ABSTRACT
Trichophyton indotineae is an emerging dermatophyte that causes severe tinea corporis and tinea cruris. Numerous cases of terbinafine- and azole-recalcitrant T. indotineae-related dermatophytosis have been observed in India over the past decade, and cases are now being recorded worldwide. Whole genome sequencing of three azole-resistant strains revealed a variable number of repeats of a 2,404 base pair (bp) sequence encoding TinCYP51B in tandem specifically at the CYP51B locus position. However, many other resistant strains (itraconazole MIC ≥0.25 µg/mL; voriconazole MIC ≥0.25 µg/mL) did not contain such duplications. Whole-genome sequencing of three of these strains revealed a variable number of 7,374 bp tandem repeat blocks harboring TinCYP51B. Consequently, two types of T. indotineae azole-resistant strains were found to host TinCYP51B in tandem sequences (type I with 2,404 bp TinCYP51B blocks and type II with 7,374 bp TinCYP51B blocks). Using the CRISPR/Cas9 genome-editing tool, the copy number of TinCYP51B within the genome of types I and II strains was brought back to a single copy. The azole susceptibility of these modified strains was similar to that of strains without TinCYP51B duplication, showing that azole resistance in T. indotineae strains is mediated by one of two types of TinCYP51B amplification. Type II strains were prevalent among 32 resistant strains analyzed using a rapid and reliable PCR test.
KEYWORDS: dermatophytes, Trichophyton indotineae, TinCYP51B, gene duplication, itraconazole, voriconazole, antifungal resistance
INTRODUCTION
The spread of antifungal resistance in dermatophytes is an emerging health problem worldwide. The growing incidence of terbinafine-resistant strains of several species of Trichophyton has been the subject of multiple reports from many countries (1 – 5). So far, in all cases, terbinafine resistance results from missense mutations in the squalene epoxidase (SQLE) gene. Given the limited number of effective antifungals currently available to treat dermatophytes, the fact that antifungal resistance is not only related to terbinafine but also to azole compounds is a major concern. Azole resistance associated with the overexpression of genes encoding multidrug transporters of the ABC family was first reported in individual cases of Trichophyton rubrum (6 – 8). Notwithstanding, azole resistance appears to be more common, especially in the emerging species Trichophyton indotineae (formerly called Trichophyton interdigitale or Trichophyton mentagrophytes type VIII) (4, 9 – 11). About 25% of T. indotineae isolated from skin dermatophytosis lesions in India were found to be less susceptible to itraconazole (ITC) and voriconazole (VRC) (10).
We recently found that the reduced susceptibility of four T. indotineae strains to azoles was mainly due to the overexpression of the CYP51B gene (TinCYP51B) encoding a sterol 14α-demethylase, a target of azole compounds such as ITC and VRC (12). The overexpression of TinCYP51B resulted from additional copies of this gene in tandem, where 100% identical 2,404 base pair (bp) block repeats harboring TinCYP51B were inserted end-to-end (12). Each block included 631 bp of the original promoter and almost the entire coding region of the sterol 14α-demethylase—only the last five codons were missing. Overexpression of the gene encoding the drug target is an effective mechanism to acquire resistance by counteracting the effects of the drug, altering the balance in favor of its target. Sterol 14α-demethylases play a crucial role in ergosterol biosynthesis in fungi, by catalyzing the oxidative removal of 14α-methyl groups from sterol precursors (13, 14)
The present study aimed to examine whether all T. indotineae strains less sensitive to azoles had the same resistance mechanism as that recently described (12). Examination of a large panel of strains deemed resistant revealed without exception overexpression of TinCYP51B, but many did not contain tandem duplications of the 2,404 bp sequence containing TinCYP51B. We found that resistance in these strains was due to extra copies of TinCYP51B by large segmental duplication of a 7,374 bp DNA fragment. Thus, two different types of tandem sequences (i.e., single-gene duplications and large segmental duplications) mediate the overexpression of TinCYP51B in T. indotineae.
RESULTS
Two different types of azole-resistant strains of T. indotineae with multiple copies of TinCYP51B
Twenty-nine T. indotineae strains deemed resistant to ITC and VRC (ITC MIC ≥ 0.25 µg/mL; VRC MIC ≥ 0.25 µg/mL; Table S1) were tested to examine whether resistance to azole in T. indotineae was always mediated by tandem duplications of 2,404 bp chromosome blocks harboring the TinCYP51B gene. PCR was performed using the P1-sense and P2-antisense primers, designed at the 3′ and 5′ ends of TinCYP51B, respectively, and genomic DNA as a target. A 956 bp DNA fragment was only to be amplified with these primers when the 2,404 bp tandem duplicates were present. The three previously characterized resistant strains TIMM200116, TIMM200118, and TIMM 200119 (Table 1; Table S1), which are known to harbor 5–7 alleles of TinCYP51B in as many 2,404 bp blocks in tandem (12), were used as positive controls. Two previously characterized susceptible strains, TIMM200114 and TIMM 200115 (Table 1; Table S1), harboring one TinCYP51B gene (12) were used as negative controls.
TABLE 1.
Phenotypic and genotypic characteristics of T. indotineae strains used in this study
| T. indotineae strains a | TBF MIC80
(μg/mL) d |
ITC MIC80
(μg/mL) d |
VRC MIC80
(μg/mL) d |
Fold expression of TinCYP51B
(mean ± SD) b |
Number of TinCYP51B alleles (mean ± SD) c |
Strain type |
|---|---|---|---|---|---|---|
| TIMM20114 | 0.125 | 0.06 | 0.015 | 1 | 1 | Azole susceptible |
| (UKJ1676/17; IFM 67092) | ||||||
| TIMM20115 | 8 | 0.06 | 0.03 | 1.0 ± 0.4 | 0.9 ± 0.2 | Azole susceptible |
| (UKJ1700/17II; IFM67093) | ||||||
| TIMM20116 | 0.125 | 0.5 | 0.5 | 34.0 ± 5.3 | 5.3 ± 0.8 | Type I |
| (UKJ1708/17; IFM 67094) | ||||||
| TIMM20118 | 8 | 0.5 | 1 | 29.8 ± 4.4 | 5.5 ± 0.6 | Type I |
| (UKJ1687/17; IFM 67096) | ||||||
| TIMM20119 | 8 | 1 | 1 | 89.9 ± 26.2 | 8.9 ± 0.6 | Type I |
| (200123/18; IFM 67097) | ||||||
| TIMM20117 | 0.125 | 0.5 | 0.5 | 9.9 ± 3.8 | 10.8 ± 0.6 | Type II |
| (200087/18; IFM 67095) | ||||||
| TIMM20120 | 0.125 | 0.25 | 1 | 12.5 ± 5.0 | 10.0 ± 1.2 | Type II |
| (250082/18) | ||||||
| TIMM20121 | 8 | 0.5 | 1 | 23.4 ± 11.6 | 14.0 ± 0.05 | Type II |
| (250084/18) | ||||||
| TIMM20122 | 8 | 0.5 | 0.5 | 22.0 ± 8.0 | 7.9 ± 0.3 | Type II |
| (250108/18) | ||||||
| TIMM20123 | 0.125 | 1 | 0.5 | 6.2 ± 2.5 | 6.4 ± 0.4 | Type II |
| (600097/19) |
All strains were from a previously published resistance study in India, with the numbering in bold (10). They were then preserved in the culture collection of Teikyo University Institute of Medical Mycology (TIMM), some of which were also preserved in the culture collection of Medical Mycology Research Center, Chiba University (IFM), through the National Bio-Resource Project, Japan (http://www.nbrp.jp/).
The results represent expression levels from three independent real-time PCR experiments. The expression levels of TinCYP51B genes were indicated as relative fold changes compared to the ΔCt mean of the data from TIMM20114 (control with a single copy of TinCYP51B). SD, standard deviation.
Data were obtained from three independent qPCR experiments using genomic DNA as a target. The TinCYP51B gene copy number of each strain was indicated as relative fold change compared to the ΔCt mean of the data from TIMM20114.
TBF, terbinafine; ITC, itraconazole; VRC, voriconazole.
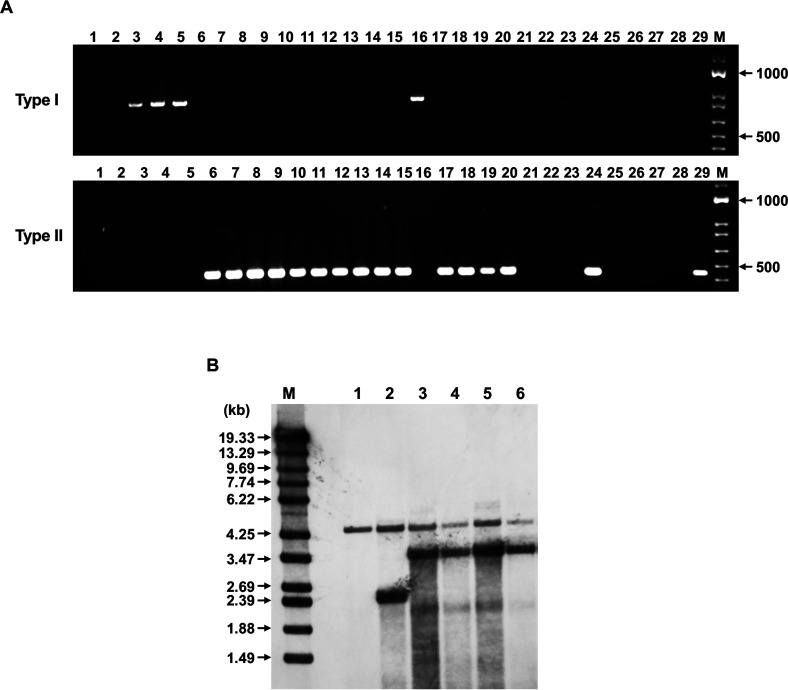
As expected, no genomic DNA amplification was obtained with the azole-susceptible control strains TIMM20114 and TIMM20115 (Fig. 1A upper panel, lanes 1–2), and an amplicon of the predicted size was obtained with TIMM20116, TIMM20118, and TIMM20119 (Fig. 1A upper panel, lanes 3–5). However, we surprisingly detected DNA amplification specific for 2,404 bp tandem duplicates in only one out of all 29 new strains included in the analysis (strain 216510/17; Fig. 1A upper panel, lane 16). No amplicon was obtained from all strains deemed susceptible to azole compounds (ITC MIC ≤ 0.25 µg/mL; VRC MIC ≤ 0.25 µg/mL; Fig. 1A upper panel, lanes 21–29). To sum up, our results pointed out toward the existence of another yet unknown mechanism of azole resistance in T. indotineae. We therefore referred to all strains for which an amplicon was obtained as “type I” azole-resistant strains and provisionally coined the term “type II” azole-resistant strains for all newly discovered resistant strains.
Fig 1.
Identification of types I and II strains in T. indotineae. (A) Types I and II strains were identified by amplification of genomic DNA with primer pairs P1–P2 and P3–P4, respectively. TIMM200114 and TIMM 200115, which are susceptible, were used as negative controls (lanes 1 and 2). Azole-resistant strains TIMM200116, TIMM200118, and TIMM 200119, which are known to harbor 5–7 alleles of CYP51B in as many 2,404 bp blocks in tandem, were used as positive controls for the type I strains (lanes 3–5). Lanes 6–29: Search for types I and II tandem sequences in the T. indotineae strains in Table S1. Lanes 6–20: Strains deemed to be resistant to azoles (ITC MIC ≥ 0.25 µg/mL; VRC MIC ≥ 0.25 µg/mL) marked by an asterisk (*) in Table S1. Strain 216510/17 was type I (lane 16). All other strains were type II. Lanes 21–29: Strains deemed to be susceptible to azoles (ITC MIC ≤ 0.25 µg/mL; VRC MIC ≤ 0.25 µg/mL) marked with a hash (#) in Table S1. Two of the later strains (UKJ 1673/17 and 250063/18) were of type II (lanes 24 and 29). (B) Southern blotting analysis of genomic DNA samples from six T. indotineae strains (TIMM20114, TIMM20118, TIMM20120, TIMM20121, TIMM20122, and TIMM20123). Genomic DNA from each strain was digested with XhoI and separated by electrophoresis on a 0.8% (w/v) agarose gel. Lanes 1–6: genomic DNA samples from TIMM20114, TIMM20118, TIMM20120, TIMM20121, TIMM20122, and TIMM20123, respectively. An internal fragment (about 410 bp) of the TinCYP51B gene was amplified by PCR with P21-P16 primers (Table S2) and used as a hybridization probe. The DNA standard fragment sizes are shown on the left.
Four type II strains, TIMM20120–TIMM20123, were chosen to further characterize the mechanism involved in azole resistance. Just as in type I strains, qRT-PCR revealed strong overexpression of TinCYP51B in these strains (Table 1). We examined whether the TIMM20120–TIMM20123 strains contained multiple copies of TinCYP51B in their genome by quantitative PCR (qPCR) using their genomic DNA as a target. The azole-susceptible strain TIMM20114, with a single copy of TinCYP51B, was used for comparison. The genomes of TIMM20120–TIMM20123 were estimated to contain 6 to 14 copies of TinCYP51B (Table 1).
The strains TIMM20120-TIMM 20123 differed from the susceptible strain TIMM20114 and the type I resistant strain TIMM20118 also in Southern blotting experiments, using a probe designed downstream of the XhoI site in the TinCYP51B open reading frame (ORF; Fig. 1B). As expected, a single 4.4 kb band was detected in the susceptible strain TIMM200114 with a single TinCYP51B gene (12). A band with the same electrophoretic mobility and a stronger 2.4 kb band were also revealed in the previously characterized type I resistant strain TIMM20118 (12), in agreement with its deposited genome sequence data (GenBank accession numbers: OK500344 and JAJVHJ000000000). The 4.4 kb band was also detected in the strains TIMM20120–TIMM20123, but there was an additional, more intense band corresponding to a 3.4 kb fragment and specific for type II strains.
Genome sequencing of type II azole-resistant strains
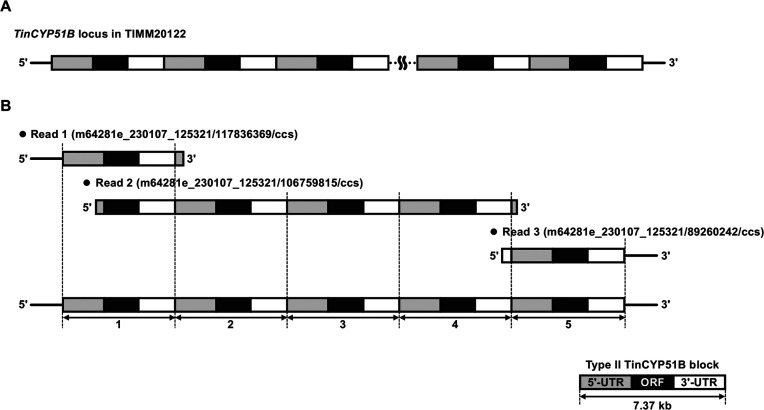
The whole genomes of the type II azole-resistant strains TIMM20121, TIMM20122, and TIMM20123 were sequenced using the PacBio sequencing technique (Table S3). Analysis of the sequences revealed that, similar to the type I resistant strains, the type II strains contained tandem duplications of the CYP51B locus. However, the duplicated blocks in the type II strains had a size of 7,374 bp instead of 2,404 bp. PacBio sequencing provided three types of reads with the TinCYP51B raw sequence data (Fig. 2): (i) reads with a DNA sequence identical to that upstream of the 7,374 bp block in TIMM20114, followed by one or two complete blocks, and ending with a partial block; (ii) reads with 1–3 complete blocks between the end and beginning of a partial block sequence; and (iii) reads starting with the end of a block, followed by 1–3 complete blocks, and ending with a DNA sequence identical to that of the unique 7,374 bp block sequence in TIMM20114. Sequences of the three read types for strains TIMM20121, TIMM20122, and TIMM20123 are given in the supplementary materials.
Fig 2.
PacBio sequencing reveals direct tandem repeats of 7,374 bp blocks in type II strain TIMM20122. (A) Scheme describing the organization of the tandem blocks within TIMM20122. The black boxes correspond to the TinCYP51B open reading frame. Upstream and downstream sequences including neighboring genes are shown by gray and white boxes, respectively. (B) Examples of the reads obtained, including reads covering the 5′ border of the duplicated regions (read m64281e_230107_125321/117836369/ccs), reads covering the 3′ border of the duplicated region (read m64281e_230107_125321/106759815/ccs), and reads localized within the duplicated region demonstrating the presence of at least five repeats in this type II strain (read m64281e_230107_125321/89260242/ccs). Sequences of these three reads are given in the supplementary materials.
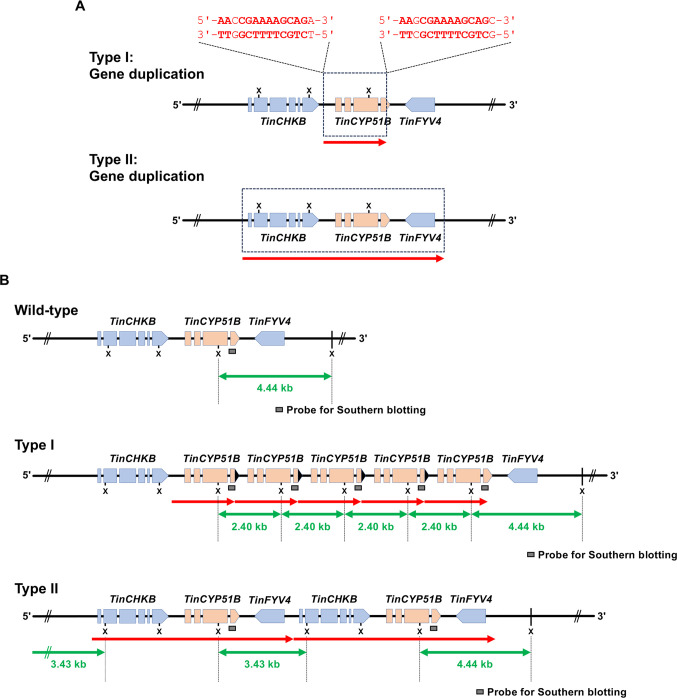
Although the duplicated blocks in type II strains were centered on TinCYP51B, they also contained two adjacent genes, TinCHKB, encoding the homolog of A. nidulans chkB (AN4279) kinase (15, 16) (upstream), and TinFYV4, homologous to Saccharomyces cerevisiae FYV4 (17) (downstream). Consequently, duplicated blocks in the type II strains were considered as large segmental duplications (Fig. 3A). Because of the large size of the duplicated blocks and the fact that there were more than five of them, the single PacBio reads could not cover the entire duplicated region. The 100% sequence identity between the blocks prevented efficient assembly of the individual reads to obtain the sequence covering an entire duplicated region (complete tandem repeat sequence) in the genomes. For strain TIMM20122, several reads covered three complete blocks of 7,374 bp, as well as parts of the two adjacent blocks, showing that this strain contains at least five blocks organized in tandem repeats. For strains TIMM20121 and TIMM20123, the PacBio sequencing revealed the presence of at least four blocks. The reads covered the 5′ and 3′ boundaries of the duplicated region for all three strains (see the supplementary materials). By combining the PacBio results with the qPCR results mentioned above, we concluded that 6 to 14 copies of the 7,374 bp block, organized in tandem repeats, were present within the genome of the type II strains TIMM20121, TIMM20122, and TIMM20123. Whatever the number of TinCYP51B alleles in the type II strains, the end-to-end 7,374 bp block sequences were consistent with the results obtained by Southern blotting analysis (Fig. 3B).
Fig 3.
Comparison of the 2,404 bp duplicated blocks within type I strains and the 7,374 bp blocks found in type II strains. Type I strains include only the TinCYP51B gene (A). Type II strains contain TinCYP51B (skin-colored boxes) and the neighboring genes TinCHKB and TinFYV4 (blue boxes) (B). The coverage of each type of block is marked with a red arrow and the sequences of the borders of the original block are also shown in red. The conserved nucleotides are marked in bold, revealing the absence of any conservation at the borders of the type II 7,374 bp duplicated blocks. The results of the PacBio sequencing results and southern blotting analysis are consistent. X, XhoI site.
Reduction of the TinCYP51B copy number to a single copy increases the susceptibility of T. indotineae to azole compounds
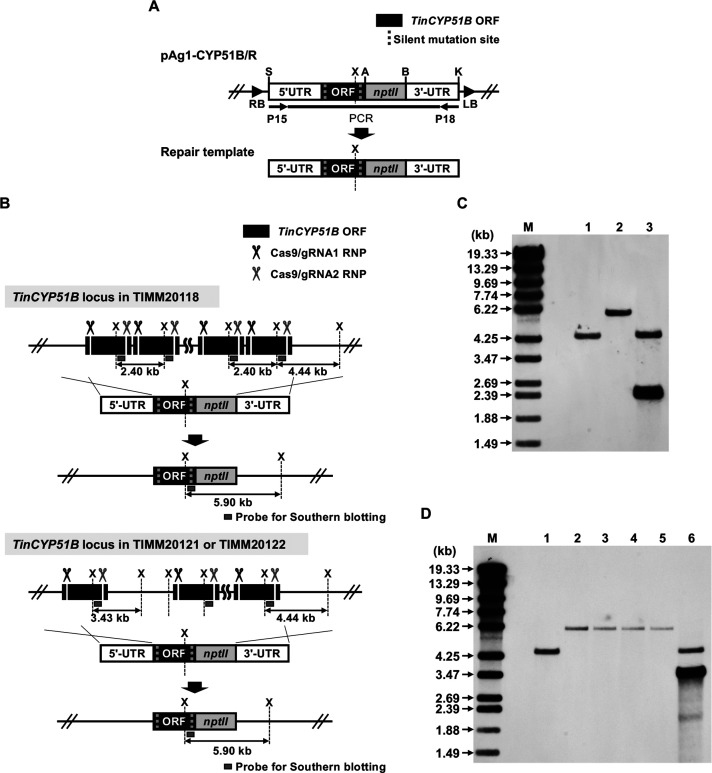
We previously showed the importance of TinCYP51B overexpression in the azole resistance of T. indotineae through RNA interference (RNAi)-mediated downregulation of the TinCYP51B gene (12). However, it was difficult to achieve complete inhibition of TinCYP51B gene expression in TinCYP51B-overexpressing strains with low azole susceptibility. Here, using the CRISPR/Cas9 genome editing tool, we attempted to reduce the copy number of the TinCYP51B gene within the genome of TinCYP51B-overexpressing strains to a single copy. For this purpose, two kinds of Cas9/single guide RNA (sgRNA) ribonucleoprotein complexes (RNPs) and the repair fragment were introduced into one type I azole-resistant strain, TIMM20118, and two type II azole-resistant strains, TIMM20121 and TIMM20122 (Fig. 4). The protoplasts/polyethylene glycol (PEG)-mediated transformation of these strains produced many G418-resistant transformants on the selective medium, 28 to 30 of which were chosen at random and tested for their growth properties on Sabouraud dextrose agar (SDA) supplemented with 2.0-µg/mL ITC.
Fig 4.
CRISPR/Cas9-mediated genetic modification of the TinCYP51B locus in the type I azole-resistant strain TIMM20118 and the type II azole-resistant strains TIMM20121 and TIMM20122. (A) Schematic representation of the binary vector pAg1-TinCYP51B/R. The nptII cassette is composed of the promoter sequence of Aspergillus nidulans tryptophan C (trpC) gene (PtrpC), nptII, and the termination sequence of A. fumigatus cgrA gene (TcgrA). The two silent mutations on the repair template cannot be cleaved by the RNP nuclease. Plasmid DNA of pAg1-TinCYP51B/R was used as a template for PCR with P15–P18 primers, to amplify the sequence (repair template) indicated by the thick line. (B) Schematic representation of the TinCYP51B locus before and after modification. The cleaved TinCYP51B locus is replaced by the repair template, resulting in the introduction of the desired modification. The replaced genomic DNA cannot be cleaved by the RNP nuclease. (C AND D) Southern blotting analysis. Aliquots of approximately 5 µg of genomic DNA from each strain were digested with XhoI and separated by electrophoresis on 0.8% (w/v) agarose gels. (C) Lane 1, TIMM20114 (azole susceptible); lane 2, 18 MM_25; lane 3, TIMM20118 (parent strain); M, DNA standard fragments (λ-EcoT14I/BglII digest). (D) Lane 1, TIMM20114 (azole susceptible); lanes 2 to 5, 21 MM_16–1, 21 MM_16–2, 21 MM_23–12, and 21 MM_23–13; lane 6, TIMM20121 (parent strain); M, DNA standard fragments. DNA standard fragment sizes are shown on the left. An internal fragment (about 410 bp) of the TinCYP51B gene was amplified by PCR with P21–P16 primers (Table S2) and used as a hybridization probe.
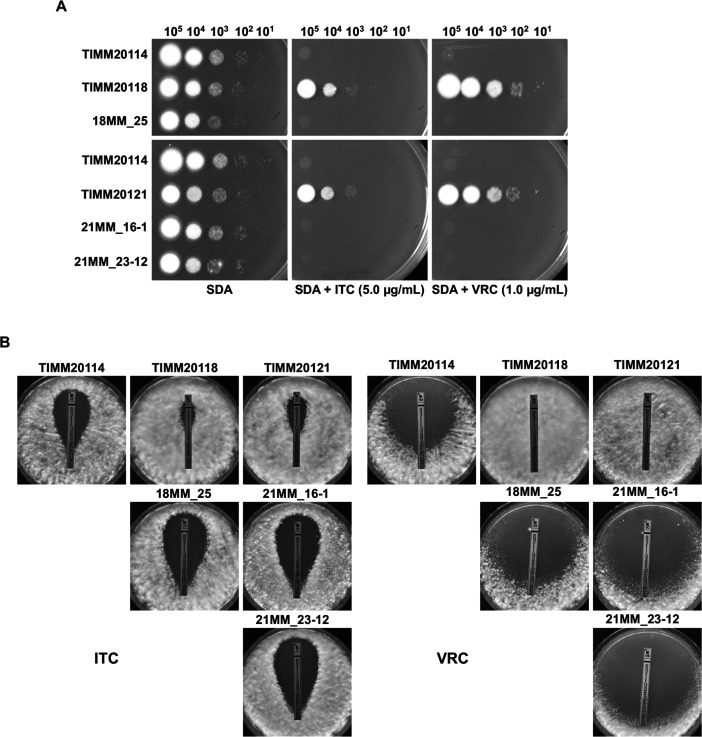
The identification of the expected clones harboring a single copy of TinCYP51B was carried out in three steps. First, since overexpression of TinCYP51B confers resistance in T. indotineae (12), we selected the transformants showing the most significant growth inhibition on ITC, which were most likely to have a reduced copy number of TinCYP51B (14 clones for TIMM20118, 12 clones for TIMM20121, and 12 clones for TIMM20122). PCR with P22–P23 primers for TIMM20118 transformants and P24–P25 primers for TIMM20121 and TIMM20122 transformants then resulted in the exclusion of, respectively, 13 clones derived from TIMM20118 and eight clones derived from TIMM20121 which still carried several copies of the TinCYP51B gene in their genome. All clones derived from TIMM20122 still contained several copies of TinCYP51B and were not further investigated. Finally, Southern blot analysis enabled us to confirm that one clone derived from TIMM20118, and four clones derived from TIMM20121 harbored one single TinCYP51B gene, as expected (Fig. 4C and D). The ITC and VRC susceptibility of these clones, assessed by serial dilution drug susceptibility tests and Etests, was similar to that of the azole-susceptible strain TIMM20114 used for comparison (Fig. 5).
Fig 5.
Evaluation of ITC and VRC susceptibility in the genetically modified T. indotineae strains by the serial dilution drug susceptibility assays and Etests. For the serial dilution drug susceptibility assays, spores from each strain were spotted at different dilutions on SDA plates, as previously described (4, 12). The plates were incubated at 28°C for 3 to 5 d.
Strains of T. indotineae with low susceptibility to azoles are mainly type II
The P3-sense and P4-antisense primers were designed 126 upstream and 309 bp downstream of the 7,374 bp block junctions in TIMM20120–TIMM20123 (Table S2). PCR reactions were performed using genomic DNA from these four strains as a target. As expected, a 435 bp amplicon was obtained for all four strains (Fig. 1A lower panel; Table 1). In total, 28 of the 32 strains with low susceptibility to azoles (MIC ITC ≥ 0.25 µg/mL; MIC VRC ≥ 0.25 µg/mL), which were not of type I, were of type II (Fig. 1A lower panel; Table S1). In conclusion, strains with low susceptibility to azoles were either types I or II, with a predominance of type II. Blocks of 7,374 bp in tandem could also be detected in T. indotineae strains that were more susceptible to ITC and VRC. Surprisingly, of the 16 strains deemed susceptible to azoles, three were found to be type II by PCR (Table S1). One of these strains, UKJ1673/17 underwent further examination and was found to contain three 7,374 bp blocks by qPCR using genomic DNA as a target (data not shown), whereas all the low susceptible type II strains contained at least five copies. These results show that strains appearing to be susceptible to azole compounds may harbor a reduced number of TinCYP51B in tandem.
DISCUSSION
Examination of a large panel of T. indotineae azole-resistant strains revealed the existence of two types of strains (types I and II) containing repeats of several TinCYP51B in tandem. Type I strains harbored TinCYP51B duplications with 2,404 bp end-to-end blocks containing only this gene (12). Type II strains revealed large segmental duplications with blocks of 7,374 bp, which contained TinCYP51B as well as the adjacent upstream and downstream genes that we named TinCHBK and TinFYV4, respectively. Whole genome sequencing could not reveal the exact number of duplications in type II strains due to technological limitations, but this number could be estimated by qPCR analysis. Here, the newly described type II strains were by far more prevalent than type I strains. In our previous study, we selected the most resistant strains based on MICs and Etests to test ITC susceptibility (Fig. 5), which is why we first characterized type I strains (12).
The case of type II strain UKJ1673/17 is interesting as it was estimated to contain only three copies of the 7,374 blocks and did not show any significantly lower susceptibility than wild-type strains containing only a single TinCYP51B gene. Therefore, in evolutionary terms, it represents an interesting link between wild-type strains with high azole susceptibility and low susceptible type II strains. These strains will be the subject of a further study.
Using the CRISPR/Cas9 genome editing tool, the copy number of TinCYP51B within the genome of the types I and II strains was brought back to a single copy. The azole resistance of these modified strains was similar to that of susceptible strains without TinCYP51B duplications, demonstrating that the reduced susceptibility of the T. indotineae strains was caused by the amplification of the TinCYP51B gene (Fig. 5). The effect of amplifying adjacent genes remains to be evaluated. The A. nidulans chkB protein kinase plays a role in protecting against UV damage in this fungus (15). S. cerevisiae FYV4 is involved in telomere length regulation (17) and is required for survival upon exposure to K1 killer toxin (18). However, as shown in Fig. 5A, a reduction of the large segmental duplications containing TinCHKB and TinFYV4 does not appear to have any significant effect on the cell growth of type II strains at least on SDA.
Short conserved sequences are missing at the borders of the duplicated blocks in type II strains
Duplications are produced by homologous recombination (HR) between directly oriented repeat sequences, insertion sequences, or transposable elements, with the intervention of a recombinase enzyme. Such a mechanism might be the cause of the duplications observed in type I strains in which short conserved sequences have been found at the borders of the duplicated blocks, called HR sites HR1 and HR2 (12). No HR sequences, such as those found at the borders of the 2,404 bp blocks in type I strains, were identified at the borders of the 7,374 bp blocks in type II strains. Duplications that lack HR sequences at the duplication block junctions arise frequently in bacteria (19). This implies that spontaneous genetic duplications can occur via mechanisms that, in the absence of any repeated sequence, can use short homologous sequences or homology-independent random end joining (20). A similar process is responsible for the tandem duplications observed in the type II strains and also seems to occur frequently in T. indotineae.
Why T. indotineae favors the duplication mechanism for azole resistance, in contrast to other dermatophyte species, remains to be clarified. This bias may be simply the result of a lack of knowledge of gene duplications, which have only recently been discovered. There is no reason why such phenomena should not occur in other dermatophytes. A large-scale survey of dermatophyte azole-resistant strains might bring some answers to this question.
Alteration in copy numbers of CYP51 genes in fungi other than T. indotineae
The copy number variation (CNV) of CYP51 genes is an effective way for fungi to acquire azole resistance (Table 2). The CNV can originate from whole chromosome duplications, as shown for chromosome 5 from Candida albicans (21, 22) and Candida glabrata (23), or for chromosome 1 from Cryptococcus neoformans (24), harboring drug target genes such as ERG11/CYP51. Large segmental duplications mediating the CNV of CYP51 genes have been observed in fungi other than T. indotineae, such as Rhynchosporium commune, a major fungal pathogen of barley, and the recently emerged multidrug-resistant Candida auris, which is the focus of intensive studies. In R. commune, the large segmental duplication of CYP51A contributes to the emergence of azole resistance (25). In C. auris, several resistance mechanisms have been described, including aneuploidy, gene copy accumulation, and new mutations in known and unknown antifungal drug resistance genes (26 – 29). For example, the segmental duplication of chromosome 1 containing ERG11, the CYP51 ortholog, and a whole chromosome 5 duplication, which contains TAC1b associated with an increased expression of ERG11, TAC1b, and CDR2, have been observed (26, 27). Gene duplications of CDR1, encoding a multidrug efflux transporter, or ERG11 were further associated with fluconazole resistance (28, 29).
TABLE 2.
Amplification of drug target genes in the literature
| Species | References | Amplified genes | Drug | Mechanism | Detection technique |
|---|---|---|---|---|---|
| Fungi | |||||
| Candida albicans | (21, 22) | ERG11/CYP51 a | Azoles | Chromosome 5 duplication (aneuploidy) | Comparative genome hybridization and karyotype analysis |
| Candida glabrata | (23) | ERG11/CYP51 | Azoles | Chromosome 5 duplication (aneuploidy) | Hybridization experiments on chromosomal blots |
| Candida auris | (26, 27) | ERG11/CYP51 | Azoles | Chromosome 5 duplication (aneuploidy) | Whole-genome sequencing (Illumina and MiSeq) |
| Candida auris | (28) | ERG11/CYP51 | Azoles | Large segmental duplication h | Whole-genome sequencing (Illumina and MiSeq) |
| Candida auris | (29) | ERG11/CYP51 and CDR1 b | Azoles | Transient gene duplication | Real-time quantitative reverse-transcription PCR (qRT-PCR) |
| Cryptococcus neoformans | (24) | ERG11/CYP51 and AFR1 b | Azoles | Chromosome 1 duplication (aneuploidy) | Comparative genome hybridization and qRT-PCR |
| Rhynchosporium commune | (25) | CYP51A | Azoles | Large segmental duplication | Whole-genome sequencing (Illumina) |
| T. indotineae | (12) | CYP51B | Azoles | Gene duplication (tandem) | Whole-genome sequencing (PacBio) and qRT-PCR |
| T. indotineae | This study | CYP51B | Azoles | Large segmental duplication (tandem) | Whole-genome sequencing (PacBio) and qRT-PCR |
| Other species | |||||
| Amaranthus tuberculatus | (30) | EPSPS c | Glyphosate | Gene duplication and chromosome duplication (aneuploidy) | Fluorescence in situ hybridization (FISH) |
| Amaranthus palmeri | (31 – 33) | EPSPS | Glyphosate | Transient gene duplication | qRT-PCR |
| Kochia scoparia | (34) | EPSPS | Glyphosate | Large segmental duplication | FISH |
| Grasses | (35 – 38) | EPSPS | Glyphosate | Transient gene duplication | qRT-PCR |
| Anopheles stephensi | (39) | GSTe2 and GSTe4 d | Dichloro-diphenyl-trichloroethane (DDT) | Large segmental duplication (tandem) | Whole-genome sequencing (Illumina) and PCR |
| E. coli | (40) | ampC e | Ampicillin | Gene duplication | β-lactamase activity and gel electrophoresis |
| Streptococcus agalactiae | (41, 42) | folP f and dfrA g | Sulphonamides and trimethoprim | Large segmental duplication (tandem) | qRT-PCR and DNA array data analysis |
Sterol 14α-demethylase gene.
Pleiotropic ABC efflux transporter genes.
5-enolpyruvylshikimate-3-phosphate synthase gene.
Glutathione S-transferase epsilon 2 and 4 genes.
β-lactamase gene.
Dihydropteroate synthase gene.
Dihydrofolate reductase gene.
Large segmental duplications contain several genes in contrast to gene duplications that contain only the target gene.
Duplications within the promoter region of the CYP51 gene and missense mutations in the coding sequence (CDS) of this gene can also lead to overexpression and confer resistance. In Aspergillus fumigatus, the overexpression of AfuCYP51A is mediated by the presence of two copies of a 34 bp tandem sequence in the AfuCYP51A promoter and by the concomitant presence of an A for T substitution at position 364 in the CDS of the gene. This nucleotide change led to the mutation Leu98His in the protein (43). A similar case is found within Blumeriella jaapii, in which PCR analyses of the region upstream of CYP51 in azole-resistant isolates indicated that DNA inserts ranging from 2.1 to 5.6 kb were present upstream of CYP51 (44).
Gene amplification, a mechanism of resistance not restricted to fungi
Amplification of the gene encoding the target molecule of a toxic chemical also occurs in plants, insects, and bacteria (Table 2). Fluorescence in situ hybridization (FISH) and qPCR have revealed that, as in the case of azole resistance in T. indotineae, the resistance of various plants against the herbicide glyphosate is mediated by amplification of the 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS) gene encoding the target of this herbicide (Table 2) (30 – 38). Glyphosate-resistant Amaranthus palmeri plants contain from fivefold to more than 160-fold copies of the EPSPS gene within their genomes compared to susceptible plants (31 – 33). In glyphosate-resistant Kochia scoparia plants, from three to more than 10 extra EPSPS copies are present as a tandem gene duplication at one locus (31, 34).
More recently, amplification of the gene encoding the target molecule of a chemical substance has also been described in Anopheles stephensi, where tandem duplication of the 3.62 kb genomic region encoding the glutathione S-transferase epsilon 2 and 4 genes was shown to be involved in DDT resistance in a strain from India. The tandem duplicated region contained two functional paralogs of GSTe2 and three functional paralogs of GSTe4 (39).
Bacteria also use gene duplications and large segmental duplications to develop resistance (19). As early as 1977, Escherichia coli K-12 mutants overproducing beta-lactamase due to repetitions of the chromosomal beta-lactamase genes were demonstrated to show ampicillin resistance (40). More recently, large segmental duplications have been described in strains of Streptococcus agalactiae resistant to sulphonamides and trimethoprim (41, 42).
Probable cause and direct identification of azole-resistant dermatophytosis by PCR
ITC is considered the main azole compound effective against dermatophytoses caused by T. indotineae (45). Fluconazole and VRC have also been used against this dermatophyte (46, 47). Other over-the-counter topical azoles, by targeting TinCYP51B, could predispose to the selection of strains with the same resistance characteristics. However, we found only two reports in PubMed on topical treatments of T. indotineae with miconazole (48, 49) under T. mentagrophytes type VIII and one report with topical clotrimazole in addition to pulsed ITC therapy (50). No reports with the use of other topical azole compounds such as econazole and oxiconazole were found. Thus, ITC, via repeated topical and systemic treatments, is probably the main contributor to the development of resistance in T. indotineae.
Resistance of T. indotineae resistant to azoles as well as to terbinafine should be monitored before starting treatment. The broth microdilution method (51) and the Etest assays are performed with T. indotineae in the same way as for yeasts and filamentous fungi. However, these tests are time-consuming to prepare and take 5–6 days to obtain MIC results after spore inoculation. An alternative way to test for azole resistance would be to carry out a direct search for tandem repeat sequences by standard PCR, using DNA extracted from cultures or directly from skin scales, as susceptibility to terbinafine can be determined by the detection of selected mutations using direct PCR and sequencing of SQLE gene amplicons (52).
MATERIALS AND METHODS
Strains and growth media
T. indotineae strains (N = 48), which had been previously characterized for susceptibility to terbinafine, ITC, and VRC, were used in this study (Table S1). The strains selected for the study of azole resistance mechanisms and the control strains are listed in Table 1. Glycerol 15% (v/v) stocks of these strains, stored at 80°C, were used for conventional culture on SDA and Sabouraud dextrose broth (Bio-Rad) at 28–30°C. Spore formation was promoted at 28°C using 1/10 SDA [0.1% (w/v) Bacto peptone (BD Bioscience) and 0.2% (w/v) dextrose (Fujifilm Wako Pure Chemical Corporation), 1.5% (w/v) Bacto agar] supplemented with 500-µg/mL cycloheximide and 50-µg/mL chloramphenicol (Fujifilm Wako Pure Chemical Corporation). Escherichia coli DH5α (Nippon Gene) was used for molecular cloning.
Chemicals
ITC (Fujifilm Wako Pure Chemical Corporation) and VRC (Tokyo Chemical Industry) were dissolved in dimethyl sulfoxide (DMSO; Fujifilm Wako Pure Chemical Corporation) to constitute stock solutions (1.0 mg/mL for less soluble compounds). Stock solutions were stored at −20°C until use.
Drug susceptibility assays
The MICs were determined using spore suspension stocks, according to the Clinical and Laboratory Standards Institute (CLSI) guidelines for the broth microdilution method (51), except for the use of SDA instead of RPMI1640 medium (4, 12). After incubation, the plates were visually evaluated and also read at 595 nm using a microtitration plate spectrophotometer (Multiskan Ascent, Thermo Fisher Scientific). The MIC80 was defined as the lowest concentration of the drug present in the wells showing a growth inhibition of ≥80% compared with the absorbance values obtained without antifungal agents.
The serial dilution drug susceptibility assays and Etest assays were performed as previously described (4, 12).
Rapid PCR identification of azole-resistant types I and II strains of T. indotineae
For rapid strain typing of azole-resistant T. indotineae by PCR, genomic DNA was extracted from the growing mycelia using the DNeasy Plant Mini Kit (Qiagen), according to the manufacturer’s protocol. PCR was performed according to a standard protocol, using P1–P2 and P3–P4 sense and antisense primers (Table S2).
Southern blotting analysis
Genomic DNA was extracted from the growing mycelia, according to a method described by Girardin et al. (53), with several minor modifications. Aliquots of 50–100 ng of the genomic DNA were used as templates for PCR. PCR was performed using PrimeSTAR HS or PrimeSTAR GXL DNA polymerases (Takara Bio). The nucleotide sequences of the primers used for PCR are listed in Table S2. For the Southern blotting analysis, aliquots of approximately 5 µg of the genomic DNA were digested with an appropriate restriction enzyme, separated by electrophoresis on 0.8% (w/v) agarose gels, and transferred onto Hybond-N+ membranes (Cytiva). Southern hybridization was performed using an ECL direct nucleic acid labeling and detection system (Cytiva), according to the manufacturer’s instructions.
Real-time quantitative PCR
For the relative quantification of the TinCYP51B gene expression in T. indotineae strains by real-time quantitative reverse-transcription PCR (qRT-PCR), total RNA extraction from the growing mycelia and first-strand cDNA synthesis were performed, as described previously for T. rubrum (4). The synthesized cDNAs were treated with DNase I (Qiagen) before use.
For the relative determination of the TinCYP51B gene copy numbers in type II azole-resistant strains by real-time quantitative PCR (qPCR), genomic DNA extraction was performed from the growing mycelia using the method described by Girardin et al. (53).
The qRT-PCR, with cDNA as a target, and the qPCR, with genomic DNA as a target, were performed using Power SYBR Green PCR master mix on a StepOne real-time PCR system (Thermo Fisher Scientific) under standard conditions, according to the manufacturer’s recommendations. The 2-ΔΔCt method was used to compare the ΔCt [threshold cycle (Ct) of the target gene (TinCYP51B) minus the Ct of the endogenous control gene (actin gene, TinACT)] values of the test strains (with unknown target gene copy numbers) with the ΔCt value of a calibrator strain (TIMM20114), which had a single copy target gene. The glyceraldehyde-3-phosphate dehydrogenase gene (TinGAPDH) was added as a target gene to confirm that there was virtually no difference in PCR reaction efficiency among the analyzed strains. The primers used to amplify TinCYP51B, TinGAPDH, and TinACT are listed in Table S2. Dissociation curves of the amplified products were plotted to confirm the absence of nonspecific products or primer dimers.
Genome sequencing and assembly
The whole genome sequencing and data analysis of the T. indotineae strains were performed by Bioengineering Lab. Co., Ltd. (Japan). Genomic DNA was extracted from the growing mycelia according to the method described by Girardin et al. (53), with several minor modifications. After the elimination of short DNA fragments (<10 kb) using the Short Read Eliminator XS kit (Circulomics), the resulting DNA was sheared to 10–20 kb on the g-TUBE (Covaris) prior to library preparation. HiFi sequencing libraries were prepared using the SMRTbell Express Template Prep Kit 2.0 (Pacific Biosciences) and bound to the sequencing polymerase enzyme using the Sequel II Binding Kit 2.0 (Pacific Biosciences), according to the manufacturer’s protocol. Shotgun genomic DNA sequence data were collected on the Sequel IIe system (Pacific Biosciences) and assembled using the IPA HiFi Genome Assembler (version 1.8.0; Pacific Biosciences).
Construction of repair templates
Repair templates harboring the E. coli neomycin phosphotransferase gene (nptII) cassette were generated using a binary vector pAg1-TinCyp51B/T (Table 3). The following two DNA fragments were generated from the genomic DNA of T. indotineae TIMM20118 by PCR: an approximately 4.32 kb fragment contains the 5-UTR and ORF of TinCYP51B using the P15-P16 primers, and an approximately 2.52 kb fragment contains the 3′-UTR of TinCYP51B using the P17–P18 primers. The silent mutations, which can’t be cleaved by the two RNP complexes (Cas9/gRNA1 RNP and Cas9/gRNA2 RNP), were introduced by overlap extension PCR and nested PCR with the primers listed in Table S2. The two obtained fragments were digested with SpeI/ApaI or BamHI/KpnI, respectively, and cloned into the corresponding restriction sites of pAg1-TinCYP51B/T, resulting in the generation of pAg1-TinCYP51B/R (Table 3; Fig. 4A).
TABLE 3.
Plasmids used in this study
| Plasmid | Description | References |
|---|---|---|
| pAg1 | A streamlined version of the binary vector pBIN19 containing sequences necessary for replication in E. coli and Agrobacterium tumefaciens (oriV and trfA), E. coli neomycin phosphotransferase gene (nptII), and the transferable DNA (T-DNA) region, with a multiple cloning site within the T-DNA region | (54) |
| pAg1-TinCyp51B/T | The 5′ UTR of the TinCYP51B gene (2.52 kb; GenBank accession no. OK539858), the nptII cassette [the promoter sequence of Aspergillus nidulans tryptophan C (trpC) gene (PtrpC; GenBank accession no. X02390), nptII, the termination sequence of Aspergillus fumigatus cgrA gene (TcgrA; GenBank accession no. EAL84894)], the 3′ UTR of the TinCYP51B gene (2.51 kb) | (12) |
| pAg1-TinCYP51B/R | The 5′ UTR and ORF of the TinCYP51B gene (4.32 kb), the nptII cassette (PtrpC, nptII, TcgrA), the 3′ UTR of the TinCYP51B gene (2.51 kb) | This study |
All primers used for the amplification of the specific DNA fragments from T. indotineae were designed based on the whole-genome sequence of T. indotineae TIMM20118 (12). The nucleotide sequences of the primers used are listed in Table S2. Where necessary, the amplified fragments were gel purified with a QIAEX II gel extraction kit (Qiagen), subcloned into HincII-digested pUC118, and sequenced.
Ribonucleoprotein (RNP) complex formation
Two 23-nt nucleotide sequences specific to the target, which contain the protospacer adjacent motifs (PAMs; 5′-NGG-3′) near the translation initiation and termination codons of the TinCYP51B ORF [5′-GCAGAAACGAGAGACAATGTCGG-3′ (− strand); 5′-TTGGCGACCCAATGGTCTCGTGG-3′ (+ strand)], were manually chosen to synthesize two Alt-R crRNAs (Integrated DNA Technologies). Two guide RNAs (gRNA1 and gRNA2) were then prepared by mixing equimolar amounts of each Alt-R crRNA and Alt-R tracrRNA (Integrated DNA Technologies) in IDT Duplex Buffer (30-mM HEPES, pH 7.5, 100-mM potassium acetate; Integrated DNA Technologies), heating to 95°C, and slowly cooling to room temperature. Two RNP complexes (Cas9/gRNA1 RNP and Cas9/gRNA2 RNP) were assembled by combining the CRISPR-Cas nuclease (HiFi Cas9 Nuclease V3, Integrated DNA Technologies) and the gRNA at a 1:1 molar ratio of gRNA and protein and incubating at room temperature for 20 min.
Fungal genetic transformation
The plasmid DNA of pAg1-TinCYP51B/R was used as a template for PCR, to amplify the sequence indicated in Fig. 4A. A pair of primers P15–P18 was used for PCR. The amplified DNA fragment was purified using the QIAquick PCR purification kit (Qiagen) and used as the repair template indicated in Fig. 4A. The repair template (2.5–5.0 µg) and two RNP complexes (55–57 nM) were then introduced into each host cell by the protoplast/PEG method, as described previously, with several minor modifications (55). After the PEG treatment, protoplasts were inoculated onto SDA supplemented with 1.2 M D-sorbitol and 1.0% (w/v) yeast extract. The plates were overlaid 24 h later with 10-mL SDA containing 250-µg/mL G418 (Fujifilm Wako Pure Chemical Corporation) and then incubated for 4–7 days. From the colonies regenerating on the selective plates, 28 to 30 clones were chosen at random and tested for their growth properties on SDA supplemented with 2.0-µg/mL ITC. Only clones showing a significantly reduced growth rate in the presence of ITC were selected and screened for their TinCYP51B loci by PCR and Southern blotting. Two pairs of the PCR primers P22–P23 and P24–P25 (Table S2) were designed to detect the type I (2,404 bp) and type II (7,374 bp) duplicated blocks within the genome of selected transformants, respectively.
ACKNOWLEDGMENTS
We thank Mineyuki Horikoshi, Atsuro Koda, and Marina Fratti for their excellent technical assistance.
This study was financially supported by Nihon Nohyaku Co., Ltd, Japan (Joint research on the mechanisms of resistance to azole compounds in dermatophytes).
The in silico analysis performed by the Swiss-Prot group of the SIB Swiss Institute of Bioinformatics was supported by the Swiss federal government through the State Secretariat for Education, Research, and Innovation (SERI).
T. Y., M. F., E. G., and M. M. designed the project and secured funding; T. Y. and M. M. conducted the molecular cloning and the production of transformants by genetic manipulations; T. Y., H. N., K. S., and M. M. carried out the drug susceptibility assays for wild-type strains and transformants by the serial dilution method, the broth microdilution method of the Clinical and Laboratory Standards Institute, or Etest assays; K. S. carried out the qPCR and qRT-PCR analysis for the TinCYP51B gene; M. F. carried out the bioinformatics analysis and gene/protein annotation; T. Y., Y-T. C., M. F., E. G., and M. M. made the manuscript, tables, and figures.
Contributor Information
Tsuyoshi Yamada, Email: tsyamada@main.teikyo-u.ac.jp.
Andreas H. Groll, University Children's Hospital Münster, Münster, Germany
DATA AVAILABILITY
The whole-genome sequences of three T. indotineae strains (TIMM20121, TIMM20122, and TIMM20123) were deposited in GenBank with the following accession numbers: JAUJAB000000000, JAUJAA000000000, and JAUIZZ000000000, respectively. The details are shown in Table S3.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/aac.00933-23.
Nucleotide sequence of the TinCYP51B locus in TIMM20121.
Nucleotide sequence of the TinCYP51B locus in TIMM20122.
Nucleotide sequence of the TinCYP51B locus in TIMM20123.
Phenotypic and genotypic characteristics of T. indotineae strains used in this study.
Oligonucleotide primers used in this study.
Overall genetic features of three T. indotineae strains.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Shen JJ, Arendrup MC, Verma S, Saunte DML. 2022. The emerging terbinafine-resistant Trichophyton epidemic: what is the role of antifungal susceptibility testing? Dermatology 238:60–79. doi: 10.1159/000515290 [DOI] [PubMed] [Google Scholar]
- 2. Yamada T, Maeda M, Alshahni MM, Tanaka R, Yaguchi T, Bontems O, Salamin K, Fratti M, Monod M. 2017. Terbinafine resistance of Trichophyton clinical isolates caused by specific point mutations in the squalene epoxidase gene. Antimicrob Agents Chemother 61:e00115-17. doi: 10.1128/AAC.00115-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Saunte DML, Hare RK, Jørgensen KM, Jørgensen R, Deleuran M, Zachariae CO, Thomsen SF, Bjørnskov-Halkier L, Kofoed K, Arendrup MC. 2019. Emerging terbinafine resistance in Trichophyton: clinical characteristics, squalene epoxidase gene mutations, and a reliable EUCAST method for detection. Antimicrob Agents Chemother 63:e01126-19. doi: 10.1128/AAC.01126-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Monod M, Feuermann M, Salamin K, Fratti M, Makino M, Alshahni MM, Makimura K, Yamada T. 2019. Trichophyton rubrum azole resistance mediated by a new ABC transporter, TruMDR3. Antimicrob Agents Chemother 63:e00863-19. doi: 10.1128/AAC.00863-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gupta AK, Cooper EA, Wang T, Polla Ravi S, Lincoln SA, Piguet V, McCarthy LR, Bakotic WL. 2023. Detection of squalene epoxidase mutations in United States patients with onychomycosis: implications for management. J Invest Dermatol:S0022-202X(23)02123-1. doi: 10.1016/j.jid.2023.04.032 [DOI] [PubMed] [Google Scholar]
- 6. Yamada T, Yaguchi T, Tamura T, Pich C, Salamin K, Feuermann M, Monod M. 2021. Itraconazole resistance of Trichophyton rubrum mediated by the ABC transporter TruMDR2. Mycoses 64:936–946. doi: 10.1111/myc.13286 [DOI] [PubMed] [Google Scholar]
- 7. Singh A, Masih A, Khurana A, Singh PK, Gupta M, Hagen F, Meis JF, Chowdhary A. 2018. High terbinafine resistance in Trichophyton interdigitale isolates in Delhi, India harbouring mutations in the squalene epoxidase gene. Mycoses 61:477–484. doi: 10.1111/myc.12772 [DOI] [PubMed] [Google Scholar]
- 8. Kano R, Kimura U, Noguchi H, Hiruma M. 2022. Clinical isolate of a multi-antifungal-resistant Trichophyton rubrum. Antimicrob Agents Chemother 66:e0239321. doi: 10.1128/aac.02393-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rudramurthy SM, Shankarnarayan SA, Dogra S, Shaw D, Mushtaq K, Paul RA, Narang T, Chakrabarti A. 2018. Mutation in the squalene epoxidase gene of Trichophyton interdigitale and Trichophyton rubrum associated with allylamine resistance. Antimicrob Agents Chemother 62:e02522-17. doi: 10.1128/AAC.02522-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ebert A, Monod M, Salamin K, Burmester A, Uhrlaß S, Wiegand C, Hipler U-C, Krüger C, Koch D, Wittig F, Verma SB, Singal A, Gupta S, Vasani R, Saraswat A, Madhu R, Panda S, Das A, Kura MM, Kumar A, Poojary S, Schirm S, Gräser Y, Paasch U, Nenoff P. 2020. Alarming India-wide phenomenon of antifungal resistance in dermatophytes: a multicentre study. Mycoses 63:717–728. doi: 10.1111/myc.13091 [DOI] [PubMed] [Google Scholar]
- 11. Tang C, Kong X, Ahmed SA, Thakur R, Chowdhary A, Nenoff P, Uhrlass S, Verma SB, Meis JF, Kandemir H, Kang Y, de Hoog GS. 2021. Taxonomy of the Trichophyton mentagrophytes/T. interdigitale species complex harboring the highly virulent, multiresistant genotype T. indotineae. Mycopathologia 186:315–326. doi: 10.1007/s11046-021-00544-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yamada T, Yaguchi T, Maeda M, Alshahni MM, Salamin K, Guenova E, Feuermann M, Monod M. 2022. Gene amplification of CYP51B: a new mechanism of resistance to azole compounds in Trichophyton indotineae. Antimicrob Agents Chemother 66:e0005922. doi: 10.1128/aac.00059-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ohba M, Sato R, Yoshida Y, Nishino T, Katsuki H. 1978. Involvement of cytochrome P-450 and a cyanide-sensitive enzyme in different steps of lanosterol demethylation by yeast microsomes. Biochem Biophys Res Commun 85:21–27. doi: 10.1016/s0006-291x(78)80005-9 [DOI] [PubMed] [Google Scholar]
- 14. Aoyama Y, Yoshida Y. 1978. The 14alpha-demethylation of lanosterol by a reconstituted cytochrome P-450 system from yeast microsomes. Biochem Biophys Res Commun 85:28–34. doi: 10.1016/s0006-291x(78)80006-0 [DOI] [PubMed] [Google Scholar]
- 15. Malavazi I, Lima JF, de Castro PA, Savoldi M, de Souza Goldman MH, Goldman GH. 2008. Genetic interactions of the Aspergillus nidulans atmAATM homolog with different components of the DNA damage response pathway. Genetics 178:675–691. doi: 10.1534/genetics.107.080879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. De Souza CP, Hashmi SB, Osmani AH, Andrews P, Ringelberg CS, Dunlap JC, Osmani SA. 2013. Functional analysis of the Aspergillus nidulans kinome. PLoS One 8:e58008. doi: 10.1371/journal.pone.0058008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gatbonton T, Imbesi M, Nelson M, Akey JM, Ruderfer DM, Kruglyak L, Simon JA, Bedalov A. 2006. Telomere length as a quantitative trait: genome-wide survey and genetic mapping of telomere length-control genes in yeast. PLoS Genet 2:e35. doi: 10.1371/journal.pgen.0020035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pagé N, Gérard-Vincent M, Ménard P, Beaulieu M, Azuma M, Dijkgraaf GJP, Li H, Marcoux J, Nguyen T, Dowse T, Sdicu A-M, Bussey H. 2003. A Saccharomyces cerevisiae genome-wide mutant screen for altered sensitivity to K1 killer toxin. Genetics 163:875–894. doi: 10.1093/genetics/163.3.875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sandegren L, Andersson DI. 2009. Bacterial gene amplification: implications for the evolution of antibiotic resistance. Nat Rev Microbiol 7:578–588. doi: 10.1038/nrmicro2174 [DOI] [PubMed] [Google Scholar]
- 20. Reams AB, Neidle EL. 2004. Gene amplification involves site-specific short homology-independent illegitimate recombination in Acinetobacter sp. strain ADP1. J Mol Biol 338:643–656. doi: 10.1016/j.jmb.2004.03.031 [DOI] [PubMed] [Google Scholar]
- 21. Coste A, Selmecki A, Forche A, Diogo D, Bougnoux M-E, d’Enfert C, Berman J, Sanglard D. 2007. Genotypic evolution of azole resistance mechanisms in sequential Candida albicans isolates. Eukaryot Cell 6:1889–1904. doi: 10.1128/EC.00151-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Morschhäuser J. 2016. The development of fluconazole resistance in Candida albicans− an example of microevolution of a fungal pathogen. J Microbiol 54:192–201. doi: 10.1007/s12275-016-5628-4 [DOI] [PubMed] [Google Scholar]
- 23. Marichal P, Vanden Bossche H, Odds FC, Nobels G, Warnock DW, Timmerman V, Van Broeckhoven C, Fay S, Mose-Larsen P. 1997. Molecular biological characterization of an azole-resistant Candida glabrata isolate. Antimicrob Agents Chemother 41:2229–2237. doi: 10.1128/AAC.41.10.2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sionov E, Lee H, Chang YC, Kwon-Chung KJ. 2010. Cryptococcus neoformans overcomes stress of azole drugs by formation of disomy in specific multiple chromosomes. PLoS Pathog 6:e1000848. doi: 10.1371/journal.ppat.1000848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Stalder L, Oggenfuss U, Mohd-Assaad N, Croll D. 2023. The population genetics of adaptation through copy number variation in a fungal plant pathogen. Mol Ecol 32:2443–2460. doi: 10.1111/mec.16435 [DOI] [PubMed] [Google Scholar]
- 26. Bing J, Hu T, Zheng Q, Muñoz JF, Cuomo CA, Huang G. 2020. Experimental evolution identifies adaptive aneuploidy as a mechanism of fluconazole resistance in Candida auris. Antimicrob Agents Chemother 65:33077664. doi: 10.1128/AAC.01466-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Carolus H, Pierson S, Muñoz JF, Subotić A, Cruz RB, Cuomo CA, Van Dijck P. 2021. Genome-wide analysis of experimentally evolved Candida auris reveals multiple novel mechanisms of multidrug resistance. mBio 12:e03333-20. doi: 10.1128/mBio.03333-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chow NA, Muñoz JF, Gade L, Berkow EL, Li X, Welsh RM, Forsberg K, Lockhart SR, Adam R, Alanio A, Alastruey-Izquierdo A, Althawadi S, Araúz AB, Ben-Ami R, Bharat A, Calvo B, Desnos-Ollivier M, Escandón P, Gardam D, Gunturu R, Heath CH, Kurzai O, Martin R, Litvintseva AP, Cuomo CA. 2020. Tracing the evolutionary history and global expansion of Candida auris using population genomic analyses. mBio 11:e03364-19. doi: 10.1128/mBio.03364-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bhattacharya S, Holowka T, Orner EP, Fries BC. 2019. Gene duplication associated with increased fluconazole tolerance in Candida auris cells of advanced generational age. Sci Rep 9:5052. doi: 10.1038/s41598-019-41513-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dillon A, Varanasi VK, Danilova TV, Koo D-H, Nakka S, Peterson DE, Tranel PJ, Friebe B, Gill BS, Jugulam M. 2017. Physical mapping of amplified copies of the 5-enolpyruvylshikimate-3-phosphate synthase gene in glyphosate-resistant Amaranthus tuberculatus. Plant Physiol 173:1226–1234. doi: 10.1104/pp.16.01427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Patterson EL, Pettinga DJ, Ravet K, Neve P, Gaines TA. 2018. Glyphosate resistance and EPSPS gene duplication: convergent evolution in multiple plant species. J Hered 109:117–125. doi: 10.1093/jhered/esx087 [DOI] [PubMed] [Google Scholar]
- 32. Gaines TA, Zhang W, Wang D, Bukun B, Chisholm ST, Shaner DL, Nissen SJ, Patzoldt WL, Tranel PJ, Culpepper AS, Grey TL, Webster TM, Vencill WK, Sammons RD, Jiang J, Preston C, Leach JE, Westra P. 2010. Gene amplification confers glyphosate resistance in Amaranthus palmeri. Proc Natl Acad Sci U S A 107:1029–1034. doi: 10.1073/pnas.0906649107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yakimowski SB, Teitel Z, Caruso CM. 2021. Defence by duplication: the relation between phenotypic glyphosate resistance and EPSPS gene copy number variation in Amaranthus palmeri. Mol Ecol 30:5328–5342. doi: 10.1111/mec.16231 [DOI] [PubMed] [Google Scholar]
- 34. Jugulam M, Niehues K, Godar AS, Koo D-H, Danilova T, Friebe B, Sehgal S, Varanasi VK, Wiersma A, Westra P, Stahlman PW, Gill BS. 2014. Tandem amplification of a chromosomal segment harboring EPSPS locus confers glyphosate resistance in Kochia scoparia. Plant Physiol 166:1200–1207. doi: 10.1104/pp.114.242826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Salas RA, Dayan FE, Pan Z, Watson SB, Dickson JW, Scott RC, Burgos NR. 2012. EPSPS gene amplification in glyphosate-resistant Italian ryegrass (Lolium perenne ssp. multiflorum) from Arkansas. Pest Manag Sci 68:1223–1230. doi: 10.1002/ps.3342 [DOI] [PubMed] [Google Scholar]
- 36. Malone JM, Morran S, Shirley N, Boutsalis P, Preston C. 2016. EPSPS gene amplification in glyphosate-resistant Bromus diandrus. Pest Manag Sci 72:81–88. doi: 10.1002/ps.4019 [DOI] [PubMed] [Google Scholar]
- 37. Ngo TD, Malone JM, Boutsalis P, Gill G, Preston C. 2018. EPSPS gene amplification conferring resistance to glyphosate in windmill grass (Chloris truncata) in Australia. Pest Manag Sci 74:1101–1108. doi: 10.1002/ps.4573 [DOI] [PubMed] [Google Scholar]
- 38. Chen J, Huang H, Zhang C, Wei S, Huang Z, Chen J, Wang X. 2015. Mutations and amplification of EPSPS gene confer resistance to glyphosate in goosegrass (Eleusine indica). Planta 242:859–868. doi: 10.1007/s00425-015-2324-2 [DOI] [PubMed] [Google Scholar]
- 39. Dykes CL, Sharma G, Behera AK, Kapoor N, Paine MJI, Donnelly MJ, Singh OP. 2022. Tandem duplication of a genomic region encoding glutathione S-transferase epsilon-2 and -4 genes in DDT-resistant Anopheles stephensi strain from India. Sci Rep 12:17872. doi: 10.1038/s41598-022-21522-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Normark S, Edlund T, Grundström T, Bergström S, Wolf-Watz H. 1977. Escherichia coli K-12 mutants hyperproducing chromosomal beta-lactamase by gene repetitions. J Bacteriol 132:912–922. doi: 10.1128/jb.132.3.912-922.1977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Brochet M, Couvé E, Zouine M, Poyart C, Glaser P. 2008. A naturally occurring gene amplification leading to sulfonamide and trimethoprim resistance in Streptococcus agalactiae. J Bacteriol 190:672–680. doi: 10.1128/JB.01357-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Brochet M, Couvé E, Zouine M, Vallaeys T, Rusniok C, Lamy MC, Buchrieser C, Trieu-Cuot P, Kunst F, Poyart C, Glaser P. 2006. Genomic diversity and evolution within the species Streptococcus agalactiae. Microbes Infect 8:1227–1243. doi: 10.1016/j.micinf.2005.11.010 [DOI] [PubMed] [Google Scholar]
- 43. Mellado E, Garcia-Effron G, Alcázar-Fuoli L, Melchers WJG, Verweij PE, Cuenca-Estrella M, Rodríguez-Tudela JL. 2007. A new Aspergillus fumigatus resistance mechanism conferring in vitro cross-resistance to azole antifungals involves a combination of cyp51A alterations. Antimicrob Agents Chemother 51:1897–1904. doi: 10.1128/AAC.01092-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ma Z, Proffer TJ, Jacobs JL, Sundin GW. 2006. Overexpression of the 14alpha-demethylase target gene (CYP51) mediates fungicide resistance in Blumeriella jaapii. Appl Environ Microbiol 72:2581–2585. doi: 10.1128/AEM.72.4.2581-2585.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Khurana A, Agarwal A, Singh A, Sardana K, Ghadlinge M, Agrawal D, Panesar S, Sethia K, Chowdhary A. 2021. Predicting a therapeutic cut-off serum level of itraconazole in recalcitrant tinea corporis and cruris-A prospective trial. Mycoses 64:1480–1488. doi: 10.1111/myc.13367 [DOI] [PubMed] [Google Scholar]
- 46. Gueneau R, Joannard B, Haddad N, Alby F, Jullien V, Schlatter J, Cotteret C, Bougnoux ME, Lanternier F, Laroche L, Delliere S, Cisternino S, Lortholary O. 2022. Extensive dermatophytosis caused by terbinafine-resistant Trichophyton indotineae, successfully treated with topical voriconazole. Int J Antimicrob Agents 60:106677. doi: 10.1016/j.ijantimicag.2022.106677 [DOI] [PubMed] [Google Scholar]
- 47. Gupta AK, Venkataraman M, Hall DC, Cooper EA, Summerbell RC. 2023. The emergence of Trichophyton indotineae: implications for clinical practice. Int J Dermatol 62:857–861. doi: 10.1111/ijd.16362 [DOI] [PubMed] [Google Scholar]
- 48. Nenoff P, Verma SB, Ebert A, Süß A, Fischer E, Auerswald E, Dessoi S, Hofmann W, Schmidt S, Neubert K, Renner R, Sohl S, Hradetzky U, Krusche U, Wenzel H-C, Staginnus A, Schaller J, Müller V, Tauer C, Gebhardt M, Schubert K, Almustafa Z, Stadler R, Fuchs A, Sitaru C, Retzlaff C, Overbeck C, Neumann T, Kerschnitzki A, Krause S, Schaller M, Walker B, Walther T, Köhler L, Albrecht M, Willing U, Monod M, Salamin K, Burmester A, Koch D, Krüger C, Uhrlaß S. 2020. Spread of terbinafine-resistant Trichophyton mentagrophytes type VIII (India) in Germany-"The tip of the iceberg?". J Fungi (Basel) 6:207. doi: 10.3390/jof6040207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Süß A, Uhrlaß S, Ludes A, Verma SB, Monod M, Krüger C, Nenoff P. 2019. Extensive tinea corporis due to a terbinafine-resistant Trichophyton mentagrophytes isolate of the Indian genotype in a young infant from Bahrain in Germany. Hautarzt 70:888–896. doi: 10.1007/s00105-019-4431-7 [DOI] [PubMed] [Google Scholar]
- 50. Kong X, Song G, Mei H, Zheng H, Tang C, de Hoog S, Li X, She X, Liu W, Liang G. 2023. The domestic isolation of terbinafine- and itraconazole-resistant Trichophyton indotineae in Chinese mainland. Mycopathologia 188:383–393. doi: 10.1007/s11046-023-00761-x [DOI] [PubMed] [Google Scholar]
- 51. Clinical and Laboratory Standards Institute . 2017. Reference method for broth dilution antifungal susceptibility testing of filamentous fungi; standard-third edition: M38. CLSI, Wayne, PA. [Google Scholar]
- 52. Blanchard G, Amarov B, Fratti M, Salamin K, Bontems O, Chang Y-T, Sabou AM, Künzle N, Monod M, Guenova E. 2023. Reliable and rapid identification of terbinafine resistance in dermatophytic nail and skin infections. J Eur Acad Dermatol Venereol. doi: 10.1111/jdv.19253 [DOI] [PubMed] [Google Scholar]
- 53. Girardin H, Latge JP. 1994. DNA extraction and quantification, p 5–9. In Molecular biology of pathogenic fungi, second edition. Telos Press. [Google Scholar]
- 54. Zhang A, Lu P, Dahl-Roshak AM, Paress PS, Kennedy S, Tkacz JS, An Z. 2003. Efficient disruption of a polyketide synthase gene (pks1) required for melanin synthesis through Agrobacterium-mediated transformation of Glarea lozoyensis. Mol Genet Genomics 268:645–655. doi: 10.1007/s00438-002-0780-4 [DOI] [PubMed] [Google Scholar]
- 55. Yamada T, Makimura K, Uchida K, Yamaguchi H. 2005. Reproducible genetic transformation system for two dermatophytes, Microsporum canis and Trichophyton mentagrophytes. Med Mycol 43:533–544. doi: 10.1080/13693780500057619 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Nucleotide sequence of the TinCYP51B locus in TIMM20121.
Nucleotide sequence of the TinCYP51B locus in TIMM20122.
Nucleotide sequence of the TinCYP51B locus in TIMM20123.
Phenotypic and genotypic characteristics of T. indotineae strains used in this study.
Oligonucleotide primers used in this study.
Overall genetic features of three T. indotineae strains.
Data Availability Statement
The whole-genome sequences of three T. indotineae strains (TIMM20121, TIMM20122, and TIMM20123) were deposited in GenBank with the following accession numbers: JAUJAB000000000, JAUJAA000000000, and JAUIZZ000000000, respectively. The details are shown in Table S3.