: Aptamer- and Peptide-Targeted MRI and CT Contrast Agents. ACS Omega 5, 22691–22701, doi: 10.1021/acsomega.0c02650 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Koudrina A et al. Exploring the Unique Contrast Properties of Aptamer–Gadolinium Conjugates in Magnetic Resonance Imaging for Targeted Imaging of Thrombi. ACS Applied Materials & Interfaces 13, 9412–9424, doi: 10.1021/acsami.0c16666 (2021). [DOI] [PubMed] [Google Scholar]
- 180.Kichkailo AS et al. Development of DNA aptamers for visualization of glial brain tumors and detection of circulating tumor cells. Mol Ther Nucleic Acids 32, 267–288, doi: 10.1016/j.omtn.2023.03.015 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Pieve CD, Perkins AC & Missailidis S Anti-MUC1 aptamers: radiolabelling with 99mTc and biodistribution in MCF-7 tumour-bearing mice. Nucl Med Biol 36, 703–710, doi: 10.1016/j.nucmedbio.2009.04.004 (2009). [DOI] [PubMed] [Google Scholar]
- 182.Liu HY, Yu X, Liu H, Wu D & She J-X Co-targeting EGFR and survivin with a bivalent aptamer-dual siRNA chimera effectively suppresses prostate cancer. Scientific Reports 6, 30346, doi: 10.1038/srep30346 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Kang H, O’Donoghue MB, Liu H & Tan W A liposome-based nanostructure for aptamer directed delivery. Chem Comm 46, 249–251, doi: 10.1039/B916911C (2010). [DOI] [PubMed] [Google Scholar]
- 184.Bouvier-Müller A & Ducongé F Application of aptamers for in vivo molecular imaging and theranostics. Adv. Drug Deliv. Rev. 134, 94–106, doi: 10.1016/j.addr.2018.08.004 (2018). [DOI] [PubMed] [Google Scholar]
- 185.Shigdar S et al. The Use of Sensitive Chemical Antibodies for Diagnosis: Detection of Low Levels of Epcam in Breast Cancer. PLoS One 8, e57613, doi: 10.1371/journal.pone.0057613 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Xiang D et al. Transforming doxorubicin into a cancer stem cell killer via EpCAM aptamer-mediated delivery. Theranostics 7, 4071–4086, doi: 10.7150/thno.20168 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Lakhin AV, Tarantul VZ & Gening LV Aptamers: problems, solutions and prospects. Acta Naturae 5, 34–43 (2013). [PMC free article] [PubMed] [Google Scholar]
- 188.Ng EW et al. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat Rev Drug Discov 5, 123–132, doi: 10.1038/nrd1955 (2006). [DOI] [PubMed] [Google Scholar]
- 189. McKeague M et al. The minimum aptamer publication standards (MAPS guidelines) for de novo aptamer selection. (2022). a. For the aptamer community to benefit from a centralized databank like the protein data bank, there needs sto be concensis guidelines for publication, here a comprehensive set of guidelikes are proposed.
- 190.Yan AC & Levy M Aptamer-Mediated Delivery and Cell-Targeting Aptamers: Room for Improvement. Nucleic Acid Ther 28, 194–199, doi: 10.1089/nat.2018.0732 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Li N et al. Technical and biological issues relevant to cell typing with aptamers. J Proteome Res 8, 2438–2448, doi: 10.1021/pr801048z (2009). [DOI] [PubMed] [Google Scholar]
- 192.Pestourie C et al. Comparison of different strategies to select aptamers against a transmembrane protein target. Oligonucleotides 16, 323–335, doi: 10.1089/oli.2006.16.323 (2006). [DOI] [PubMed] [Google Scholar]
- 193.Spill F et al. Controlling uncertainty in aptamer selection. Proc Natl Acad Sci U S A 113, 12076–12081, doi: 10.1073/pnas.1605086113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Stock C et al. Migration of human melanoma cells depends on extracellular pH and Na+/H+ exchange. J Physiol 567, 225–238, doi: 10.1113/jphysiol.2005.088344 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Chi CN, Engstrom A, Gianni S, Larsson M & Jemth P Two conserved residues govern the salt and pH dependencies of the binding reaction of a PDZ domain. J Biol Chem 281, 36811–36818, doi: 10.1074/jbc.M607883200 (2006). [DOI] [PubMed] [Google Scholar]
- 196.Eaton BE, Gold L & Zichi DA Let’s get specific: the relationship between specificity and affinity. Chem Biol 2, 633–638, doi: 10.1016/1074-5521(95)90023-3 (1995). [DOI] [PubMed] [Google Scholar]
- 197.Shangguan D, Cao ZC, Li Y & Tan W Aptamers evolved from cultured cancer cells reveal molecular differences of cancer cells in patient samples. Clin Chem 53, 1153–1155, doi: 10.1373/clinchem.2006.083246 (2007). [DOI] [PubMed] [Google Scholar]
- 198.Freage L, Jamal D, Williams NB & Mallikaratchy PR A Homodimeric Aptamer Variant Generated from Ligand-Guided Selection Activates the T Cell Receptor Cluster of Differentiation 3 Complex. Mol Ther Nucleic Acids 22, 167–178, doi: 10.1016/j.omtn.2020.08.016 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Batool S et al. Dimerization of an aptamer generated from Ligand-guided selection (LIGS) yields a high affinity scaffold against B-cells. Biochim Biophys Acta Gen Subj 1863, 232–240, doi: 10.1016/j.bbagen.2018.10.006 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Lee JF, Hesselberth JR, Meyers LA & Ellington AD Aptamer database. Nucleic Acids Res 32, D95–100, doi: 10.1093/nar/gkh094 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Li J, Ma X, Li X & Gu J PPAI: a web server for predicting protein-aptamer interactions. BMC Bioinformatics 21, 236, doi: 10.1186/s12859-020-03574-7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Thodima V, Pirooznia M & Deng Y RiboaptDB: a comprehensive database of ribozymes and aptamers. BMC Bioinformatics 7 Suppl 2, S6, doi: 10.1186/1471-2105-7-S2-S6 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Bourlai E. J. M. a. T . Biclustering for ssDNA aptamer motif prototypes. IEEE-EMBS International Conference on Biomedical and Health Informatics (BHI), 292–295, doi: 10.1109/BHI.2016.7455892 (2016). [DOI] [Google Scholar]
- 204.Cruz-Toledo J et al. Aptamer Base: a collaborative knowledge base to describe aptamers and SELEX experiments. Database (Oxford) 2012, bas006, doi: 10.1093/database/bas006 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Qi S et al. Strategies to manipulate the performance of aptamers in SELEX, post-SELEX and microenvironment. Biotechnology Advances 55, 107902, doi: 10.1016/j.biotechadv.2021.107902 (2022). [DOI] [PubMed] [Google Scholar]
- 206.Kolm C et al. DNA aptamers against bacterial cells can be efficiently selected by a SELEX process using state-of-the art qPCR and ultra-deep sequencing. Sci Rep 10, 20917, doi: 10.1038/s41598-020-77221-9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Wei X, Ma P, Imran Mahmood K, Zhang Y & Wang Z A review: Construction of aptamer screening methods based on improving the screening rate of key steps. Talanta 253, 124003, doi: 10.1016/j.talanta.2022.124003 (2023). [DOI] [Google Scholar]
- 208.Gordon CKL et al. Click-Particle Display for Base-Modified Aptamer Discovery. ACS Chem Biol 14, 2652–2662, doi: 10.1021/acschembio.9b00587 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Wang J et al. Multiparameter Particle Display (MPPD): A Quantitative Screening Method for the Discovery of Highly Specific Aptamers. Angew Chem Int Ed Engl 56, 744–747, doi: 10.1002/anie.201608880 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Hung LY, Wang CH, Hsu KF, Chou CY & Lee GB An on-chip Cell-SELEX process for automatic selection of high-affinity aptamers specific to different histologically classified ovarian cancer cells. Lab Chip 14, 4017–4028, doi: 10.1039/c4lc00587b (2014). [DOI] [PubMed] [Google Scholar]
- 211.Lee S et al. A cross-contamination-free SELEX platform for a multi-target selection strategy. BioChip Journal 7, 38–45, doi: 10.1007/s13206-013-7106-y (2013). [DOI] [Google Scholar]
- 212.Cox JC et al. Automated selection of aptamers against protein targets translated in vitro: from gene to aptamer. Nucleic Acids Res 30, e108, doi: 10.1093/nar/gnf107 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Hunniger T, Wessels H, Fischer C, Paschke-Kratzin A & Fischer M Just in time-selection: A rapid semiautomated SELEX of DNA aptamers using magnetic separation and BEAMing. Analytical chemistry 86, 10940–10947, doi: 10.1021/ac503261b (2014). [DOI] [PubMed] [Google Scholar]
- 214.Townshend B, Xiang JS, Manzanarez G, Hayden EJ & Smolke CD A multiplexed, automated evolution pipeline enables scalable discovery and characterization of biosensors. Nat Commun 12, 1437, doi: 10.1038/s41467-021-21716-0 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Chinnappan R et al. In vitro selection of DNA aptamers and their integration in a competitive voltammetric biosensor for azlocillin determination in waste water. Analytica Chimica Acta 1101, 149–156, doi: 10.1016/j.aca.2019.12.023 (2020). [DOI] [PubMed] [Google Scholar]
- 216.Hong KL & Sooter LJ In Vitro Selection of a Single-Stranded DNA Molecular Recognition Element against the Pesticide Fipronil and Sensitive Detection in River Water. International Journal of Molecular Sciences 19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Bashir A et al. Machine learning guided aptamer refinement and discovery. Nat Commun 12, 2366, doi: 10.1038/s41467-021-22555-9 ().2021 a. The future of aptamer selections is trending toward machine learning and artificial intelligence, this work provides an example of the potential these technologies hold to solve the bottle-neck of selection and post selection processing.
- 218.Perez Tobia J et al. Machine Learning Directed Aptamer Search from Conserved Primary Sequences and Secondary Structures. ACS Synthetic Biology 12, 186–195, doi: 10.1021/acssynbio.2c00462 (2023). [DOI] [PubMed] [Google Scholar]
- 219.Moussa S et al. Diversifying Design of Nucleic Acid Aptamers Using Unsupervised Machine Learning. The Journal of Physical Chemistry B 127, 62–68, doi: 10.1021/acs.jpcb.2c05660 (2023). [DOI] [PubMed] [Google Scholar]
- 220.Iwano N, Adachi T, Aoki K, Nakamura Y & Hamada M Generative aptamer discovery using RaptGen. Nature Computational Science 2, 378–386, doi: 10.1038/s43588-022-00249-6 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Lee SJ, Cho J, Lee B-H, Hwang D & Park J-W Design and Prediction of Aptamers Assisted by In Silico Methods. Biomedicines 11 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Sun D et al. Computational tools for aptamer identification and optimization. TrAC Trends in Analytical Chemistry 157, 116767, doi: 10.1016/j.trac.2022.116767 (2022). [DOI] [Google Scholar]
- 223. Liu R, McConnell EM, Li J & Li Y Advances in functional nucleic acid based paper sensors. Journal of Materials Chemistry B 8, 3213–3230, doi: 10.1039/C9TB02584G (2020). a. A comprehensive review of aptamer applications for environmental monitoring sorted into examples of water, soil, and air monitoring.
- 224.Strack RL, Disney MD & Jaffrey SR A superfolding Spinach2 reveals the dynamic nature of trinucleotide repeat-containing RNA. Nat Methods 10, 1219–1224, doi: 10.1038/nmeth.2701 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Song W et al. Imaging RNA polymerase III transcription using a photostable RNA-fluorophore complex. Nat Chem Biol 13, 1187–1194, doi: 10.1038/nchembio.2477 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Dolgosheina EV et al. RNA mango aptamer-fluorophore: a bright, high-affinity complex for RNA labeling and tracking. ACS Chem Biol 9, 2412–2420, doi: 10.1021/cb500499x (2014). [DOI] [PubMed] [Google Scholar]
- 227.Guan B & Zhang X Aptamers as Versatile Ligands for Biomedical and Pharmaceutical Applications. Int J Nanomedicine 15, 1059–1071 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Afrasiabi S, Pourhajibagher M, Raoofian R, Tabarzad M & Bahador A Therapeutic applications of nucleic acid aptamers in microbial infections. J Biomed Sci 27, 6, doi: 10.1186/s12929-019-0611-0 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Zou X, Wu J, Gu J, Shen L & Mao L Application of Aptamers in Virus Detection and Antiviral Therapy. Front Microbiol 10, 1462, doi: 10.3389/fmicb.2019.01462 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230. Bates PJ, Laber DA, Miller DM, Thomas SD & Trent JO Discovery and development of the G-rich oligonucleotide AS1411 as a novel treatment for cancer. Exp Mol Pathol 86, 151–164, doi: 10.1016/j.yexmp.2009.01.004 (2009). a. As one of the aptamers that has been investigated in clinical trials, and widely applied for cancer applications, this paper is required reading for any aptamer researcher.
- 231.Kwon H-M et al. An RNA Aptamer That Specifically Binds to the Glycosylated Hemagglutinin of Avian Influenza Virus and Suppresses Viral Infection in Cells. PLoS One 9, e97574, doi: 10.1371/journal.pone.0097574 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Bagalkot V, Farokhzad OC, Langer R & Jon S An aptamer-doxorubicin physical conjugate as a novel targeted drug-delivery platform. Angew Chem Int Ed Engl 45, 8149–8152, doi: 10.1002/anie.200602251 (2006). [DOI] [PubMed] [Google Scholar]
- 233.Hicke BJ et al. Tumor targeting by an aptamer. J Nucl Med 47, 668–678 (2006). [PubMed] [Google Scholar]
- 234.Chu TC, Twu KY, Ellington AD & Levy M Aptamer mediated siRNA delivery. Nucleic Acid Res 34, e73–e73, doi: 10.1093/nar/gkl388 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Esposito CL et al. Multifunctional Aptamer-miRNA Conjugates for Targeted Cancer Therapy. Mol Ther 22, 1151–1163, doi: 10.1038/mt.2014.5 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Kim D, Jeong YY & Jon S A Drug-Loaded Aptamer–Gold Nanoparticle Bioconjugate for Combined CT Imaging and Therapy of Prostate Cancer. ACS Nano 4, 3689–3696, doi: 10.1021/nn901877h (2010). [DOI] [PubMed] [Google Scholar]
- 237.Neubacher S & Hennig S RNA Structure and Cellular Applications of Fluorescent Light-Up Aptamers. Angewandte Chemie International Edition 58, 1266–1279, doi: 10.1002/anie.201806482 2019(). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


