ABSTRACT
KEYWORDS: tecovirimat, mpox, human monkeypox virus, genomic surveillance, drug resistance
LETTER
Tecovirimat, also known as TPOXX or ST-246, is a drug available for the treatment of mpox. Tecovirimat targets the conserved orthopoxvirus VP37 protein (also known as F13) required for extracellular virus particle generation (1, 2). Multiple VP37 mutations associated with tecovirimat resistance have been reported within the current global mpox outbreak in immunocompromised individuals with advanced HIV infection (3 – 5). In many of these cases, resistance mutation heterogeneity was observed following tecovirimat exposure, suggesting resistance emerged under selective pressure during treatment.
To monitor circulating monkeypox virus (MPXV) within California, a genomic surveillance network was established whereby clinical and commercial laboratories provided positive specimens for whole-genome sequencing using an amplicon-based protocol and subsequent analysis (6 – 9). Through this surveillance, 11 mpox cases were identified in southern California with the same tecovirimat resistance-associated mutation (Table 1): a three-nucleotide deletion in the vaccinia virus Copenhagen F13L gene homolog (OPG057) resulting in asparagine removed from position 267 in the VP37 protein (VP37:N267del) (5) (https://www.fda.gov/emergency-preparedness-and-response/mcm-issues/fda-mpox-response#therapeutics). VP37:N267del was the only tecovirimat resistance-associated mutation detected in identified specimens and had allele frequencies greater than 89% in all instances, suggesting infections may have occurred with predominantly mutant virus. Phenotypic testing in vitro (3 – 5) confirmed tecovirimat resistance in ten identified specimens with EC50 values ranging from 1.488 to 3.977 µM, corresponding to an 85- to 230-fold change compared to wild-type isolates.
TABLE 1.
Patient and laboratory information for southern California MPXV specimens with the VP37:N267del mutation
| Case | Symptom onset date | No. JYNNEOS doses a | Tecovirimat received b ? | Specimen c | Specimen collection date | Specimen source | Reference genome coverage | Mean sequencing depth | GISAID d ID | GenBank ID | SRA e ID |
|---|---|---|---|---|---|---|---|---|---|---|---|
| A | 2022–11-28 | 0/2 | No | A-1 | 2022–12-14 | Foot | 79.79% | 4760.8× | EPI_ISL_16997471 | OQ503823.2 | SRR23580583 |
| B | 2022–11-29 | 0/2 | No | B-1 | 2022–12-02 | Unknown | 97.51% | 557.22× | EPI_ISL_16997450 | OQ503816.2 | SRR23580591 |
| C | 2022–12-01 | 0/2 | No | C-1 | 2022–12-14 | Left upper arm | 97.51% | 828.74× | EPI_ISL_16997457 | OQ503819.2 | SRR23580587 |
| D | 2022–12-13 | 0/2 | No | D-1 | 2022–12-19 | Right ankle | 98.56% | 1016.0× | EPI_ISL_16679232 | OQ330999.2 | SRR23238058 |
| D-2 | 2022–12-19 | Left wrist | 79.41% | 2240.4× | EPI_ISL_16679231 | OQ330998.2 | SRR23238059 | ||||
| D-3 | 2022–12-19 | Right breast | 79.50% | 2505.4× | EPI_ISL_16679233 | OQ331000.2 | SRR23238056 | ||||
| E | 2022–12-19 | 0/2 | No | E-1 | 2023–01-02 | Unknown | 98.65% | 777.32× | EPI_ISL_16930177 | OQ547883.2 | SRR24451328 |
| F | 2023–01-02 | 2/2 | No | F-1 | 2023–01-08 | Unknown | 97.63% | 3281.2× | EPI_ISL_17211332 | OQ644784.2 | SRR23893269 |
| G | 2023–01-04 | 0/2 | No | G-1 | 2023–01-06 | Unknown | 96.85% | 409.70× | EPI_ISL_16997466 | OQ503826.2 | SRR23580580 |
| G-2 | 2023–01-06 | Left buttock | 98.88% | 756.89× | EPI_ISL_16930180 | OQ547884.2 | SRR24451327 | ||||
| H | 2023–01-04 | 1/2 | No | H-1 | 2023–01-10 | Unknown | 96.16% | 624.38× | EPI_ISL_16997441 | OQ503830.2 | SRR23580575 |
| I | 2023–01-06 | 0/2 | No | I-1 | 2023–01-10 | Left buttock | 95.39% | 345.58× | EPI_ISL_16997402 | OQ547899.2 | SRR23730906 |
| J | 2023–01-08 | 2/2 | No | J-1 | 2023–01-13 | Mouth | 99.03% | 895.48× | EPI_ISL_17206617 | OQ991943.1 | SRR24451324 |
| K | 2023–01-09 | 0/2 | No | K-1 | 2023–01-10 | Right side face | 98.95% | 816.41× | EPI_ISL_16930183 | OQ547885.2 | SRR24451326 |
JYNNEOS doses received 14+ days before symptom onset.
Tecovirimat received before specimen collection.
All specimens were lesion swabs.
GISAID, Global Initiative for Sharing All Influenza Data.
SRA, Sequence Read Archive.
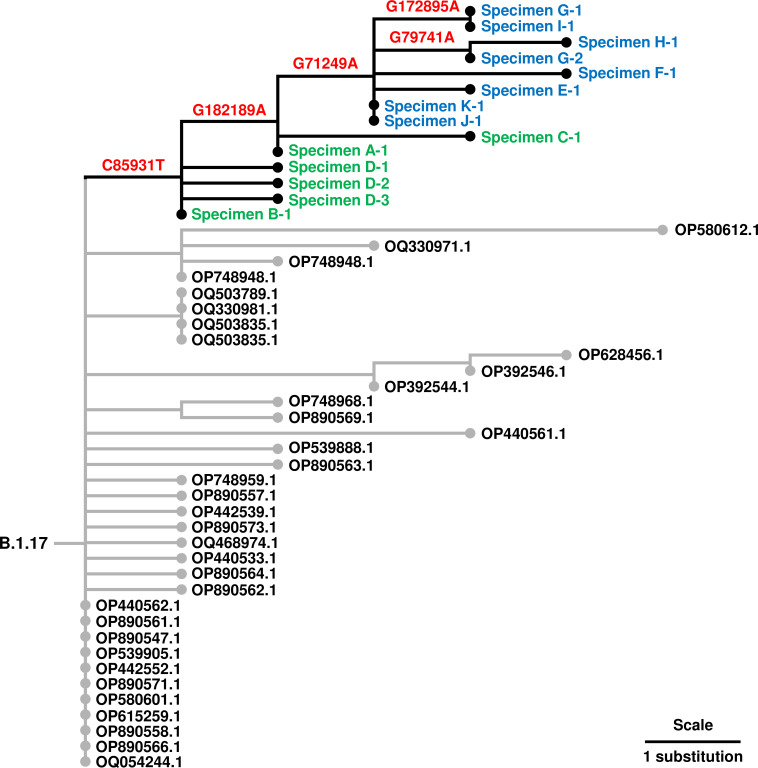
Of the 11 identified VP37:N267del cases, 10 involved males and 1 a female. Symptom onset ranged from late November 2022 to early January 2023 (Table 1). Four patients had conditions associated with reduced immune function, including HIV infection. Previously, tecovirimat resistance within the current outbreak was observed in immunocompromised individuals with severe mpox disease (3 – 5). No patients in this study required hospitalization, suggesting resistance alone does not result in increased virulence. Two patients were fully vaccinated with two doses of JYNNEOS vaccine, and one patient received only a single dose more than a month before symptom onset. For all cases, tecovirimat was not received at any time prior to specimen collection. Seven cases were epidemiologically linked to a group sex event, indicating person-to-person transmission. Supporting their relatedness, phylogenetic analysis showed that all identified VP37:N267del specimens cluster within the B.1.17 lineage of Clade IIb (10), and the seven cases with confirmed epidemiologic links cluster further (Fig. 1). Whether any identified cases involved exposure to mpox patients undergoing tecovirimat treatment is unknown.
Fig 1.
Phylogeny of southern California MPXV specimens with VP37:N267del mutations. Phylogenetic tree obtained from UShER (10) containing southern California MPXV specimens with VP37:N267del mutations (labeled in blue and green) and their relationship to other B.1.17 sequences available from GenBank with accession numbers labeled in black. VP37:N267del specimens labeled in blue have confirmed epidemiologic links, while those in green have no known links. Substitution mutations relative to the Clade IIb, lineage B.1 reference sequence identified using Nextclade v2.11.0 (MPXV_USA_2022_MA001 in NC_063383 coordinates, or pseudo_ON563414) (8) associated with branches are in red. Phylogenetic tree was visualized using Iroki (11). Sequencing of VP37:N267del specimens was performed on Illumina platforms (MiSeq and NextSeq 1000) using an amplicon-based protocol (6), and consensus sequences were generated using the TheiaCov_Illumina_PE v2.3.2 workflow on Terra.bio (7) with a minimum sequencing depth of 10 reads at each position. Analysis using iVar v1.3.1 (9) with a minimum allele frequency of 10% determined that VP37:N267del allele frequencies were greater than 89% in all identified specimens and was the only tecovirimat resistance-associated mutation detected.
These findings demonstrate community transmission of an MPXV variant with a tecovirimat resistance-associated mutation (VP37:N267del) in southern California. Person-to-person transmission without tecovirimat exposure suggests this mutation does not carry a significant fitness cost, which is consistent with in vitro replication levels being similar between wild type and tecovirimat-resistant F13L mutants in other orthopoxviruses (2). This differs from mutations conferring resistance to cidofovir/brincidofovir (that targets viral DNA polymerase) being linked to reduced fitness in vaccinia virus (12 – 15). This work emphasizes the important need for mpox patients receiving tecovirimat to take steps to prevent transmission of potentially resistant virus. Additionally, this study showcases the value of genomic surveillance in detecting emerging resistance within outbreaks.
ACKNOWLEDGMENTS
We gratefully acknowledge the work of our public health laboratory partners and the California Department of Public Health Mpox Response Team, especially Monica Haw, Chantha Kath, Shiffen Getabetcha, Robbie Snyder, and Kathy Harriman. The authors would also like to thank Briar Lucero for technical assistance.
This work was supported by the Epidemiology and Laboratory Capacity for Prevention and Control of Emerging Infectious Diseases cooperative agreements of the Centers for Disease Control and Prevention awarded to the County of Los Angeles Department of Public Health (grant 6 NU50CK000498) and the California Department of Public Health (grant 5 NU50CK000539). A.K. was supported by the Global Infectious Disease Epidemiology Training Grant 2T32AI052073-16A1 from the National Institutes of Health.
The County of Los Angeles Department of Public Health Institutional Review Board (IRB) determined that this study met the criteria for not research per 45 CFR 46.102(e). Therefore, IRB review was not required.
Use of trade names and commercial sources is for identification only and does not imply endorsement by the U.S. Department of Health and Human Services or the County of Los Angeles Department of Public Health. The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention or their institutions, the County of Los Angeles Department of Public Health, the California Department of Public Health, or the California Health and Human Services Agency.
Contributor Information
Nicole M. Green, Email: nicgreen@ph.lacounty.gov.
Miguel Angel Martinez, IrsiCaixa Institut de Recerca de la Sida, Badalona, Barcelona, Spain .
REFERENCES
- 1. Yang G, Pevear DC, Davies MH, Collett MS, Bailey T, Rippen S, Barone L, Burns C, Rhodes G, Tohan S, Huggins JW, Baker RO, Buller RLM, Touchette E, Waller K, Schriewer J, Neyts J, DeClercq E, Jones K, Hruby D, Jordan R. 2005. An orally bioavailable antipoxvirus compound (ST-246) inhibits extracellular virus formation and protects mice from lethal orthopoxvirus challenge. J Virol 79:13139–13149. doi: 10.1128/JVI.79.20.13139-13149.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Duraffour S, Lorenzo MM, Zöller G, Topalis D, Grosenbach D, Hruby DE, Andrei G, Blasco R, Meyer H, Snoeck R. 2015. ST-246 is a key antiviral to inhibit the viral F13L phospholipase, one of the essential proteins for orthopoxvirus wrapping. J Antimicrob Chemother 70:1367–1380. doi: 10.1093/jac/dku545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Alarcón J, Kim M, Terashita D, Davar K, Garrigues JM, Guccione JP, Evans MG, Hemarajata P, Wald-Dickler N, Holtom P, Garcia Tome R, Anyanwu L, Shah NK, Miller M, Smith T, Matheny A, Davidson W, Hutson CL, Lucas J, Ukpo OC, Green NM, Balter SE. 2023. An Mpox-related death in the United States. N Engl J Med 388:1246–1247. doi: 10.1056/NEJMc2214921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Garrigues JM, Hemarajata P, Karan A, Shah NK, Alarcón J, Marutani AN, Finn L, Smith TG, Gigante CM, Davidson W, Wynn NT, Hutson CL, Kim M, Terashita D, Balter SE, Green NM. 2023. Identification of tecovirimat resistance-associated mutations in human monkeypox virus - Los Angeles County. Antimicrob Agents Chemother 67. doi: 10.1128/aac.00568-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Smith TG, Gigante CM, Wynn NT, Matheny A, Davidson W, Yang Y, Condori RE, O’Connell K, Kovar L, Williams TL, Yu YC, Petersen BW, Baird N, Lowe D, Li Y, Satheshkumar PS, Hutson CL. 2023. Resistance to anti-orthopoxviral drug tecovirimat (TPOXX®) during the 2022 Mpox outbreak in the US. Public and global health. doi: 10.1101/2023.05.16.23289856 [DOI] [Google Scholar]
- 6. Chen NFG, Chaguza C, Gagne L, Doucette M, Smole S, Buzby E, Hall J, Ash S, Harrington R, Cofsky S, Clancy S, Kapsak CJ, Sevinsky J, Libuit K, Park DJ, Hemarajata P, Garrigues JM, Green NM, Sierra-Patev S, Carpenter-Azevedo K, Huard RC, Pearson C, Incekara K, Nishimura C, Huang JP, Gagnon E, Reever E, Razeq J, Muyombwe A, Borges V, Ferreira R, Sobral D, Duarte S, Santos D, Vieira L, Gomes JP, Aquino C, Savino IM, Felton K, Bajwa M, Hayward N, Miller H, Naumann A, Allman R, Greer N, Fall A, Mostafa HH, McHugh MP, Maloney DM, Dewar R, Kenicer J, Parker A, Mathers K, Wild J, Cotton S, Templeton KE, Churchwell G, Lee PA, Pedrosa M, McGruder B, Schmedes S, Plumb MR, Wang X, Barcellos RB, Godinho FMS, Salvato RS, Ceniseros A, Breban MI, Grubaugh ND, Gallagher GR, Vogels CBF. 2023. Development of an amplicon-based sequencing approach in response to the global emergence of Mpox. PLoS Biol 21:e3002151. doi: 10.1371/journal.pbio.3002151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Libuit KG, Doughty EL, Otieno JR, Ambrosio F, Kapsak CJ, Smith EA, Wright SM, Scribner MR, Petit III RA, Mendes CI, Huergo M, Legacki G, Loreth C, Park DJ, Sevinsky JR. 2023. Accelerating bioinformatics implementation in public health. Microbial Genomics 9. doi: 10.1099/mgen.0.001051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Aksamentov I, Roemer C, Hodcroft EB, Neher RA. 2021. Nextclade: clade assignment, mutation calling and quality control for viral genomes. JOSS 6:3773. doi: 10.21105/joss.03773 [DOI] [Google Scholar]
- 9. Grubaugh ND, Gangavarapu K, Quick J, Matteson NL, De Jesus JG, Main BJ, Tan AL, Paul LM, Brackney DE, Grewal S, Gurfield N, Van Rompay KKA, Isern S, Michael SF, Coffey LL, Loman NJ, Andersen KG. 2019. An amplicon-based sequencing framework for accurately measuring intrahost virus diversity using primalseq and iVar. Genome Biol 20:8. doi: 10.1186/s13059-018-1618-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Turakhia Y, Thornlow B, Hinrichs AS, De Maio N, Gozashti L, Lanfear R, Haussler D, Corbett-Detig R. 2021. Ultrafast sample placement on existing tRees (UShER) enables real-time phylogenetics for the SARS-CoV-2 pandemic. Nat Genet 53:809–816. doi: 10.1038/s41588-021-00862-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Moore RM, Harrison AO, McAllister SM, Polson SW, Wommack KE. 2020. Iroki: automatic customization and visualization of phylogenetic trees. PeerJ 8:e8584. doi: 10.7717/peerj.8584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Smee DF, Wandersee MK, Bailey KW, Hostetler KY, Holy A, Sidwell RW. 2005. Characterization and treatment of cidofovir-resistant vaccinia (WR strain) virus infections in cell culture and in mice. Antivir Chem Chemother 16:203–211. doi: 10.1177/095632020501600306 [DOI] [PubMed] [Google Scholar]
- 13. Andrei G, Gammon DB, Fiten P, De Clercq E, Opdenakker G, Snoeck R, Evans DH. 2006. Cidofovir resistance in vaccinia virus is linked to diminished virulence in mice. J Virol 80:9391–9401. doi: 10.1128/JVI.00605-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kornbluth RS, Smee DF, Sidwell RW, Snarsky V, Evans DH, Hostetler KY. 2006. Mutations in the E9L polymerase gene of cidofovir-resistant vaccinia virus strain WR are associated with the drug resistance phenotype. Antimicrob Agents Chemother 50:4038–4043. doi: 10.1128/AAC.00380-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Becker MN, Obraztsova M, Kern ER, Quenelle DC, Keith KA, Prichard MN, Luo M, Moyer RW. 2008. Isolation and characterization of cidofovir resistant vaccinia viruses. Virol J 5:58. doi: 10.1186/1743-422X-5-58 [DOI] [PMC free article] [PubMed] [Google Scholar]