Abstract
Almost 40 years since the discovery of microtubule dynamic instability, the molecular mechanisms underlying microtubule dynamics remain an area of intense research interest. The “standard model” of microtubule dynamics implicates a “cap” of GTP-bound tubulin dimers at the growing microtubule end as the main determinant of microtubule stability. Loss of the GTP-cap leads to microtubule “catastrophe,” a switch-like transition from microtubule growth to shrinkage. However, recent studies, using biochemical in vitro reconstitution, cryo-EM, and computational modeling approaches, challenge the simple GTP-cap model. Instead, a new perspective on the mechanisms of microtubule dynamics is emerging. In this view, highly dynamic transitions between different structural conformations of the growing microtubule end – which may or may not be directly linked to the nucleotide content at the microtubule end – ultimately drive microtubule catastrophe.
Keywords: cytoskeleton, GTP-cap, microtubule catastrophe, microtubule dynamic instability, microtubules
INTRODUCTION: MICROTUBULE DYNAMICS CAN BE EXPLAINED BY THE GTP-CAP MODEL
Microtubules are dynamic polymers found in all eukaryotic cells. As one of the major components of the cytoskeleton, microtubules are essential for a number of dynamic cellular processes: from forming the mitotic spindle and axoneme, to acting as tracks for intracellular transport by molecular motor proteins. Microtubules are stiff, hollow tubes typically containing 13 protofilaments formed through self-assembly of αβ-tubulin heterodimers that join longitudinally in a head-to-tail fashion.[1-3] This arrangement results in structurally distinct polymer ends, with α-tubulin exposed at one end (termed the minus end), and β-tubulin exposed at the other (termed the plus end).[4,5] Notably, both microtubule ends exhibit stochastic transitions between phases of growth and shrinkage, a behavior known as microtubule dynamic instability (Figure 1A,B).[6] However, the biochemical and structural polarity of microtubule polymers gives rise to distinct polymerization dynamics of the two ends, with plus ends typically growing faster and being more dynamic than minus ends. Over the years, many key principles of dynamic instability have been identified[3,7,8]; however, some fundamentals of the process are still not understood.
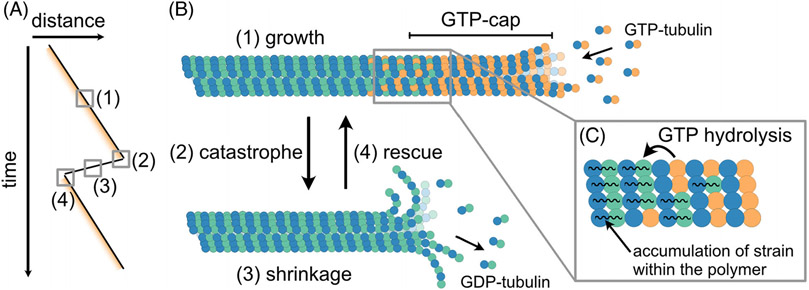
FIGURE 1.
Microtubule dynamics can be explained by the GTP-cap model. (A) A “kymograph” schematic of microtubule end position over time. Yellow shading represents the “GTP-cap.” Microtubule ends switch between phases of growth (1) and shrinkage (3) through transitions called “catastrophe” (2) and “rescue” (4). (B) Microtubules grow by the addition of GTP-tubulin heterodimers onto their ends. After incorporation into the microtubule polymer, GTP in the β-subunit of tubulin undergoes hydrolysis after a delay, leaving a “cap” of GTP-tubulin at the growing end. (C) GTP-hydrolysis induces a conformational change resulting in strain build-up within the GDP-polymer lattice (C). Loss of the protective GTP-cap from the microtubule end exposes the unstable GDP-tubulin lattice causing microtubule catastrophe – the switch into a phase of rapid microtubule shrinkage.
The original discovery of microtubule dynamic instability almost four decades ago led to the GTP-cap model as an explanation for the observed characteristics of microtubule dynamics.[6] Tubulin is a GTPase, and thus has the ability to bind and hydrolyze GTP. Microtubules grow by the addition of GTP-associated αβ-tubulin heterodimers to the polymer ends. Once a dimer is incorporated into the microtubule lattice and covered by another incoming dimer, the GTP in the β-subunit is able to undergo hydrolysis.[9,10] GTP hydrolysis induces a number of conformational changes within the tubulin dimer, building strain within the polymer, and resulting in an unstable microtubule lattice composed of tubulin dimers with GDP in their β-subunit (referred to as “GDP-tubulin”) (Figure 1C).[11-16] However, given that GTP hydrolysis does not occur instantaneously but after a delay, a “cap” of GTP-tubulin dimers remains at the growing end.[9,11,17] This GTP-cap is thought to be a stabilizing structure, protecting a growing microtubule from “catastrophe” – the switch into a phase of rapid shrinkage.[6,18,19]
The GTP-cap model suggests a direct link between the microtubule growth rate and microtubule stability. Namely, the size of the protective GTP-cap is the result of the balance between the rate of GTP-tubulin incorporation at a growing microtubule end (i.e., the microtubule growth rate), and the rate of subsequent GTP hydrolysis within the polymer (Figure 1B,C). Assuming that the GTP hydrolysis rate is constant, the faster the microtubule grows, the larger its GTP-cap will be. A larger GTP-cap would presumably be harder to lose, and thus a faster-growing microtubule is expected to be more stable against catastrophe.[6] Indeed, a multitude of studies with purified tubulin in vitro established that increasing the tubulin concentration leads to a simultaneous increase in microtubule growth rate and a suppression of catastrophe, consistent with the idea that faster-growing microtubules possess larger, more protective GTP-caps.[20-24] However, confirming that a larger GTP-cap correlates with suppression of catastrophe and thus increased microtubule lifetime was a decades-long challenge because of the inability to directly visualize the GTP-cap on a dynamic microtubule end. It would not be until a proxy for the GTP-cap on dynamic microtubules was established that the correlation between the GTP-cap size, microtubule growth rate, and microtubule catastrophe could be tested.
EB PROTEINS MARK THE GTP-CAP
In recent years, EB proteins have emerged as a tool for studying the GTP-cap on a growing microtubule. EBs constitute a highly conserved family of microtubule-associated proteins (MAPs), widely recognized for their comet-shaped localization at growing microtubule ends (Figure 2).[25-28] As such, EBs are commonly used to visualize and track the dynamic microtubule network in cells.[29-31] The ability of EBs to autonomously recognize the growing microtubule end was established using in vitro reconstitution with purified Mal3, an EB homologue from fission yeast.[32] Importantly, this study found that Mal3 recognizes both growing microtubule plus and minus ends, but does not localize to shrinking or static ends. Thus, the microtubule feature recognized by EBs is only present on tips of growing microtubules.
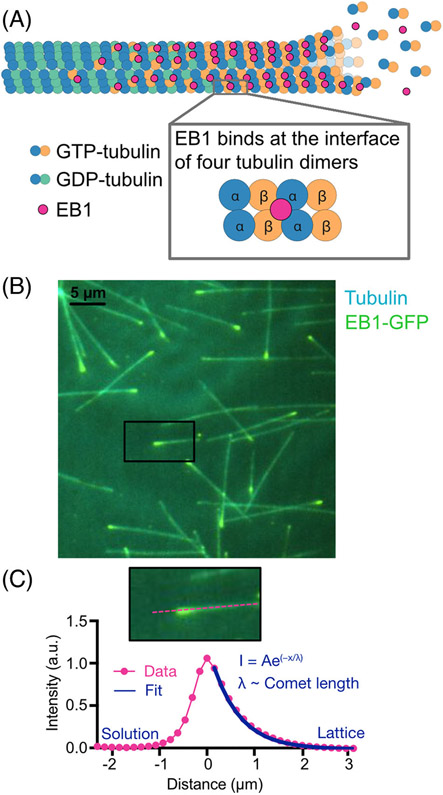
FIGURE 2.
EB localization on growing microtubule ends serves as a proxy for the GTP-cap. (A) EBs bind at the interface of four tubulin dimers, in a location that facilitates EB’s recognition of the nucleotide state of tubulin within the microtubule lattice. Due to its nucleotide sensitivity, EB localization can be used to mark the GTP-cap. (B) Total-internal-reflection-fluorescence image of EB1-GFP localization on ends of growing microtubules in vitro. (C) The fluorescent EB1-GFP signal resembles a comet-like shape, exponentially decaying along the microtubule lattice. Fluorescent comet profiles are averaged together then fit to an exponential decay function and the decay constant is used as a proxy for the length of the GTP-cap.
The first clue about EBs’ tip-tracking mechanism came from another reconstitution study, demonstrating that human EB1 preferentially binds microtubule lattices assembled with GMPCPP, a GTP-analogue, over the dynamically grown GDP-lattice extensions.[33] This observation suggested that EB1 directly recognizes the nucleotide state of tubulin within the microtubule lattice. Subsequent cryo-electron microscopy (EM) studies using GTPɣS, another GTP-analogue, established that EBs bind between two microtubule protofilaments, at the interface of four tubulin dimers, in a region close to the exchangeable GTP-binding site (Figure 2A).[14,16,34] This binding across four tubulin dimers allows EBs to sense the structural changes induced by GTP hydrolysis within the tubulin dimers in the microtubule lattice. Most recently, studies employing a tubulin mutant that cannot hydrolyze GTP provided further support that EBs preferentially bind to the GTP-cap.[35]
In addition to enabling nucleotide sensing, EB localization at the interface of four tubulin dimers may promote EBs’ direct effects on microtubule dynamics. Cryo-EM structural studies found that EB-decorated microtubules adopt a compacted configuration, considered to be associated with a post-GTP-hydrolysis state.[16] Thus, EBs may directly influence the rate of GTP hydrolysis. Furthermore, increasing the concentration of Mal3 within a sub-saturating regime (below 100 nM) was reported to decrease the length of Mal3 comets, consistent with the model that EBs increase the GTP-hydrolysis rate, and thus decrease the size of the GTP-cap.[36] Notably, EBs’ primary effect on microtubule dynamics in vitro is an increase in microtubule catastrophe frequency,[37,38] which may be a consequence of an overall decrease in the GTP-cap size.
A recent study employing 3D single-molecule diffusion simulations suggested that EB tip-tracking is facilitated by increased binding to open and tapered microtubule end structures, which may be present during microtubule growth.[39] Additionally, EB binding to the outer surface of curved and straight tubulin sheets at microtubule ends was reported using cryo-EM tomography approaches.[40] Although the exact details and affinities of EBs for distinct structural features that may be present at microtubule ends are still not fully understood, the exponentially decaying profile of EB comet intensity along the microtubule lattice is now widely accepted as a bona fide marker of the decaying GTP-cap on growing microtubule ends.[35]
THE GTP-CAP SIZE IS NOT THE SOLE DETERMINANT OF THE MICROTUBULE LIFETIME
With EB comets in hand as a proxy for the GTP-cap, measurements of microtubule dynamics in saturating EB conditions over a range of tubulin concentrations established that the length of the GTP-cap indeed increases with increasing tubulin concentration,[32] correlating with an increase in microtubule growth rate and a simultaneous suppression of microtubule catastrophe.[20,41] Specifically, the EB-comet size was found to scale linearly with the microtubule growth rate over a range of tubulin concentrations, reaching lengths of 600 nm or more at the highest tubulin concentrations tested, while microtubule catastrophe frequency exhibited a sharp decrease as a function of the EB-comet size.[41] While early studies using GMPCPP, a slowly hydrolyzable GTP analogue, reported that even a small GTP-like cap may be sufficient to protect the microtubule against catastrophe,[19,42] these recent EB-comet measurements showed that microtubule stability (in the absence of MAPs) increases for larger GTP-caps, with catastrophe frequency becoming negligible for EB comets larger than 300 nm (corresponding to a GTP-cap of about 40 tubulin layers).[41] In addition, high-temporal resolution studies of EB localization during the transition to microtubule catastrophe found that EB is gradually lost prior to the onset of catastrophe, supporting the stabilizing role of the GTP-cap in microtubule dynamics.[34,43] One of the studies postulated that there is a specific minimum threshold of EB density on the microtubule end that must be reached prior to catastrophe, indicating that the GTP-cap must decrease to a critical size before it will no longer protect a microtubule against catastrophe.[43] Taken together, these observations suggested that the size of the GTP-cap is the main determinant of the microtubule lifetime, and that the GTP-cap must be kept small for microtubules to undergo dynamic instability.
Microtubule minus ends are more stable than plus ends, despite having smaller GTP-caps
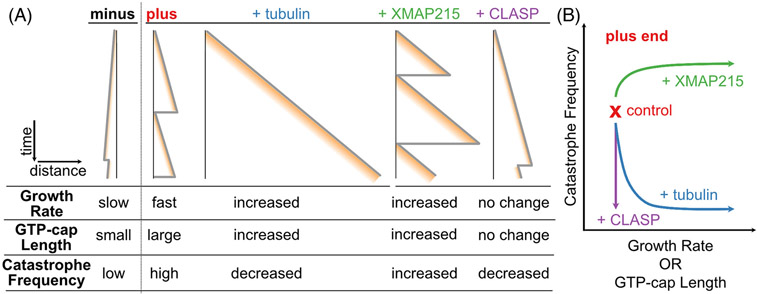
The concept that the GTP-cap size determines microtubule stability is predominantly based on in vitro studies focusing only on microtubule plus-end dynamics. Microtubule plus ends are clearly dynamic in cells, while minus ends are often anchored at microtubule organizing centers, obscuring their dynamics and regulation.[44] Nevertheless, early in vitro reconstitution studies established that minus ends also undergo dynamic instability.[20,45] Interestingly, the characteristics of minus-end dynamics are clearly distinct from those at plus ends: at a given tubulin concentration, minus ends grow slower, yet typically exhibit longer lifetimes compared to their plus end counterparts (Figure 3A).[20,45,46] The finding that minus ends have low catastrophe frequency in spite of their slow growth rates raises the possibility that minus ends have inherently larger GTP-caps, perhaps due to a distinct GTP-hydrolysis rate. However, a recent study using EB-comet measurements at both microtubule ends found that the GTP-cap length at minus ends scales with the microtubule growth rate in the same way it does at plus ends, suggesting that the rate of GTP hydrolysis at the two ends is the same.[46] In other words, a microtubule end growing at a specific growth rate (achieved at different ends using different tubulin concentrations) has the same GTP-cap length, regardless of whether it is a plus or a minus end. Yet, this same-sized GTP-cap is more protective at microtubule minus ends than at plus ends. The authors found that the enhanced microtubule stability at minus ends stems from a lower GTP-tubulin off-rate at the minus end compared to the plus end, rendering the GTP-cap at minus ends more stable. This finding presents a clear example of the GTP-cap size not being the sole determinant of microtubule stability.
FIGURE 3.
The GTP-cap size is not the sole determinant of the microtubule lifetime. (A) Schematic time–distance plots (kymographs) of microtubule end dynamics and corresponding GTP-cap sizes in different polymerization conditions. Microtubule minus ends grow slower than plus ends and have a smaller GTP-cap, but undergo catastrophe less frequently.[46] With the addition of tubulin, microtubule plus ends display increased growth rate and GTP-cap sizes, as well as decreased catastrophe frequency, consistent with the canonical model of microtubule stability. In contrast, the addition of XMAP215 or CLASP breaks the canonical relationship between microtubule growth and catastrophe. XMAP215 increases microtubule growth rate, GTP-cap size and catastrophe frequency.[41] CLASP suppresses microtubule catastrophe without increasing the microtubule growth rate or the GTP-cap size.[74] (B) Functional relationship between microtubule growth rate, GTP-cap ength, and catastrophe frequency at the microtubule plus end in different polymerization conditions.
XMAP215 promotes microtubule catastrophe despite increasing the GTP-cap size
While early in vitro studies mainly focused on the dynamic instability of microtubules assembled with purified tubulin alone, microtubule dynamics in cells are dramatically modulated by a multitude of MAPs. Some of the most prominent MAPs belong to the class of TOG-domain proteins, characterized by containing at least one tumor overexpression gene (TOG) domain.[47-51] TOG domains are known for their ability to directly bind tubulin dimers,[52] and they often function in ordered arrays within the protein.[53] The best-studied TOG-domain proteins belong to the XMAP215 and CLASP protein families, which play important roles in cell division, cell migration, and neuronal development.[54-56]
Proteins from the XMAP215 family are potent microtubule polymerases, with conserved functions in promoting microtubule assembly. Depletion of XMAP215 family proteins results in characteristically small mitotic spindles.[57] The family is named after Xenopus microtubule assembly protein of 215 kDa (XMAP215), which on its own accelerates microtubule growth rates by an order of magnitude in vitro.[58-60] XMAP215’s polymerase activity relies on an array of TOG domains, with the minimal polymerase unit containing TOG1 and TOG2,[61] which preferentially bind to a curved conformation of tubulin characteristic of tubulin dimers in solution.[52,62] XMAP215’s effects on microtubule growth are further enhanced with EB1, achieving growth rates in vitro that match the fast microtubule growth observed in cells.[38] While EB1 and XMAP215 do not interact directly, their synergy is realized through allosteric effects on the microtubule lattice, with EB1 likely promoting the formation of lateral bonds between tubulin dimers and facilitating the straightening of protofilaments into the microtubule lattice.[16,34] Given the preferential binding of the polymerase TOG domains to curved tubulin dimers, protofilament straightening facilitates faster unbinding of XMAP215 from a lattice-incorporated tubulin dimer, ultimately speeding up XMAP215’s overall polymerase activity.[38,63]
Surprisingly, both in the presence and absence of EB1, XMAP215 accelerates microtubule growth rates without increasing microtubule lifetime (Figure 3), contrary to what would be expected based on the GTP-cap model.[38,41,58] A logical hypothesis is that XMAP215 is able to decrease the GTP-cap size, by for example increasing the GTP-hydrolysis rate, resulting in fast microtubules with small protective caps. However, an investigation of microtubules polymerized with matching growth rates, achieved using either XMAP215 or high tubulin concentrations, revealed that both the amount of EB1 and the average EB1 comet lengths are not smaller in the presence of XMAP215.[41] Thus, XMAP215 does not decrease the GTP-cap size. Instead, XMAP215 was found to promote irregularities in microtubule growth, causing large fluctuations in growth rate, as well as tapered and curled microtubule end configurations (Figure 4).[41] Interestingly, high-spatiotemporal-resolution tracking of microtubule ends and their EB1 comets prior to catastrophe demonstrated that, in the presence of XMAP215, microtubules transition to catastrophe more abruptly, with faster instantaneous growth rates and higher EB1 content than previously established with tubulin alone.[41,43] These results underscore the lack of a universal minimal GTP-cap size threshold needed for catastrophe. Moreover, these findings point to the relevance of structural aspects of microtubule end beyond the GTP-tubulin content, further indicating that the GTP-cap size is not the sole determinant of microtubule stability.
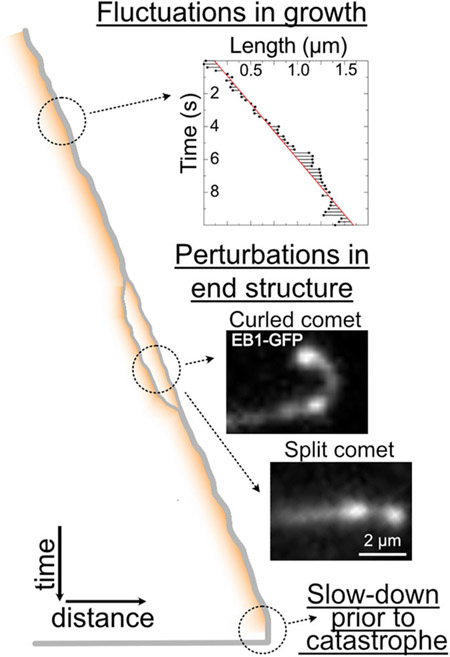
FIGURE 4.
Light microscopy provides hints into dynamic changes at growing microtubule ends. A time–distance schematic (kymograph) of a growing microtubule end. (Top) Microtubule ends can exhibit significant fluctuations in end position over time.[83,84] (©2021 Farmer et al. Inset originally published in the Journal of Cell Biology. https://doi.org/10.1083/jcb.202012144) (Middle) Observations of growing microtubule ends reveal occurrences of microtubule end splitting, curling and repair.[41,73,123] (Bottom) Microtubules typically exhibit a slow-down in growth accompanied by the loss of the EB-comet prior to catastrophe.[34,41,124]
CLASPs promote microtubule lifetime without increasing the GTP-cap size
The members of the cytoplasmic linker-associated protein (CLASP) family also rely on their TOG domains to regulate individual microtubule dynamics and spatial architecture of microtubule networks.[56,64-66] CLASPs localize to the tips of microtubules at the leading edge of migrating cells and are necessary for proper cell migration.[66-68] In cell division, CLASPs are found at kinetochores and along the mitotic spindle, and depletion of CLASPs results in severe spindle defects.[69,70] Additionally, CLASPs promote microtubule nucleation and density at microtubule organizing centers, including both the centrosome and the Golgi.[71,72]
In vitro studies have demonstrated that CLASPs suppress catastrophe frequency and promote rescue during periods of growth, resulting in overall microtubule stabilization.[73-75] Such stabilizing effects could be a consequence of larger GTP-caps, achieved either by an increase in microtubule growth rate or a decrease in the GTP-hydrolysis rate. Notably, measurements of microtubule dynamics found that CLASPs can modulate catastrophe and rescue without inducing significant changes in the average growth or shrinkage rates (Figure 3).[73,74] Measurements of EB comets in the presence of CLASPs are complicated by the fact that CLASPs directly interact with EBs to achieve microtubule end-localization in cells.[56] Experiments using EB1delC – a truncated EB1 construct that recognizes the nucleotide state of tubulin, but lacks the C-terminal CLASP-binding domain – showed that CLASP2 does not increase the amount of EB1delC at growing microtubule ends.[74] Consistent results were obtained by investigating the comets of Bim1, a budding yeast EB homologue, in the presence of an isolated CLASP TOG domain that on its own regulates microtubule dynamics.[76] Thus, CLASPs do not increase the GTP-cap size to suppress microtubule catastrophe.
When microtubules are grown in the absence of MAPs, microtubule catastrophe is typically preceded by a slowdown in microtubule growth rate, with a vast majority of growth slowdowns resulting in catastrophe. In contrast, the majority of microtubules experiencing a growth slowdown in the presence of CLASP transition back to robust growth, rather than undergoing catastrophe.[77] An investigation of EB comets in the presence of CLASP further revealed an increased frequency of EB-comet-repair events, in which a subset of lagging protofilaments catches up to the leading protofilaments to form a single, repaired, EB comet (Figure 4).[73] These observations suggest that CLASP stabilizes incomplete lattices, allowing the microtubule to repair itself,[73] instead of undergoing a catastrophe as a result of a disrupted end.
The molecular mechanisms behind CLASPs’ stabilization of microtubules are still not understood. The finding that a single CLASP TOG domain can reproduce the regulatory effects of the full-length protein refutes the hypothesis that CLASPs simply tether unpolymerized tubulin dimers to microtubule ends, making them available for protection against catastrophe or promotion of rescue.[73,76] Instead, CLASPs must stabilize the microtubule by directly acting at an inter- and/or intra-dimer level. However, it should be noted that these effects have to be temporary or occur at a protofilament level, since they do not affect the overall rate of microtubule shrinkage.[74,76] Despite CLASPs’ exact molecular mechanisms remaining elusive, it is clear that CLASPs do not stabilize microtubules by increasing the GTP-cap size.
XMAP215 and CLASPs present just two examples of microtubule regulators that break the canonical relationship between microtubule growth rate and microtubule lifetime. Although many regulators of microtubule dynamics induce either simultaneous increase in growth rate and lifetime (such as e.g., tau[78,79]) or simultaneous suppression of growth rate and increase in catastrophe frequency (such as Op18/Stathmin[80,81]), there are still others for which this relationship between growth and lifetime is not maintained.[82] To what extent their effects may rely on the modulation of the GTP-cap size remains largely unknown.
THE DYNAMIC MICROTUBULE END STRUCTURE IS THE ULTIMATE DETERMINANT OF MICROTUBULE STABILITY
Microtubule growth fluctuations and aging add complexity to the growing microtubule end
The realization that the mean GTP-cap size is not the primary determinant of microtubule lifetime raises different possibilities. For example, the GTP-cap could fluctuate so rapidly that it can be easily lost, even when very large. Alternatively, the actual stabilizing structure at the growing microtubule end might be much smaller, and not equivalent to the GTP-tubulin region recognized by EBs.
Significant fluctuations in microtubule growth at the nanoscale have been detected using high-resolution optical tweezers approaches.[83-85] These measurements led to the idea that the size of the GTP-cap itself may rapidly fluctuate, potentially due to a large GTP-tubulin off-rate during microtubule growth.[86] Subsequent measurements of microtubule growth using both optical tweezers and total-internal-reflection-fluorescence (TIRF) microscopy reported rapid millisecond-scale tubulin on-off kinetics, suggesting that, like the tubulin on-rate, the tubulin off-rate also increases with the concentration of tubulin in solution.[87] These fast rates of single-molecule tubulin dimer kinetics have recently been contested by new measurements using interferometric scattering microscopy with gold nanoparticle-labeled tubulin.[88,89] Here, the authors reported two distinct, albeit slower tubulin off-rates, corresponding to different sites of tubulin association at the microtubule end distinguished by the number of longitudinal and lateral bonds formed. Regardless of the exact rates of tubulin on/off kinetics, the effects of microtubule growth fluctuations are likely to be important for the size of the GTP-cap and overall microtubule stability.
An additional piece of the microtubule dynamics puzzle comes in the form of microtubule aging. Namely, lifetime distributions of microtubules grown with tubulin alone in vitro do not fit a single exponential decay, which would be predicted by purely stochastic, random occurrences of microtubule catastrophe.[90,91] Rather, microtubule catastrophe is more likely for microtubules that have been growing for several minutes than those that have just started to grow. This finding suggests that the microtubule “age” is somehow recorded, and that temporal transitions, presumably occurring at the growing microtubule tip over a span of minutes, ultimately result in tip destabilization and microtubule catastrophe.
What could be the nature of microtubule aging? Theoretical and computational models of microtubule dynamics over the years have provided some exciting hypotheses. Although earlier, analytically tractable single-filament models of dynamic instability have not been able to recapitulate microtubule aging, a simple multi-protofilament generalization has captured the observed characteristics of microtubule aging.[92] In this model, a critical number of individual microtubule protofilaments need to be destabilized to trigger microtubule catastrophe. Specifically, the authors proposed that destabilization of individual protofilaments is a consequence of exposing a GDP-tubulin subunit at the protofilament end, which would occur by dissociation of terminal GTP-tubulin subunits. Once exposed, the terminal GDP-tubulin would impede further association of incoming GTP-tubulin dimers, rendering the protofilament end “damaged.” Importantly, this model clearly distinguished the stabilizing feature of the microtubule end from the GTP-cap; once a GDP-subunit is exposed at the end, the GTP content of subunits buried in the lattice below it becomes irrelevant. This concept that GDP-tubulin plays an important role at the growing microtubule end has gained further recent support.[93,94] Of note, a new study of microtubule growth using interference reflection microscopy reported that GTP hydrolysis leads to a significant increase in growth fluctuations when compared to the microtubule growth in which GTPase activity is absent.[95] Supported by computational modeling, the authors proposed that exposure of GDP-tubulin at the growing end “poisons” the microtubule growth, and may serve as a precursor to catastrophe.
A model in which exposure of terminal GDP-tubulin leads to microtubule aging and eventually results in catastrophe could explain the observed effects of several MAPs. XMAP215’s potent polymerase activity is typically attributed to an increase in the tubulin on-rate, although promotion of growth could to some extent also be a consequence of suppression of an otherwise large tubulin off-rate.[96] However, a reduction in the tubulin off-rate should result in suppression of growth fluctuations in the presence of XMAP215, which is contrary to experimental observations.[41] On the other hand, some degree of heterogeneity in individual protofilament growth rates is expected if XMAP215 molecules act processively on a single-protofilament level,[60] especially if the amounts of XMAP215 are not saturating. Additionally, XMAP215’s ability to tightly bind and remove terminal GTP-like tubulin subunits in the absence of soluble tubulin[60,97] could manifest itself in an occasional removal of GTP-tubulin even in conditions of growth, leading to a GDP-tubulin exposure. If the accumulation of a few terminal GDP-tubulin protofilaments is sufficient for catastrophe, this could explain why, in the presence of XMAP215, microtubules undergo catastrophe with an overall larger GTP-tubulin content[41] – GTP-tubulin subunits buried in the tubulin lattice might not be relevant to overall polymer stability in these highly fluctuating growth conditions. Conversely, an anti-catastrophe factor like CLASP might be able to stabilize the transiently poisoned GDP-end just enough to facilitate the terminal nucleotide exchange and binding of an incoming GTP-tubulin dimer to rescue the microtubule end from an impending catastrophe.[75,77]
Specific conformational changes at the growing microtubule end have been implicated in additional models of microtubule aging and catastrophe. Observations of fluorescent tubulin suggested that growing microtubule tips are tapered rather than blunt, and that the degree of tip taper increases over time. This observation was supported by a computational model which predicted variability in individual protofilament lengths, and suggested that microtubule aging is a consequence of the progressive evolution of microtubule end tapering.[98] Another model proposed that extension of interprotofilament cracks into the GDP microtubule lattice leads to microtubule catastrophe.[99,100] Further insight was gained by a coarse-grained Brownian dynamics approach, which explicitly simulated individual tubulin–tubulin interactions while incorporating thermal fluctuations and mechanics of individual protofilaments.[101] This approach allowed the investigation of not only fluctuations in lengths, but also curvature of individual protofilaments. The authors proposed that the slow evolution of stochastic and reversible microtubule end configurations involving increasing numbers of protofilament curls eventually results in microtubule catastrophe. As such, this model also challenged the notion that the sheer size of the GTP-cap determines microtubule stability, although it was restricted to short (~second) temporal scales, and could thus not directly predict experimentally measured microtubule lifetimes.
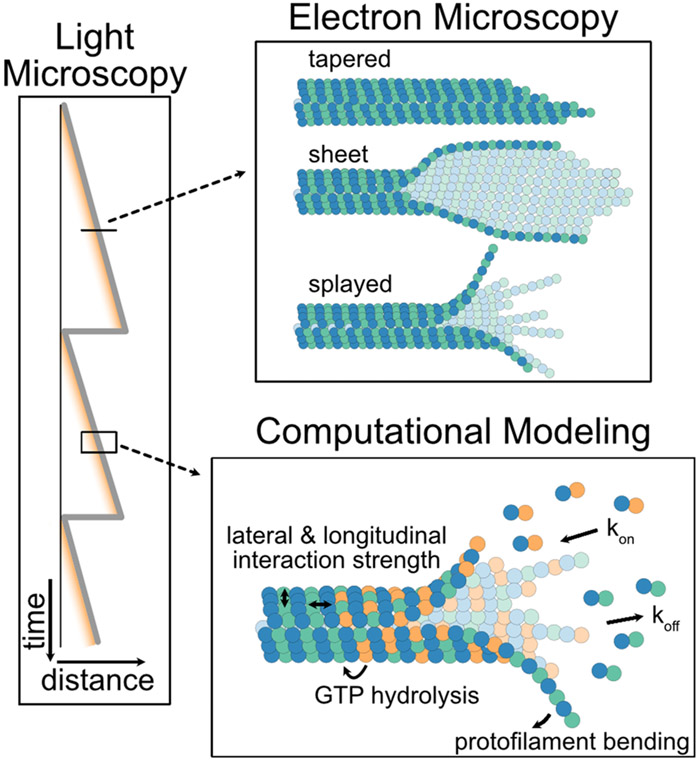
While microtubule aging presents a new set of conundrums, it is clear that resolving a high-spatiotemporal-resolution structure of the dynamic microtubule end is essential if we are to fully understand the molecular mechanisms of microtubule dynamics. Careful examinations using light imaging approaches and improved image analysis techniques have provided recent evidence of dramatic changes in the microtubule end structure over time, including microtubule end tapering, protofilament splitting, and repair (Figure 4),[39,41,73,98,102] as well as allowed measurements of tubulin polymerization kinetics at a single-molecule level.[88,89] Nevertheless, investigation of the microtubule end at the ultra-structural level still requires high-resolution EM approaches.
High-resolution imaging and computational modeling approaches provide novel insights into mechanisms of microtubule dynamics
Recent advances in cryo-EM imaging have yielded critical insights into the heterogeneity of microtubule end structures (Figure 5). While there is now an overall consensus that growing microtubule ends are certainly not blunt, the extent to which individual protofilaments are laterally connected and curved during growth is a subject of an ongoing debate.[103] Early cryo-EM studies suggested that protofilaments at the growing end are relatively straight, but could vary in length, resulting in a “tapered” end.[104] Other studies reported laterally connected subsets of protofilaments forming curved “sheets” at growing microtubule ends.[40,105,106] Finally, recent applications of 3D cryo-electron tomography suggested that growing microtubule ends display disconnected and individually curved protofilaments,[107] similar to the end configurations well accepted for shrinking microtubules.[104,108]
FIGURE 5.
A multi-method approach is necessary to elucidate the role of dynamic microtubule end conformations in microtubule stability. Light microscopy approaches provide both spatial and temporal information over large timescales. Electron microscopy approaches are used to gain high-spatial-resolution insights into fixed microtubule end conformations.[105,106,111] Computational approaches bridge the molecular to polymer scales, allowing simulation of molecular interactions through space and time using a variety of assumptions and input parameters.[109]
The main outstanding challenge lies in connecting these fixed, high-resolution snapshots to the live, dynamic transitions at microtubule ends. For example, given that EBs localize to the interface of four tubulin dimers, observations of split EB comets suggest that, at least in some circumstances, laterally connected subsets of protofilaments do exist on growing microtubule ends. This disconnect between the static ultra-structural and the dynamic low-resolution view of the microtubule tip is bridged by the continuous development of new computational modeling strategies (Figure 5).[109] Recent refinements of dynamic instability models using a variety of coarse-grained and multiscale computational approaches further support the role of 3D microtubule end geometry in the underlying mechanism of microtubule dynamics. Moreover, the mechanical aspects of individual protofilament geometries are increasingly appreciated.[110,111] Of note, a recent study using Brownian dynamics explained how thermal fluctuations drive straightening of curved, flexible protofilaments to promote formation of lateral protofilament interactions, thus supporting microtubule polymerization.[111] On the other hand, individual protofilaments must also be sufficiently stiff to support the established role of microtubule depolymerization in force generation.[103,112] Exploring a range of protofilament bending stiffnesses, this computational model is consistent with the flared microtubule end morphology. Furthermore, based on their model, the authors proposed that GTP hydrolysis triggers changes in either lateral bond strength or protofilament stiffness to drive microtubule catastrophe.[111]
Finally, with the increasing computational capabilities, molecular dynamics (MD) simulations that traditionally addressed atomistic-level details on nanosecond scales are becoming more informative for the processes of microtubule dynamics occurring over much larger timescales. MD approaches are particularly powerful for incorporating new high-resolution details of tubulin dimer structures obtained using cryo-EM approaches.[101,113-117] A recent study, employing atomistic-level MD simulations of the complete microtubule plus-end tip, explored the effect of GTP and GDP states on the elasticity and splaying dynamics of microtubule ends.[118] Interestingly, these simulations support the existence of metastable, laterally connected protofilament clusters, akin to sheet-like structures observed by cryo-EM.[106] Whether the durations of these simulations span time scales sufficiently long to detect potential protofilament splaying is not clear. Nevertheless, this study exemplifies that there is still a lot unknown about the dynamic structural transitions at the microtubule end that ultimately drive catastrophe.
OUTLOOK
After 40 years of active research on microtubule dynamics, the molecular mechanisms underlying microtubule dynamic instability remain an exciting and intense field of study. New details arising from state-of-the-art structural, computational, and biochemical reconstitution approaches are continuously woven into our understanding of microtubule dynamics. While the GTP-cap has been deemed the determinant of microtubule stability for decades, we now understand that the nucleotide content at the microtubule end can rapidly fluctuate. More importantly, an appreciation for the incredibly dynamic changes in microtubule end configurations has emerged. To what extent these configurations are directly linked to the nucleotide state of tubulin remains unknown; however, these dynamic transitions at the microtubule end are now recognized as the ultimate driver of microtubule catastrophe.
There is, however, still much to be learned. The majority of structural and computational studies have been focused on the dynamics of microtubule plus ends in the absence of any MAPs. Encompassing microtubule minus ends, as well as the effects of individual regulatory proteins, is an essential next step for the models of microtubule dynamics. Furthermore, our understanding of the mechanisms of microtubule catastrophe in cells where ensembles of regulatory proteins drive microtubule dynamics is particularly limited. Only a small number of recent studies have begun to address the high-resolution end structure and the size of the GTP-cap in cells.[107,119,120] Yet microtubules display a wide range of dynamics in cellular processes, including migration, polarization, and differentiation.[121,122] Future studies investigating the molecular mechanisms that drive microtubule dynamics in myriad cellular contexts will be required for the full understanding of the dynamic microtubule network in cells.
ACKNOWLEDGMENTS
We thank the members of the Zanic laboratory for discussions and feedback. We acknowledge the support by National Institutes of Health grant R35GM119552 to M.Z.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
DATA AVAILABILITY STATEMENT
Data sharing not applicable – no new data generated.
REFERENCES
- 1.Tilney LG, Bryan J, Bush DJ, Fujiwara K, Mooseker MS, Murphy DB, & Snyder DH (1973). Microtubules: Evidence for 13 protofilaments. Journal of Cell Biology, 59(2), 267–275. 10.1083/jcb.59.2.267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Margolis RL, & Wilson L (1978). Opposite end assembly and disassembly of microtubules at steady state in vitro. Cell, 13(1), 1–8. 10.1016/0092-8674(78)90132-0 [DOI] [PubMed] [Google Scholar]
- 3.Desai A, & Mitchison TJ (1997). Microtubule polymerization dynamics. Annual Review of Cell and Developmental Biology, 13(1), 83–117. 10.1146/annurev.cellbio.13.1.83 [DOI] [PubMed] [Google Scholar]
- 4.Fan J, Griffiths AD, Lockhart A, Cross RA, & Amos LA (1996). Microtubule minus ends can be labelled with a phage display antibody specific to alpha-tubulin. Journal of Molecular Biology, 259(3), 325–330. 10.1006/jmbi.1996.0322 [DOI] [PubMed] [Google Scholar]
- 5.Nogales E, Whittaker M, Milligan RA, & Downing KH (1999). High-resolution model of the microtubule. Cell, 96(1), 79–88. 10.1016/S0092-8674(00)80961-7 [DOI] [PubMed] [Google Scholar]
- 6.Mitchison TJ, & Kirschner MW (1984). Dynamic instability of microtubule growth. Nature, 310, 237–242. [DOI] [PubMed] [Google Scholar]
- 7.Kirschner M, & Mitchison T (1986). Beyond self-assembly: From microtubules to morphogenesis. Cell, 45(3), 329–342. 10.1016/0092-8674(86)90318-1 [DOI] [PubMed] [Google Scholar]
- 8.Brouhard GJ, & Rice LM (2018). Microtubule dynamics: An interplay of biochemistry and mechanics. Nature Reviews Molecular Cell Biology, 19(7), 451–463. 10.1038/s41580-018-0009-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carlier M-F, & Pantaloni D (1981). Kinetic analysis of guanosine 5′-triphosphate hydrolysis associated with tubulin polymerization. Biochemistry, 20(7), 1918–1924. 10.1021/bi00510a030 [DOI] [PubMed] [Google Scholar]
- 10.Nogales E, Wolf SG, & Downing KH (1998). Structure of the ab-tubulin dimer by electron crystallography. Nature, 391, 199–204. 10.1007/1-4020-3920-4 [DOI] [PubMed] [Google Scholar]
- 11.David-Pfeuty T, Erickson HP, & Pantaloni D (1977). Guanosinet-riphosphatase activity of tubulin associated with microtubule assembly. Proceedings of the National Academy of Sciences of the United States of America, 74(12), 5372–5376. 10.1073/pnas.74.12.5372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.MacNeal RK, & Purich DL (1978). Stoichiometry and role of GTP hydrolysis in bovine neurotubule assembly. Journal of Biological Chemistry, 253(13), 4683–4687. [PubMed] [Google Scholar]
- 13.Carlier M-F, Didry D, Simon C, & Pantaloni D (1989). Mechanism of GTP hydrolysis in tubulin polymerization: Characterization of the kinetic intermediate microtubule-GDP-Pi using phosphate analogues. Biochemistry, 28, 1783–1791. 10.1021/bi00430a054 [DOI] [PubMed] [Google Scholar]
- 14.Alushin GM, Lander GC, Kellogg EH, Zhang R, Baker D, & Nogales E (2014). High-resolution microtubule structures reveal the structural transitions in ab-tubulin upon GTP hydrolysis. Cell, 157(5), 1117–1129. 10.1016/j.cell.2014.03.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geyer EA, Burns A, Lalonde BA, Ye X, Piedra F-A, Huffaker TC, & Rice LM (2015). A mutation uncouples the tubulin conformational and GTPase cycles, revealing allosteric control of microtubule dynamics. ELife, 4, e10113. 10.7554/eLife.10113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang R, Alushin GM, Brown A, & Nogales E (2015). Mechanistic origin of microtubule dynamic instability and its modulation by EB proteins. Cell, 162(4), 849–859. 10.1016/j.cell.2015.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weisenberg RC, Deery WJ, & Dickinson PJ (1976). Tubulin-nucleotide interactions during the polymerization and depolymerization of microtubules. Biochemistry, 15(19), 4248–4254. 10.1021/bi00664a018 [DOI] [PubMed] [Google Scholar]
- 18.Hyman AA, Salser S, Drechsel DN, Unwin N, & Mitchison TJ (1992). Role of GTP hydrolysis in microtubule dynamics : Information from a slowly hydrolyzable analogue, GMPCPP. Molecular Biology of the Cell, 3, 1155–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caplow M, & Shanks J (1996). Evidence that a single monolayer tubulin-GTP cap is both necessary and sufficient to stabilize microtubules. Molecular Biology of the Cell, 7(4), 663–675. 10.1091/MBC.7.4.663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walker RA, O’Brien ET, Pryer NK, Soboeiro MF, Voter WA, Erickson HP, & Salmon ED (1988). Dynamic instability of individual microtubules. The Journal of Cell Biology, 107, 1437–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Drechsel DN, Hyman AA, Cobb MH, & Kirschner MW (1992). Modulation of the dynamic instability of tubulin assembly by the microtubule-associated protein tau. Molecular Biology of the Cell, 3(10), 1141–1154. 10.1091/mbc.3.10.1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fygenson DK, Braun E, & Libchaber A (1994). Phase diagram of microtubules. Physical Review E, 50(2), 1579–1588. 10.1103/PhysRevE.50.1579 [DOI] [PubMed] [Google Scholar]
- 23.Gardner MK, Zanic M, Gell C, Bormuth V, & Howard J (2011). Depolymerizing kinesins Kip3 and MCAK shape cellular microtubule architecture by differential control of catastrophe. Cell, 147(5), 1092–1103. 10.1016/j.cell.2011.10.037 [DOI] [PubMed] [Google Scholar]
- 24.Arpaǧ G, Lawrence EJ, Farmer VJ, Hall SL, & Zanic M (2020). Collective effects of XMAP215, EB1, CLASP2, and MCAK lead to robust microtubule treadmilling. Proceedings of the National Academy of Sciences of the United States of America, 117(23), 12847–12855. 10.1073/pnas.2003191117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Su L-K, Burrell M, Hill DE, Gyuris J, Brent R, Wiltshire R, Trent J, Vogelstein B, & Kinzler KW (1995). APC binds to the novel protein EB1. Cancer Research, 55, 2972–2977. [PubMed] [Google Scholar]
- 26.Akhmanova A, & Steinmetz MO (2008). Tracking the ends: A dynamic protein network controls the fate of microtubule tips. Nature Reviews Molecular Cell Biology, 9(4), 309–322. 10.1038/nrm2369 [DOI] [PubMed] [Google Scholar]
- 27.Nehlig A, Molina A, Rodrigues-Ferreira S, Honoré S, & Nahmias C (2017). Regulation of end-binding protein EB1 in the control of microtubule dynamics. Cellular and Molecular Life Sciences, 74(13), 2381–2393. 10.1007/s00018-017-2476-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mustyatsa VV, Boyakhchyan AV, Ataullakhanov FI, & Gudimchuk NB (2017). EB-family proteins: Functions and microtubule interaction mechanisms. Biochemistry (Moscow), 82(7), 791–802. 10.1134/S0006297917070045 [DOI] [PubMed] [Google Scholar]
- 29.Tirnauer JS, O’Toole E, Berrueta L, Bierer BE, & Pellman D (1999). Yeast Bim1p promotes the G1-specific dynamics of microtubules. Journal of Cell Biology, 145(5), 993–1007. 10.1083/jcb.145.5.993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mimori-Kiyosue Y, Shiina N, & Tsukita S (2000). The dynamic behavior of the APC-binding protein EB1 on the distal ends of microtubules. Current Biology, 10(14), 865–868. 10.1016/S0960-9822(00)00600-X [DOI] [PubMed] [Google Scholar]
- 31.Piehl M, & Cassimeris L (2003). Organization and dynamics of growing microtubule plus ends during early mitosis. Molecular Biology of the Cell, 14, 916–925. 10.1091/mbc.E02 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bieling P, Laan L, Schek H, Munteanu EL, Sandblad L, Dogterom M, Brunner D, & Surrey T (2007). Reconstitution of a microtubule plus-end tracking system in vitro. Nature, 450(7172), 1100–1105. 10.1038/nature06386 [DOI] [PubMed] [Google Scholar]
- 33.Zanic M, Stear JH, Hyman AA, & Howard J (2009). EB1 recognizes the nucleotide state of tubulin in the microtubule lattice. PLoS ONE, 4(10), 1–5. 10.1371/journal.pone.0007585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maurer SP, Fourniol FJ, Bohner G, Moores CA, & Surrey T (2012). EBs recognize a nucleotide-dependent structural cap at growing microtubule ends. Cell, 149(2), 371–382. 10.1016/j.cell.2012.02.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roostalu J, Thomas C, Cade NI, Kunzelmann S, Taylor IA, & Surrey T (2020). The speed of GTP hydrolysis determines GTP cap size and controls microtubule stability. ELife, 9, 1–22. 10.7554/eLife.51992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maurer SP, Bieling P, Cope J, Hoenger A, & Surrey T (2011). GTPyS microtubules mimic the growing microtubule end structure recognized by end-binding proteins (EBs). Proceedings of the National Academy of Sciences of the United States of America, 108(10), 3988–3993. 10.1073/pnas.1014758108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vitre B, Coquelle FM, Heichette C, Garnier C, Chrétien D, & Arnal I (2008). EB1 regulates microtubule dynamics and tubulin sheet closure in vitro. Nature Cell Biology, 10(4), 415–421. 10.1038/ncb1703 [DOI] [PubMed] [Google Scholar]
- 38.Zanic M, Widlund PO, Hyman AA, & Howard J (2013). Synergy between XMAP215 and EB1 increases microtubule growth rates to physiological levels. Nature Cell Biology, 15(5), 688–693. 10.1038/ncb2744 [DOI] [PubMed] [Google Scholar]
- 39.Reid TA, Coombes C, Mukherjee S, Goldblum RR, White K, Parmar S, Mcclellan M, Zanic M, Courtemanche N, & Gardner MK (2019). Structural state recognition facilitates tip tracking of EB1 at growing microtubule ends. ELife, 1–32. 10.1101/636092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guesdon A, Bazile F, Buey RM, Mohan R, Monier S, García RR, Angevin M, Heichette C, Wieneke R, Tampé R, Duchesne L, Akhmanova A, Steinmetz MO, & Chrétien D (2016). EB1 interacts with outwardly curved and straight regions of the microtubule lattice. Nature Cell Biology, 18, 1102–1108. 10.1038/ncb3412 [DOI] [PubMed] [Google Scholar]
- 41.Farmer V, Arpağ G, Hall S, & Zanic M (2021). XMAP215 promotes microtubule catastrophe by disrupting the growing microtubule end. Journal of Cell Biology, 220(10), 1–13. 10.1101/2020.12.29.424748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Drechsel DN, & Kirschner MW (1994). The minimum GTP cap required to stabilize microtubules. Current Biology, 4(12), 1053–1061. 10.1016/S0960-9822(00)00243-8 [DOI] [PubMed] [Google Scholar]
- 43.Duellberg C, Cade NI, Holmes D, & Surrey T (2016). The size of the EB cap determines instantaneous microtubule stability. ELife, 5, e13470. 10.7554/eLife.13470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Akhmanova A, & Steinmetz MO (2015). Control of microtubule organization and dynamics: Two ends in the limelight. Nature Reviews Molecular Cell Biology, 16(12), 711–726. 10.1038/nrm4084 [DOI] [PubMed] [Google Scholar]
- 45.Horio T, & Hotani H (1986). Visualization of the dynamic instability of individual micortubules by dark-field microscopy. Nature, 320, 646–650. 10.1038/323646a0 [DOI] [PubMed] [Google Scholar]
- 46.Strothman C, Farmer V, Arpağ G, Rodgers N, Podolski M, Norris S, Ohi R, & Zanic M (2019). Microtubule minus-end stability is dictated by the tubulin off-rate. The Journal of Cell Biology, 218(9), 2841–2853. 10.1083/jcb.201905019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Charrasse S, Mazel M, Taviaux S, Berta P, Chow T, & Larroque C (1995). Characterization of the cDNA and pattern of expression of a new gene over-expressed in human hepatomas and colonic tumors. European Journal of Biochemistry, 234(2), 406–413. 10.1111/j.1432-1033.1995.406_b.x [DOI] [PubMed] [Google Scholar]
- 48.Charrasse S, Schroeder M, Gauthier-Rouviere C, Ango F, Cassimeris L, Gard DL, & Larroque C (1998). The TOGp protein is a new human microtubule-associated protein homologous to the Xenopus XMAP215. Journal of Cell Science, 111(10), 1371–1383. [DOI] [PubMed] [Google Scholar]
- 49.Slep KC (2009). The role of TOG domains in microtubule plus end dynamics. Biochemical Society Transactions, 37(5), 1002–1006. 10.1042/BST0371002 [DOI] [PubMed] [Google Scholar]
- 50.Slep KC (2018). A cytoskeletal symphony: Owed to TOG. Developmental Cell, 46(1), 5–7. 10.1016/j.devcel.2018.06.010 [DOI] [PubMed] [Google Scholar]
- 51.Farmer VJ, & Zanic M (2021). TOG-domain proteins. Current Biology, 31, R499–R501. 10.1016/j.cub.2021.01.039 [DOI] [PubMed] [Google Scholar]
- 52.Ayaz P, Ye X, Huddleston P, Brautigam CA, & Rice LM (2012). A TOG: αβ-Tubulin complex structure reveals conformation-based mechanisms for a microtubule polymerase. Science, 337(6096), 857–860. 10.1126/science.1221698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Byrnes AE, & Slep KC (2017). TOG–tubulin binding specificity promotes microtubule dynamics and mitotic spindle formation. The Journal of Cell Biology, 216(6), 1641–1657. http://jcb.rupress.org/content/early/2017/05/15/jcb.201610090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gard DL, Becker BE, & Josh Romney R (2004). MAPping the eukaryotic tree of life: Structure, function, and evolution of the MAP215/Dis1 family of microtubule-associated proteins. International Review of Cytology, 239, 179–272. 10.1016/S0074-7696(04)39004-2 [DOI] [PubMed] [Google Scholar]
- 55.Al-Bassam J,&Chang F (2011). Regulation of microtubule dynamics by TOG-domain proteins XMAP215/Dis1 and CLASP. Trends in Cell Biology, 21(10), 604–614. 10.1016/j.tcb.2011.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lawrence EJ, Zanic M, & Rice LM (2020). CLASPs at a glance. Journal of Cell Science, 133(8), jcs243097. 10.1242/jcs.243097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cullen CF, Deák P, Glover DM, & Ohkura H (1999). mini spindles: A gene encoding a conserved microtubule-associated protein required for the integrity of the mitotic spindle in Drosophila. Journal of Cell Biology, 146(5), 1005–1018. 10.1083/jcb.146.5.1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vasquez RJ, Gard DL, & Cassimeris L (1994). XMAP from Xenopus eggs promotes rapid plus end assembly of microtubules and rapid microtubule polymer turnover. Journal of Cell Biology, 127(4), 985–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gard DL, & Kirschner MW (1987). A microtubule-associated protein from Xenopus eggs that specifically promotes assembly at the plus-end. Journal of Cell Biology, 105(5), 2203–2215. 10.1083/jcb.105.5.2203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brouhard GJ, Stear JH, Noetzel TL, Al-Bassam J, Kinoshita K, Harrison SC, Howard J, & Hyman AA (2008). XMAP215 is a processive microtubule polymerase. Cell, 132(1), 79–88. 10.1016/j.cell.2007.11.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Widlund PO, Stear JH, Pozniakovsky A, Zanic M, Reber S, Brouhard GJ, Hyman AA, & Howard J (2011). XMAP215 polymerase activity is built by combining multiple tubulin-binding TOG domains and a basic lattice-binding region. Proceedings of the National Academy of Sciences of the United States of America, 108(7), 2741–2746. 10.1073/pnas.1016498108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Al-Bassam J, Larsen NA, Hyman AA, & Harrison SC (2007). Crystal Structure of a TOG Domain: Conserved Features of XMAP215/Dis1-Family TOG Domains and Implications for Tubulin Binding. Structure, 15(3), 355–362. 10.1016/j.str.2007.01.012 [DOI] [PubMed] [Google Scholar]
- 63.Grimaldi AD, Zanic M, & Kaverina I (2015). Encoding the microtubule structure: Allosteric interactions between the microtubule +TIP complex master regulators and TOG-domain proteins. Cell Cycle, 14(9), 1375–1378. 10.1080/15384101.2015.1026521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lemos CL, Sampaio P, Maiato H, Costa M, Omel’yanchuk LV, Liberal V, & Sunkel CE (2000). Mast, a conserved microtubule-associated protein required for bipolar mitotic spindle organization. EMBO Journal, 19(14), 3668–3682. 10.1093/emboj/19.14.3668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Inoue YH, Avides MDC, Shiraki M, Deak P, Yamaguchi M, Nishimoto Y, Matsukage A, & Glover DM (2000). Orbit, a novel microtubule-associated protein essential for mitosis in 3Drosophila melanogaster. Journal of Cell Biology, 149(1), 153–166. 10.1083/jcb.149.1.153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Akhmanova A, Hoogenraad CC, Drabek K, Stepanova T, Dortland B, Verkerk T, Vermeulen W, Burgering BM, De Zeeuw CI, Grosveld F, & Galjart N (2001). Clasps are CLIP-115 and -170 associating proteins involved in the regional regulation of microtubule dynamics in motile fibroblasts. Cell, 104, 923–935. [DOI] [PubMed] [Google Scholar]
- 67.Wittmann T, & Waterman-Storer CM (2005). Spatial regulation of CLASP affinity for microtubules by Rac1 and GSK3β in migrating epithelial cells. Journal of Cell Biology, 169(6), 929–939. 10.1083/jcb.200412114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Drabek K, van Ham M, Stepanova T, Draegestein K, van Horssen R, Sayas CL, Akhmanova A, Ten Hagen T, Smits R, Fodde R, Grosveld F, & Galjart N (2006). Role of CLASP2 in microtubule stabilization and the regulation of persistent motility. Current Biology, 16(22), 2259–2264. 10.1016/j.cub.2006.09.065 [DOI] [PubMed] [Google Scholar]
- 69.Maiato H, Fairley EAL, Rieder CL, Swedlow JR, Sunkel CE, & Earnshaw WC (2003). Human CLASP1 is an outer kinetochore component that regulates spindle microtubule dynamics. Cell, 113(7), 891–904. 10.1016/S0092-8674(03)00465-3 [DOI] [PubMed] [Google Scholar]
- 70.Maiato H, Khodjakov A, & Rieder CL (2005). Drosophila CLASP is required for the incorporation of microtubule subunits into fluxing kinetochore fibres. Nature Cell Biology, 7(1), 42–47. 10.1038/ncb1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Efimov A, Kharitonov A, Efimova N, Loncarek J, Miller PM, Andreyeva N, Gleeson P, Galjart N, Maia ARR, McLeod IX, Yates JR, Maiato H, Khodjakov A, Akhmanova A, & Kaverina I (2007). Asymmetric CLASP-dependent nucleation of noncentrosomal microtubules at the trans-Golgi network. Developmental Cell, 12(6), 917–930. 10.1016/j.devcel.2007.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Miller PM, Folkmann AW, Maia ARR, Efimova N, Efimov A, & Kaverina I (2009). Golgi-derived CLASP-dependent microtubules control Golgi organization and polarized trafficking in motile cells. Nature Cell Biology, 11(9), 1069–1080. 10.1038/ncb1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Aher A, Kok M, Sharma A, Rai A, Olieric N, Rodriguez-Garcia R, Katrukha EA, Weinert T, Olieric V, Kapitein LC, Steinmetz MO, Dogterom M, & Akhmanova A (2018). CLASP suppresses microtubule catastrophes through a single TOG domain. Developmental Cell, 46(1), 40–58.e8. 10.1016/j.devcel.2018.05.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lawrence EJ, Arpag G, Norris SR, & Zanic M (2018). Human CLASP2 specifically regulates microtubule catastrophe and rescue. Molecular Biology of the Cell, 29(10), 1168–1177. 10.1091/mbc.E18-01-0016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lawrence EJ, & Zanic M (2019). Rescuing microtubules from the brink of catastrophe: CLASPs lead the way. Current Opinion in Cell Biology, 56, 94–101. 10.1016/j.ceb.2018.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Majumdar S, Kim T, Chen Z, Munyoki S, Tso S-C, Brautigam CA, & Rice LM (2018). An isolated CLASP TOG domain suppresses microtubule catastrophe and promotes rescue. Molecular Biology of the Cell, (11), 1497–1503. 10.1111/cbdd.13316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mahserejian SM, Scripture JP, Mauro AJ, Lawrence EJ, Jonasson EM, Murray KS, Li J, Gardner M, Alber M, Zanic M, & Goodson HV (2022). Quantification of microtubule stutters: Dynamic instability behaviors that are strongly associated with catastrophe. Molecular Biology of the Cell, 33(3), ar22. 10.1091/mbc.E20-06-0348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Panda D, Samuel JC, Massie M, Feinstein SC, & Wilson L (2003). Differential regulation of microtubule dynamics by three- and four-repeat tau: Implications for the onset of neurodegenerative disease. Proceedings of the National Academy of Sciences of the United States of America, 100(16), 9548–9553. 10.1073/pnas.1633508100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ramirez-Rios S, Denarier E, Prezel E, Vinit A, Stoppin-Mellet V, Devred F, Barbier P, Peyrot V, Sayas CL, Avila J, Peris L, Andrieux A, Serre L, Fourest-Lieuvin A, & Arnal I (2016). Tau antagonizes end-binding protein tracking at microtubule ends through a phosphorylation-dependent mechanism. Molecular Biology of the Cell, 27(19), 2924–2934. 10.1091/mbc.E16-01-0029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sobel A, Boutterin MC, Beretta L, Chneiweiss H, Doye V, & Peyro-Saint-Paul H (1989). Intracellular substrates for extracellular signaling. Journal of Biological Chemistry, 264(7), 3765–3772. 10.1016/s0021-9258(19)84915-3 [DOI] [PubMed] [Google Scholar]
- 81.Belmont LD, & Mitchison TJ (1996). Identification of a protein that interacts with tubulin dimers and increases the catastrophe rate of microtubules. Cell, 84(4), 623–631. 10.1016/S0092-8674(00)81037-5 [DOI] [PubMed] [Google Scholar]
- 82.Bowne-Anderson H, Hibbel A, & Howard J (2015). Regulation of microtubule growth and catastrophe: Unifying theory and experiment. Trends in Cell Biology, 25(12), 769–779. 10.1016/j.tcb.2015.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kerssemakers JWJ, Munteanu EL, Laan L, Noetzel TL, Janson ME, & Dogterom M (2006). Assembly dynamics of microtubules at molecular resolution. Nature, 442(7103), 709–712. 10.1038/nature04928 [DOI] [PubMed] [Google Scholar]
- 84.Schek HT, Gardner MK, Cheng J, Odde DJ, & Hunt AJ (2007). Microtubule assembly dynamics at the nanoscale. Current Biology, 17(17), 1445–1455. 10.1016/j.cub.2007.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gardner MK, Hunt AJ, Goodson HV,&Odde DJ (2008). Microtubule assembly dynamics: New insights at the nanoscale. Current Opinion in Cell Biology, 20(1), 64–70. 10.1016/j.ceb.2007.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Howard J, & Hyman AA (2009). Growth, fluctuation and switching at microtubule plus ends. Nature Reviews Molecular Cell Biology, 10(8), 569–574. 10.1038/nrm2713 [DOI] [PubMed] [Google Scholar]
- 87.Gardner MK, Charlebois BD, Janosi IM, Howard J, Hunt AJ, & Odde DJ (2011). Rapid microtubule self-assembly kinetics. Cell, 146(4), 582–592. 10.1016/j.cell.2011.06.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mickolajczyk KJ, Geyer EA, Kim T, Rice LM, & Hancock WO (2019). Direct observation of individual tubulin dimers binding to growing microtubules. Proceedings of the National Academy of Sciences of the United States of America, 116(15), 7314–7322. 10.1101/418053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cleary JM,& Hancock WO (2021). Review Molecular mechanisms underlying microtubule growth dynamics. Current Biology, 31(10), R560–R573. 10.1016/j.cub.2021.02.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Odde DJ, Cassimeris L,& Buettner HM (1995). Kinetics of microtubule catastrophe assessed by probabilistic analysis. Biophysical Journal, 69(3), 796–802. 10.1016/S0006-3495(95)79953-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gardner MK, Zanic M, & Howard J (2013). Microtubule catastrophe and rescue. Current Opinion in Cell Biology, 25(1), 14–22. 10.1016/j.ceb.2012.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bowne-Anderson H, Zanic M, Kauer M, & Howard J (2013). Microtubule dynamic instability: A new model with coupled GTP hydrolysis and multistep catastrophe. BioEssays, 35(5), 452–461. 10.1002/bies.201200131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Piedra F-A, Kim T, Garza ES, Geyer EA, Burns A, Ye X, Rice LM, & Reck-Peterson SL (2016). GDP-to-GTP exchange on the microtubule end can contribute to the frequency of catastrophe. Molecular Biology of the Cell, 27, 3515–3525. 10.1091/mbc.E16-03-0199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kim T, & Rice LM (2019). Long-range, through-lattice coupling improves predictions of microtubule catastrophe. Molecular Biology of the Cell, 30(12), 1451–1462. 10.1091/mbc.E18-10-0641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cleary JM, Kim T, Cook ASI, McCormick LA, Hancock WO, & Rice LM (2022). Measurements and simulations of microtubule growth imply strong longitudinal interactions and reveal a role for GDP on the elongating end. ELife, 121(3), 521a. 10.7554/eLife.75931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Howard J, & Hyman AA (2003). Dynamics and mechanics of the microtubule plus end. Nature, 422(6933), 753–758. 10.1038/nature01600 [DOI] [PubMed] [Google Scholar]
- 97.Shirasu-Hiza M, Coughlin P, & Mitchison TJ (2003). Identification of XMAP215 as a microtubule-destabilizing factor in Xenopus egg extract by biochemical purification. Journal of Cell Biology, 161(2), 349–358. 10.1083/jcb.200211095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Coombes CE, Yamamoto A, Kenzie MR, Odde DJ, & Gardner MK (2013). Evolving tip structures can explain age-dependent microtubule catastrophe. Current Biology, 23(14), 1342–1348. 10.1016/j.cub.2013.05.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Margolin G, Gregoretti IV, Cickovski TM, Li C, Shi W, Alber MS, & Goodson HV (2012). The mechanisms of microtubule catastrophe and rescue: Implications from analysis of a dimer-scale computational model. Molecular Biology of the Cell, 23(4), 642–656. 10.1091/mbc.E11-08-0688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li C, Li J, Goodson HV,& Alber MS (2014). Microtubule dynamic instability: The role of cracks between protofilaments. Soft Matter, 10(12), 2069–2080. 10.1039/c3sm52892h [DOI] [PubMed] [Google Scholar]
- 101.Zakharov P, Gudimchuk NB, Voevodin V, Tikhonravov A, Ataullakhanov FI, & Grishchuk EL (2015). Molecular and mechanical causes of microtubule catastrophe and aging. Biophysical Journal, 109(12), 2574–2591. 10.1016/j.bpj.2015.10.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Demchouk AO, Gardner MK, & Odde DJ (2011). Microtubule tip tracking and tip structures at the nanometer scale using digital fluorescence microscopy. Cellular and Molecular Bioengineering, 4(2), 192–204. 10.1007/s12195-010-0155-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gudimchuk NB,& McIntosh JR (2021). Regulation of microtubule dynamics, mechanics and function through the growing tip. Nature Reviews Molecular Cell Biology, 22(12), 777–795. 10.1038/s41580-021-00399-x [DOI] [PubMed] [Google Scholar]
- 104.Mandelkow E-M, Mandelkow E, & Milliganll RA (1991). Microtubule dynamics and microtubule caps: A time-resolved cryo- electron microscopy study. The Journal of Cell Biology, 114(5), 977–991. 10.1083/jcb.114.5.977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chrétien D, Fuller SD, & Karsenti E (1995). Structure of growing microtubule ends: Two-dimensional sheets close into tubes at variable rates. Journal of Cell Biology, 129(5), 1311–1328. 10.1083/jcb.129.5.1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Atherton J, Stouffer M, Francis F, & Moores CA (2018). Microtubule architecture in vitro and in cells revealed by cryo-electron tomography. Acta Crystallographica Section D: Structural Biology, 74(6), 572–584. 10.1107/S2059798318001948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mcintosh JR, Toole EO, Morgan G, Austin J, Ulyanov E, Ataullakhanov FI, & Gudimchuk NB (2018). Microtubules grow by the addition of bent guanosine triphosphate tubulin to the tips of curved protofilaments. Journal of Cell Biology, 217(8), 2691–2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Simon JR, & Salmon ED (1990). The structure of microtubule ends during the elongation and shortening phases of dynamic instability examined by negative-stain electron microscopy. Journal of Cell Science, 96(Pt 4), 668–673. 10.1109/TSP.2003.822287 [DOI] [PubMed] [Google Scholar]
- 109.Hemmat M, Castle BT,& Odde DJ (2018). Microtubule dynamics: Moving toward a multi-scale approach. Current Opinion in Cell Biology, 50, 8–13. 10.1016/j.ceb.2017.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Michaels TCT, Feng S, Liang H, & Mahadevan L (2020). Mechanics and kinetics of dynamic instability. ELife, 9, e54077. 10.7554/eLife.54077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gudimchuk NB, Ulyanov EV, Toole EO, Page CL, Vinogradov DS, Morgan G, Li G, Moore JK, Szczesna E, Roll-mecak A, Ataullakhanov FI, & Mcintosh JR (2020). Mechanisms of microtubule dynamics and force generation examined with computational modeling and electron cryotomography. Nature Communications, 11(1), 3765. 10.1038/s41467-020-17553-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Grishchuk EL, Molodtsov MI, Ataullakhanov FI, & McIntosh JR (2005). Force production by disassembling microtubules. Nature, 438(7066), 384–388. 10.1038/nature04132 [DOI] [PubMed] [Google Scholar]
- 113.Tong D,& Voth GA (2020). Microtubule simulations provide insight into the molecular mechanism underlying dynamic instability. Biophysical Journal, 118(12), 2938–2951. 10.1016/j.bpj.2020.04.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Fedorov VA, Orekhov PS, Kholina EG, Zhmurov AA, Ataullakhanov FI, Kovalenko IB, & Gudimchuk NB (2019). Mechanical properties of tubulin intra- and inter-dimer interfaces and their implications for microtubule dynamic instability. PLOS Computational Biology, 15(8), e1007327. 10.1371/journal.pcbi.1007327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bollinger JA, & Stevens MJ (2018). Catastrophic depolymerization of microtubules driven by subunit shape change. Soft Matter, 1748–1752. 10.2018/SM/C7SM02033C [DOI] [PubMed] [Google Scholar]
- 116.Ayoub AT, Klobukowski M, & Tuszynski JA (2015). Detailed per-residue energetic analysis explains the driving force for microtubule disassembly. PLoS Computational Biology, 11(6), 1–21. 10.1371/journal.pcbi.1004313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Molodtsov MI, Ermakova EA, Shnol EE, Grishchuk EL, McIntosh JR, & Ataullakhanov FI (2005). A molecular-mechanical model of the microtubule. Biophysical Journal, 88(5), 3167–3179. 10.1529/biophysj.104.051789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Igaev M, & Grubmuller H (2022). Bending-torsional elasticity and energetics of the plus-end microtubule tip. Proceedings of the National Academy of Sciences of the United States of America, 121(3), 113a. 10.1016/j.bpj.2021.11.2143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Seetapun D, Castle BT, McIntyre AJ, Tran PT, & Odde DJ (2012). Estimating the microtubule GTP cap size in vivo. Current Biology, 22(18), 1681–1687. 10.1016/j.cub.2012.06.068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Mustyatsa VV, Kostarev AV, Tvorogova AV, Ataullakhanov FI, Gudimchuk NB, & Vorobjev IA (2019). Fine structure and dynamics of EB3 binding zones on microtubules in fibroblast cells. Molecular Biology of the Cell, 30, 2105–2114. 10.1091/mbc.e18-11-0723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Rusan NM, Fagerstrom CJ, Yvon A-MC, & Wadsworth P (2001). Cell cycle-dependent changes in microtubule dynamics in living cells expressing green fluorescent protein-alpha tubulin. Molecular Biology of the Cell, 12(4), 971–980. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=32280&tool=pmcentrez&rendertype=abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Akhmanova A, & Kapitein LC (2022). Mechanisms of microtubule organization in differentiated animal cells. Nature Reviews Molecular Cell Biology, 23, 541–558. 10.1038/s41580-022-00473-y [DOI] [PubMed] [Google Scholar]
- 123.Doodhi H, Prota AE, Kapitein LC, Akhmanova A, & Steinmetz MO (2016). Termination of protofilament elongation by eribulin induces lattice defects that promote microtubule catastrophes. Current Biology, 26, 1713–1721. 10.1016/j.cub.2016.04.053 [DOI] [PubMed] [Google Scholar]
- 124.Maurer SP, Fourniol FJ, & Hoenger A (2014). Seeded microtubule growth for cryoelectron microscopy of end-binding proteins. Methods in Molecular Biology, 1136, 247–260. 10.1007/978-1-4939-0329-0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing not applicable – no new data generated.