Abstract
Gene expression in murine dendritic cells (DCs) infected with green fluorescent protein-expressing Salmonella enterica serovar Typhimurium BRD509 was studied by mRNA differential display. Infected DCs were sorted from uninfected cells by flow cytometry. The mRNA expression patterns of infected and uninfected cells revealed a number of differentially expressed transcripts, which included the macrophage-derived chemokine (MDC). Up-regulation of MDC transcription in infected DCs was confirmed by Northern blotting, and the kinetics of MDC expression was examined by real-time reverse transcription-PCR, with which 31- and 150-fold increases were detected at 2 and 6 h postinfection, respectively. The increased release by DCs of MDC into culture media was detected by an enzyme-linked immunosorbent assay. The biological activity of MDC was investigated in in vitro and in vivo assays. In vitro, supernatants from S. enterica serovar Typhimurium-infected DCs were chemoattractive to T cells, and neutralization of MDC in these supernatants inhibited T-cell migration. Passive transfer of anti-MDC antibody to mice infected with BRD509 revealed that neither growth of the bacterium nor resistance of the mice to reinfection was affected and that in vivo inhibition of MDC did not affect T-cell responses, as measured by the gamma interferon ELISPOT method 3 days after challenge infection.
Salmonella enterica causes a variety of localized and systemic diseases, depending on the host and bacterial strain involved (35). S. enterica serovar Typhi, which causes human typhoid fever, remains a health threat for people worldwide, and there are more than 16 million cases and 600,000 deaths annually (17). S. enterica serovar Typhimurium infection of mice, which shares many features of human S. enterica serovar Typhi infection, is a broadly used and well-characterized animal model for human typhoid fever (47). Following oral administration of S. enterica serovar Typhimurium to mice, the bacteria penetrate the intestinal mucosa through invasion of M cells of Peyer's patches (7, 19) and migrate via the lymph nodes to the spleen and liver, where they reside intracellularly within macrophages and replicate within specialized vacuoles (7, 34). As the interactions between S. enterica serovar Typhimurium and macrophages are thought to play a central role in determining disease outcome, there have been numerous studies describing the features of such interactions from various viewpoints, such as the virulence mechanisms employed by the bacterium to kill cells (6, 10, 32, 48) and the responses of macrophages to the invading bacterium (36, 49).
Recent studies have described the interactions between S. enterica serovar Typhimurium and dendritic cells (DCs). DCs, like macrophages, are antigen-presenting cells which play a central role in linking innate immunity and adaptive immunity (3). However, unlike macrophages, DCs have a unique ability to induce antigen-specific primary T-cell activation (2). Salmonella invades and survives within both human and murine DCs (28, 40, 41, 51), and Jantsch et al. (18) reported that in bone marrow-derived DCs, intracellular S. enterica serovar Typhimurium represents a static, nondividing population, suggesting that DCs fail to kill this intracellular pathogen. Given the migratory capacity of DCs (30) and their presence in Peyer's patches, DCs are likely to serve as an efficient dissemination vehicle for Salmonella out of the mucosal site (22), a thesis supported by studies of Rescigno et al., which showed that DCs mediate Salmonella invasion (33). Bone marrow-derived DCs, as well as freshly isolated DCs from the spleen and mesenteric lymph nodes, can process Salmonella and present bacterial antigens to specific CD4+ and CD8+ T cells (41-43, 53). Although both DCs and macrophages phagocytose Salmonella and present the processed bacterial proteins, their roles in initiating and sustaining immune responses are probably different (52). It has been suggested that upon S. enterica serovar Typhimurium infection, macrophages act more as key effectors than as response initiators, while in contrast, DCs are the principal antigen-presenting cells involved in the priming of naïve T cells (49).
Compared with the extensive investigations of the interaction between S. enterica serovar Typhimurium and macrophages, only limited data are available on the effect of S. enterica serovar Typhimurium on gene expression by DCs. For example, Rosenberger et al. studied the expression of nearly 600 genes after S. enterica serovar Typhimurium infection of a murine macrophage cell line by using microarrays (36); however, to our current knowledge, no such study has been performed with S. enterica serovar Typhimurium-infected DCs.
In order to identify de novo-expressed genes which may be involved in the response of DCs to S. enterica serovar Typhimurium infection and to expand the current knowledge of gene expression profiles in this infection model without a specific focus on any single category of genes, in this study differential display was used to compare mRNA samples from S. enterica serovar Typhimurium-infected and uninfected DCs. A green fluorescent protein (GFP)-expressing S. enterica serovar Typhimurium strain was constructed to facilitate sorting of infected DCs from uninfected bystander DCs. By comparing S. enterica serovar Typhimurium-infected DCs with uninfected control DCs, the macrophage-derived chemokine (MDC) gene was found to be up-regulated during infection. Murine MDC was previously known as ABCD-1 (39) and recently was designated CCL22 (55). MDC is classified as a CC chemokine and is a chemoattractant for DCs, macrophages, NK cells, and T cells (8, 9, 14, 37, 39). The receptor identified to date for MDC is CCR4 (16), which is expressed on the Th2 subset of mature CD4+ T cells (4, 38). The role of MDC in the biology of murine Salmonella infection was studied further.
MATERIALS AND METHODS
Salmonella strains.
The Salmonella strains used in this study were the virulent strain S. enterica serovar Typhimurium SL1344 and an aroA aroD mutant of S. enterica serovar Typhimurium SL1344 (BRD509, a kind gift from G. Dougan, Imperial College, London, United Kingdom). Recombinant S. enterica serovar Typhimurium expressing GFP was generated by transformation of BRD509 with plasmid pGF3, which resulted in BRD509/pGF3 (streptomycin and ampicillin resistant). Plasmid pGF3 was constructed by replacing the C fragment of tetanus toxin from the pTETtac4 plasmid (12) with a PCR-amplified GFP-encoding fragment from plasmid pAJGFP21 (a gift from A. Jo, The University of Melbourne, Melbourne, Australia).
Mice.
For all experiments, male, 6-to-8-week old C57BL/6 mice were used. All mice were bred and housed at the animal facility of The University of Melbourne Department of Microbiology and Immunology. All animal experiments were approved by The University of Melbourne Animal Ethics and Experimentation Committee and complied with the Prevention of Cruelty to Animals Act (1986) and the NHMRC Australian Code of Practice for the Care and Use of Animals for Scientific Purposes (1997).
Cell cultures.
Bone marrow-derived DCs were generated as described previously (25) from C57BL/6 mice (animal care facility, Department of Microbiology and Immunology, The University of Melbourne). The DC culture medium was RPMI 1640 supplemented with 10% fetal calf serum, 10% granulocyte-macrophage colony-stimulating factor-containing cell supernatant, 100 U of penicillin G sodium per ml, 100 μg of streptomycin sulfate per ml, 2 mM l-glutamine, and 50 μM 2-mercaptoethanol. On day 8 of culture, nonadherent and semiadherent cells were harvested, which routinely yielded a population with more than 90% CD11chigh cells. Splenic T cells were generated by the method of Julius et al. (20). CD4+ T cells were enriched by depletion of CD8+ T cells by using anti-mouse CD8 Dynabeads (Dynal, Oslo, Norway) according to manufacturer's instructions. To activate cells, 2 × 106 CD4+ T cells resuspended in 1 ml of T-cell culture medium were seeded into each well of a 24-well plate, which was precoated with 200 μl of a 10-μg/ml solution of purified anti-CD3 antibody (BD Bioscience, San Jose, Calif.), and were incubated at 37°C and 5% CO2 overnight (37). The T-cell culture medium was RPMI 1640 supplemented with 10% fetal calf serum, 100 U of penicillin G sodium per ml, 100 μg of streptomycin sulfate per ml, 2 mM l-glutamine, 1 mM pyruvate, and 50 μM 2-mercaptoethanol.
DC infection and sorting.
S. enterica serovar Typhimurium strain BRD509/GF3 was grown overnight without shaking until the absorbance at 600 nm was 0.5 to 0.8. DCs were infected at a multiplicity of infection of 20:1 in DC culture medium for 2 h at 37°C in the presence of 5% CO2. When a longer period of infection (more than 2 h) was required, DCs were washed twice in phosphate-buffered saline (PBS) to remove extracellular bacteria and cultured with fresh DC culture medium supplemented with 100 μg of gentamicin per ml to inhibit extracellular bacterial growth (31). To prepare cell samples used for differential display, the infected and uninfected immature DCs were sorted with a MoFlo cell sorter (DakoCytomation Glostrup, Denmark) by using the phenotypes GFP+ MHC-IIlow and MHC-IIlow, respectively.
Differential display.
A differential display analysis was carried out as described previously (23, 24), with minor modifications. Briefly, total RNA was isolated from uninfected and infected DCs by using the Wizard Plus SV total RNA isolation system (Promega, Madison, Wis.) according to the manufacturer's instructions. First-strand cDNA was synthesized for either infected or uninfected DCs by using the following three anchored primers: 3RA (5′ GCA AGC TTT TTT TTT TTA 3′), 3RC (5′ GCA AGC TTT TTT TTT TTC 3′), and 3RG (5′ GCA AGC TTT TTT TTT TTG 3′). Each reverse transcription mixture contained 6 μl of total RNA (0.2 μg), 12 μl of 5× Moloney murine leukemia virus reverse transcriptase buffer (Promega), 4.8 μl of deoxynucleoside triphosphates (dNTPs) (250 μM each), 28.2 μl of sterile water, and one of the three anchored primers (6 μl of 2 μM 3RA, 3RC, or 3RG). The mixture was preheated to 65°C for 5 min and then incubated at 37°C for 10 min. Three microliters of a 200-U/μl solution of Moloney murine leukemia virus reverse transcriptase (Promega) was added, and the mixture was incubated at 37°C for 50 min, which was followed by a final step of heating at 75°C for 5 min. The newly synthesized cDNA was PCR amplified by using the following nine arbitrary primers: 5AP1 (5′ CGG GAA GCT TAT CTT GAT TGC C 3′), 5AP2 (5′ CGG GAA GCT TAT AGG TGA CCG T 3′), 5AP3 (5′ CGG GAA GCT TAT CTT TGG TCA G 3′), 5AP4 (5′ CGG GAA GCT TAT TAC AAG GAC G 3′), 5AP5 (5′ CGG GAA GCT TAT TGG CAT TGC A 3′), 5AP6 (5′ CGG GAA GCT TAT CCA AGC ATG G 3′), 5AP7 (5′ CGG GAA GCT TAT TCC TGT GTG A 3′), 5AP8 (5′ CGG GAA GCT TAT CGA GAC TAG C 3′), and 5AP9 (5′ CGG GAA GCT TAT GCT AGC AGA C 3′). These primers were separately paired with one of the three anchored primers (3RA, 3RC, or 3RG). Each 20-μl PCR mixture contained 2 μl of 10× PCR buffer, 1.2 μl of MgCl2 (25 mM), 1.6 μl of dNTPs (25 μM each), 0.2 μl of a 5-U/μl solution of Taq DNA polymerase, 0.4 μl of α-35S-labeled dATP (10 mCi/ml; Amersham), and 8.6 μl of sterile water, as well as an anchored primer (2 μl of a 2 μM solution of one of the three anchored primers [3RA, 3RC, or 3RG]), an arbitrary primer (2 μl of a 2 μM solution of one of the nine arbitrary primers [5AP1 to 5AP9]), and a cDNA sample (2 μl). The PCR conditions were as follows: 10 cycles of 30 s at 94°C, 2 min at 41°C, and 30 s at 72°C, followed by 25 cycles of 15 s at 94°C, 30 s at 41°C, and 30 s at 72°C and a final incubation at 72°C for 8 min.
Twenty microliters of each PCR product was mixed with 4 μl of gel loading buffer (Promega), incubated at 95°C for 5 min, and immediately placed on ice for 5 min. The products were subsequently separated on a 6% polyacrylamide gel in Tris-borate-EDTA buffer for 3 h at 60 W. The gel was dried and exposed to X-ray film overnight at −70°C or scanned with a phosphorimager (FLA-3000; Fuji Film, Tokyo, Japan).
Differentially expressed bands were gel purified, reamplified with the appropriate anchored and arbitrary primers, and either sequenced directly by using the corresponding arbitrary primers or sequenced after they were cloned into the pGEM-T easy vector (Promega). Sequencing was performed with a Big Dye Ready Reaction kit (Applied Biosystems, Foster City, Calif.). Electrophoresis and base calling were conducted at the DNA sequencing laboratory at the Australian Genome Research Facility (Melbourne, Victoria, Australia). The sequencing data were analyzed by using the Sequencher 3.0 software (Genecodes Corporation Ann Arbor, Mich.), and similarity comparisons were assessed by using a BLAST search against the GenBank database (http://www.ncbi.nlm.nih.gov/BLAST/).
Northern blot analysis.
For uninfected and infected DCs, total RNA was electrophoresed by using a method modified from the method of Goda and Minton (13). Briefly, 0.2 μg of RNA was denatured by heating at 75°C for 5 min, immediately loaded onto a 1% agarose gel (supplemented with 20 mM guanidine thiocyanate), and electrophoresed for 2 h at 65 V. RNA was transferred to a Hybond N+ nylon membrane (Amersham, Alesbury, United Kingdom) by using a downward silica gel transfer system as described previously (45, 46) and was cross-linked to the membrane by UV light treatment at 1,200 μW for 45 s by using a Spectrolinker XL-1000 UV cross-linker (Spectronics Corporation, Westbury, N.Y.). Radioactively labeled probes were generated in a 40-μl mixture which comprised 2 μl of DNA template (purified MDC or β-actin gene PCR products), 4 μl of 10× PCR buffer, 2 μl of MgCl2 (25 mM), 4 μl of dNTPs (250 μM each), 0.4 μl of Taq DNA polymerase, 2 μl of each of the forward and reverse primers (2 μM each) (for MDC, forward primer 5′ AGC CTC AGC TGA CAG CAT ATG GGA 3′ and reverse primer 5′ GAC CTT CAG GAC ATG CAT GGG CAG T 3′; for the β-actin gene, forward primer 5′ ACT ATT GGC AAC GAG CGG TTC CGA T 3′ and reverse primer 5′ TAC TTG CGC TCA GGA GGA GCA ATG A 3′), 2 μl of [α-32P]dATP (10 mCi/ml; Amersham), and sterile water. The PCR conditions consisted of an initial incubation at 94°C for 1.5 min, followed by 30 cycles of 94°C for 30 s, 56°C for 30 s, and 72°C for 30 s and a final incubation at 72°C for 8 min. The labeled probes were purified to remove unincorporated nucleotides by using Micro Bio-Spin P-30 chromatography columns (Bio-Rad, Hercules, Calif.) according to the manufacturer's instructions. Probe hybridization was performed at 68°C in the presence of 5 ml of hybridization solution (7% sodium dodecyl sulfate, 0.18 M Na2HPO4, 0.07 M NaH2PO4; pH 7.2) overnight. After hybridization, the membranes were washed twice under high-stringency conditions (0.1× SSC-0.1% sodium dodecyl sulfate at 68°C for 15 min [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate]). Membranes were rehybridized with labeled the β-actin gene probe after removal of the previous bound MDC probe. For analyses, membrane images were obtained by exposure to X-ray film overnight at −70°C or the membranes were scanned with a phosphorimager.
Real-time RT-PCR.
Real-time reverse transcription (RT)-PCR was performed by using the protocol of Applied Biosystems (SDS Compendium 7700, version 4.0), with minor modifications. The primers and probe for MDC were designed by using the Primer Express software (Applied Biosystems) and were purchased from Applied Biosystems (forward primer 5′ GAG TTC TTC TGG ACC TCA AAA TCC 3′, reverse primer 5′ TCT CGG TTC TTG ACG GTT ATC A 3′, and probe 5′ AAG CCT GGC GTT GTT 3′). Briefly, cDNA was synthesized as described previously. For MDC, each 25-μl (final volume) real-time RT-PCR mixture consisted of 12.5 μl of 2× TaqMan Universal PCR master mixture (Applied Biosystems), 0.75 μl of each of the primers (forward and reverse; 10 μM each), and 0.5 μl of probe (10 μM). For GAPDH (the endogenous control), each 25-μl (final volume) reaction mixture consisted of 12.5 μl of 2× TaqMan Universal PCR master mixture, 0.25 μl of a solution containing the forward and reverse primers (10 μM each), and 0.25 μl of probe (20 μM). For all samples (MDC and GAPDH), either a standard containing 1 μl of quantified DNA (purified PCR products of MDC and GAPDH), a negative control containing sterile water (no-template control), or a test sample containing cDNA were set up, and each sample was tested in duplicate. The PCR conditions were 2 min at 50°C and 10 min at 95°C, followed by 45 cycles of 15 s at 95°C and 60 s at 58°C. The reactions were performed with an ABI Prism 7700 sequence detection system (Applied Biosystems). The data were collected and analyzed further by using the Sequence Detector 1.7 software (Applied Biosystems). Standard curves were generated by using serially diluted standard control samples, and the input amounts of unknown test samples were calculated by using these standard curves. Finally, the expression level of MDC was normalized to the expression level of GAPDH.
Detection of MDC by ELISA.
To detect the expression level of secreted MDC in supernatants of S. enterica serovar Typhimurium-infected and uninfected DCs, a sandwich enzyme-linked immunosorbent assay (ELISA) was carried out. Briefly, a 96-well Maxisorp immunoplate (Nunc A/S, Roskilde, Denmark) was coated (100 μl/well) overnight at 4°C with a 1-μg/ml solution of capture antibody (rat anti-mouse MDC antibody; R&D Systems, Minneapolis, Minn.). Following washes, the plate was incubated either with 100-μl portions of fivefold serial dilutions (starting at 25 ng/ml) of recombinant mouse MDC/CCL22 (rmMDC) (used as standards; R&D Systems) or with supernatants of S. enterica serovar Typhimurium-infected and uninfected DCs. The detection antibody (rabbit anti-MDC antiserum, generated in this study) was diluted 1:100 with PBS and added at a concentration of 100 μl/well. The absorbance at 492 nm of each sample was measured with a Titertek Multiskan plate reader (Labsystems Multiskan, Helsinki, Finland).
Chemotaxis assay.
To examine the migration of T cells induced by various media, a chemotaxis assay was performed. rmMDC and supernatants from S. enterica serovar Typhimurium-infected and uninfected DCs were diluted to appropriate concentrations with culture medium. When required, rat anti-mouse MDC antibody (R&D Systems) and rabbit anti-MDC antiserum (raised in this study) were added to the medium described above in order to specifically neutralize MDC. Culture medium alone was used as the background migration control. Five hundred microliters of each sample was added in duplicate to the lower wells of 5-μm-pore-size, polycarbonate, Transwell chambers in a 24-well plate (Corning Costar, Corning, N.Y.), and the plate was then equilibrated by incubating it at 37°C in the presence of 5% CO2 for 15 min. After equilibration, 100 μl of a suspension containing 1.5 × 106 activated CD4+ T cells per ml was loaded into the upper wells of the Transwell chambers, and the plate was incubated at 37°C in the presence of 5% CO2 for 2 h. After incubation, cells from the lower wells were collected by centrifugation at 1,800 × g for 5 min and resuspended in 50 μl of culture medium. Cells were stained with trypan blue, and viable cells were enumerated by microscopy.
Immunization protocol.
Immunization of mice was performed as described previously (49, 50). S. enterica serovar Typhimurium strains BRD509 and SL1344 were grown overnight statically at 37°C to an optical density at 600 nm of 0.5 to 0.8 and diluted with PBS. For immunization, mice were either orally inoculated with 1010 CFU of BRD509 or intravenously inoculated with 100 CFU of bacteria. For challenge, mice were orally inoculated with 107 CFU of wild-type SL1344.
Use of anti-MDC antibodies in vivo.
Passive transfer of anti-MDC antiserum was performed as described by Matsukawa et al. (29). Briefly, mice were injected intraperitoneally (i.p.) with 0.5 ml of rabbit anti-MDC antiserum. This anti-MDC antiserum was generated by immunization of rabbits with glutathione S-transferase (GST)-fused recombinant murine MDC protein produced in this study (data not shown). The specificity of this antiserum was confirmed by Western blotting, and the titer of the antiserum was determined by direct ELISA (data not shown). The MDC neutralization capability of this antiserum was determined by a chemotaxis assay, which showed that this antiserum could inhibit MDC-induced T-cell migration at a dilution of 1:50. As a control, mice were injected i.p. with 0.5 ml of rabbit anti-GST antiserum.
Viable counts of S. enterica serovar Typhimurium in organs.
Viable counts of bacteria in organs at various times were determined as described previously (49, 50). Briefly, the number of bacteria in each organ was determined by plating serial dilutions of organ homogenates on Luria-Bertani agar plates supplemented with appropriate antibiotics.
IFN-γ ELISPOT assay.
The gamma interferon (IFN-γ) ELISPOT assay was performed essentially as described by Wijburg et al. (50) in order to examine the effect of MDC neutralization on Salmonella-specific IFN-γ-secreting cells.
RESULTS
Infection and sorting of DCs with S. enterica serovar Typhimurium expressing GFP.
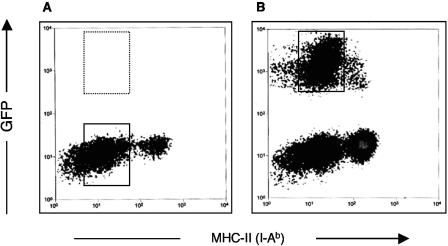
The use of GFP-expressing S. enterica serovar Typhimurium BRD509/GF3 to infect DCs enabled strict definition of infected cell populations and thus minimized the potential for artifacts generated by bystander cell populations. Following infection, BRD509/GF3-infected DCs were stained with anti-MHC-II (I-Ab) antibody and analyzed by flow cytometry (Fig. 1). It was apparent that the majority of infected (GFP+) DCs expressed low levels of major histocompatibility complex II (MHC-II) (i.e., were immature DCs). In contrast, only a small proportion of infected (GFP+) DCs expressed high levels of MHC-II (i.e., were mature DCs). In order to acquire the most homogeneous cell population for differential display, S. enterica serovar Typhimurium-infected immature DCs were sorted by using the GFP+ MHC-IIlow phenotype, and uninfected immature DCs, expressing an equivalent level of MHC-II, were sorted and used as control samples (Fig. 1).
FIG. 1.
Flow cytometric cell sorting of DCs after S. enterica serovar Typhimurium infection. DCs were either mock infected (A) or infected with GFP-expressing S. enterica serovar Typhimurium strain BRD509/GF3 for 6 h (B). Uninfected DCs and Salmonella-infected DCs were sorted with a flow cytometric cell sorter by using the phenotypes MHC-II low (box in panel A) and GFP + MHC-II low (box in panel B), respectively.
Increased transcription of MDC in DCs following S. enterica serovar Typhimurium infection.
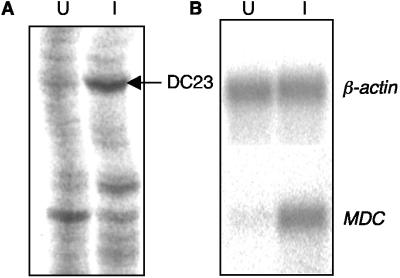
Total RNA was isolated from S. enterica serovar Typhimurium-infected and uninfected immature DCs at 6 h after (mock) infection. This time was chosen mainly because preliminary experiments (results not shown) suggested that this period of time was sufficient for signaling to occur and for effector molecule mRNA to be expressed. The RNA was reverse transcribed into cDNAs and then amplified and compared by using the differential display technique. Three anchored primers and nine arbitrary primers were used to amplify the resultant cDNAs, and the PCR products obtained from infected and uninfected DCs were compared side by side on a polyacrylamide gel. Differentially expressed gene candidates were determined by the presence or absence of bands or by changes in the intensity of the bands. A total of 21 differentially expressed bands were identified, and the DNA was recovered and reamplified by PCR. A portion of one differential display gel is shown in Fig. 2A. One band (DC23) which was up-regulated in infected DCs was reamplified by using primers 5AP7 and 3RC, and the resultant PCR product was further sequenced by using primer 5AP7. Comparing the sequencing data with the GenBank database revealed that this differential display product was 100% identical to the MDC gene (GenBank accession number AF076596).
FIG. 2.
Up-regulation of MDC in DCs during S. enterica serovar Typhimurium infection. (A) Differential display was used to identify differentially expressed genes in DCs following S. enterica serovar Typhimurium BRD509/GF3 infection. A portion of a differential display gel is shown. The arrow indicates the position of a differentially displayed band whose intensity was reduced in uninfected DCs (lane U) compared with infected DCs (lane I). Subsequent sequencing and a BLAST search against the GenBank database assigned this differential display product to the MDC gene. (B) Northern blotting was used to confirm the increased expression of MDC. RNA was extracted from sorted uninfected and infected DCs, hybridized with the MDC-specific DNA probe, and then washed and rehybridized with the β-actin gene probe. The hybridization pattern was analyzed with a phosphorimager.
To confirm the apparent increased transcription from MDC, the MDC transcript was analyzed in a Northern blot. A 171-bp PCR product was amplified by using MDC gene-specific primers with the incorporation of [α-32P]dATP (data not shown). This radioactively labeled PCR product was hybridized to total RNA obtained from infected and uninfected DCs. The β-actin gene was used as an internal control, and the same blot was rehybridized after removal of the MDC probe. The results showed that RNA samples from infected and uninfected DCs hybridized to the β-actin gene probe with approximately the same intensity. However, the intensity of the MDC hybridization signal in the RNA sample obtained from infected DCs was much stronger than the intensity of the signal in the RNA obtained from uninfected DCs (Fig. 2B). The relative levels of expression of MDC compared to the expression of the β-actin gene (determined by dividing the hybridization signal of MDC by the hybridization signal of β-actin gene) were 0.42 and 0.07 for infected and uninfected DCs, respectively, indicating that there was 6.0-fold up-regulation of MDC mRNA in infected DCs.
Expression of the MDC gene and protein in DCs following S. enterica serovar Typhimurium infection.
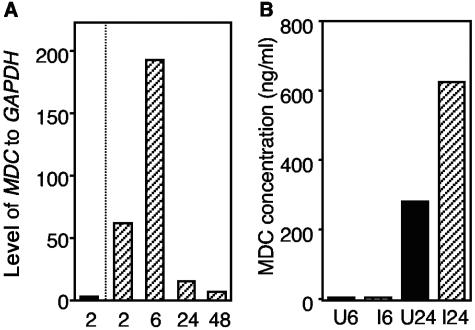
Real-time RT-PCR was used to investigate the kinetic expression of the MDC gene in DCs during S. enterica serovar Typhimurium infection. DCs were infected with S. enterica serovar Typhimurium strain BRD509/GF3, and infected immature DCs were sorted at different times. Uninfected immature DCs were sorted after 2 h of incubation and used as a control for basal MDC expression. The relative expression of MDC to GAPDH in uninfected DCs and infected DCs at 2, 6, 24, and 48 h postinfection is shown in Fig. 3A, and the data indicate that there was a 31-fold increase in the level of MDC transcription in infected DCs at 2 h postinfection.
FIG. 3.
MDC expression in DCs. (A) Real-time RT-PCR was used to study the kinetic expression of the MDC gene in DCs following S. enterica serovar Typhimurium infection and sorting of infected and uninfected cells. The data are the relative expression levels of MDC compared to the expression levels of GAPDH in uninfected DCs which were sorted 2 h after mock infection (solid bar) or in S. enterica serovar Typhimurium-infected DCs sorted at different times postinfection (in hours) (cross-hatched bars). The results of one of two experiments are shown, and the results were similar in the different experiments. (B) MDC protein expression as determined by an ELISA (detection limit, <0.04 ng/ml). At 6 and 24 h after infection and sorting, culture supernatants from uninfected DCs (solid bars) and infected DCs (cross-hatched bars) were collected, and the amounts of MDC in the supernatants were analyzed. U6 and U24, uninfected cells at 6 and 24 h postinfection, respectively; I6 and I24, infected cells at 6 and 24 h postinfection, respectively.
Having found transcriptional up-regulation of MDC, we then investigated the MDC protein levels in DC culture supernatants. A sandwich ELISA was performed to determine the levels of secreted MDC derived from culture supernatants from Salmonella-infected and uninfected DCs collected at 6 and 24 h postinfection. Serial dilutions of recombinant MDC/CCL22 (R&D Systems) were used to generate a standard curve. The results show that the concentration of the MDC protein was low but detectable in the supernatants of infected and uninfected DCs at 6 h postinfection (6.3 ng/ml for uninfected DCs and 4.8 ng/ml for infected DCs), while at 24 h postinfection the supernatant from infected DCs contained a larger amount of secreted MDC protein (626 ng/ml) than the supernatant from uninfected DCs contained (281 ng/ml) (Fig. 3B).
Salmonella-infected DCs induced T-cell migration.
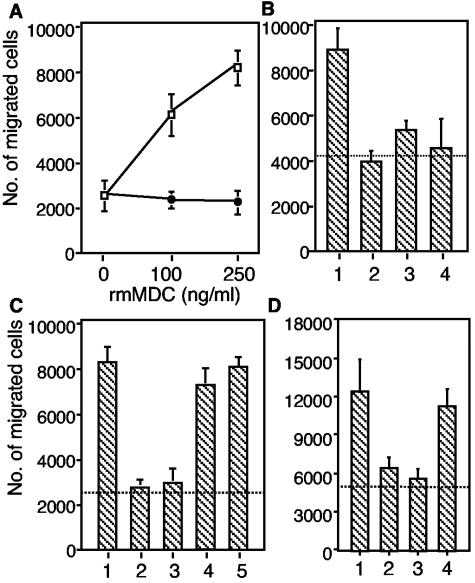
Because MDC is chemoattractive to T cells, macrophages, DCs, and NK cells (8, 9, 14, 37, 39), the ability of secreted MDC to induce T-cell migration was examined with a chemotaxis assay. As a positive control, recombinant MDC/CCL22 (R&D Systems) induced T-cell migration in a dose-dependent manner, which was completely inhibited by rat anti-mouse MDC neutralizing antibody (R&D Systems) at a concentration of 10 μg/ml (Fig. 4A). Supernatants of infected and uninfected DC cultures were diluted 1:5 with DC culture medium and were used to induce T-cell migration. The results showed that both supernatants elicited T-cell migration compared with the background control. Furthermore, the number of cells migrating in response to the supernatant of Salmonella-infected DCs was higher than the number of cells migrating in response to the supernatant of uninfected DCs (Fig. 4B), indicating that the supernatant of infected DCs is more chemoattractive to T cells than the supernatant of uninfected DCs. To determine whether MDC contributed to the induction of T-cell migration, supernatants were incubated with anti-MDC neutralizing antibody (R&D Systems) before they were used to induce T-cell migration. As shown in Fig. 4B, anti-MDC antibody inhibited supernatant-induced T-cell migration, suggesting that MDC was probably the major functional chemoattractant of T cells in these DC-derived supernatants.
FIG. 4.
MDC-induced chemotaxis of CD4 + T cells. Purified CD4 + cells (1.5 × 10 5 cells) were added to the upper wells of a microchamber, and medium was added to the lower chamber. The data are the means and standard errors for duplicate samples. (A) Migratory responsiveness of CD4 + T cells to increasing doses of rmMDC (□) and in the presence of 10 μg of rat anti-mouse MDC antibody per ml (•). (B) Migration of CD4 + T cells induced by MDC in the DC culture supernatant. Bar 1, infected DC supernatant diluted 1:5; bar 2, infected DC supernatant diluted 1:5 plus anti-mouse MDC; bar 3, uninfected DC supernatant diluted 1:5; bar 4, uninfected DC supernatant diluted 1:5 plus anti-mouse MDC. (C) Migration of CD4 + T cells was inhibited by rabbit anti-MDC antiserum but not by the control antiserum. Bar 1, 250 ng of rmMDC per ml; bar 2, rmMDC plus anti-MDC antiserum diluted 1:5; bar 3, rmMDC plus anti-MDC antiserum diluted 1:50; bar 4, rmMDC plus control antiserum diluted 1:5; bar lane 5, rmMDC plus control antiserum diluted 1:50. (D) Bar 1, infected DC supernatant diluted 1:5; bar 2, infected DC supernatant diluted 1:5 plus anti-mouse MDC; bar 3, infected DC supernatant diluted 1:5 plus anti-MDC antiserum diluted 1:10; bar 4, infected DC supernatant diluted 1:5 plus control anti-GST antiserum diluted 1:10. The dotted lines indicate the background migration observed in medium-only control samples.
The chemotaxis assay was also used to test the biological activity of rabbit anti-MDC antiserum generated in this study. The results showed that rabbit anti-MDC antiserum inhibited rmMDC-induced T-cell migration at dilutions of 1:5 and 1:50. In contrast, the control antiserum (rabbit anti-GST antiserum) did not inhibit rmMDC-induced T-cell migration (Fig. 4C). In addition, rabbit anti-MDC antiserum, but not the control antiserum, inhibited T-cell migration induced with supernatant from infected DCs (Fig. 4D). The various background levels of T-cell migration in individual experiments were likely due to the use of different culture media.
Effect of MDC neutralization on the anti-S. enterica serovar Typhimurium immune response.
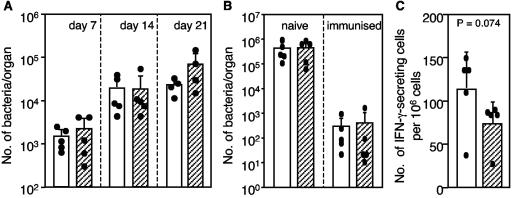
To examine the biological function of MDC in vivo, rabbit anti-MDC antiserum was injected i.p. into mice. The effect of MDC neutralization on bacterial growth was examined (Fig. 5A). Mice were injected i.p. with 0.5 ml of the anti-MDC antiserum or the control antiserum, and 1 day later each mouse was inoculated intravenously with attenuated S. enterica serovar Typhimurium strain BRD509. In order to completely block MDC function, further injections of antiserum (0.5 ml) were administered on days 7 and 14. To determine the bacterial loads at different times after immunization, spleens were collected, and the number of bacteria was enumerated by viable counting. The results showed that the bacterial loads in the spleens in both groups (control antiserum- and anti-MDC antiserum-treated mice) increased from day 7 to day 21. The mean bacterial numbers in the anti-MDC antiserum-treated mice were greater than the numbers in the control antiserum-treated mice; however, the difference was not statistically different. Next, the effect of passive transfer of anti-MDC antiserum on the clearance of wild-type S. enterica serovar Typhimurium SL1344 in naïve and immunized mice was evaluated. Mice were immunized with BRD509 by using a regimen known to elicit protective immune responses (50). One day prior to challenge, naïve or immunized mice were treated with anti-MDC antiserum or control antiserum. On days 3 and 5 postchallenge, the Peyer's patches, mesenteric lymph nodes, spleen, and liver of each mouse were collected, and the number of S. enterica serovar Typhimurium SL1344 cells was determined by viable counting. Figure 5B shows the bacterial counts in the spleens of naïve and immunized mice on day 5 postchallenge. The bacterial load in the naïve mice was approximately 1,000-fold higher than the bacterial load in the immunized mice, indicating that the immunized mice were able to control the challenge infection. No statistically significant difference (as determined by the Mann-Whitney U test) was observed in the number of bacteria detected when anti-MDC antiserum-treated mice and control antiserum-treated mice were compared for either the naïve or immunized animals. Similar patterns were observed in the other organs and tissues that were examined (data not shown).
FIG. 5.
Effects of MDC-specific antiserum on in vivo anti-S. enterica serovar Typhimurium immune responses. Groups of five mice were injected i.p. with 0.5 ml of rabbit anti-MDC antiserum (cross-hatched bars) or isotype control anti-GST antiserum (open bars). The bars and error bars indicate the means and standard deviations, respectively, for each group of mice, and the solid dots indicate the values for individual mice. (A) On day zero, mice were injected i.p. with antiserum, and on day 1 mice were immunized intravenously with 100 CFU of S. enterica serovar Typhimurium BRD509. On days 7 and 14, two more injections of antiserum were administered. The bacterial load in each mouse spleen was determined by plating serial dilutions of spleen homogenates. (B) Naïve and BRD509-immunized mice were injected i.p. with antiserum 1 day prior to oral challenge infection with wild-type S. enterica serovar Typhimurium SL1344. On day 5 after the challenge, the bacterial load in each mouse spleen was determined by viability counting. (C) Effect of MDC neutralization on the number of IFN-γ-secreting cells present in the spleens of BRD509-immunized mice 3 days after challenge infection with SL1344 as determined by an ELISPOT assay. The nonparametric Mann-Whitney U test was used for statistical analysis.
The effect of MDC neutralization on the number of S. enterica serovar Typhimurium-specific IFN-γ-secreting cells (presumably T cells) in the spleen was also determined. The numbers of IFN-γ-producing splenocytes present in the anti-MDC antiserum- and control antiserum-treated BRD509-immunized mice were determined by using an ELISPOT assay 3 days after challenge with wild-type S. enterica serovar Typhimurium SL1344. As Fig. 5C shows, the anti-MDC antiserum-treated mice had a lower average number of IFN-γ-secreting cells in the spleen (73 ± 27 cells/106 cells) than the control antiserum-treated mice had (113 ± 45 cells/106 cells), although again the difference was not statistically significant (P = 0.074, as determined by the Mann-Whitney U test).
DISCUSSION
In this study, we used GFP-expressing S. enterica serovar Typhimurium to infect DCs and then used flow cytometry to sort infected DCs from uninfected DCs. This was done because infection of DCs with Salmonella does not guarantee infection of each individual DC and only a proportion of DCs in the culture are infected, while the remaining DCs are noninfected bystanders. These two cell populations may be functionally different; e.g., they may differ in their antigen presentation capabilities (52, 54) and thus differ in their gene expression profiles. In addition, DCs have two distinct developmental stages (immature and mature). It is well established that immature DCs are more effective at antigen capture and processing, whereas mature DCs are the major antigen-presenting cells (2, 3). The different functions of immature and mature DCs are almost certainly reflected by differences in the gene expression profiles. Thus, the use of flow cytometry to selectively sort uninfected and infected DCs which express the same level of MHC-II not only reduced the potential interference derived from sorting manipulation but also facilitated the identification of the bacterial infection-related genes. The finding that the majority of infected DCs were immature DCs, characterized by MHC-IIlow expression, supported the hypothesis that immature DCs are more phagocytic than mature DCs or are more readily invaded by S. enterica serovar Typhimurium.
Although the up-regulation of MDC mRNA was initially identified in DCs infected with an attenuated Salmonella strain, further study by real-time RT-PCR showed that wild-type Salmonella infection of DCs can also induce a similar increased level of MDC mRNA (data not shown). This finding supports previous microarray studies which revealed increased transcription of chemokines, such as macrophage inflammatory protein 1α (MIP-1α), in Salmonella-infected cells (36) and a potential role for these important mediators in host immunity. The successful activation of an effective host immune response requires communication between immune cells during their activation and during their mobilization to the site of infection (26). The chemokine system plays a crucial role in this process. For example, T-cell migration can be promoted by T-cell-attracting chemokines secreted by DCs (11), which can enhance the interaction between DCs and relatively rare antigen-specific T cells.
In mice, high levels of MDC mRNA expression were found in activated splenic B cells and DCs (39). In studies of human cells, DCs, B cells, and macrophages produced MDC constitutively, while NK cells, monocytes, and CD4+ T cells produced MDC upon stimulation (1). The stimulators for MDC expression include lipopolysaccharide, interleukin-4 (IL-4), IL-13, IL-1, and tumor necrosis factor alpha (1, 5), while the inhibitors include IFN-γ and IL-12 (5, 15). The study described here is the first study to show that S. enterica serovar Typhimurium infection can induce MDC expression by DCs. Moreover, in an attempt to examine the kinetic expression of MDC, we found that MDC transcription was increased as early as 2 h postinfection and declined to basal levels 48 h later. In order to determine whether the up-regulation of MDC at the mRNA level leads to up-regulation of MDC protein expression, we quantified secreted MDC in culture supernatants of S. enterica serovar Typhimurium-infected and uninfected DCs by ELISA. The results showed that the secreted MDC protein was present at an increased level in the supernatant of infected DCs compared with the supernatant of uninfected DCs at 24 h postinfection but not at an early time (6 h postinfection). This unsynchronous up-regulation of MDC mRNA and protein may reflect the lag between mRNA expression and protein secretion. Nonetheless, these results together suggested that MDC may play a role in the host's anti-Salmonella immune response.
Within macrophages, Salmonella infection can induce the up-regulation of chemokines, such as MIP-1α, MIP-1β, and MIP-2α (36). Given the essential roles that DCs and macrophages play in immune responses, we hypothesized that by increasing the expression and secretion of MDC, DCs attract T cells and enhance the cell-cell contact between DCs and T cells to effect an anti-Salmonella immune response. To examine this hypothesis, we investigated the function of MDC in vitro and in vivo. By using a chemotaxis assays, we found that culture supernatants of uninfected and infected DCs induced the migration of activated CD4+ T cells but not the migration of naïve T cells (data not shown) and that the supernatant of infected DCs was more chemoattractive than the supernatant of uninfected DCs. Moreover, neutralization of MDC by the MDC-specific antibody completely abrogated the chemoattractive capacity of the supernatants. These results suggested that secreted MDC in DC supernatants was responsible for the observed T-cell migration and that Salmonella infection-induced increased transcription of MDC could lead to more potent recruitment of activated T cells.
In the past few years, in vivo studies have suggested that MDC plays an important role in immunity. For example, Kikuchi and Crystal reported that MDC chemoattracts T cells in vivo (21), and antigen-specific T cells can rapidly acquire MDC responsiveness in vivo after subcutaneous injection of antigen (44). A pivotal role for MDC was found in mice during experimental sepsis induced by cecal ligation and puncture (29). In this model, administration of MDC conferred protection to mice; it increased the animal survival rate, reduced the bacterial load, and enhanced bacterial clearance. Conversely, neutralization of endogenous MDC by administration of anti-MDC antibody decreased the recruitment of peritoneal macrophages and increased the bacterial load. Considering these previously described findings and our findings, we hypothesized that neutralization of endogenous MDC in Salmonella-infected mice might exacerbate the infection or at least might not have a beneficial effect. The rabbit anti-MDC antiserum raised in this study was thoroughly tested before administration to mice for its ability to neutralize MDC in vitro. This anti-MDC antiserum not only inhibited T-cell migration induced by recombinant murine MDC (i.e., MDC produced in E. coli) but also completely blocked T-cell migration induced by supernatant obtained from S. enterica serovar Typhimurium-infected DCs. We investigated the effect of MDC neutralization on the in vivo growth of attenuated S. enterica serovar Typhimurium, the clearance of virulent S. enterica serovar Typhimurium from immune animals, and the number of S. enterica serovar Typhimurium-specific IFN-γ-secreting cells. We report here that no statistically significant difference was observed between the treatment and control groups for any of these measures. The failure of the passively transferred rabbit anti-MDC antiserum to inhibit MDC-mediated immune phenomena could be explained in a number of ways. First, MDC may play only a minor, if any, role in vivo in anti-Salmonella immunity. Second, the biological function of MDC may be redundant. Redundancy of chemokines has been reported previously (27), and the absence of MDC may be compensated for by other chemokines. Third, we cannot rule out the possibility of incomplete neutralization of MDC in vivo. It is not unlikely that the rabbit antiserum may not reach the local in vivo sites where Salmonella-infected DCs reside in high enough concentrations. During this experiment we found that increased levels of MDC were present in the sera of infected mice compared with the levels in normal mice (mean levels, 0.18 and 0.08 ng/ml, respectively), but attempts to show that there was a reduction in the level of the chemokine in mice which received the MDC-specific rabbit sera were hindered by the presence of high levels of circulating rabbit antibodies. It may be necessary to develop MDC gene knockout mice to more fully understand the role of this chemokine in the murine model of S. enterica serovar Typhimurium immunity.
The type of study reported here is important in the postgenomic era, in which the functionality of the numerous gene products identified in transcriptome and proteome analyses must now be established. Studies to date indicate that the immune response to Salmonella in the murine model is the complex result of the interaction of many cellular and soluble factors, and only through systematic analysis of putative mediators will a more complete understanding of the individual components be established.
Acknowledgments
This project was carried out with the financial support of an NHMRC program grant (The Australian Bacterial Pathogenesis Program) and the Australian Government's Cooperative Research Centres program (CRC for Vaccine Technology). Odilia Wijburg is a Doherty Fellow of the NHMRC.
Editor: S. H. E. Kaufmann
REFERENCES
- 1.Andrew, D. P., M. S. Chang, J. McNinch, S. T. Wathen, M. Rihanek, J. Tseng, J. P. Spellberg, and C. G. Elias, 3rd. 1998. STCP-1 (MDC) CC chemokine acts specifically on chronically activated Th2 lymphocytes and is produced by monocytes on stimulation with Th2 cytokines IL-4 and IL-13. J. Immunol. 161:5027-5038. [PubMed] [Google Scholar]
- 2.Banchereau, J., F. Briere, C. Caux, J. Davoust, S. Lebecque, Y. J. Liu, B. Pulendran, and K. Palucka. 2000. Immunobiology of dendritic cells. Annu. Rev. Immunol. 18:767-811. [DOI] [PubMed] [Google Scholar]
- 3.Banchereau, J., and R. M. Steinman. 1998. Dendritic cells and the control of immunity. Nature 392:245-252. [DOI] [PubMed] [Google Scholar]
- 4.Bonecchi, R., G. Bianchi, P. P. Bordignon, D. D'Ambrosio, R. Lang, A. Borsatti, S. Sozzani, P. Allavena, P. A. Gray, A. Mantovani, and F. Sinigaglia. 1998. Differential expression of chemokine receptors and chemotactic responsiveness of type 1 T helper cells (Th1s) and Th2s. J. Exp. Med. 187:129-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonecchi, R., S. Sozzani, J. T. Stine, W. Luini, G. D'Amico, P. Allavena, D. Chantry, and A. Mantovani. 1998. Divergent effects of interleukin-4 and interferon-gamma on macrophage-derived chemokine production: an amplification circuit of polarized T helper 2 responses. Blood 92:2668-2671. [PubMed] [Google Scholar]
- 6.Brennan, M. A., and B. T. Cookson. 2000. Salmonella induces macrophage death by caspase-1-dependent necrosis. Mol. Microbiol. 38:31-40. [DOI] [PubMed] [Google Scholar]
- 7.Carter, P. B., and F. M. Collins. 1974. The route of enteric infection in normal mice. J. Exp. Med. 139:1189-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang, M., J. McNinch, C. Elias, 3rd, C. L. Manthey, D. Grosshans, T. Meng, T. Boone, and D. P. Andrew. 1997. Molecular cloning and functional characterization of a novel CC chemokine, stimulated T cell chemotactic protein (STCP-1) that specifically acts on activated T lymphocytes. J. Biol. Chem. 272:25229-25237. [DOI] [PubMed] [Google Scholar]
- 9.Chantry, D., A. J. DeMaggio, H. Brammer, C. J. Raport, C. L. Wood, V. L. Schweickart, A. Epp, A. Smith, J. T. Stine, K. Walton, L. Tjoelker, R. Godiska, and P. W. Gray. 1998. Profile of human macrophage transcripts: insights into macrophage biology and identification of novel chemokines. J. Leukoc. Biol. 64:49-54. [DOI] [PubMed] [Google Scholar]
- 10.Chen, L. M., K. Kaniga, and J. E. Galan. 1996. Salmonella spp. are cytotoxic for cultured macrophages. Mol. Microbiol. 21:1101-1115. [DOI] [PubMed] [Google Scholar]
- 11.Cyster, J. G. 1999. Chemokines and cell migration in secondary lymphoid organs. Science 286:2098-2102. [DOI] [PubMed] [Google Scholar]
- 12.Fairweather, N. F., S. N. Chatfield, A. J. Makoff, R. A. Strugnell, J. Bester, D. J. Maskell, and G. Dougan. 1990. Oral vaccination of mice against tetanus by use of a live attenuated Salmonella carrier. Infect. Immun. 58:1323-1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goda, S. K., and N. P. Minton. 1995. A simple procedure for gel electrophoresis and northern blotting of RNA. Nucleic Acids Res. 23:3357-3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Godiska, R., D. Chantry, C. J. Raport, S. Sozzani, P. Allavena, D. Leviten, A. Mantovani, and P. W. Gray. 1997. Human macrophage-derived chemokine (MDC), a novel chemoattractant for monocytes, monocyte-derived dendritic cells, and natural killer cells. J. Exp. Med. 185:1595-1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iellem, A., L. Colantonio, S. Bhakta, S. Sozzani, A. Mantovani, F. Sinigaglia, and D. D'Ambrosio. 2000. Inhibition by IL-12 and IFN-alpha of I-309 and macrophage-derived chemokine production upon TCR triggering of human Th1 cells. Eur. J. Immunol 30:1030-1039. [DOI] [PubMed] [Google Scholar]
- 16.Imai, T., D. Chantry, C. J. Raport, C. L. Wood, M. Nishimura, R. Godiska, O. Yoshie, and P. W. Gray. 1998. Macrophage-derived chemokine is a functional ligand for the CC chemokine receptor 4. J. Biol. Chem. 273:1764-1768. [DOI] [PubMed] [Google Scholar]
- 17.Ivanhoff, B. 1995. Typhoid fever, global situation and W.H.O. recommendations. Southeast Asian J. Trop. Med. Public Health 26(Suppl. 2):1-6. [Google Scholar]
- 18.Jantsch, J., C. Cheminay, D. Chakravortty, T. Lindig, J. Hein, and M. Hensel. 2003. Intracellular activities of Salmonella enterica in murine dendritic cells. Cell. Microbiol. 5:933-945. [DOI] [PubMed] [Google Scholar]
- 19.Jensen, V. B., J. T. Harty, and B. D. Jones. 1998. Interactions of the invasive pathogens Salmonella enterica serovar Typhimurium, Listeria monocytogenes, and Shigella flexneri with M cells and murine Peyer's patches. Infect. Immun. 66:3758-3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Julius, M. H., E. Simpson, and L. A. Herzenberg. 1973. A rapid method for the isolation of functional thymus-derived murine lymphocytes. Eur. J. Immunol. 3:645-649. [DOI] [PubMed] [Google Scholar]
- 21.Kikuchi, T., and R. G. Crystal. 2001. Antigen-pulsed dendritic cells expressing macrophage-derived chemokine elicit Th2 responses and promote specific humoral immunity. J. Clin. Investig. 108:917-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knodler, L. A., and B. B. Finlay. 2001. Salmonella and apoptosis: to live or let die? Microbes Infect. 3:1321-1326. [DOI] [PubMed] [Google Scholar]
- 23.Liang, P., L. Averboukh, and A. B. Pardee. 1993. Distribution and cloning of eukaryotic mRNAs by means of differential display: refinements and optimization. Nucleic Acids Res. 21:3269-3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liang, P., and A. B. Pardee. 1992. Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science 257:967-971. [DOI] [PubMed] [Google Scholar]
- 25.Lutz, M. B., N. Kukutsch, A. L. Ogilvie, S. Rossner, F. Koch, N. Romani, and G. Schuler. 1999. An advanced culture method for generating large quantities of highly pure dendritic cells from mouse bone marrow. J. Immunol. Methods 223:77-92. [DOI] [PubMed] [Google Scholar]
- 26.Mackay, C. R. 2001. Chemokines: immunology's high impact factors. Nat. Immunol. 2:95-101. [DOI] [PubMed] [Google Scholar]
- 27.Mantovani, A. 1999. The chemokine system: redundancy for robust outputs. Immunol. Today 20:254-257. [DOI] [PubMed] [Google Scholar]
- 28.Marriott, I., T. G. Hammond, E. K. Thomas, and K. L. Bost. 1999. Salmonella efficiently enter and survive within cultured CD11c+ dendritic cells initiating cytokine expression. Eur. J. Immunol. 29:1107-1115. [DOI] [PubMed] [Google Scholar]
- 29.Matsukawa, A., C. M. Hogaboam, N. W. Lukacs, P. M. Lincoln, H. L. Evanoff, and S. L. Kunkel. 2000. Pivotal role of the CC chemokine, macrophage-derived chemokine, in the innate immune response. J. Immunol. 164:5362-5368. [DOI] [PubMed] [Google Scholar]
- 30.Mellman, I., and R. M. Steinman. 2001. Dendritic cells: specialized and regulated antigen processing machines. Cell 106:255-258. [DOI] [PubMed] [Google Scholar]
- 31.Michetti, P., N. Porta, M. J. Mahan, J. M. Slauch, J. J. Mekalanos, A. L. Blum, J. P. Kraehenbuhl, and M. R. Neutra. 1994. Monoclonal immunoglobulin A prevents adherence and invasion of polarized epithelial cell monolayers by Salmonella enterica serovar Typhimurium. Gastroenterology 107:915-923. [DOI] [PubMed] [Google Scholar]
- 32.Monack, D. M., B. Raupach, A. E. Hromockyj, and S. Falkow. 1996. Salmonella enterica serovar Typhimurium invasion induces apoptosis in infected macrophages. Proc. Natl. Acad. Sci. USA 93:9833-9838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rescigno, M., M. Urbano, B. Valzasina, M. Francolini, G. Rotta, R. Bonasio, F. Granucci, J. P. Kraehenbuhl, and P. Ricciardi-Castagnoli. 2001. Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat. Immunol. 2:361-367. [DOI] [PubMed] [Google Scholar]
- 34.Richter-Dahlfors, A., A. M. Buchan, and B. B. Finlay. 1997. Murine salmonellosis studied by confocal microscopy: Salmonella enterica serovar Typhimurium resides intracellularly inside macrophages and exerts a cytotoxic effect on phagocytes in vivo. J. Exp. Med. 186:569-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosenberger, C. M., A. J. Pollard, and B. B. Finlay. 2001. Gene array technology to determine host responses to Salmonella. Microbes Infect. 3:1353-1360. [DOI] [PubMed] [Google Scholar]
- 36.Rosenberger, C. M., M. G. Scott, M. R. Gold, R. E. Hancock, and B. B. Finlay. 2000. Salmonella enterica serovar Typhimurium infection and lipopolysaccharide stimulation induce similar changes in macrophage gene expression. J. Immunol. 164:5894-5904. [DOI] [PubMed] [Google Scholar]
- 37.Ross, R., X. L. Ross, H. Ghadially, T. Lahr, J. Schwing, J. Knop, and A. B. Reske-Kunz. 1999. Mouse langerhans cells differentially express an activated T cell-attracting CC chemokine. J. Investig. Dermatol. 113:991-998. [DOI] [PubMed] [Google Scholar]
- 38.Sallusto, F., D. Lenig, C. R. Mackay, and A. Lanzavecchia. 1998. Flexible programs of chemokine receptor expression on human polarized T helper 1 and 2 lymphocytes. J. Exp. Med. 187:875-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schaniel, C., E. Pardali, F. Sallusto, M. Speletas, C. Ruedl, T. Shimizu, T. Seidl, J. Andersson, F. Melchers, A. G. Rolink, and P. Sideras. 1998. Activated murine B lymphocytes and dendritic cells produce a novel CC chemokine which acts selectively on activated T cells. J. Exp. Med. 188:451-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schoppet, M., H. I. Huppertz, A. Simm, and A. Bubert. 2000. Infection of dendritic cells by Enterobacteriaceae. Med. Microbiol. Immunol 188:191-196. [DOI] [PubMed] [Google Scholar]
- 41.Svensson, M., C. Johansson, and M. J. Wick. 2000. Salmonella enterica serovar Typhimurium-induced maturation of bone marrow-derived dendritic cells. Infect. Immun. 68:6311-6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Svensson, M., B. Stockinger, and M. J. Wick. 1997. Bone marrow-derived dendritic cells can process bacteria for MHC-I and MHC-II presentation to T cells. J. Immunol. 158:4229-4236. [PubMed] [Google Scholar]
- 43.Svensson, M., and M. J. Wick. 1999. Classical MHC class I peptide presentation of a bacterial fusion protein by bone marrow-derived dendritic cells. Eur. J. Immunol. 29:180-188. [DOI] [PubMed] [Google Scholar]
- 44.Tang, H. L., and J. G. Cyster. 1999. Chemokine up-regulation and activated T cell attraction by maturing dendritic cells. Science 284:819-822. [DOI] [PubMed] [Google Scholar]
- 45.Tang, S. L., S. Nuttall, K. Ngui, C. Fisher, P. Lopez, and M. Dyall-Smith. 2002. HF2: a double-stranded DNA tailed haloarchaeal virus with a mosaic genome. Mol. Microbiol. 44:283-296. [DOI] [PubMed] [Google Scholar]
- 46.Tarasov, V. Y., M. G. Pyatibratov, S. L. Tang, M. Dyall-Smith, and O. V. Fedorov. 2000. Role of flagellins from A and B loci in flagella formation of Halobacterium salinarum. Mol. Microbiol. 35:69-78. [DOI] [PubMed] [Google Scholar]
- 47.Tsolis, R. M., R. A. Kingsley, S. M. Townsend, T. A. Ficht, L. G. Adams, and A. J. Baumler. 1999. Of mice, calves, and men. Comparison of the mouse typhoid model with other Salmonella infections. Adv. Exp. Med. Biol. 473:261-274. [PubMed] [Google Scholar]
- 48.Watson, P. R., A. V. Gautier, S. M. Paulin, A. P. Bland, P. W. Jones, and T. S. Wallis. 2000. Salmonella enterica serovars Typhimurium and Dublin can lyse macrophages by a mechanism distinct from apoptosis. Infect. Immun. 68:3744-3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wijburg, O. L., C. P. Simmons, N. van Rooijen, and R. A. Strugnell. 2000. Dual role for macrophages in vivo in pathogenesis and control of murine Salmonella enterica var. Typhimurium infections. Eur. J. Immunol. 30:944-953. [DOI] [PubMed] [Google Scholar]
- 50.Wijburg, O. L., N. Van Rooijen, and R. A. Strugnell. 2002. Induction of CD8+ T lymphocytes by Salmonella enterica serovar Typhimurium is independent of Salmonella pathogenicity island 1-mediated host cell death. J. Immunol. 169:3275-3283. [DOI] [PubMed] [Google Scholar]
- 51.Yrlid, U., M. Svensson, C. Johansson, and M. J. Wick. 2000. Salmonella infection of bone marrow-derived macrophages and dendritic cells: influence on antigen presentation and initiating an immune response. FEMS Immunol. Med. Microbiol. 27:313-320. [DOI] [PubMed] [Google Scholar]
- 52.Yrlid, U., M. Svensson, A. Kirby, and M. J. Wick. 2001. Antigen-presenting cells and anti-Salmonella immunity. Microbes Infect. 3:1239-1248. [DOI] [PubMed] [Google Scholar]
- 53.Yrlid, U., and M. J. Wick. 2002. Antigen presentation capacity and cytokine production by murine splenic dendritic cell subsets upon Salmonella encounter. J. Immunol. 169:108-116. [DOI] [PubMed] [Google Scholar]
- 54.Yrlid, U., and M. J. Wick. 2000. Salmonella-induced apoptosis of infected macrophages results in presentation of a bacteria-encoded antigen after uptake by bystander dendritic cells. J. Exp. Med. 191:613-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zlotnik, A., and O. Yoshie. 2000. Chemokines: a new classification system and their role in immunity. Immunity 12:121-127. [DOI] [PubMed] [Google Scholar]