Abstract
The presence of a highly conserved nahAc allele among phylogenetically diverse bacteria carrying naphthalene-catabolic plasmids provided evidence for in situ horizontal gene transfer at a coal tar-contaminated site (J. B. Herrick, K. G. Stuart-Keil, W. C. Ghiorse, and E. L. Madsen, Appl. Environ. Microbiol. 63:2330–2337, 1997). The objective of the present study was to identify and characterize the different-sized naphthalene-catabolic plasmids in order to determine the probable mechanism of horizontal transfer of the nahAc gene in situ. Filter matings between naphthalene-degrading bacterial isolates and their cured progeny revealed that the naphthalene-catabolic plasmids were self-transmissible. Limited interstrain transfer was also found. Analysis of the restriction fragment length polymorphism (RFLP) patterns indicated that catabolic plasmids from 12 site-derived isolates were closely related to each other and to the naphthalene-catabolic plasmid (pDTG1) of Pseudomonas putida NCIB 9816-4, which was isolated decades ago in Bangor, Wales. The similarity among all site-derived naphthalene-catabolic plasmids and pDTG1 was confirmed by using the entire pDTG1 plasmid as a probe in Southern hybridizations. Two distinct but similar naphthalene-catabolic plasmids were retrieved directly from the microbial community indigenous to the contaminated site in a filter mating by using a cured, rifampin-resistant site-derived isolate as the recipient. RFLP patterns and Southern hybridization showed that both of these newly retrieved plasmids, like the isolate-derived plasmids, were closely related to pDTG1. These data indicate that a pDTG1-like plasmid is the mobile genetic element responsible for transferring naphthalene-catabolic genes among bacteria in situ. The pervasiveness and persistence of this naphthalene-catabolic plasmid suggest that it may have played a role in the adaptation of this microbial community to the coal tar contamination at our study site.
Microbial adaptation to environmental change may be facilitated by transfer and sorting of genetic material between and within naturally occurring populations (35). Horizontal gene transfer (HGT) is a major mechanism for microorganisms to acquire new metabolic traits in new combinations. As such, HGT has important implications in the spread of antibiotic resistance (8, 42), in assessment of the risk of releasing genetically engineered organisms (17, 41, 69), and in the effectiveness of microbially mediated control of environmental contamination (36, 52, 65). The occurrence and impact of HGT in naturally occurring microbial communities have been documented by using a variety of complementary experimental procedures that fall into two general categories: retrospective and mechanistic.
In the retrospective approach, HGT is inferred after the discovery of consistent phylogenetic patterns of particular genes, operons, or plasmids which contrast with those of their hosts. One far-reaching example of the retrospective phylogenetic approach involves the functional and evolutionary relationships among oxygenase enzymes that have been established by examining sequence similarities and operon structures in homologous genes coding for aromatic hydrocarbon metabolism (23, 65, 66, 72). Narrower, often site-specific examination of recently isolated bacteria and their genes has also shown or suggested HGT. In this regard, Fulthorpe et al. (19) found that several genes involved in 2,4-dichlorophenoxyacetic acid (2,4-D) metabolism in 32 bacteria from diverse locations were arranged in mosaics and concluded that recombination had played a major role in the evolution of 2,4-D-catabolic bacteria. In a related study, when Ka et al. (30) examined 2,4-D-degrading bacteria from eight different field plots by using catabolic gene probes, they found that a large group of taxonomically diverse isolates shared the tfdA, tfdB, tfdC, and tfdD genes on mobile plasmids. A closer examination of the plasmids from Alcaligenes paradoxus 2811P and Pseudomonas pickettii 712 isolates revealed that they carried nearly identical 2,4-D-catabolic plasmids, indicating that HGT had occurred (31). HGT was also inferred when Matheson et al. (40) sequenced a chromosomally located tfdA gene in a Burkholderia sp. isolated in Michigan and found 99.5% sequence similarity to a chromosomal gene in a Burkholderia strain isolated in Oregon. Furthermore, Rosselló-Mora et al. (49) examined 11 strains of naphthalene-degrading bacteria from contaminated sites in Spain and, after finding that consistent hybridization patterns for the nahA, nahG, and nahH gene probes were shared among four different genomovars of P. stutzeri hosts, concluded that HGT had occurred.
Mechanistic approaches to HGT have sought evidence for mobilization of particular genes between donor and recipient populations in experiments of controlled duration and location. The major variables in these experimental systems have been the gene(s) of interest (e.g., catabolic genes or those involved in resistance to heavy metals or antibiotics), the donors and recipients (defined or undefined), and the setting (laboratory or field). Permutations of these variables have included the use of defined donor and recipient bacteria added to laboratory-incubated environmental samples (3, 20, 34, 43, 68, 73); unknown donors from environmental samples and defined recipients incubated in the laboratory (59, 63, 64); defined donors and recipients added and retrieved from field sites (3, 39, 43, 71); and defined donors commingled with undefined, naturally occurring recipient microbial communities in laboratory (16, 24, 68) or field (44, 76) experiments.
Herrick et al. (26) recently reported in situ horizontal transfer of a naturally occurring naphthalene-catabolic gene (nahAc) between naturally occurring bacteria isolated from a coal tar waste-contaminated field site. Partial sequencing of the nahAc gene showed that identical alleles were shared among seven taxonomically diverse hosts (as determined by partial 16S rRNA gene sequences). Furthermore, large (70- to 88-kb) plasmids that hybridized to nahAc gene probes were shared by 12 naphthalene-degrading, site-derived isolates. Intracellular gene rearrangements were suggested because the plasmids were of different sizes and because the nahAc gene was located on the chromosome or on both the chromosome and plasmid of selected isolates. The present study was designed to further characterize the nature of the mobile genetic element and its mechanism of transfer between the contaminated-site-derived bacteria.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
Naphthalene-degrading strains isolated from the sediment of the seep region of a coal tar-contaminated site are designated Cg- and have been described previously (26). The naphthalene catabolic plasmids found within these strains are designated pCg-. The archetypal naphthalene degraders Pseudomonas putida PpG7 (12) and P. putida NCIB 9816-4 (15) were included in the study for comparison. All strains and sizes of plasmids are listed in Table 1. All naphthalene-degrading strains were maintained on mineral salts basal medium (MSB) with naphthalene vapor as the sole carbon source (MSB-N) (60). Rifampin (Sigma) was added to the medium at 300 μg/ml from a filter-sterilized 20-mg/ml stock in methanol as needed. Any traces of methanol that may not have volatilized from the medium did not support detectable growth of sediment bacteria. Cycloheximide (Sigma) was added as needed to the medium at 100 μg/ml from a filter-sterilized 10-mg/ml stock. Stock solutions of cycloheximide were initially prepared in 50% ethanol, but it was determined after an initial screen that the ethanol was serving as a carbon source for sediment bacteria. Thereafter, cycloheximide stock solutions were prepared in water.
TABLE 1.
Naphthalene-degrading bacterial strains and plasmids used in this study
| Strain abbreviationa | Strain identityb | Origin | Naphthalene plasmid | Plasmid size (kb)c | Reference(s) |
|---|---|---|---|---|---|
| PpG7 | Pseudomonas putida PpG7 | Berkeley, Calif. | NAH7 | 77 | 12 |
| NCIB9816-4 | Pseudomonas putida NCIB 9816-4 | Bangor, Wales | pDTG1 | 81 | 15, 54 |
| Cg1 | Pseudomonas putida Cg1 | Glens Falls, N.Y. | pCg1 | 88 | 26 |
| Cg2 | Pseudomonas fluorescens Cg2 | Glens Falls, N.Y. | pCg2 | 76 | 26 |
| Cg4 | Gram-negative rod Cg4 | Glens Falls, N.Y. | pCg4 | 70 | 26 |
| Cg5 | Pseudomonas fluorescens Cg5 | Glens Falls, N.Y. | pCg5 | 78 | 26 |
| Cg7 | Pseudomonas strain Cg7 | Glens Falls, N.Y. | pCg7a and b | 78 and 96 | 26 |
| Cg8 | Gram-negative rod Cg8 | Glens Falls, N.Y. | pCg8 | 76 | 26 |
| Cg9 | Pseudomonas fluorescens Cg9 | Glens Falls, N.Y. | pCg9 | 76 | 26 |
| Cg11 | Pseudomonas mendocina Cg11 | Glens Falls, N.Y. | pCg11 | 86 | 26 |
| Cg12 | Pseudomonas strain Cg12 | Glens Falls, N.Y. | pCg12 | 76 | 26 |
| Cg15 | Gram-negative rod Cg15 | Glens Falls, N.Y. | pCg15 | 86 | 26 |
| Cg16 | Pseudomonas strain Cg16 | Glens Falls, N.Y. | pCg16 | 76 | 26 |
| Cg21 | Gram-negative rod Cg21 | Glens Falls, N.Y. | pCg21 | 76 | 26 |
Abbreviations referring to plasmid-cured strains have the suffix C added. Corresponding strains that are both cured and resistant to rifampin have the suffix CR added. After successful matings, the corresponding transconjugants were assigned a T as suffix.
All Glens Falls isolates are identified by multiple substrate utilization patterns in the Biolog identification system (26).
Determined by pulsed-field gel electrophoresis.
Curing.
Naphthalene-degrading strains were grown in tubes containing 3 ml of Luria-Bertani (LB) broth (51) and subcultured (1% transfer) into fresh medium every other day. Sampling for cured colonies continued periodically for approximately 9 months, at which time naphthalene-negative colonies from all Cg strains were found. Curing was verified by the absence of large plasmids relative to the presence of the plasmid in the original parent strain.
ERIC REP-PCR fingerprinting.
The identities of cured and parent strains were compared by using the following enterobacterial repetitive intergenic consensus (ERIC) sequence PCR primers: ERIC-1R, 5′-ATGTAAGCTCCTGGGGATTCAC-3′; ERIC-2, 5′-AAGTAAGTGACTGGGGTGAGCG-3′ (25, 70). The ERIC repetitive extragenic palindrome (REP)-PCR amplification procedure was as follows. A 10-μl volume of sterile water was added to each PCR tube and inoculated with cells on the tip of a sterile platinum inoculating needle. The cells were lysed by being heated to 95°C for 5 min. PCR cocktail (37.5 μl containing 50 mM KCl, 10 mM Tris [pH 8.8], 1.5 mM MgCl2, 0.1 mg of bovine serum albumin per ml, 0.05% Tween 20, 2.0 μM each primer, and 1 U of Taq polymerase) and 2 drops of sterile light mineral oil were added to each tube. The tubes were heated to 80°C (for “hot start”), and 50 μM (final concentration) deoxynucleoside triphosphates were added to each tube. Cycling on a thermal cycler (MJ Research, Watertown, Mass.) was carried out as follows: 95°C for 7 min (1 cycle); 94°C for 1 min, 52°C for 1 min, and 65°C for 6 min (34 cycles); 65°C for 16 min (1 cycle).
Amplification of nahAc.
Cured strains were also tested for amplification of the nahAc gene with the following PCR primers: Cg1 upper, 5′-GCCCCAACGGTGAACTGC-3′ (this study, corresponding to positions 326 to 343 of nahAc [56]) and nahAc5, 5′-GGAGGTCATTTGCAAGCCTG-3′ (26). PCR amplification was carried out in a 50-μl volume with the thermal cycler and reagents as previously described (26) but with primer concentrations of 0.8 μM and a cycling regime of 95°C for 5 min (1 cycle); 94°C for 1 min, 58°C for 1 min, and 72°C for 1 min (35 cycles); 72°C for 5 min (1 cycle).
Selection of rifampin-resistant recipient strains.
Spontaneous rifampin-resistant cured strains were selected on LB plates containing a rifampin gradient (0 to 100 μg/ml) (13). Colonies able to grow at the highest rifampin concentration were picked and transferred onto LB plates containing successively higher concentrations of rifampin, up to 500 μg/ml. ERIC REP-PCR was performed as above to confirm that the identities of rifampin-resistant cured strains matched the original strains.
Conjugal filter matings.
Recipient cells (109, grown overnight in LB broth amended with 200 μg of rifampin per ml) were harvested and washed with phosphate-buffered saline (PBS) (51). To the same tube, 109 donor cells (grown overnight in LB broth) were added and harvested. The cells were resuspended in 50 μl of PBS and placed on a sterile, 0.22-μm-pore-size, 25-mm-diameter Millipore filter on an LB plate. The plates were incubated overnight, agar side down, in the dark at 22°C. The filters were removed and vortexed in 1 ml of PBS for 30 s to remove cells. Transconjugants were enumerated on MSB-N plates amended with rifampin. Controls containing donor cells only were enumerated on MSB-N plates, and recipient cells were enumerated on LB plates amended with rifampin. Both donor and recipient controls were also plated onto MSB-N plates amended with rifampin to account for spontaneous rifampin resistance and/or experimental errors.
Plasmid characterization and Southern hybridizations.
Plasmids were isolated by a modification of the alkali lysis method of Anderson and McKay (2, 55). Because we had no success in isolating plasmids by other procedures attempted (32, 46), we describe our isolation protocol here. Cultures grown overnight (or until turbid) in 50 ml of LB broth at 23°C were harvested, washed with Tris-EDTA (TE, pH 8.0), and resuspended in 10 ml of 6.7% sucrose–50 mM Tris–20 mM EDTA (pH 8) in a 35-ml Oakridge tube. RNase A (15 μl of a 10-mg/ml solution) and 25 mg of dry lysozyme were added, and the contents of the tubes were gently mixed. After an incubation for 5 min at 37°C, 400 μl of 0.5 M disodium EDTA, 1.25 ml of 0.25 M EDTA-50 mM Tris (pH 8.0), and 1.0 ml of 20% sodium dodecyl sulfate–50 mM Tris–20 mM EDTA (pH 8) were all added, with gentle mixing between additions. The tubes were incubated at 37°C for 10 min (until lysis) and then incubated at 55°C for 30 min with proteinase K (100 μl of a 20-mg/ml solution). Freshly prepared 3 N NaOH (0.8 ml) was added, and the contents of the tubes were mixed gently for 10 min. Tris-HCl (pH 7.0) (1.3 ml of a 2 M solution) was then added, and the contents of the tubes were again mixed gently for 3 min. Sodium acetate (pH 5.2) (0.55 ml of a 3 M solution) and 5 M NaCl (1.65 ml) were added, and the contents of the tubes were mixed gently. A phenol-chloroform extraction was performed, and the aqueous phase was removed with a truncated pipette tip and reextracted again with chloroform to remove residual phenol. An equal volume of isopropanol (approximately 16 ml) was added to the separated aqueous phase, and DNA was precipitated overnight at −20°C. DNA was pelleted by centrifuging at 10,000 × g for 30 min. The air-dried pellet was resuspended in 200 μl of sterile water for direct plasmid detection by standard gel electrophoresis (15 μl on a 0.7% agarose gel) or in 1 ml of TE (pH 7.0) for further purification on a Qiagen anion-exchange column. For the latter procedure, 4 ml of QBT buffer (750 mM NaCl, 50 mM morpholinepropanesulfonic acid [MOPS; pH 7.0], 15% ethanol, 0.15% Triton X-100) was added, and the tubes were centrifuged at 12,000 × g for 5 min. The supernatant was applied to a Qiagen plasmid midi-tip and purified as specified by the manufacturer for purification of plasmid DNA prepared by other methods. Plasmid DNA (17.5 μl of the final 200-μl preparation) was digested with restriction enzymes overnight under the reaction conditions recommended by the supplier (Gibco). Restriction fragments were separated on a 0.7% agarose gel in Tris-acetate-EDTA at 3 to 4 V/cm. The gels were stained with SYBR Green (Molecular Probes, Eugene, Oreg.) for 40 min and visualized under 300-nm UV illumination. A probe for Southern hybridization was prepared by digesting pDTG1 with BamHI and labeling by nick translation with digoxigenen-dUTP, as specified in the Genius system user’s guide, version 3.0 (Boehringer Mannheim). Southern hybridization was performed with nylon membranes (Micron Separations Inc., Westboro, Mass.) at 65°C overnight, and the membranes were washed with 0.5× wash buffer at 65°C (Boehringer Mannheim).
Exogenous isolation of plasmid from seep sediment.
Recipient cells (5 × 108) were harvested and washed as above for conjugal filter matings. Donor cells were obtained from seep sediment as follows. A 100-ml volume of 0.1% Na4P2O7 (pH 7.2) was added to 10 g of seep sediment (sampled 7 months before the experiment and stored at 4°C) and blended in a Waring blender for two 30-s intervals (4, 28). The soil was allowed to settle for 5 min, after which 0.5 ml of the supernatant was added to the microcentrifuge tube containing the recipient cells and centrifuged for 5 min. The pellet was resuspended in 50 μl of PBS, and the filter mating proceeded as above. Transconjugants were enumerated on MSB-N plates amended with rifampin and cycloheximide. Plates containing potential transconjugant colonies were incubated under a mixture of naphthalene and indole vapors for 2 days, after which the plates were examined for the presence of indigo colonies (14, 29). These indigo-positive colonies were tested for growth with and without naphthalene on MSB plates amended with rifampin and cycloheximide (prepared in water). Confirmation that transconjugant colonies arose from the recipient used was obtained by comparing the ERIC REP-PCR fingerprints. Detection of large plasmids and amplification of nahAc gene in indigo-positive, naphthalene-positive colonies were carried out as above.
RESULTS
Development of recipients for conjugation experiments. (i) Curing.
All naphthalene-degrading bacteria were repeatedly transferred in LB broth for several months. During this time, 10 of the 12 seep strains (strains Cg1, Cg2, Cg4, Cg5, Cg8, Cg9, Cg11, Cg12, Cg16, and Cg21), as well as the archetypal naphthalene degraders NCIB9816-4 and PpG7, were cured of their large plasmids. Large supercoiled plasmids were detected in parent naphthalene-degrading strains but were absent in the plasmid isolations from the respective cured strains (data not shown). Confirmation that each of the cured strains was, in fact, derived from each parent strain was provided by comparing the ERIC REP-PCR fingerprints of the cured and original parent strains (data not shown). Loss of plasmid correlated with a loss in ability to grow with naphthalene as the sole carbon source. Concurrent loss of the nahAc gene was verified by lack of PCR amplification of a 407-bp fragment of nahAc in the cured strains, while the parent naphthalene-degrading strains all contained the amplifiable nahAc gene (data not shown). The remaining two seep-derived strains, Cg7 and Cg15, also lost the ability to grow on naphthalene during this curing regime. However, these strains retained smaller plasmids (data not shown) and are the subject of another study.
(ii) Selection for rifampin resistance in cured strains.
By successively growing spontaneous resistant mutants on increasing concentrations of rifampin, strains that were resistant to rifampin (MIC of greater than 500 μg/ml) were obtained (data not shown). Confirmation that each of the rifampin-resistant cured strains was derived from each parent strain was again provided by comparing the ERIC REP-PCR fingerprints of the rifampin-resistant cured and original parent strains (data not shown). These rifampin-resistant cured strains, designated Cg1.CR, Cg2.CR, Cg4.CR, Cg5.CR, Cg8.CR, Cg9.CR, Cg12.CR, Cg16.CR, Cg21.CR, PpG7.CR, and NCIB9816-4.CR, were then used as recipients in mating experiments. The REP-PCR pattern for the cured strain Cg11 did not match its parent; therefore, additional experiments with this bacterium were abandoned.
Self-transmissibility of naphthalene plasmids.
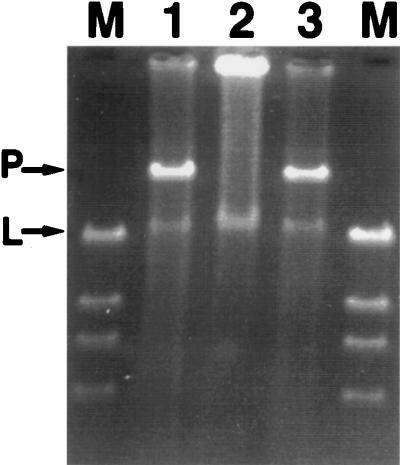
Filter matings between eight of the nine naphthalene-degrading seep isolates and their respective cured, rifampin-resistant progeny yielded transconjugant colonies which were able to grow on MSB-N amended with rifampin (Table 2). Conjugation was considered successful when the number of transconjugant colonies was at least 3 orders of magnitude greater than the number of spontaneous resistant donor colonies on the same selective medium. Thus, of the 11 intrastrain matings performed, 9 were successful (all but NCIB9816-4 and Cg5). Intrastrain matings for Cg5 and for NCIB9816-4 were attempted four and five times, respectively, before being considered unsuccessful. One donor, Cg21, was able to transfer the naphthalene-catabolic plasmid to three other hosts (Table 2). No transconjugants were found in other interstrain matings tested (Table 2). The presence of a new plasmid in the transconjugants was verified by alkali lysis and gel electrophoresis. Figure 1 shows plasmid profiles for the donor, recipient, and transconjugant for strain Cg12. Analogous profiles were obtained for all successful matings. Data in Table 2 and Fig. 1 clearly demonstrate that the naphthalene-catabolic plasmids in bacteria isolated from our contaminated study site were self-transmissible.
TABLE 2.
Results of laboratory filter matings between plasmid-containing, naphthalene-degrading strains and cured, rifampin-resistant recipient strainsa
| Donorb | Recipient | No. of transconjugants per donor | Spontaneous resistance frequencyc |
|---|---|---|---|
| PpG7 | PpG7.CR | 10−2 | 10−6 |
| NCIB9816-4 | NCIB9816-4.CRd | 10−7e | 10−8 |
| NCIB9816-4 | Cg21.CR | 10−6e | 10−6 |
| Cg1 | Cg1.CR | 10−3 | 10−8 |
| Cg1 | NCIB9816-4.CR | 10−8e | 10−8 |
| Cg2 | Cg2.CR | 10−4 | 10−7 |
| Cg2 | Cg12.CR | 10−8e | 10−8 |
| Cg4 | Cg4.CRf | 10−4 | 10−7 |
| Cg4 | Cg2.CR | 10−8e | 10−9 |
| Cg5 | Cg5.CRg | 10−6e | 10−8 |
| Cg8 | Cg8.CR | 10−4 | 10−7 |
| Cg9 | Cg9.CRf | 10−6 | 10−9 |
| Cg12 | Cg12.CR | 10−5 | 10−8 |
| Cg16 | Cg16.CR | 10−4 | 10−8 |
| Cg21 | Cg21.CR | 10−3 | 10−7 |
| Cg21 | Cg1.CR | 10−5 | 0 |
| Cg21 | Cg8.CR | 10−5 | 0 |
| Cg21 | NCIB9816-4.CR | 10−6 | 0 |
All matings attempted once, unless otherwise noted.
For strain designation, see Table 1.
Number of spontaneous antibiotic-resistant colonies per donor.
Mating attempted five times. pDTG1 was previously shown to be self-transmissible (10−6/donor) (45).
A mating was considered unsuccessful if there was less than a 103-fold difference between the number of transconjugants and the number of spontaneous antibiotic-resistant donors.
One of three matings successful.
Mating attempted four times.
FIG. 1.
Gel electrophoresis profiles for donor, recipient, and transconjugant bacteria. Data, shown here for strain Cg12, were obtained for all successful matings listed in Table 2. Partially purified plasmids were separated on a 0.7% agarose gel. P, Covalently closed supercoiled plasmid; L, linear genomic DNA; M, lambda DNA cut with HindIII. Lanes: 1, Cg12 (donor); 2, Cg12.CR (recipient); 3, Cg12.T (transconjugant).
Analysis of RFLP patterns.
To characterize the self-transmissible naphthalene plasmids, we isolated them from each of their hosts by a modification of the alkali lysis method of Anderson and McKay (2, 55) and then purified them on a Qiagen anion-exchange column. Digestion of the naphthalene catabolic plasmids with restriction enzymes BamHI, HindIII, SmaI, and XhoI revealed a pattern of restriction fragment lengths common to plasmids from all of our site-derived bacteria and pDTG1 (data not shown for BamHI, SmaI, and XhoI, but see the description of Southern hybridization of pDTG1 to HindIII-digested site-derived plasmids in Fig. 2 [below]). This common pattern was unlike the restriction fragment length polymorphism (RFLP) pattern for the canonical naphthalene catabolic plasmid, NAH7. Table 3 lists the sizes of the restriction fragments common to all seep-derived naphthalene-catabolic plasmids and pDTG1, as determined by comparing the distance of band migration to the migration of a size standard (lambda DNA digested with HindIII). RFLP patterns generated for NAH7 (SmaI and HindIII) and pDTG1 (BamHI, HindIII, and XhoI) agreed well with previously published results (53, 54, 74). Some of the plasmids (pDTG1, pCg1, pCg4, pCg5, pCg7, pCg11, and pCg15) contained one or two restriction fragments that were slightly altered (shifted, missing, or additional) from the RFLP pattern shared among the naphthalene plasmids (Table 3; Fig. 2 [see below]). These alterations in restriction fragment lengths correlated well with differences in the overall size of the plasmids (Table 1).
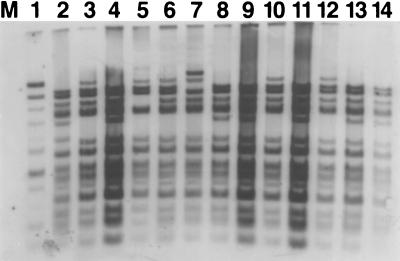
FIG. 2.
Southern hybridization of pDTG1 to naphthalene-catabolic plasmids residing in bacteria isolated from coal tar waste-contaminated sediments. The plasmids were digested with HindIII and electrophoresed on a 0.7% agarose gel. M, lambda DNA cut with HindIII. Digested plasmids from each strain were isolated and loaded into the following lanes: 1, PpG7; 2, NCIB9816-4; 3, Cg1; 4, Cg2; 5, Cg4; 6, Cg5; 7, Cg7; 8, Cg8; 9, Cg9; 10, Cg11; 11, Cg12; 12, Cg15; 13, Cg16; 14, Cg21.
TABLE 3.
Sizes of the restriction fragments common to all naphthalene-catabolic plasmids from site-derived isolates and P. putida NCIB9816-4a
| Size of fragment (kb) with restriction enzyme:
| ||||
|---|---|---|---|---|
| SmaI | BamHI | XhoI | HindIII | |
| >23.0b | >23.0b | >23.0b | 16.1c | 3.3 |
| 11.6 | 23.0 | 10.9 | 13.2 | 2.5 |
| 7.2 | 20.0 | 6.0 | 11.2 | 2.0d |
| 5.8 | 9.3 | 5.2 | 9.8 | 1.6 |
| 5.0 | 5.5 | 3.9 | 8.6 | 1.3 |
| 3.4 | 4.0 | 2.4 | 7.8 | 1.2 |
| 1.9 | 1.9 | 6.4c | 1.0 | |
| 1.5 | 4.5 | 0.9 | ||
| 1.4 | 3.4 | 0.8 | ||
See the text for minor variations from this pattern for certain plasmids.
Resolution of standard agarose gels (0.7%) did not allow accurate sizing of fragments greater than 23 kb.
Plasmids contain either the 16.1-kb fragment (Cg1, Cg4, Cg5, Cg7, Cg11, Cg15) or the 6.4-kb fragment (PpNCIB9816-4, Cg2, Cg8, Cg9, Cg12, Cg15, Cg21).
Doublet.
Southern hybridization of pDTG1 to naphthalene-catabolic plasmids from field-site derived bacteria.
To confirm similarities between our site-derived naphthalene-catabolic plasmids and pDTG1, the entire pDTG1 plasmid was labeled and used as a probe in Southern hybridizations to three of the four restriction digests described above. All of the restriction fragments found in the common plasmid pattern hybridized to pDTG1 with the same high intensity (Fig. 2, lanes 3 to 14; here an HindIII digestion is shown); pDTG1 self-hybridization is shown in Fig. 2, lane 2. Hybridization of the entire pDTG1 to NAH7 showed variable intensity (lane 1). In strain Cg7, an additional large restriction fragment of approximately 21 kb was also found (largest band in lane 7). This is consistent with our previous finding that strain Cg7 contains two closely related naphthalene-catabolic plasmids that differ in size by approximately 20 kb (Table 1) (26). Besides the two fragment sizes that differ in HindIII digests of the naphthalene-catabolic plasmids (Table 3), other variations in the banding patterns in Fig. 2 include a possible shift in the 4.5-kb fragment to 5.5 kb in pDTG1 (lane 2), a deleted 9.8-kb band in pCg4 (lane 5), and a possible shift in the 13.2-kb fragment to an approximately 11-kb fragment in strain Cg7 (lane 7). When our site-derived naphthalene plasmids were digested (BamHI and SmaI) and probed with pDTG1, high-intensity hybridization signals again confirmed a pattern of restriction fragments common to all our site-derived naphthalene plasmids (Table 3 and data not shown).
Exogenous isolation of plasmids from seep sediment.
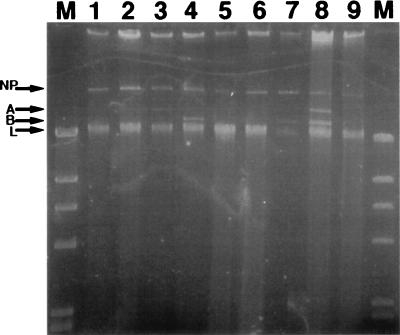
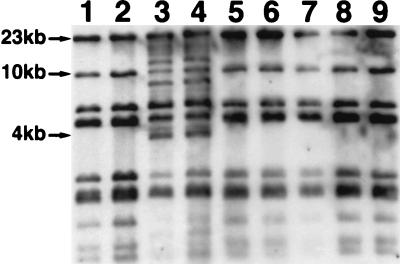
Microorganisms indigenous to sediment from our Glens Falls, N.Y., study site were obtained by homogenizing the sediment in a blender with 0.1% Na4P2O7. These cells from the sediment homogenate were used as donors in filter matings with all nine of the cured, rifampin-resistant, seep-derived recipient strains. Microorganisms in aliquots of sediment homogenate plated onto transconjugant-selective medium (no added recipient bacteria) failed to grow, indicating that the rifampin effectively inhibited native naphthalene-degrading bacteria. Potential transconjugants were screened in indole vapor to detect the activity of a dioxygenase gene in transforming indole to indigo (14, 29). Only when Cg9.CR served as a recipient were any indigo-colored putative transconjugant colonies observed. These 12 indigo colonies, all derived from Cg9.CR (as verified by ERIC REP-PCR [data not shown]), were picked and restreaked onto fresh transconjugant-selective medium, after which only 8 transconjugant colonies continued to grow. All the transconjugant strains were found to contain the nahAc gene by PCR amplification (data not shown). A large plasmid comparable in size to pCg9 (corresponding to band NP in Fig. 3, lane 1) was found in all of these transconjugant strains (lanes 2 through 9). Furthermore, additional smaller plasmids, bands A and B, were found either singly (lanes 3, 4, and 7) or together (lane 8) in four of the transconjugants. After Southern transfer, the plasmids in the gel shown in Fig. 3 were hybridized to the pDTG1 probe. This revealed that only the largest plasmid band found in each transconjugant (corresponding to band NP in Fig. 3, lanes 1 to 9) gave a positive hybridization signal (data not shown). Once confident that the uncharacterized plasmids carried by transconjugant strains would not hybridize to pDTG1, we digested total plasmid extractions from the eight transconjugant strains with XhoI, separated the fragments by electrophoresis, and performed Southern analysis with the pDTG1 probe (Fig. 4). All eight exogenously isolated plasmids (Fig. 4, lanes 2 to 9) shared the same pattern of nine restriction fragments found in pCg9 (lane 1). Transconjugant naphthalene-catabolic plasmids in lanes 3 and 4 (Fig. 4) contained additional restriction fragments of approximately 12, 8.5, and 4 kb. Longer exposure of the film shown in Fig. 4 revealed that the low-molecular-weight bands in lanes 1, 2, 4, 5, 6, 8, and 9 were also present in lanes 3 and 7 (data not shown). In addition to the XhoI restriction fragments common to all site-derived plasmids and pDTG1, pCg5 and pCg7 each have one XhoI restriction fragment of approximately 12 and 7 kb, respectively (data not shown). However, none of the naphthalene-catabolic plasmids previously isolated from our site-derived strains have the same XhoI RFLP pattern seen in the transconjugant naphthalene-catabolic plasmids in lanes 3 and 4 (Fig. 4). Thus, in addition to retrieving the same naphthalene-catabolic plasmid previously isolated from our site-derived strains, our exogenous plasmid isolation procedure has retrieved a novel variant of this plasmid. Data in Fig. 4 clearly show that the microorganisms indigenous to our coal tar-contaminated site possess the self-transmissible naphthalene-catabolic plasmid previously found in site-derived isolates and that its transmission to site-derived recipient strains can be rapidly induced under laboratory conditions.
FIG. 3.
Plasmids retrieved from eight transconjugant colonies after filter matings between indigenous sediment microorganisms and recipient strain Cg9.CR. Undigested, covalently closed supercoiled plasmid extractions were run on 0.7% agarose gel. NP, naphthalene-catabolic plasmid; A, uncharacterized plasmid; B, another uncharacterized plasmid; L, linear genomic DNA. Plasmids were isolated from strain Cg9 (original) (lane 1) and transconjugant colonies (lanes 2 to 9).
FIG. 4.
Southern hybridization of pDTG1 to naphthalene-catabolic plasmids retrieved from sediment microorganisms after filter matings with recipient strain Cg9.CR. The plasmids were cut with XhoI and run on a 0.7% agarose gel. Plasmids were isolated from parent strain Cg9 (lane 1) and transconjugant colonies (lanes 2 to 9).
DISCUSSION
The presence of a highly conserved nahAc allele among diverse bacteria carrying naphthalene-catabolic plasmids provided evidence for in situ horizontal gene transfer at a coal tar-contaminated site (26). Because the plasmids differed slightly in size (Table 1) and an isolate from another region of the same contaminated field site carried the same nahAc allele (the Cg1 allele [26]) on its chromosome, the mechanism of HGT was uncertain. Among the possible mechanisms were the following: (i) individual genes may have been recruited and transferred between replicons on gene cassettes or transposons (1, 64, 67); (ii) genes may have been transferred between bacteria via conjugation if the genes were located on self-transmissible conjugative transposons or plasmids (41, 50); and (iii) portions of the genome not otherwise organized on a mobile element may have been transferred via the mechanisms of transformation or transduction (38).
This study determined that all of the seep-derived naphthalene catabolic plasmids that we tested were self-transmissible (Table 2). This is consistent with previous reports that all other naphthalene-catabolic plasmids examined to date from pseudomonad strains are also self-transmissible (75). In addition, we found limited interstrain transfer of the naphthalene-catabolic plasmid among our seep-derived isolates (Fig. 1; Table 2). RFLP analysis of these plasmids indicated that they are all closely related to each other and to the naphthalene-catabolic plasmid (pDTG1) of P. putida NCIB 9816-4, which was isolated decades ago in Bangor, Wales (Fig. 2; Table 3). Similarity among all site-derived naphthalene-catabolic plasmids and pDTG1 was confirmed by using the entire pDTG1 as a probe in Southern hybridizations (Fig. 2). The same naphthalene-catabolic plasmid and a related variant were retrieved directly from the microbial community indigenous to the contaminated-site sediment by using a cured, rifampin-resistant seep isolate as a recipient in laboratory filter matings (Fig. 3 and 4). This entire plasmid is therefore a mobile genetic element responsible for transferring naphthalene-catabolic genes among bacteria in situ at this site, most probably via conjugation.
To date, all of the naphthalene-catabolic plasmids examined with respect to incompatibility fall into the IncP9 or IncP7 groups (5, 75) and have been isolated from fluorescent Pseudomonas spp. (rRNA group I). Plasmids from these two incompatibility groups are not considered to have a broad host range (5, 47). The incompatibility group of pDTG1 has not been determined, but another naphthalene-catabolic plasmid from P. putida NCIB 9816-3 belongs to the IncP9 incompatibility group (75).
The exogenous plasmid isolation technique, in which plasmids are directly captured into an appropriate recipient bacterium without the prior cultivation of the plasmids’ original host, has been successfully applied in the isolation of mercury resistance plasmids (7, 18, 37, 48, 59), 2,4-D-catabolic plasmids (63), and mobilizing plasmids (27, 61, 68). In our study, two closely related naphthalene-catabolic plasmids, as well as at least two other uncharacterized plasmids, were retrieved in the eight transconjugant colonies we obtained by the exogenous isolation technique (Fig. 3 and 4). Like the isolate-derived plasmids, these newly retrieved naphthalene-catabolic plasmids were both closely related to pDTG1 (Fig. 4). Other exogenous plasmid isolation experiments have retrieved a group of plasmids which varied in RFLP patterns but were still related to each other (37, 59, 63, 68). Different researchers, using this plasmid capture technique, have also reported a predominance of one plasmid type from a single isolation experiment (18, 27, 48). Although all eight exogenously isolated naphthalene-catabolic plasmids examined in the present study were obtained from a single filter mating experiment, subsequent exogenous plasmid isolations from seep sediment with the same recipients under different incubation conditions also obtained only naphthalene-catabolic plasmids closely related to pDTG1 (data not shown). Thus, after examining two types of plasmid sources (naphthalene-degrading isolates and plasmid capture from the sediment community), we have been able to isolate only variants of pDTG1 from our site. In contrast, when Alcaligenes eutrophus strains deficient in 2,4-D degradation were used to recover 2,4-D-degradative plasmids from contaminated soil, plasmids representing two incompatibility groups were isolated (63).
Unlike the present study, in which similarity to a previously examined plasmid was found, all of the mercury-resistant plasmids exogenously isolated by Dahlberg et al. (7) differed from previously described mercury-resistant plasmids, as gauged by hybridization to inc/rep probes. A unique aspect of this marine study (7) was that plasmid recovery was successful when a mating medium (artificial seawater) was used without addition of high concentrations of nutrients.
It should be noted that the success of exogenous plasmid isolation experiments is dependent on an appropriate recipient bacterium to act as a genetic sink for capture of transmissible plasmids. Naphthalene-catabolic plasmids whose host range did not include our recipient bacteria could not have been retrieved. It is possible that microorganisms native to our study site contain a greater diversity of naphthalene-catabolic genes (and the mobile genetic elements on which they may reside) than we are detecting. In support of this idea, a gram-positive (putatively Arthrobacter sp.) naphthalene degrader has been isolated from contaminated seep sediment (25). Based on negative results from both PCR amplification and hybridization experiments, the nahAc gene in this gram-positive bacterium shows no homology to the nahAc of P. putida PpG7, which differs by 5% from the Cg1 allele of nahAc (25). The use of recipient bacteria from genera other than the rRNA type I Pseudomonas used here (such as the gram-positive isolate) may allow the exogenous isolation of different naphthalene-catabolic plasmids from our site.
The results of our exogenous plasmid isolation experiment indicate the long-term persistence of this naphthalene-catabolic plasmid in our coal tar-contaminated site. The seep-derived strains which carried the plasmid were originally isolated from a sediment sample taken from the field in July 1992, whereas the exogenous plasmid isolation procedure was performed on sediment which had been sampled from the study site more than 4 years later. Other reports support in situ persistence of plasmids. In a study of exogenously isolated mercury-resistant plasmids, Lilley et al. (37) isolated and characterized 79 plasmids from the phytosphere of field-grown sugar beets. Three of the five types of plasmids (grouped on the basis of RFLP patterns) persisted over the course of 3 consecutive years, despite the absence of mercury contamination (37). Top et al. (63) also noted that one of the 2,4-D-catabolic plasmids isolated in their study had persisted in the treated soil for at least 3 years.
Despite the above-discussed methodological limitations, we have gathered evidence for in situ persistence and horizontal transfer of a single naphthalene-catabolic plasmid type in our coal tar-contaminated field site. This suggests that the coal tar contamination has applied selective pressure for the proliferation of this plasmid. This selective pressure could act by at least three different mechanisms. The coal tar contamination may have acted specifically to induce the transfer of pDTG1. Such substrate-specific active transfer has been found to be the case for transfer of tetracycline resistance determinants (50), as well as for conjugal transfer of opine-catabolizing Ti plasmids of Agrobacterium tumefaciens (21). However, other studies examining the transfer of plasmids encoding resistance to mercury (33), other heavy metals (9), or naphthalene catabolism (11) have failed to find any evidence for increased transfer of the mobile genetic element in the presence of their respective substrates.
Alternatively, carbon input associated with the coal tar contamination could have caused an increase in the total amount of conjugation among certain bacterial populations. Because conjugation is dependent on cell density (57), population growth would enhance conjugation rates simply because of greater cell-to-cell contact. In support of this, some researchers have observed a general increase in the number of plasmid-containing bacteria in environments contaminated with organic pollutants compared with that in pristine sites (6, 36, 61). The coal tar contamination in our study site may have increased the specific growth rate of native microorganisms. Enhanced growth rate has been correlated with an increase in the plasmid transfer rate (58); for example, soils amended with nutrients have shown increased plasmid transfer relative to unamended soils (22, 34, 41, 62).
However, we speculate that a third mechanism, selection for growth of transconjugants containing pDTG1, may also be operating at our field site. Rather than increasing the actual plasmid transfer frequency, the selection may be acting on the particular catabolic genes which reside on pDTG1, possibly because the encoded enzymes have a kinetic or affinity advantage over other catabolic alleles at the ambient naphthalene concentrations. It has been noted that under strong selective pressure of high substrate levels, one type of catabolic gene can predominate (often in diverse bacterial hosts) or exclude other catabolic alleles (10, 30, 45).
ACKNOWLEDGMENTS
This research was supported by the Air Force Office of Scientific Research grant F49620-95-0346 and USDA/Hatch grant 189434.
We are grateful to R. Garen for gel image preparation. We also acknowledge two anonymous reviewers for suggesting improvements to the manuscript.
REFERENCES
- 1.Amabile-Cuevas C F. Gene flux and antibiotic resistance. In: Amabile-Cuevas C F, editor. Origin, evolution, and spread of antibiotic resistance genes. Boca Raton, Fla: CRC Press, Inc.; 1993. pp. 24–76. [Google Scholar]
- 2.Anderson D G, McKay L L. Simple and rapid method for isolating large plasmid DNA from lactic streptococci. Appl Environ Microbiol. 1983;46:549–552. doi: 10.1128/aem.46.3.549-552.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bale M J, Fry J C, Day M J. Plasmid transfer between strains of Pseudomonas aeruginosa on membrane filters attached to river stones. J Gen Microbiol. 1987;133:3099–3107. doi: 10.1099/00221287-133-11-3099. [DOI] [PubMed] [Google Scholar]
- 4.Balkwill D L, Ghiorse W C. Characterization of subsurface bacteria associated with two shallow aquifers in Oklahoma. Appl Environ Microbiol. 1985;50:580–588. doi: 10.1128/aem.50.3.580-588.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boronin A M. Diversity and relationships of Pseudomonas plasmids. In: Galli E, Silver S, Witholt B, editors. Pseudomonas: molecular biology and biotechnology. Washington, D.C: American Society for Microbiology; 1992. pp. 329–340. [Google Scholar]
- 6.Campbell J I A, Jacobsen C S, Sorensen J. Species variation and plasmid incidence among fluorescent Pseudomonas strains isolated from agricultural and industrial soils. FEMS Microbiol Ecol. 1995;18:51–62. [Google Scholar]
- 7.Dahlberg C, Linberg C, Torsvik V L, Hermansson M. Conjugative plasmids isolated from bacteria in marine environments show various degrees of homology to each other and are not closely related to well-characterized plasmids. Appl Environ Microbiol. 1997;63:4692–4697. doi: 10.1128/aem.63.12.4692-4697.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davies J. Inactivation of antibiotics and the dissemination of resistance genes. Science. 1994;264:375–382. doi: 10.1126/science.8153624. [DOI] [PubMed] [Google Scholar]
- 9.de Rore H, Top E, Houwen F, Mergeay M, Verstraete W. Evolution of heavy metal resistant transconjugants in a soil environment with a concomitant selective pressure. FEMS Microbiol Ecol. 1994;14:263–273. [Google Scholar]
- 10.Dunbar J, White S, Forney L. Genetic diversity through the looking glass: effect of enrichment bias. Appl Environ Microbiol. 1997;63:1326–1331. doi: 10.1128/aem.63.4.1326-1331.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duncan K E, Ferguson N, Istock C A. Fitnesses of a conjugative plasmid and its host bacteria in soil microcosms. Mol Biol Evol. 1995;12:1012–1021. [Google Scholar]
- 12.Dunn N W, Gunsalus I C. Transmissible plasmid coding early enzymes of naphthalene oxidation in Pseudomonas putida. J Bacteriol. 1973;114:974–979. doi: 10.1128/jb.114.3.974-979.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eisenstadt E, Carlton B C, Brown B J. Gene mutation. In: Gerhardt P, et al., editors. Methods for general and molecular bacteriology. Washington, D.C: American Society for Microbiology; 1994. pp. 297–316. [Google Scholar]
- 14.Ensley B D, Ratzkin B J, Osslund T D, Simon M J, Wackett L P, Gibson D T. Expression of naphthalene oxidation genes in Escherichia coli results in the biosynthesis of indigo. Science. 1983;222:167–169. doi: 10.1126/science.6353574. [DOI] [PubMed] [Google Scholar]
- 15.Evans W C, Fernley H N, Griffiths E. Oxidative metabolism of phenanthrene and anthracene by soil Pseudomonads. Biochem J. 1965;95:819–831. doi: 10.1042/bj0950819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Focht D D, Searles D B, Koh S-C. Genetic exchange in soil between introduced chlorobenzoate degraders and indigenous biphenyl degraders. Appl Environ Microbiol. 1996;62:3910–3913. doi: 10.1128/aem.62.10.3910-3913.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fry J C, Day M J. Plasmid transfer and release of genetically engineered bacteria in nature: a discussion and summary. In: Fry J C, Day M J, editors. Bacterial genetics in natural environments. New York, N.Y: Chapman & Hall; 1990. pp. 243–250. [Google Scholar]
- 18.Fry J C, Day M J. Plasmid transfer in the epilithon. In: Fry J C, Day M J, editors. Bacterial genetics in natural environments. New York, N.Y: Chapman & Hall; 1990. pp. 55–80. [Google Scholar]
- 19.Fulthorpe R R, McGowan C, Maltseva O V, Holben W E, Tiedje J M. 2,4-Dichlorophenoxyacetic acid-degrading bacteria contain mosaics of catabolic genes. Appl Environ Microbiol. 1995;61:3274–3281. doi: 10.1128/aem.61.9.3274-3281.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fulthorpe R R, Wyndham R C. Transfer and expression of the catabolic plasmid pBR60 in wild bacterial recipients in a freshwater ecosystem. Appl Environ Microbiol. 1991;57:1546–1553. doi: 10.1128/aem.57.5.1546-1553.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fuqua W C, Winans S C. A luxR-luxI type regulatory system activates Agrobacterium Ti plasmid conjugal transfer in the presence of a plant tumor metabolite. J Bacteriol. 1994;176:2796–2806. doi: 10.1128/jb.176.10.2796-2806.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goetz A, Smalla K. Manure enhances plasmid mobilization and survival of Pseudomonas putida introduced in to field soil. Appl Environ Microbiol. 1997;63:1980–1986. doi: 10.1128/aem.63.5.1980-1986.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harayama S, Kok M, Neidle E L. Functional and evolutionary relationships among diverse oxygenases. Annu Rev Microbiol. 1992;46:565–601. doi: 10.1146/annurev.mi.46.100192.003025. [DOI] [PubMed] [Google Scholar]
- 24.Henschke R B, Schmidt F R J. Plasmid mobilization from genetically engineered bacteria to members of the indigenous soil microflora in situ. Curr Microbiol. 1990;20:105–110. [Google Scholar]
- 25.Herrick J B. Detection, divergence, and phylogeny of a naphthalene dioxygenase gene and naphthalene-degrading bacteria native to a coal-tar contaminated site. Ph.D. thesis. Ithaca, N.Y: Cornell University; 1995. [Google Scholar]
- 26.Herrick J B, Stuart-Keil K G, Ghiorse W C, Madsen E L. Natural horizontal transfer of a naphthalene dioxygenase gene between bacteria native to a coal tar-contaminated field site. Appl Environ Microbiol. 1997;63:2330–2337. doi: 10.1128/aem.63.6.2330-2337.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hill K E, Weightman A J, Fry J C. Isolation and screening of plasmids from the epilithon which mobilize recombinant plasmid pD10. Appl Environ Microbiol. 1992;58:1292–1300. doi: 10.1128/aem.58.4.1292-1300.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holben W E. Isolation and purification of bacterial DNA from soil. In: Weaver R W, Angle J S, Bottomley P S, editors. Methods of soil analysis, part 2. Microbiological and biochemical properties. Madison, Wis: Soil Science Society of America; 1995. pp. 727–751. [Google Scholar]
- 29.Jenkins R O, Dalton H. The use of indole as a spectrophotometric assay substrate for toluene dioxygenase. FEMS Microbiol Lett. 1985;30:227–232. [Google Scholar]
- 30.Ka J O, Holben W E, Tiedje J M. Genetic and phenotypic diversity of 2,4-dichlorophenoxyacetic acid (2,4-D)-degrading bacteria isolated from 2,4-D-treated field soils. Appl Environ Microbiol. 1994;60:1106–1115. doi: 10.1128/aem.60.4.1106-1115.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ka J O, Tiedje J M. Integration and excision of a 2,4-dichlorophenoxyacetic acid-degradative plasmid in Alcaligenes paradoxus and evidence of its natural intergeneric transfer. J Bacteriol. 1994;176:5284–5289. doi: 10.1128/jb.176.17.5284-5289.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kado C I, Liu S T. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981;145:1365–1373. doi: 10.1128/jb.145.3.1365-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kinkle B K, Sadowsky M J, Schmidt E L, Koskinen W C. Plasmids pJP4 and r68.45 can be transferred between populations of bradyrhizobia in nonsterile soil. Appl Environ Microbiol. 1993;59:1762–1766. doi: 10.1128/aem.59.6.1762-1766.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klingmüller W. Plasmid transfer in natural soil: a case by case study with nitrogen-fixing Enterobacter. FEMS Microbiol Ecol. 1991;85:107–116. [Google Scholar]
- 35.Lawrence J G. Selfish operons and speciation by gene transfer. Trends Microbiol. 1997;5:355–359. doi: 10.1016/S0966-842X(97)01110-4. [DOI] [PubMed] [Google Scholar]
- 36.Leahy J G, Colwell R R. Microbial degradation of hydrocarbons in the environment. Microbiol Rev. 1990;54:305–315. doi: 10.1128/mr.54.3.305-315.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lilley A K, Bailey M J, Day M J, Fry J C. Diversity of mercury resistance plasmids obtained by exogenous isolation from the bacteria of sugar beet in three successive years. FEMS Microbiol Ecol. 1996;20:211–227. [Google Scholar]
- 38.Lorenz M G, Wackernagel W. Bacterial gene transfer by natural genetic transformation in the environment. Microbiol Rev. 1994;58:563–602. doi: 10.1128/mr.58.3.563-602.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marcinek H, Wirth R, Muscholl-Silberhorn A, Gauer M. Enterococcus faecalis gene transfer under natural conditions in municipal sewage water treatment plants. Appl Environ Microbiol. 1998;64:626–632. doi: 10.1128/aem.64.2.626-632.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matheson V G, Forney L J, Suwa Y, Nakatsu C H, Sexstone A J, Holben W E. Evidence for acquisition in nature of a chromosomal 2,4-dichlorophenoxyacetic acid/alpha-ketoglutarate dioxygenase gene by different Burkholderia spp. Appl Environ Microbiol. 1996;62:2457–2463. doi: 10.1128/aem.62.7.2457-2463.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller R V, Levy S B. Horizontal gene transfer in relation to environmental release of genetically engineered microorganisms. In: Levy S B, Miller R V, editors. Gene transfer in the environment. New York, N.Y: McGraw-Hill Book Co.; 1989. pp. 405–416. [Google Scholar]
- 42.Nikolich M P, Hong G, Shoemaker N B, Salyers A A. Evidence for natural horizontal transfer of tetQ between bacteria that normally colonize humans and bacteria that normally colonize livestock. Appl Environ Microbiol. 1994;60:3255–3260. doi: 10.1128/aem.60.9.3255-3260.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O’Morchoe S B, Ogunseitan O, Sayler G S, Miller R V. Conjugal transfer of R68.45 and FP5 between Pseudomonas aeruginosa strains in a freshwater environment. Appl Environ Microbiol. 1988;54:1923–1929. doi: 10.1128/aem.54.8.1923-1929.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peters M, Heinaru E, Talpsep E, Wand H, Stottmeister U, Heinaru A. Acquisition of a deliberately introduced phenol degradation operon, pheBA, by different indigenous Pseudomonas species. Appl Environ Microbiol. 1997;63:4899–4906. doi: 10.1128/aem.63.12.4899-4906.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pettigrew C A, Breen A, Corcoran C, Sayler G S. Chlorinated biphenyl mineralization by individual populations and consortia of freshwater bacteria. Appl Environ Microbiol. 1990;56:2036–2045. doi: 10.1128/aem.56.7.2036-2045.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qiagen. Purification of cosmids and very low copy-number plasmids. Qiagen plasmid handbook. Chatworth, Calif: Qiagen, Inc.; 1995. [Google Scholar]
- 47.Ramos-Gonzalez M-I, Duque E, Ramos J L. Conjugational transfer of recombinant DNA in cultures and in soils: host range of Pseudomonas putida TOL plasmid. Appl Environ Microbiol. 1991;57:3020–3027. doi: 10.1128/aem.57.10.3020-3027.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rochelle P A, Fry J C, Day M J. Factors affecting conjugal transfer of plasmids encoding mercury resistance from pure cultures and mixed natural suspensions of epilithic bacteria. J Gen Microbiol. 1989;135:409–424. doi: 10.1099/00221287-135-2-409. [DOI] [PubMed] [Google Scholar]
- 49.Rosselló-Mora R A, Lalucat J, Garcia-Valdes E. Comparative biochemical and genetic analysis of naphthalene degradation among Pseudomonas stutzeri strains. Appl Environ Microbiol. 1994;60:966–972. doi: 10.1128/aem.60.3.966-972.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Salyers A A, Shoemaker N B, Stevens A M, Li L-Y. Conjugative transposons: an unusual and diverse set of integrated gene transfer elements. Microbiol Rev. 1995;59:579–590. doi: 10.1128/mr.59.4.579-590.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 52.Sayler G S, Hooper S W, Layton A C, King J M H. Catabolic plasmids of environmental and ecological significance. Microb Ecol. 1990;19:1–20. doi: 10.1007/BF02015050. [DOI] [PubMed] [Google Scholar]
- 53.Serdar C M. Plasmid involvement in the metabolism of naphthalene and parathion by Pseudomonas strains. Ph.D. thesis. Austin: University of Texas at Austin; 1985. [Google Scholar]
- 54.Serdar C M, Gibson D T. Isolation and characterization of altered plasmids in mutant strains of Pseudomonas putida NCIB 9816. Biochem Biophys Res Commun. 1989;164:764–771. doi: 10.1016/0006-291x(89)91525-8. [DOI] [PubMed] [Google Scholar]
- 55.Siering P L. Application of molecular approaches to investigate the role of microorganisms in manganese cycling in wetland environments. Ph.D. thesis. Ithaca, N.Y: Cornell University; 1996. [Google Scholar]
- 56.Simon M J, Osslund T D, Saunders R, Ensley B D, Suggs S, Harcourt A, Suen W C, Cruden D L, Gibson D T, Zylstra G J. Sequences of genes encoding naphthalene dioxygenase in Pseudomonas putida strains G7 and NCIB 9816-4. Gene. 1993;127:31–37. doi: 10.1016/0378-1119(93)90613-8. [DOI] [PubMed] [Google Scholar]
- 57.Simonsen L. Dynamics of plasmid transfer on surfaces. J Gen Microbiol. 1990;136:1001–1008. doi: 10.1099/00221287-136-6-1001. [DOI] [PubMed] [Google Scholar]
- 58.Smets B F, Rittmann B E, Stahl D A. The specific growth rate of Pseudomonas putida PAW1 influences the conjugal transfer rate of the TOL plasmid. Appl Environ Microbiol. 1993;59:3430–3437. doi: 10.1128/aem.59.10.3430-3437.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smit E, Wolters A, van Elsas J D. Self-transmissible mercury resistant plasmids with gene-mobilizing capacity in soil bacterial populations: influence of wheat roots and mercury addition. Appl Environ Microbiol. 1998;64:1210–1219. doi: 10.1128/aem.64.4.1210-1219.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stanier R Y, Palleroni N J, Douderoff M. The aerobic pseudomonads: a taxonomic study. J Gen Microbiol. 1966;43:159–271. doi: 10.1099/00221287-43-2-159. [DOI] [PubMed] [Google Scholar]
- 61.Top E, de Smet I, Verstraete W, Dijkmans R, Mergeay M. Exogenous isolation of mobilizing plasmids from polluted soils and sludges. Appl Environ Microbiol. 1994;60:831–839. doi: 10.1128/aem.60.3.831-839.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Top E, Mergeay M, Springael D, Verstraete W. Gene escape model transfer of heavy metal resistance genes from Escherichia coli to Alcaligenes eutrophus on agar plates and in soil samples. Appl Environ Microbiol. 1990;56:2471–2479. doi: 10.1128/aem.56.8.2471-2479.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Top E M, Holben W E, Forney L J. Characterization of diverse 2,4-dichlorophenoxyacetic acid-degradative plasmids isolated from soil by complementation. Appl Environ Microbiol. 1995;61:1691–1698. doi: 10.1128/aem.61.5.1691-1698.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Top E M, Maltseva O V, Forney L J. Capture of a catabolic plasmid that encodes only 2,4-dichlorophenoxyacetic acid:alpha-ketoglutaric acid dioxygenase (TfdA) by genetic complementation. Appl Environ Microbiol. 1996;62:2470–2476. doi: 10.1128/aem.62.7.2470-2476.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van der Meer J R. Evolution of novel metabolic pathways for the degradation of chloroaromatic compounds. Antonie Leeuwenhoek. 1997;71:159–178. doi: 10.1023/a:1000166400935. [DOI] [PubMed] [Google Scholar]
- 66.van der Meer J R, deVos W M, Harayama S, Zehnder A J B. Molecular mechanisms of genetic adaptation to xenobiotic compounds. Microbiol Rev. 1992;56:677–694. doi: 10.1128/mr.56.4.677-694.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van der Meer J R, Zehnder A J B, de Vos W M. Identification of a novel composite transposable element, Tn5280, carrying chlorobenzene dioxygenase genes of Pseudomonas sp. strain P51. J Bacteriol. 1991;173:7077–7083. doi: 10.1128/jb.173.22.7077-7083.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van Elsas J D, McSpadden Gardener B B, Wolters A C, Smit E. Isolation, characterization, and transfer of cryptic gene-mobilizing plasmids in the wheat rhizosphere. Appl Environ Microbiol. 1998;64:880–889. doi: 10.1128/aem.64.3.880-889.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.van Overbeek L S, van Veen J A, van Elsas J D. Induced reporter gene activity, enhanced stress resistance, and competitive ability of a genetically modified Pseudomonas fluorescens strain released into a field plot planted with wheat. Appl Environ Microbiol. 1997;63:1965–1973. doi: 10.1128/aem.63.5.1965-1973.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Versalovic J, Koeuth T, Lupski J R. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991;19:6823–6832. doi: 10.1093/nar/19.24.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Williams H G, Day M J, Fry J C, Stewart G J. Natural transformation in river epilithon. Appl Environ Microbiol. 1996;62:2994–2998. doi: 10.1128/aem.62.8.2994-2998.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Williams P A, Sayers J R. The evolution of pathways for aromatic hydrocarbon oxidation in Pseudomonas. Biodegradation. 1994;5:195–217. doi: 10.1007/BF00696460. [DOI] [PubMed] [Google Scholar]
- 73.Wyndham R C, Nakatsu C, Peel M, Cashore A, Ng J, Szilagyi F. Distribution of the catabolic transposon Tn5271 in a groundwater bioremediation system. Appl Environ Microbiol. 1994;60:86–93. doi: 10.1128/aem.60.1.86-93.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yen K M, Gunsalus I C. Plasmid gene organization: naphthalene/salicylate oxidation. Proc Natl Acad Sci USA. 1982;79:874–878. doi: 10.1073/pnas.79.3.874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yen K M, Serdar C M. Genetics of naphthalene catabolism in Pseudomonads. Crit Rev Microbiol. 1988;15:247–268. doi: 10.3109/10408418809104459. [DOI] [PubMed] [Google Scholar]
- 76.Zhou J-Z, Tiedje J M. Gene transfer from a bacterium injected into an aquifer to an indigenous bacterium. Mol Ecol. 1995;4:613–618. doi: 10.1111/j.1365-294x.1995.tb00261.x. [DOI] [PubMed] [Google Scholar]