Abstract
The toluene/o-xylene monooxygenase cloned from Pseudomonas stutzeri OX1 displays a very broad range of substrates and a very peculiar regioselectivity, because it is able to hydroxylate more than one position on the aromatic ring of several hydrocarbons and phenols. The nucleotide sequence of the gene cluster coding for this enzymatic system has been determined. The sequence analysis revealed the presence of six open reading frames (ORFs) homologous to other genes clustered in operons coding for multicomponent monooxygenases found in benzene- and toluene-degradative pathways cloned from Pseudomonas strains. Significant similarities were also found with multicomponent monooxygenase systems for phenol, methane, alkene, and dimethyl sulfide cloned from different bacterial strains. The knockout of each ORF and complementation with the wild-type allele indicated that all six ORFs are essential for the full activity of the toluene/o-xylene monooxygenase in Escherichia coli. This analysis also shows that despite its activity on both hydrocarbons and phenols, toluene/ o-xylene monooxygenase belongs to a toluene multicomponent monooxygenase subfamily rather than to the monooxygenases active on phenols.
Bacterial enzymatic systems able to oxidize toluene include dioxygenases (43), monooxygenases that oxidize the methyl group (42), and several monooxygenases that catalyze the hydroxylation of the aromatic ring. Among the latter, toluene-2-monooxygenase from Burkholderia (Pseudomonas) cepacia G4 (40) and Pseudomonas sp. strain JS150 (18), toluene-3-monooxygenase from Burkholderia (Pseudomonas) pickettii PKO1 (31), and toluene-4-monooxygenase from Pseudomonas mendocina KR1 (44) have been particularly studied. These enzymes are multicomponent complexes that display a certain regioselectivity for hydroxylation and, usually, a broad range of substrates. For most of these systems, biochemical and/or genetic studies are available (3, 18, 28, 33, 45, 46). Other multicomponent monooxygenases, active on phenols, were identified in phenol-degrading Pseudomonas and Acinetobacter strains (9, 15, 29, 30). Among the monooxygenase systems that recognize toluene, only those from B. cepacia G4 (T2MO) and Pseudomonas sp. strain JS150 (Tb2MO) (18, 39) were able to hydroxylate phenols. Interestingly, the sequence analysis of Tb2MO-encoding genes showed that this system is more similar to phenol hydroxylases than to the other sequenced monooxygenases (18).
Pseudomonas stutzeri OX1 metabolizes o-xylene, in addition to toluene, via a novel catabolic pathway in which toluene/ o-xylene monooxygenase initially hydroxylates toluene, yielding a mixture of o-, m-, and p-cresol, and o-xylene, producing 2,3-dimethylphenol (2,3-DMP) and 3,4-DMP (2). Toluene/ o-xylene monooxygenase is also responsible for the further hydroxylation of the phenolic intermediates arising from the hydrocarbons to (di)methylcatechols (Fig. 1A), and in this respect, it resembles toluene-2-monooxygenase encoded by B. cepacia G4 and Pseudomonas sp. strain JS150 (18, 39).
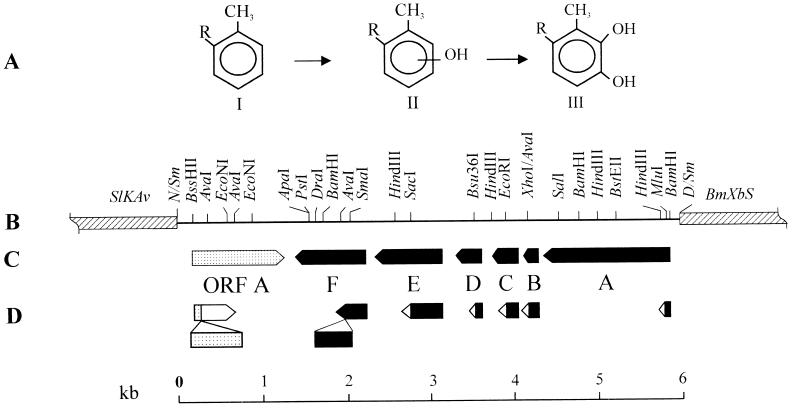
FIG. 1.
Reactions (A) catalyzed by and genetic map (B and C) of the P. stutzeri OX1 locus coding for toluene/o-xylene monooxygenase. (A) I, R=H, toluene; R=CH3, o-xylene; II, R=H, o-, m-, or p-cresol; R=CH3, 2,3- or 3,4-DMP; III, R=H, 3-methylcatechol (4-methylcatechol can also be formed from cresols); R=CH3, 3,4-dimethylcatechol. (B) Restriction endonuclease map of the DNA fragment expressing toluene/o-xylene monooxygenase activity (pBZ1260). Striped boxes represent the vector polylinker with relevant cloning sites. Sl, SalI; K, KpnI; Av, AvaI; N, NotI; Sm, SmaI; D, DraI; Bm, BamHI; Xb, XbaI; S, SalI. (C) The boxes indicate the ORFs identified (also see Table 2), and the points of the arrows indicate the direction of the transcription. In panel D, the mutations introduced in each ORF in turn are schematized (further details are reported in Materials and Methods).
In a previous study, the locus coding for toluene/o-xylene monooxygenase (tou, for toluene/o-xylene utilization) was cloned from the chromosome of P. stutzeri OX1 and mapped to a 6-kb DraI-NotI fragment (2). Based on the locus size and the number of different polypeptides coded for by the region, we postulated that toluene/o-xylene monooxygenase was a multicomponent enzyme. To confirm this, we set out to determine its nucleotide sequence and to show that each open reading frame (ORF) found in the locus was essential for full activity of the monooxygenase.
MATERIALS AND METHODS
Bacteria, plasmids, and general procedures.
Escherichia coli DH5α (14) was grown at 37°C in Luria broth (LB) or in M9 salts medium (19) supplemented with 10 mM malate. Kanamycin and ampicillin were used in selective media at 50 and 100 μg/ml, respectively. Induction of the lacIq-regulated lac promoter of plasmids based on pGEM3Z (Promega) or pVLT33 (5) was performed by addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 1 mM. E. coli cells were transformed with plasmid DNAs by electroporation (8). Plasmid preparations and all DNA manipulations were carried out according to standard procedures (37).
A summary of the plasmids used in this work is given in Table 1. To generate the plasmid pMM3356, which is both compatible with pGEM3Z and carries the entire set of tou genes, the 0.4-kb BssHII-EcoNI fragment downstream of the tou genes was deleted from pBZ1260 (2) (see Fig. 1B for the restriction map), giving rise to plasmid pMZ1256; the insert was then transferred into pVLT33 as a KpnI-XbaI cassette to give pMM3356. To facilitate the mutagenesis procedure, the insert of pMZ1256 was also cloned in pSP72 (Promega), producing plasmid pMZ1257. With the exception of touF, all of the tou genes were mutated by frameshift, with cutting and blunting of the protruding ends of restriction endonuclease sites present inside the genes. touA, touD, and touE were mutated by elimination of the MluI, Bsu36I, and SacI sites of pMZ1256 or pMZ1257. The corresponding inserts were then transferred into pVLT33 as a KpnI-XbaI cassette to give pMM3301, pMM3304, and pMM3305. A touB mutation was obtained by elimination of the XhoI restriction site in pMM3356. The resulting plasmid was pMM3302. The plasmid pMM3303, which carries a mutated touC, was obtained by elimination of the EcoRI site inside the gene upon cloning of the KpnI-XbaI cassette containing the tou genes in a pVLT33 vector previously deprived of its EcoRI site. The plasmid pMM3306, which carries a deletion in touF, was obtained by elimination of the Apa-SmaI fragment downstream of touE in pMM3356.
TABLE 1.
Characteristics of the plasmids used in this study
| Plasmid | Relevant featuresa | Source or reference |
|---|---|---|
| pGEM-3Z | Apr, cloning vector | Promega Corp. |
| pBZ1260 | AprtouABCDEF orfA, 6-kb NotI-DraI fragment of the P. stutzeri OX1 DNA | 2 |
| pMZ1256 | AprtouABCDEF, DraI-EcoNI fragmentb | This work |
| pMZ1201 | AprtouA, DraI-AvaI fragmentb | This work |
| pMZ9002 | AprtouB, SalI-EcoRI fragmentb | This work |
| pMZ1203 | AprtouC, obtained by cloning of the PCR amplification product | This work |
| pMZ1204 | AprtouD, HindIII-HindIII fragmentb | This work |
| pMZ1205 | AprtouE, obtained by cloning of the PCR amplification product | This work |
| pMZ1006 | AprtouF, HindIII-EcoNI fragmentb | This work |
| pVLT33 | Kmr, cloning vector, compatible with pGEM-3Z | 5 |
| pMM3356 | Kmr, as pMZ1256, transferred in pVLT33 as a KpnI-XbaI cassette | This work |
| pMM3301 | Kmr, same insert as pMM3356, touA mutant by blunting of the MluI site | This work |
| pMM3302 | Kmr, same insert as pMM3356, touB mutant by blunting of the XhoI site | This work |
| pMM3303 | Kmr, same insert as pMM3356, touC mutant by blunting of the EcoRI site | This work |
| pMM3304 | Kmr, same insert as pMM3356, touD mutant by blunting of the Bsu36I site | This work |
| pMM3305 | Kmr, same insert as pMM3356, touE mutant by blunting of the SacI site | This work |
| pMM3306 | KmrtouF mutant, DraI-SmaI fragmentb | This work |
Apr, ampicillin resistance; Kmr, kanamycin resistance.
Restriction sites refer to the pBZ1260 map shown in Fig. 1B.
A series of pGEM3Z-based plasmids expressing tou genes separately were generated as follows. Plasmid pMZ1201, which expresses touA, was obtained by deletion in pMZ1256 of a KpnI-AvaI fragment containing the other tou genes. Plasmid pMZ9002, which expresses touB, was obtained by deletion of a 3.8-kb EcoRI fragment in pBZ9047 (2). pMZ1203 and pMZ1205, which express touC and touE, respectively, were obtained by PCR amplification of touC with primers 5′-TGAGCGTGATTTTGTTA-3′ and 5′-CCGCCACATCCCCCGCC-3′ and amplification of touE with primers 5′-CCTTGGGCTTAGACCGG-3′ and 5′-CAGAAAAGAAACCATTG-3′, cloning of the corresponding amplified fragments in pGEMT (Promega), and then transfer of the inserts into pGEM3Z. Plasmid pMZ1204, which expresses touD, was constructed by subcloning the 1.1-kb HindIII-HindIII fragment from pMZ1256 into pGEM3Z. pMZ1006, which expresses touF, was obtained by deletion of the KpnI-EcoNI fragment from pBZ1035 (2).
DNA sequencing and sequence analyses.
Plasmid templates for DNA sequencing were isolated by use of purification kits purchased from Macherey-Nagel-Düren or Qiagen. Nucleotide sequence was determined directly from plasmid pBZ1260 or its derivatives (2) by the dideoxy chain termination technique (38), with the Deaza G/AT7Sequencing Mixes kit according to the supplier’s instructions (Pharmacia Biotech), [α-35S]dATP and T7, SP6, or specific synthetic primers.
The sequence was analyzed with the Genetics Computer Group (GCG; Madison, Wis.) software package, version 7.3 (6). The National Center for Biotechnology Information BLAST program (1) was used to search a nonredundant peptide sequence database (GenBank CDS translations plus PDB plus Swiss-Prot plus SPupdate plus PIR).
Phylogenetic trees showing the relationships between the large and small subunits of the terminal hydroxylase component of several multicomponent monooxygenases were obtained with the program PHYLO_WIN (13), which uses the maximum parsimony algorithms contained in the PHYLIP package (9a).
Enzyme assays.
The rates at which E. coli cells metabolized toluene, o-xylene, m-cresol, and 2,3-DMP were determined by monitoring changes in phenolic compound concentration in the medium with a colorimetric assay developed previously (2). Briefly, a culture grown in minimal medium M9 in the presence of 10 mM malate and 1 mM IPTG was washed twice in 0.1 M phosphate buffer (pH 7.2) and suspended in the same buffer to obtain A600 ≅ 2. Glucose (final concentration, 5 mM) and 70 μl of 3% (vol/vol) toluene or o-xylene in N,N-dimethylformamide, 50 μl of 40 mM m-cresol, or 70 μl of 40 mM 2,3-DMP in water was added to 20 ml of cell suspension. At 5, 10, and 15 min after incubation at 30°C with the substrate, 1-ml samples were collected, and the phenol concentration in the medium was determined. Specific activities were reported as nanomoles of produced or disappeared phenolic compound per minute per milligram of cell proteins.
Analysis of plasmid-encoded polypeptides.
E. coli cells were grown at 37°C in LB until A600 = 0.6 and then supplemented with 1 mM IPTG. One-milliliter samples were collected prior to induction and then at 30, 60, 90, and 180 min after the addition of IPTG. The cells were harvested by centrifugation at 14,000 × g, resuspended in 50 to 100 μl of sodium dodecyl sulfate (SDS) gel loading buffer (50 mM Tris-HCl [pH 6.8], 100 mM dithiothreitol, 2% SDS, 0.1% bromophenol blue, 10% glycerol), boiled for 3 min, and centrifuged at 14,000 × g for 3 min. Ten to fifteen microliters of the supernatants was analyzed by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) (37). The sizes of the polypeptides were determined with Sigma Chemical Co. calibration kits SDS-6H, SDS-7, and MW-SDS-17S.
Nucleotide sequence accession number.
The nucleotide sequence reported here and the amino acid sequences derived from translation of the tou genes have been submitted to the EMBL data bank under accession no. AJ005663.
RESULTS
Nucleotide sequence and sequence analysis of the toluene/o-xylene monooxygenase locus.
The locus coding for toluene/o-xylene monooxygenase was previously mapped to a 6-kb DraI-NotI fragment of a P. stutzeri OX1 chromosome (2) (Fig. 1B). To further characterize the organization and the structure of the genes encoding the toluene/o-xylene monooxygenase, we determined the nucleotide sequence of this tou (toluene/o-xylene utilization) locus. Translation of the sequence in all of the reading frames possible revealed a cluster of six ATG-starting ORFs, preceded by a potential ribosome binding site and designated touABCDEF (Fig. 1C and Table 2), in the direction the transcription of the locus was shown to occur (2). Near the stop codon of touF, a potential dyad symmetry structure resembling a rho factor-independent terminator was present. The average G+C content was approximately 50%, which was low compared with that of the chromosomes of Pseudomonas species (60 to 66%) (24).
TABLE 2.
Sequence analysis of the P. stutzeri OX1 toluene/o-xylene monooxygenase locus
| Coordinates | tou ORF | Amino acid no. | Molecular mass (kDa) of the product
|
|
|---|---|---|---|---|
| Expected | Estimateda | |||
| 210–1706 | touA | 498 | 57.6 | 58 |
| 1742–2002 | touB | 86 | 10.3 | 9 |
| 2017–2355 | touC | 112 | 12.5 | 12 |
| 2542–2874 | touD | 110 | 12.3 | 27 |
| 2905–3897 | touE | 330 | 38.3 | 38 |
| 3979–4977 | touF | 332 | 36.6 | 40 |
| 5130–6158b | orfA | 342 | 38.8 | NDc |
SDS-PAGE.
Reverse sequence.
ND, not determined.
Comparison of the six predicted tou polypeptide sequences with those of a nonredundant peptide sequence database revealed a high degree of similarity with the six subunits of toluene and benzene multicomponent monooxygenases from B. pickettii PKO1 (3), P. mendocina KR1 (45, 46), B. cepacia AA1 (23), and Pseudomonas aeruginosa JI104 (21). In addition to those in toluene/benzene multicomponent monooxygenases, lower but significant similarities to polypeptides from other enzyme systems were also found, most notably, the phenol hydroxylases from Pseudomonas (15, 29, 30) and Acinetobacter (9) strains, the toluene/benzene-2-monooxygenase (BMO) from Pseudomonas sp. strain JS150 (18), the soluble methane monooxygenases from different methanotrophs (4, 26, 41), the epoxidase from Nocardia corallina B-276 (36), and dimethyl sulfide oxygenase from Acinetobacter sp. strain 20B (16) (Table 3).
TABLE 3.
Pairwise comparisons of amino acid sequences translated from touABCDEF with those of similar proteins
| tou gene | Similar gene | Organisma | Degradative pathway for substrate | % Identityb | % Similarityb | Reference |
|---|---|---|---|---|---|---|
| touA | bmoA | P. aeruginosa JI104 | Benzene | 73.1 | 77.9 | 21 |
| tbuA1 | B. pickettii PKO1 | Toluene | 68.0 | 75.5 | 3 | |
| tmoA | P. mendocina KR1 | Toluene | 66.8 | 75.1 | 46 | |
| tbhA | B. cepacia AA1 | Toluene | 67.6 | 72.7 | 23 | |
| tbmD | Pseudomonas sp. strain JS150 | Toluene | 27.3 | 37.0 | 18 | |
| dmpN | P. putida CF600 | Phenol | 27.6 | 37.5 | 30 | |
| mmoX | M. capsulatus (Bath) | Methane | 27.2 | 33.7 | 41 | |
| dsoD | Acinetobacter sp. strain 20B | Dimethyl sulfide | 30.1 | 38.8 | 16 | |
| amoC | N. corallina B-276 | Alkene | 25.1 | 32.4 | 36 | |
| touB | bmoB | P. aeruginosa JI104 | Benzene | 50.0 | 66.3 | 21 |
| tbuU | B. pickettii PKO1 | Toluene | 52.3 | 63.9 | 3 | |
| tmoB | P. mendocina KR1 | Toluene | 53.6 | 69.1 | 46 | |
| tbhB | B. cepacia AA1 | Toluene | 35.2 | 43.6 | 23 | |
| touC | bmoC | P. aeruginosa JI104 | Benzene | 64.8 | 76.5 | 21 |
| tbuB | B. pickettii PKO1 | Toluene | 52.2 | 63.9 | 3 | |
| tmoC | P. mendocina KR1 | Toluene | 57.1 | 70.5 | 46 | |
| tbhC | B. cepacia AA1 | Toluene | 57.6 | 66.6 | 23 | |
| touD | bmoD1 | P. aeruginosa JI104 | Benzene | 56.8 | 64.2 | 21 |
| tbuV | B. pickettii PKO1 | Toluene | 58.6 | 70.2 | 3 | |
| tmoD | P. mendocina KR1 | Toluene | 63.1 | 70.8 | 46 | |
| tbhD | B. cepacia AA1 | Toluene | 54.8 | 63.4 | 23 | |
| tbmC | Pseudomonas sp. strain JS150 | Toluene | 38.2 | 46.1 | 18 | |
| dmpM | P. putida CF600 | Phenol | 34.4 | 43.3 | 30 | |
| mmoB | M. capsulatus (Bath) | Methane | 27.4 | 36.8 | 41 | |
| dsoC | Acinetobacter sp. strain 20B | Dimethyl sulfide | 31.4 | 43.8 | 16 | |
| touE | tbuA2 | B. pickettii PKO1 | Toluene | 65.6 | 72.3 | 3 |
| tbhE | B. cepacia AA1 | Toluene | 58.2 | 63.1 | 23 | |
| tmoE | P. mendocina KR1 | Toluene | 56.2 | 63.3 | 46 | |
| tbmB | Pseudomonas sp. strain JS150 | Toluene | 22.7 | 31.7 | 18 | |
| dmpL | P. putida CF600 | Phenol | 27.1 | 36.4 | 30 | |
| dsoB | Acinetobacter sp. strain 20B | Dimethyl sulfide | 23.8 | 33.1 | 16 | |
| amoA | N. corallina B-276 | Alkene | 24.9 | 34.1 | 36 | |
| touF | tbuC | B. pickettii PKO1 | Toluene | 40.9 | 51.1 | 3 |
| tbhF | B. cepacia AA1 | Toluene | 39.4 | 46.5 | 23 | |
| tmoF | P. mendocina KR1 | Toluene | 35.3 | 44.8 | 45 | |
| tbmF | Pseudomonas sp. strain JS150 | Toluene | 30.2 | 45.1 | 18 | |
| dmpP | P. putida CF600 | Phenol | 28.0 | 39.7 | 30 | |
| mmoC | M. capsulatus (Bath) | Methane | 23.4 | 33.3 | 41 | |
| dsoF | Acinetobacter sp. strain 20B | Dimethyl sulfide | 27.9 | 38.1 | 16 | |
| amoD | N. corallina B-276 | Alkene | 30.4 | 41.6 | 36 |
Only one representative strain is reported for substrates other than toluene.
The percentages of identity and similarity were obtained from overall pairwise comparisons of amino acid sequences by using the GCG program GAP with a gap weight of 30 and a gap length weight of 0.1.
TouA, TouD, TouE, and TouF were similar to proteins found in several multicomponent monooxygenases. In particular, TouA and TouE were similar to the large and small oxygenase subunits. The large subunit of the multicomponent monooxygenases is characterized by a region resembling a dinuclear iron binding ligand identified by a pair of fixed-space domains with the amino acid sequence Asp-Glu-X-Arg-His (10, 11). In TouA, this motif was found between amino acids 130 and 137 and 230 and 234 and was also perfectly conserved (multialignment not shown). TouF was similar to the NADH-ferredoxin oxidoreductase components of several mono- and dioxygenase systems. These peptides possess an N-terminal region similar to that of the chloroplast-type ferredoxins, in which Cys (Cys37, -42, -45, and -77 in TouF) and Gly (Gly 40 and -52 in TouF), involved in the coordination of two iron atoms of the [2Fe-2S] cluster, are conserved. The C-terminal region resembles oxidoreductases from bacterial, yeast, plant, and human origins. In this region of TouF, we found four conserved regions (amino acids 141 to 154, 166 to 196, 208 to 245, and 289 to 318) involved in NADH or FADH interaction (27, 45).
In contrast, the only similarity TouB showed was to a small peptide found exclusively in benzene/toluene multicomponent monooxygenases. In the case of toluene-4-monooxygenase, it was shown to take part in the terminal hydroxylase component (33).
TouC shared the highest degree of similarity with polypeptides in benzene/toluene multicomponent monooxygenases. However, the region of TouC between amino acids 40 and 70 was perfectly alignable with a domain present in a large number of proteins categorized as ferredoxin types, in which two conserved Cys residues followed by His residues (Cys45, His47, and Cys64, His67 in TouC), putatively involved in the coordination of a Rieske-type iron-sulfur cluster, are present (25, 32).
The translation of the DNA sequence downstream of touABCDEF revealed, on the opposite strand, the presence of an ORF, named ORF A, potentially coding for a polypeptide with a size of 38.8 kDa (Fig. 1C and Table 2). This ORF was similar to some transposase genes found in Alcaligenes eutrophus IS1086 (7) (73.7% similarity) and Escherichia hermannii IS30 (22) (59.8% similarity).
Characterization of the polypeptides coded for by touABCDEF and complementation analysis.
The product of each tou gene was analyzed by SDS-PAGE. As expected from sequencing data (Table 2), the molecular masses of TouA, TouB, TouC, and TouE were estimated to be 58, 9, 12, and 38 kDa, respectively. The estimated molecular mass of TouF was 40 kDa. This value was slightly higher than expected and might be due to the acidic negatively charged N-terminal region, which can retard migration, as previously observed with other chloroplast-type ferredoxins (17). The estimated molecular mass of TouD was 27 kDa, more than twice the expected mass. A similar overestimation of molecular mass in SDS-PAGE was also observed with the TmoC peptide (46). The low molecular mass along with the acidity of the peptide (pI 4.45) (20) or the incomplete reduction of a dimeric form might account for this result.
To provide genetic evidence that each Tou polypeptide was involved in the formation of the toluene/o-xylene monooxygenase complex, a single mutation was introduced into each of the tou genes, and its effect on the enzymatic activity was investigated. We previously suggested (2) that this enzymatic complex was also responsible for the second step of the toluene/o-xylene catabolic pathway, which is the formation of catechols from the phenolic intermediates. The mutant gene clusters were thus assayed both for phenolic compound production from toluene or o-xylene and for phenolic compound consumption.
The enzymatic system encoded by the wild-type touABCDEF gene cluster was able to hydroxylate both hydrocarbons and phenols (Table 4). Decreasing levels of activity were observed when the substrates, in the following order, were toluene, m-cresol, o-xylene, and 2,3-DMP, suggesting that the activity is affected more by the presence of a second methyl group than by that of a hydroxyl group. The mutations in touA, touB, touD, and touE led to a complete loss of activity. The mutation in touC did not completely abolish monooxygenase activity and affected the oxidation of hydrocarbons more than that of phenolic compounds, with 93 to 95% and 68 to 80% reductions, respectively. Finally, as was also observed for the TmoF subunit of P. mendocina KR1 (45), the deletion of touF caused a reduction in enzymatic activity ranging from 26 to 73%, depending on the substrate (Table 4).
TABLE 4.
Complementation between individually cloned tou genes and the tou gene clusters carrying the corresponding mutation
| Plasmid(s)a | tou genesb | IPTG addition | Toluene/o-xylene monooxygenase sp act (nmol min−1 mg of protein−1) on substrate
|
|||
|---|---|---|---|---|---|---|
| Step Ic
|
Step IId
|
|||||
| Toluene | o-Xylene | m-Cresol | 2,3-DMP | |||
| pMM3356 | ABCDEF | + | 10.00 | 4.71 | 6.25 | 3.50 |
| − | 2.30 | 0.00 | 2.30 | 2.00 | ||
| pMM3301 | ABCDEF | + | 0.00 | 0.00 | 0.34 | 0.00 |
| − | 0.00 | 0.00 | 0.00 | 0.00 | ||
| pMM3302 | ABCDEF | + | 0.00 | 0.00 | 0.00 | 0.00 |
| − | 0.00 | 0.00 | 0.00 | 0.00 | ||
| pMM3303 | ABCDEF | + | 0.72 | 0.24 | 1.28 | 1.12 |
| − | 0.00 | 0.00 | 0.00 | 0.00 | ||
| pMM3304 | ABCDEF | + | 0.00 | 0.00 | 0.00 | 0.00 |
| − | 0.00 | 0.00 | 0.00 | 0.00 | ||
| pMM3305 | ABCDEF | + | 0.00 | 0.00 | 0.00 | 0.00 |
| − | 0.00 | 0.00 | 0.00 | 0.00 | ||
| pMM3306 | ABCDEF | + | 4.31 | 1.30 | 4.65 | 1.40 |
| − | 0.00 | 0.00 | 0.00 | 0.96 | ||
| pMM3301, pMZ1201 | ABCDEF + A | + | 2.23 | 1.23 | 1.57 | 2.47 |
| − | 0.00 | 0.00 | 0.00 | 0.70 | ||
| pMM3302, pMZ9002 | ABDEF + B | + | 4.16 | 0.75 | 2.61 | 0.83 |
| − | 0.00 | 0.00 | 0.00 | 0.00 | ||
| pMM3303, pMZ1203 | ABCDEF + C | + | 2.94 | 0.78 | 5.42 | 2.08 |
| − | 0.00 | 0.00 | 0.00 | 0.00 | ||
| pMM3304, pMZ1204 | ABCDEF + D | + | 11.47 | 2.87 | 5.05 | 2.35 |
| − | 0.67 | 0.32 | 0.84 | 0.71 | ||
| pMM3305, pMZ1205 | ABCDEF + E | + | 5.52 | 1.83 | 6.94 | 2.79 |
| − | 0.89 | 0.26 | 0.77 | 0.69 | ||
| pMM3306, pMZ1006 | ABCDEF + F | + | 6.45 | 3.00 | 6.45 | 2.62 |
| − | 2.20 | 0.00 | 0.00 | 1.40 | ||
Construction of each plasmid is described in Materials and Methods. The host was E. coli DH5α.
The mutated genes are indicated in boldface.
Conversion of the specified hydrocarbon into a phenolic compound.
Consumption of the specified phenolic compound.
The frameshift mutations introduced in each tou gene could exert some degree of polarity on downstream genes. To rule out the possibility that the mutant phenotypes were due exclusively to a polar effect, complementation tests between plasmids carrying the single mutations and those carrying only one of the tou genes were performed. As shown in Table 4, each mutation could be complemented by the corresponding wild-type tou gene, rescuing 16 to 100% of the activity levels measured in pMM3356-carrying cells. The incomplete complementation observed in some cases might be due to a polar effect of the mutation in the gene cluster or to a disproportion in plasmid copy number. In accordance with the activity levels measured with the wild-type system, the percentages of the activity levels rescued in complementation assays were higher with monomethylated substrates than with the dimethylated ones.
DISCUSSION
The nucleotide sequence of the locus coding for P. stutzeri OX1 toluene/o-xylene monooxygenase revealed six ORFs, designated touABCDEF, which showed relevant similarities to the subunits of several enzymatic complexes involved in the monooxygenation of aromatic compounds. Each gene found in the locus was shown to be essential for the full enzymatic activity. These results provide genetic evidence that toluene/o-xylene monooxygenase, the enzyme responsible for the initial steps of toluene and o-xylene catabolism in P. stutzeri OX1, is a multicomponent monooxygenase.
The gene cluster encoding the P. stutzeri toluene/o-xylene monooxygenase has a GC ratio similar to, and the same gene arrangement as, the tbu, tmo, tbh, and bmo operons of the toluene monooxygenases from B. pickettii PKO1 (3), P. mendocina KR1 (45, 46), B. cepacia AA1 (23), and P. aeruginosa JI104 (21). These data, together with the presence of a putative transposase (ORF A), suggest that these genes might have been recently acquired by gene transfer from other bacteria. Further investigations are required to confirm this hypothesis.
Comparison of Tou polypeptides with those belonging to more-characterized systems led us to hypothesize for them a role in a four-component monooxygenase.
TouF and TouC may represent the components of the electron transport chain. TouF is presumably necessary for NADH oxidation and for the transport of the two reduction equivalents to the central Rieske-type ferredoxin (TouC). ORFs having the Rieske-type motif or ferredoxin-like motifs were found in virtually all of the aromatic compound-hydroxylating complexes. In the two-component systems, the NADH-ferredoxin reductase activity is due to a single component (i.e., XylA or BenC) that probably evolved from the fusion of an NADH reductase with a ferredoxin (27). In three- or four-component systems, a Rieske-type ferredoxin that transfers the electrons from the NADH reductase to the terminal oxygenase is present. The Rieske-type ferredoxin was found to be essential for reconstruction of NADH-dependent catalytic activity of T4MO in vitro, by mediating electron transfer between the reductase and the hydroxylase (33). In the cloned P. stutzeri OX1 monooxygenase, the knockout of either touF or touC did not lead to a complete loss of activity. Due to their role, it may be suggested that both functions can be at least partially accomplished by host proteins.
For polypeptides such as TouD, a regulatory function of the catalysis has been postulated (3, 12, 33, 35) but has not yet been demonstrated. In the case of DmpM protein from Pseudomonas putida CF600, its interaction with both the hydroxylase component and phenol has been suggested (35). Pikus et al. (33) demonstrated that TmoD is a high-affinity component of the T4MO complex rather than a subunit of the hydroxylase and suggested that it may have a role related to catalysis. Consistent with these hypotheses, in our in vivo experiments, the knockout of the touD gene led to a complete loss of activity, and complementation with the wild-type touD made it possible to rescue 60 to 100% of the wild-type activity.
TouA, TouB, and TouE may represent the three peptides constituting the catalytic subunit of the enzymatic complex. Indeed, the knockout of each of the corresponding genes led to a complete loss of activity with every substrate. A three-polypeptide terminal oxygenase was also found in the P. putida CF600 phenol hydroxylase (34) and the B. cepacia G4 T2MO complex (28), but TouB and similar peptides seem to characterize the monooxygenases active on toluene and benzene.
Further proof that toluene/o-xylene monooxygenase from P. stutzeri OX1 is closely related to toluene monooxygenases comes from phylogenetic analysis (not shown) of the large and small subunits of the terminal hydroxylase component of several multicomponent monooxygenases. In fact, the TouA and TouE proteins from P. stutzeri OX1 are included in the same group as BMO, T3MO, and T4MO from P. aeruginosa JI104 (21), B. pickettii PKO1 (3), B. cepacia AA1 (23), and P. mendocina KR1 (46), which could be defined as a toluene subfamily. Especially in the case of the large subunit, similar subgroups can be recognized for phenol and methane monooxygenases. The only toluene monooxygenase which appears to be an exception is Tb2MO from Pseudomonas sp. strain JS150, which, despite its activity on hydrocarbons, was previously found to be more similar to phenol monooxygenases than to toluene monooxygenases (18).
Based on the genetic analysis, the P. stutzeri OX1 toluene/o-xylene monooxygenase can be considered to belong to a toluene monooxygenase subfamily; however, it is peculiar from a biochemical point of view. In fact, in comparison to the other toluene monooxygenases, it displays a broader range of substrates, recognizing both hydrocarbons and phenols, and a more relaxed regioselectivity of aromatic ring hydroxylation, being able to hydroxylate more than one position on both natural and nonnatural substrates (2). It thus combines the specificity and the regioselectivity of all of the enzymes belonging to the toluene monooxygenase subfamily and of Tb2MO from Pseudomonas sp. strain JS150 (18), T2MO from B. cepacia G4 (28, 40), and multicomponent phenol hydroxylases (9, 15, 29, 30).
Despite their genetic similarity, the enzymatic systems belonging to the toluene monooxygenase subfamily are thus shown to display different regioselectivities and different substrate specificities. Further efforts are under way to isolate determinants that affect their biochemical properties.
ACKNOWLEDGMENTS
This work was supported by the Ministero dell’Università e della Ricerca Scientifica e Tecnologica and by Piano Nazionale Biotecnologie Vegetali, MIRAAF (Rome).
We are grateful to F. Bolognese for technical support, M. Pinti for collaboration with the experimental work, J. R. Valverde for computer assistance with sequence analysis, and I. Cases and V. de Lorenzo for inspiring discussions.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Bertoni G, Bolognese F, Galli E, Barbieri P. Cloning of the genes for and characterization of the early stages of toluene and o-xylene catabolism in Pseudomonas stutzeri OX1. Appl Environ Microbiol. 1996;62:3704–3711. doi: 10.1128/aem.62.10.3704-3711.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Byrne A M, Kukor J J, Olsen R H. Sequence analysis of the gene cluster encoding toluene-3-monooxygenase from Pseudomonas pickettii PKO1. Gene. 1995;154:65–70. doi: 10.1016/0378-1119(94)00844-i. [DOI] [PubMed] [Google Scholar]
- 4.Cardy D L N, Laidler V, Salmond G P C, Murrell J C. Molecular analysis of the methane monooxygenase (MMO) gene cluster of Methylosinus trichosporium OB3b. Mol Microbiol. 1991;5:335–342. doi: 10.1111/j.1365-2958.1991.tb02114.x. [DOI] [PubMed] [Google Scholar]
- 5.de Lorenzo V, Eltis L, Kessler B, Timmis K N. Analysis of Pseudomonas gene products using lacIq/Ptrp-lac plasmids and transposons that confer conditional phenotypes. Gene. 1993;123:17–24. doi: 10.1016/0378-1119(93)90533-9. [DOI] [PubMed] [Google Scholar]
- 6.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dong Q, Sadouk A, van der Lelie D, Taghavi S, Ferhat A, Nuyten J M, Borremans B, Mergeay M, Toussaint A. Cloning and sequencing of IS1086, an Alcaligenes eutrophus insertion element related to IS30 and IS4351. J Bacteriol. 1992;174:8133–8138. doi: 10.1128/jb.174.24.8133-8138.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dower W J, Miller J F, Ragsdale C W. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 1988;16:6125–6145. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ehrt S, Schirmer F, Hillen W. Genetic organization, nucleotide sequence and regulation of expression of genes encoding phenol hydroxylase and catechol 1,2-dioxygenase in Acinetobacter calcoaceticus NCIB8250. Mol Microbiol. 1995;18:13–20. doi: 10.1111/j.1365-2958.1995.mmi_18010013.x. [DOI] [PubMed] [Google Scholar]
- 9a.Felsenstein J. PHYLIP (phylogenetic inference package), version 3.57c. Seattle: Department of Genetics, University of Washington; 1993. [Google Scholar]
- 10.Fox B G, Shanklin J, Sommerville C, Münck E. Stearoyl-acyl carrier protein delta 9-desaturase from Ricinus communis is a diiron-oxo protein. Proc Natl Acad Sci USA. 1993;90:2486–2490. doi: 10.1073/pnas.90.6.2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fox B G, Surerus K K, Münck E, Lipscomb J D. Evidence for a μ-oxobridged binuclear iron cluster in the hydroxylase component of methane monooxygenase. J Biol Chem. 1988;263:10553–10556. [PubMed] [Google Scholar]
- 12.Froland W A, Andersson K K, Lee S K, Liu Y, Lipscomb J D. Methane monooxygenase component B and reductase alter the regioselectivity of the hydroxylase component-catalyzed reactions. A novel role for protein-protein interactions in an oxygenase mechanism. J Biol Chem. 1992;267:17588–17597. [PubMed] [Google Scholar]
- 13.Galtier N, Gouy M, Gautier C. SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. CABIOS. 1996;12:543–548. doi: 10.1093/bioinformatics/12.6.543. [DOI] [PubMed] [Google Scholar]
- 14.Hanahan D. Techniques for transformation of E. coli. In: Glover D M, editor. DNA cloning. The practical approach. Vol. 1. Oxford, United Kingdom: IRL Press, Ltd.; 1985. pp. 109–136. [Google Scholar]
- 15.Herrmann H, Muller C, Schmidt I, Mahnke J, Petruschka L, Hahnke K. Localization and organization of phenol degradation genes of Pseudomonas putida strain H. Mol Gen Genet. 1995;247:240–246. doi: 10.1007/BF00705655. [DOI] [PubMed] [Google Scholar]
- 16.Horinouchi M, Kasuga K, Nojiri H, Yamane H, Omori T. Cloning and characterization of genes encoding an enzyme which oxidizes dimethyl sulfide in Acinetobacter sp. strain 20B. FEMS Microbiol Lett. 1997;155:99–105. doi: 10.1111/j.1574-6968.1997.tb12692.x. [DOI] [PubMed] [Google Scholar]
- 17.Huisman J G, Mooreman A F M, Verkley F N. In vitro synthesis of chloroplast ferredoxin as a high molecular weight precursor in a cell-free protein synthesizing system from wheat germs. Biochem Biophys Res Commun. 1978;82:1121–1131. doi: 10.1016/0006-291x(78)90303-0. [DOI] [PubMed] [Google Scholar]
- 18.Johnson G R, Olsen R H. Nucleotide sequence analysis of genes encoding a toluene/benzene-2-monooxygenase from Pseudomonas sp. strain JS150. Appl Environ Microbiol. 1995;61:3336–3346. doi: 10.1128/aem.61.9.3336-3346.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kahn M, Kolter R, Thomas C M, Figurski D, Meyer R, Remaut E, Helinski D R. Plasmid cloning vehicles derived from plasmid ColE1, F, RK6 and RK2. Methods Enzymol. 1979;68:268–280. doi: 10.1016/0076-6879(79)68019-9. [DOI] [PubMed] [Google Scholar]
- 20.Kaufmann E, Geisler N, Weber K. SDS-PAGE strongly overestimates the molecular masses of the neurofilament proteins. FEBS Lett. 1984;170:81–84. doi: 10.1016/0014-5793(84)81373-3. [DOI] [PubMed] [Google Scholar]
- 21.Kitayama A, Suzuki E, Kawakami Y, Nagamune T. Gene organization and low regiospecificity in aromatic-ring hydroxylation of a benzene monooxygenase of Pseudomonas aeruginosa JI104. J Ferment Bioeng. 1996;82:421–425. [Google Scholar]
- 22.Lawrence J G, Ochman H, Hartl D L. The evolution of insertion sequences within enteric bacteria. Genetics. 1992;131:9–20. doi: 10.1093/genetics/131.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma, Y., and D. S. Herson. 1997. GenBank accession no. AF001356.
- 24.Mandel M. Deoxyribonucleic acid base composition in the genus Pseudomonas. J Gen Microbiol. 1966;43:273–292. doi: 10.1099/00221287-43-2-273. [DOI] [PubMed] [Google Scholar]
- 25.Mason J R, Cammack R. The electron-transport proteins of hydroxylating bacterial dioxygenase. Annu Rev Microbiol. 1992;46:277–305. doi: 10.1146/annurev.mi.46.100192.001425. [DOI] [PubMed] [Google Scholar]
- 26.McDonald I R, Uchiyama H, Kambe S, Yagi O, Murrell J C. The soluble methane monooxygenase gene cluster of the trichloroethylene-degrading methanotroph Methylocystis sp. strain M. Appl Environ Microbiol. 1997;63:1898–1904. doi: 10.1128/aem.63.5.1898-1904.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neidle E L, Hartnett C, Ornston L N, Bairoch A, Rekik M, Harayama S. Nucleotide sequence of the Acinetobacter calcoaceticus benABC genes for benzoate 1,2-dioxygenase reveal evolutionary relationships among multicomponent oxygenases. J Bacteriol. 1991;173:5385–5395. doi: 10.1128/jb.173.17.5385-5395.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Newmann L M, Wackett L P. Purification and characterization of toluene-2-monooxygenase from Burkholderia cepacia G4. Biochemistry. 1995;34:14066–14076. doi: 10.1021/bi00043a012. [DOI] [PubMed] [Google Scholar]
- 29.Ng L C, Shingler V, Sze C C, Poh C L. Cloning and sequences of the first eight genes of the chromosomally encoded (methyl)phenol degradation pathway from Pseudomonas putida P35X. Gene. 1994;151:29–36. doi: 10.1016/0378-1119(94)90629-7. [DOI] [PubMed] [Google Scholar]
- 30.Nordlund I, Powlowski J, Shingler V. Complete nucleotide sequence and polypeptide analysis of multicomponent phenol hydroxylase from Pseudomonas sp. strain CF600. J Bacteriol. 1990;172:6826–6833. doi: 10.1128/jb.172.12.6826-6833.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Olsen R H, Kukor J J, Kaphammer B. A novel toluene-3-monooxygenase pathway cloned from Pseudomonas pickettii PKO1. J Bacteriol. 1994;176:3749–3756. doi: 10.1128/jb.176.12.3749-3756.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Otaka E, Ooi T. Examination of protein sequence homologies. V. New perspectives on evolution between bacterial and chloroplast-type ferredoxins inferred from sequence evidence. J Mol Evol. 1989;29:246–254. doi: 10.1007/BF02100208. [DOI] [PubMed] [Google Scholar]
- 33.Pikus J D, Studts J M, Achim C, Kauffmann K E, Münk E, Steffan R J, McClay K, Fox B G. Recombinant toluene-4-monooxygenase: catalytic and Mössbauer studies of the purified di-iron and Rieske components of a four-protein complex. Biochemistry. 1996;35:9106–9119. doi: 10.1021/bi960456m. [DOI] [PubMed] [Google Scholar]
- 34.Powlowski J, Shingler V. Genetics and biochemistry of phenol degradation by Pseudomonas sp. CF600. Biodegradation. 1994;5:219–236. doi: 10.1007/BF00696461. [DOI] [PubMed] [Google Scholar]
- 35.Qian H, Edlund U, Powlowsky J, Shingler V, Sethson I. Solution structure of phenol hydroxylase protein component P2 determined by NMR spectroscopy. Biochemistry. 1997;36:495–504. doi: 10.1021/bi9619233. [DOI] [PubMed] [Google Scholar]
- 36.Saeki H, Furuhashi K. Cloning and characterization of a Nocardia corallina B-276 gene cluster encoding alkene monooxygenase. J Ferment Bioeng. 1994;78:339–406. [Google Scholar]
- 37.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 38.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shields M S, Montgomery S O, Chapman P J, Cuskey S M, Pritchard P H. Novel pathway of toluene catabolism in the trichloroethylene-degrading bacterium G4. Appl Environ Microbiol. 1989;55:1624–1629. doi: 10.1128/aem.55.6.1624-1629.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shields M S, Montgomery S O, Cuskey S M, Chapman P J, Pritchard P H. TOM, a new aromatic degradative plasmid from Burkholderia (Pseudomonas) cepacia G4. Appl Environ Microbiol. 1995;61:1352–1356. doi: 10.1128/aem.61.4.1352-1356.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stainthorpe A C, Lees V, Salmond G P C, Dalton H, Murrell J C. The methane monooxygenase gene cluster of Methylococcus capsulatus (Bath) Gene. 1990;91:27–34. doi: 10.1016/0378-1119(90)90158-n. [DOI] [PubMed] [Google Scholar]
- 42.Suzuki M, Hayakawa T, Shaw J P, Rekik M, Harayama S. Primary structure of xylene monooxygenase: similarities to and differences from the alkane hydroxylation system. J Bacteriol. 1991;173:1690–1695. doi: 10.1128/jb.173.5.1690-1695.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wackett L P. Toluene dioxygenase from Pseudomonas putida F1. Methods Enzymol. 1990;188:39–45. doi: 10.1016/0076-6879(90)88010-8. [DOI] [PubMed] [Google Scholar]
- 44.Whited G M, Gibson D T. Toluene-4-monooxygenase, a three-component enzyme system that catalyzes the oxidation of toluene to p-cresol in Pseudomonas mendocina KR1. J Bacteriol. 1991;173:3010–3016. doi: 10.1128/jb.173.9.3010-3016.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yen K-M, Karl M R. Identification of a new gene, tmoF, in the Pseudomonas mendocina KR1 gene cluster encoding toluene-4-monooxygenase. J Bacteriol. 1992;174:7253–7261. doi: 10.1128/jb.174.22.7253-7261.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yen K-M, Karl M R, Blatt L M, Simon M J, Winter R B, Fausset P R, Lu H S, Harcourt A A, Chen K K. Cloning and characterization of a Pseudomonas mendocina KR1 gene cluster encoding toluene-4-monooxygenase. J Bacteriol. 1991;173:5315–5327. doi: 10.1128/jb.173.17.5315-5327.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]