Abstract
Group B streptococci (GBS) usually behave as commensal organisms that asymptomatically colonize the gastrointestinal and urogenital tracts of adults. However, GBS are also pathogens and the leading bacterial cause of life-threatening invasive disease in neonates. While the events leading to transmission and disease in neonates remain unclear, GBS carriage and level of colonization in the mother have been shown to be significant risk factors associated with invasive infection. Surface antigens represent ideal vaccine targets for eliciting antibodies that can act as opsonins and/or inhibit colonization and invasion. Using a genetic screen for exported proteins in GBS, we identified a gene, designated lrrG, that encodes a novel LPXTG anchored surface antigen containing leucine-rich repeat (LRR) motifs found in bacterial invasins and other members of the LRR protein family. Southern blotting showed that lrrG was present in all GBS strains tested, representing the nine serotypes, and revealed the presence of an lrrG homologue in Streptococcus pyogenes. Recombinant LrrG protein was shown in vitro to adhere to epithelial cells in a dose-dependent manner, suggesting that it may function as an adhesion factor in GBS. More importantly, immunization with recombinant LrrG elicited a strong immunoglobulin G response in CBA/ca mice and protected against lethal challenge with virulent GBS. The data presented in this report suggest that this conserved protein is a highly promising candidate antigen for use in a GBS vaccine.
Streptococcus agalactiae, or Lancefield group B streptococci (GBS), is a predominant cause of infections in newborn infants and neonates, with mortality rates as high as 30% in preterm infants (47, 71). GBS are also being increasingly associated with invasive disease in the adult population, such as pregnant women, older persons, or individuals with underlying chronic illnesses (15, 74). The bacteria are commonly found in the gastrointestinal tract of adult humans without any symptoms, but colonization of the rectovaginal compartment of pregnant women is associated with an increased risk of neonatal infection. In the United States, prevention strategies are based on intrapartum administration of antibiotics to mothers who might be considered at risk, e.g., due to early delivery, high intrapartum temperature, and previous history of neonatal GBS infection, or to pregnant women shown to be heavily colonized with GBS before gestation (8). Although these two approaches, often used in combination in clinical practice, have significantly reduced the incidence of neonatal GBS sepsis and meningitis (25, 71, 73), invasive disease due to GBS infection remains the leading cause of neonatal morbidity and mortality in the United States (11, 70). In addition, intrapartum prohylaxis is unlikely to be effective against late-onset GBS disease and would probably not be effective against GBS-related stillbirths and preterm delivery (13, 64). With a reported increase in macrolide resistance in clinical isolates of GBS, efforts have shifted toward vaccination as an attractive alternative for disease prevention (11, 13, 24, 74).
Current progress toward the development of an effective GBS vaccine has primarily focused on the use of the bacterial capsule as a primary immunogen, with nine serotypes having been identified so far. Recent population studies of serotype distributions suggest that a capsular polysaccharide (CPS) vaccine would need to contain capsule types Ia, Ib, II, III, V, VI, and VIII in order to prevent the majority of GBS infections in North America and Japan (23, 39, 43).
Initial vaccination studies with pure preparations of type Ia, II, and III CPSs in humans were disappointing in that only 40 and 60% of individuals developed significant increases in capsule type-specific antibodies (3). However, studies carried out in the 1990s showed that conjugation of the polysaccharide to various protein carriers led to the stimulation of T-cell-dependent antigenic recognition and profound enhancement of the immunogenicity of CPS vaccines (5, 6, 60, 61)
An alternative strategy for protection against GBS infection would be to develop a vaccine with a conserved surface protein antigen. Surface antigens represent good candidates for vaccines, as antibodies directed against these targets have the potential to interfere with bacterial virulence and also to promote opsonophagocytosis through binding to γFc receptors on phagocytes (17, 44). In contrast to CPSs, protein antigens have the potential to elicit protective T-cell-dependent antibody responses and long-lasting immunity without the need for conjugation to other molecules. Several GBS surface proteins have already been investigated as potential vaccine candidates; these include Rib, the alpha and beta subunits of the C protein, Sip, Fbs, C5a peptidase, and the R proteins (2, 12, 18, 20, 50, 80). While these antigens, with the exception of C5a peptidase and Sip, are capable of eliciting antibodies in mice and conferring a degree of protection against lethal challenge with GBS, they are not conserved among all serotypes. The alpha subunit of the C protein, for example, is present in only approximately 50% of all clinical isolates, while Rib is expressed in the majority of serotype III strains but rarely in strains of other serotypes (16, 80). Furthermore, both the alpha subunit of the C protein and Rib possess repeating peptide sequences that show strain-to-strain variations in the numbers of repeats, something which can influence their immunogenic properties (20, 34, 38, 45, 46, 54).
Here we report the characterization of a gene, designated lrrG, that encodes a novel leucine-rich repeat (LRR) surface protein that is conserved among all serotypes of GBS. Furthermore, when tested as a vaccine antigen, LrrG was shown to be immunogenic and capable of eliciting protection against experimental infection with GBS.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial stains and plasmid used in this study are listed in Table 1. Streptococcal isolates were grown in Todd-Hewitt medium (Oxoid, Basingstoke, United Kingdom) or on Todd-Hewitt blood agar plates supplemented with 5% (vol/vol) defibrinated horse blood (TCS Microbiology) at 37°C. Escherichia coli strain DH5α served as a host strain for cloning purposes, and E. coli strain BL21λ(DE3) (Stratagene) was used for protein overexpression. E. coli was grown at 37°C under aeration in Luria broth, and clones carrying plasmid pET28b (Novagen) were selected in the presence of kanamycin (50 μg ml−1).
TABLE 1.
Bacterial strains and plasmid used in this study
| Strain or plasmid | Relevant characteristic(s)a | Source or reference |
|---|---|---|
| Strains | ||
| S. agalactiae | ||
| 97/0099 | Type III encapsulated GBS isolate from the blood of a septic newborn | Public Health Laboratory Service, Colindale, London, United Kingdom |
| 515a | Type 1a | 4 |
| A909 | Type 1a | 40 |
| SB35 | Type 1b | 80 |
| H36B | Type 1b | 40 |
| 18RS21 | Type II | 31 |
| 1954/92 | Type II | 41 |
| 118/158 | Type II | 41 |
| BS29 | Type II | 41 |
| BM110 | Type III | 55 |
| BS30 | Type III | 80 |
| M781 | Type III | 62 |
| 3139 (1/82) | Type IV | 26 |
| 1169-NT | Type V | 86 |
| GBS VI | Type VI | Jitka Motlovab |
| 7271 | Type VII | 36 |
| JM9 | Type VIII | 37 |
| S. pneumoniae VH14 | Type 14 strain | 2a |
| S. pyogenes NCTC 8198 | M1 strain | American Type Culture Collection |
| E. coli | ||
| DH5α | General cloning strain | Stratagene |
| BL21 λ(DE3) | F−ompT hsdSB (rB− mB−) gal dcm (DE3) | Stratagene |
| Plasmid pET28b | Vector for overexpressing His-tagged proteins with a bacteriophage T7 promoter; Kmr | Novagen |
Kmr, kanomycin resistance.
The strain kindly provided by Jitka Motlova originated from the Czech National Collection of Type Cultures, Prague, Czech Republic, and was verified to be serotype VI GBS.
DNA manipulations.
Routine molecular biology techniques for PCR amplification and cloning were performed as described previously (68). Vent DNA polymerase (New England Biolabs [NEB], Beverly, Mass.) was used for standard PCR, while rTth DNA polymerase (Applied Biosystems, Warrington, United Kingdom) was used for long-range PCR according to the manufacturer's instructions. DNA sequencing was carried out as a service at the Department of Genetics, University of Cambridge, and was performed by use of an ABI automated sequencing machine with BigDye chemistry (Applied Biosystems). Chromosomal DNA was isolated from streptococcal strains as described by Madoff et al. (46). Plasmid DNA was isolated from E. coli by using plasmid miniprep columns (Qiagen) and from Lactococcus lactis by using a modified Qiagen plasmid miniprep procedure (85). DNA restriction and modification enzymes were used according to the manufacturer's recommendations (NEB). E. coli cells were routinely transformed by heat shock following CaCl2 treatment (68).
Bioinformatic searches.
BLAST searches of all predicted open reading frames (ORFs) were performed by using a BLASTP search of amino acid similarities to sequences in the GenBank nonredundant protein database (1). Alignments were carried out by using CLUSTAL W (http://www.ebi.ac.uk/clustalw/) (83). In addition to BLAST similarity searches, functional domains were tentatively identified by searching for similarities in the InterPro database of protein families (http://www.ebi.ac.uk/interpro). Regions containing repeats were identified by using Prospero (http://www.well.ox.ac.uk/rmott/ARIADNE/prospero). SignalP (http://www.cbs.dtu.dk/services/SignalP-2.0) was used for the prediction of signal peptide regions (57).
Southern hybridization.
For Southern hybridization, genomic DNA (5 μg) from each streptococcal strain was digested with approximately 20 U of EcoRV (NEB), separated by conventional electrophoresis, and transferred to a positively charged nitrocellulose membrane (Hybond N+; Amersham) by alkaline transfer as described previously (68). Southern blots were hybridized with a digoxigenin (DIG)-labeled lrrG DNA probe that was obtained by PCR with DIG-dUTP (Roche Diagnostics) as well as primers having the sequences 5′-ATGACAAAAAAACATCTTAAAACG and 5′-TTGCGGCCGCTTTTCTTGCTCGTTTTCC, which were designed to amplify the complete ORF of lrrG, including the native signal peptide region. Hybridization was conducted overnight at 65°C with 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) containing 50 mM sodium phosphate, 0.02% sodium dodecyl sulfate (SDS), 1% blocking reagent (Boehringer Mannheim), and 0.1% N-lauroylsarcosine. Blots were washed initially at room temperature with 2× SSC-0.1% SDS and then with 0.5× SSC-0.1% SDS at 68°C. Detection of gene-specific hybridized bands was carried out by chemiluminescence.
Cloning and expression of recombinant LrrG and its derivatives.
Plasmid pET28b was used for the synthesis of a carboxyl-terminal hexahistidine-tagged LrrG fusion protein and its truncated versions, which were constructed as follows. For LrrG, a DNA fragment encoding the mature form of LrrG lacking the native signal peptide region was PCR amplified from GBS strain 97/0099 chromosomal DNA with primers having the sequences 5′-CATGCCATGGTATATGGATTAGAAAGAGAGGAATC and 5′-TTGCGGCCGCTTTTCTTGCTCGTTTTCC (underlining indicates either a NcoI or a NotI restriction enzyme site). The amino terminus-encoding region of lrrG (encoding truncated derivative LrrGnt) was PCR amplified with primers having the sequences 5′-CATGCCATGGTATATGGATTAGAAAGAGAGGAATC and 5′-TTGCGGCCGCTACTTCACTAAGGGCATTATCC, while the carboxyl terminus-encoding region of lrrG (truncated derivative LrrGct) was amplified with primers having the sequences 5′-CCCATGGTCTTACCGCCAAATTTACAG and 5′-TTGCGGCCGCTTTTCTTGCTCGTTTTCC. Amplification of the repeat-encoding region of lrrG (truncated derivative LrrGcr) was performed with primers having the sequences 5′-CCCATGGGAATTAATAAGTTATCTCAAACAT and 5′-TTGCGGCCGCCTCTTTTTCCAAGCGCTTAAC. aroD (encoding the GBS intracellular control protein) was amplified with primers having the sequences 5′-CATGCCATGGCAAAAATAGTAGTACCAGTAATGCCTC and 5′-TTGCGGCCGCCTCTGAAATAGTAATTTGTCCG. rib (encoding surface protein Rib) was amplified with primers having the sequences 5′-CCCATGGCTGAAGTAATTTCAGGAAGTGC and 5′-TTGCGGCCGCATCCTCTTTTTTCTTAGAAACAGATAA. These primers were designed to include either a NcoI or a NotI restriction enzyme site in order to facilitate cloning into plasmid vector pET28b.
Amplified products were purified by using a Qiagen PCR purification kit, digested, and ligated with NcoI-NotI-digested pET28b DNA before being transformed into expression host E. coli BL21λ(DE3). The identity of the cloned DNA fragment was verified by DNA sequencing. LrrG, LrrGnt, LrrGcr, and Rib were purified under nondenaturing conditions by metal affinity chromatography. In brief, 500-ml cultures of E. coli BL21λ(DE3) containing a particular clone were grown in Luria broth with kanamycin to an optical density at 600 nm of between 0.5 and 0.6 before the addition of isopropyl-β-d-thiogalactopyranoside (IPTG) (final concentration, 1 mM). Cultures were typically allowed to grow for a further 3 h, at which point cell pellets were collected by centrifugation and stored at −20°C. In order to purify His-tagged proteins, cell pellets were resuspended in 10 ml of lysis buffer (50 mM NaH2PO4 [pH 7.5], 500 mM NaCl, 30 mM imidazole, 10% glycerol) containing 1 mg of lysozyme/ml, 1 mM phenylmethylsulfonyl fluoride, and an EDTA-free protease inhibitor cocktail tablet (Roche). Cell suspensions were incubated for 30 min on ice, disrupted by sonication (MSE Soniprep), and centrifuged at 12,000 × g for 20 min at 4°C to remove insoluble materials. The clear supernatant was mixed with 1.5 ml of a 50% slurry of Ni2+ and nitrilotriacetic acid resin (Qiagen) at 4°C under gentle shaking for 2 h and loaded into a polypropylene column. After unbound proteins were washed away, recombinant proteins were eluted in elution buffer (0.5 M imidazole, 50 sodium phosphate [pH 7.5], 300 mM NaCl, 10% glycerol). LrrGct and AroD were purified under denaturing conditions. Induced cell pellets were resuspended in 10 ml of denaturation buffer (8 M urea, 0.1 M sodium phosphate [pH 7.5], 300 mM NaCl, 10% glycerol). Cells were incubated overnight at 40°C, and lysates were centrifuged at 12,000 × g for 20 min at 4°C. The supernatant was mixed with Ni2+-nitrilotriacetic acid resin as previously described. The resin was washed with a gradient of 8 to 0 M urea in 0.1 M sodium phosphate (pH 7.5)-300 mM NaCl-10% glycerol, and recombinant proteins were eluted in elution buffer. Eluate fractions were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE); those containing pure proteins were pooled, dialyzed against phosphate-buffered saline (PBS; pH 7.0) containing 10% glycerol, and stored in aliquots at −80°C until required for use.
Polyclonal antibodies to LrrG.
Antisera were raised in female New Zealand White rabbits (6 to 8 weeks old) by subcutaneous (s.c.) immunization with recombinant LrrG. The first dose (100 μg/0.5 ml of PBS) was emulsified in 0.5 ml of complete Freund's adjuvant, and the subsequent doses (days 28 and 63; 75 μg) were emulsified in 0.5 ml of incomplete Freund's adjuvant. Blood was collected approximately 2 weeks after the last dose and allowed to clot overnight at 4°C, and sera were collected after centrifugation.
Preparation of GBS subcellular fractions and SDS-PAGE.
Subcellular fractions of GBS were obtained as described by Kling et al. (34) with the following modifications. Mid-exponential-phase cultures of GBS in Todd-Hewitt broth (50 ml) were centrifuged at 3,000 × g for 10 min to pellet the bacteria. Supernatants were passed through 0.22-μm-pore-size filters (Millipore) before the addition of trichloroacetic acid (10% [wt/vol]) in order to precipitate secreted proteins. The remaining bacterial pellets were treated with mutanolysin to release cell wall proteins into the supernatants, which were collected following centrifugation. The remaining cell pellets were subjected to vigorous sonication (MSE Soniprep) and centrifuged at 14,000 × g to further separate the cytosolic proteins from insoluble cell envelope material. All protein samples were resuspended in SDS-PAGE sample buffer and heated at 100°C for 3 min in order to solubilize proteins prior to SDS-PAGE analysis (12% [wt/vol] polyacrylamide gels) by the method described by Sambrook et al. (68).
Western blotting.
Proteins were size separated by SDS-PAGE and transferred to nitrocellulose membranes by using a semidry transfer chamber (Bio-Rad Laboratories, Hercules, Calif.) for 25 min at 15 V. Following transfer, the membranes were incubated at 4°C overnight in a 5% (wt/vol) solution of dehydrated milk in PBS and then incubated for 2 h with rabbit anti-LrrG (1:1,000) diluted in PBS containing 0.05% Tween 20 (PBST). The membranes then were washed three times (for 5 min each time) with PBST before being incubated with alkaline phosphatase-labeled anti-rabbit immunoglobulin G (IgG; Sigma) (1:30,000) diluted in PBST. Unbound antibodies were removed by washing the membranes five times (for 3 min each time) with PBST before the addition of 5-bromo-4-chloro-3-indolylphosphate-nitroblue tetrazolium substrate solution. The developing reaction was stopped by washing the membranes with copious amounts of water. The membranes then were air dried and stored in the dark.
Vaccination experiments.
Groups of 6- to 7-week-old CBA/ca mice (Harlan Ltd., Bicester, United Kingdom) were immunized s.c. with 25 μg of recombinant protein mixed 1:1 with alum adjuvant (Pierce) in a final volume of 0.1 ml. A separate group of mice were immunized with 25 μg of bovine serum albumin (BSA) (Sigma). Four weeks later, mice were given a booster. In order to analyze immune responses, blood samples were collected from the tail artery of each mouse at 3 and 6 weeks after the initial immunization. Three weeks after the booster, mice were challenged intraperitoneally with 0.5 ml of a standard inoculum (see below) of serotype III GBS strain 97/0099 diluted in Todd-Hewitt broth and pretitrated to correspond to a lethal challenge dose that would result in all unvaccinated mice succumbing to a lethal infection. To prepare the standard inoculum, GBS strain 97/0099 was serially passaged in mice to enhance virulence, inoculated into Todd-Hewitt broth containing 20% heat-inactivated fetal calf serum, and grown to late exponential phase before being frozen at −80°C in aliquots. To estimate the lethal challenge dose, groups of eight mice were injected intraperitoneally with dilutions of the standard inoculum and observed every 2 h over a 7-day period for mice that had succumbed to infection. Mice showing substantial symptoms of infection were humanely killed at the recorded time of observation. Although the number of mice in each group was relatively small, an estimate of the lethal dose of GBS can be determined; such estimation is aided by the fact that the lethal challenge dose typically falls within a narrow range of bacterial concentrations (1 × 106 to 5 × 106). The same monitoring procedures were used in the GBS challenge experiments with vaccinated mice. All protocols were approved by and carried out according to United Kingdom Home Office regulations.
Collection of sera.
Blood samples were collected from the tail artery of each mouse at 21 and 42 days after the initial immunization and were allowed to clot overnight at 4°C. Each sample was applied to a SeraSieve column (Hughes & Hughes Ltd.) and centrifuged at 10,000 × g for 3 min to remove any remaining coagulated blood. Collected serum samples were stored in aliquots at −20°C.
ELISA detection of antigen-specific serum antibodies.
As described previously (65), enzyme-linked immunosorbent assay (ELISA) plates were coated with 3 μg of recombinant LrrG/well and blocked with 3% dehydrated milk solution in PBST. Sera were tested in duplicate by using a twofold dilution series from an initial dilution of 1:50 to a final dilution of 1:107. Duplicate samples of a 1:50 dilution of preimmune serum were also included as controls on each ELISA plate. Anti-mouse IgG, IgG1, and IgG2a antibodies conjugated to alkaline phosphatase (Southern Biotechnology Associates, Birmingham, Ala.) (1:3,000) were applied before development with p-nitrophenyl phosphate substrate (Acros Organics) (1 mg/ml) in diethanolamine (Acros Organics) buffer. Plates were examined at 405 nm by using a microplate reader (Titertek Multiscan MCC 340), and the end-point titer was calculated as the dilution resulting in the same optical density as that determined for a 1:50 dilution of pooled preimmune sera.
Analysis of LrrG-epithelial cell interactions.
Recombinant LrrG and its truncated derivatives were assayed for their relative affinities for binding to lung epithelial cell line HEp-2 (ATCC CCL) and cervical epithelial cell line ME180 (ATCC HTB33). Cells were grown at 37°C in a humidified atmosphere containing 5% CO2 and were maintained in Dulbecco's modified Eagle medium supplemented with 10% fetal calf serum, 4 mM l-glutamine, 10 mM HEPES, 10 U of penicillin/ml, and 10 μg of streptomycin/ml. For adherence assays, cells were used to seed 96-well plates (Becton Dickinson) and were grown until the cell monolayers reached confluence (typically 1 to 2 days). Prior to incubation with recombinant LrrG, the cell monolayers were washed with fresh medium, fixed with 0.2% glutaraldehyde, and blocked with PBS containing 3% milk powder for 1 h at 37°C. Preliminary experiments performed without fixation of the cells were shown to produce similar results. Various amounts of LrrG, LrrGcr, LrrGnt, and LrrGct proteins were added to individual wells and incubated for 2 h at 37°C. The AroD protein of GBS was also included as a negative control to monitor nonspecific interactions between proteins and host cells, as it is known to be a highly conserved intracellular bacterial enzyme involved in the biosynthesis of aromatic amino acids. After thorough washing with PBS-Ca2+, anti-His-IgG (Amersham) (1:1,000) was added to the wells, and the plates were incubated for 1 h at room temperature. Alkaline phosphatase-conjugated goat anti-mouse IgG secondary antibody (Southern Biotechnology Associates) (1:3,000) was used to detect mouse anti-His-IgG bound to adherent recombinant LrrG and its truncated derivatives. Plates were developed by the addition of p-nitrophenyl phosphate substrate solution, and the absorbance at 405 nm was measured by using a microplate reader.
Statistical analyses.
All statistical analyses were carried by using the Minitab 10.5 Xtra Power statistical software package. For ELISA data, antigen-specific antibody titers were expressed as geometric mean titers based on readings for duplicate samples. For survival data, the statistical significance of survival times of vaccinated groups was determined by using the Mann-Whitney U test.
Nucleotide sequence accession number.
The genomic sequence was assigned accession number AY909605 in the GenBank database.
RESULTS
Cloning and sequence analysis of lrrG.
A partial gene fragment containing an LRR domain characteristic of the large group of LRR proteins of Listeria (35, 49) was one of many gene fragments previously identified in screens for surface proteins with export-specific reporter vectors similar to those described previously (22, 63). When this work was carried out, the complete genome sequence of GBS was not published; therefore, a genome walking approach was used to obtain the complete ORF of lrrG. This process involved combining digested GBS genomic DNA with an appropriately digested adapter DNA (multiple cloning site of pUC19) in a ligation mixture. The ligation mixture then was used as a template in long-range PCR to amplify DNA fragments present upstream or downstream of the originally identified GBS sequence essentially as described previously (33). The resulting sequence contig revealed a 3,168-bp ORF, designated lrrG, which was preceded by a potential ribosome-binding site (GAAAGG) and putative −10 (AAATAT) and −35 (TATACT) promoter elements upstream of lrrG. The predicted coding sequence of LrrG comprises 1,056 amino acids with a predicted molecular mass of 118.1 kDa and a pI of 8.34 (Fig. 1).
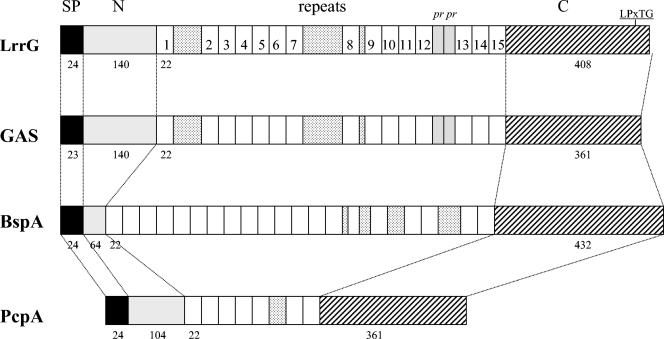
FIG. 1.
Alignment of the amino acid sequence of the LrrG protein from S. agalactiae (97/0099) with LRR proteins from S. pyogenes (GAS; hypothetical protein), B. forsythus (surface antigen BspA), and S. pneumoniae (PcpA). SP, signal peptide region; N, amino terminus; pr, partial repeat; C, carboxyl terminus; LPxTG, LPXTG cell wall anchoring motif. The individual repeats for LrrG are numbered. Numerals below the maps indicate the number of amino acid residues in each protein domain.
BLASTP analysis of the amino acid sequence identified strong homology (71% identity and 81% similarity over the full length of LrrG) to an as-yet-uncharacterized hypothetical surface protein from Streptococcus pyogenes (accession no. AAK33772). Protein sequence homology was also observed between LrrG and the previously characterized BpsA LRR surface antigen (24% identity and 38% similarity over the full length of LrrG) of Bacteroides forsythus (76). LrrG also shows similarity to PcpA, a choline-binding surface antigen found in Streptococcus pneumoniae (69). Interestingly, homology to this surface antigen resides in three overlapping blocks of sequence, the first of which shares 28% identity and 44% similarity with residues 111 to 286 of LrrG. The second region of homology, between residues 204 and 585, shares 21% identity and 43% similarity with PcpA, while the third region, spanning residues 233 to 301, shares 32% identity and 55% similarity with PcpA (Fig. 1). A similar pattern of low-level homology was also observed between LrrG and another LRR protein, TpLRR (28% identity and 42% similarity overall), located in the outer membrane of Treponema pallidum (78). Through the use of various bioinformatic approaches as well as visual analysis of the entire amino acid sequence of LrrG, 15 full-length LRRs and 2 partial LRRs (Fig. 2) were identified within the amino-terminal and central portions of the protein (residues 165 to 647). Most, if not all, of the LRRs have been proposed to play a role in protein-protein interactions (28), and in bacteria, the LRR is typically 20 to 24 amino acid residues long and contains the consensus sequence XLXXIXXLPXXLXXIXXAFXX (35). In addition, a putative integrin-binding arginine-glycine-aspartic acid (RGD) motif (14) located at position 143 of LrrG could play a role in attachment to host cells.
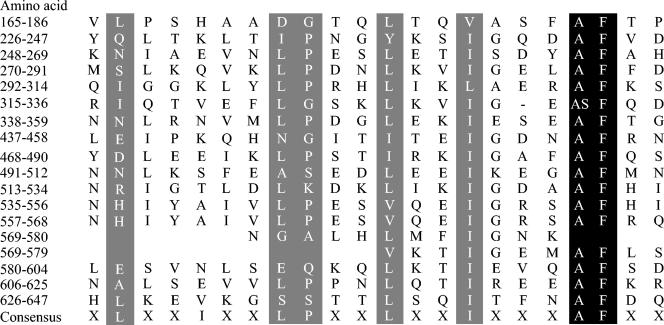
FIG. 2.
Alignment of the 22-amino-acid repeats of the LrrG protein and the consensus LRR protein motif. The amino acid positions of the repeats are indicated on the left. Amino acid residues are indicated in the sequences by the one-letter code and are included if present at a particular position in more than half of the repeats. Amino acid positions with identical or similar amino acid substitutions within the LrrG repeat region and LRR consensus sequences are highlighted.
The genetic organization of the chromosomal lrrG locus in strain 97/0099 was also investigated by genome walking and DNA sequencing. The lrrG gene and its putative promoter were situated between nrdA and a downstream ORF encoding a putative protein. In the published genome sequences of two GBS strains (19, 82), this genetic organization is conserved, and nrdA is annotated as encoding a ribonucleoside diphosphate reductase.
LrrG gene conservation.
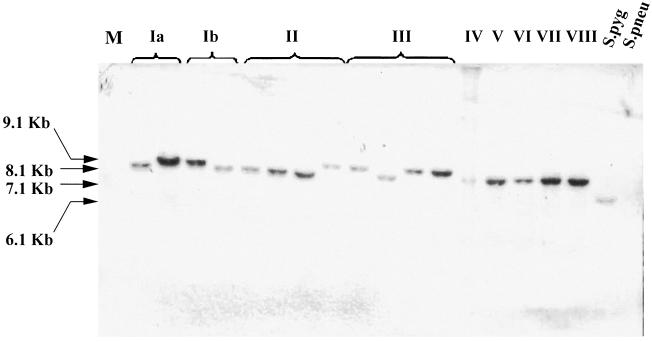
At present, nine immunologically distinct CPS serotypes of GBS have been identified as being clinically significant (13, 23, 39, 43). In order to assess the level of conservation of the LrrG gene among such relevant serotypes of GBS, genomic DNA from a panel of 17 GBS clinical isolates, including at least 1 strain of each serotype, was subjected to Southern blot analysis with the full-length lrrG gene as a DNA probe. In addition, a clinical isolate of S. pneumoniae (serotype 14) and S. pyogenes M1 were included in the analysis. Southern blots were hybridized with an lrrG gene probe amplified by PCR from GBS serotype III strain 97/0099. DNA fragments varying in size from 6.1 to 9.1 kb were detected in all clinical isolates and all possible serotypes of GBS (Fig. 3), indicating that the lrrG gene was present in all of these isolates. As a control, Southern blot analysis was carried out with a bca gene probe (bca encodes the serotype Ia-, Ib-, and II-specific C protein alpha subunit), and a band was detected only in serotype Ia, Ib, and II isolates, as expected (data not shown). Interestingly, the lrrG gene probe also hybridized to a single 6.1-kb EcoRV-digested genomic DNA fragment in S. pyogenes (Fig. 3). The intensity of the S. pygoenes DNA fragment detected by Southern blotting compared favorably with those of its GBS counterparts, suggesting that a homologue of lrrG with high sequence similarity is present in S. pyogenes. This suggestion was confirmed by subsequent interrogation of the gene database, which identified an S. pyogenes gene encoding a hypothetical protein containing LRRs; CLUSTAL W analysis revealed 75.1% identity at the nucleotide level with lrrG. In contrast, no homologous genomic DNA fragments in S. pneumoniae were detected by Southern blotting with the lrrG gene probe, indicating significant variations between the DNA sequences of the pneumococcal choline-binding protein A gene (pcpA) and lrrG (32.82% nucleotide identity over the entire length of lrrG).
FIG. 3.
Southern blot hybridization of chromosomal DNAs from various streptococcal species with the lrrG gene. Lanes (left to right): Ia, S. agalactiae 515a and S. agalactiae A909; Ib, S. agalactiae SB35 and S. agalactiae H36B; II, S. agalactiae 18RS21, S. agalactiae 1954/92, S. agalactiae 118/158, and S. agalactiae BS29; III, S. agalactiae BM110, S. agalactiae BS30, S. agalactiae 97/0099, and S. agalactiae M781; IV, S. agalactiae 3139; V, S. agalactiae 1169-NT; VI, S. agalactiae GBS6; VII, S. agalactiae 7271; VIII, S. agalactiae JM9; S.pyg, S. pyogenes M1; S.pneu, S. pneumoniae VH14; M, 1-kb molecular weight markers. The sizes of the chromosomal DNA fragments hybridizing with the lrrG gene are indicated on the left. Hybridization with the DIG-labeled probe was carried out at 42°C.
A single 3-kb lrrG gene fragment was successfully amplified from all but one GBS strain (serotype III M781) by PCR with primers targeting the extreme N- and C-terminal sequences encoded by the lrrG gene from strain 97/0099 (see Materials and Methods). In GBS strain M781, two DNA fragments, of approximately 1.6 and 1.0 kb, were amplified by PCR, but the reasons for the irregularity observed with this strain remain unknown. The 3-kb fragments amplified from the strains appeared uniform in length, suggesting that the numbers of LRRs do not vary, as is the case with some other repeat-containing proteins of GBS, such as Rib and the C protein alpha subunit. However, a deletion of a single LRR would be difficult to detect by agarose gel electrophoresis, whereas a difference in length of 132 bp (i.e., two repeats) would be discernible. Given that there was no evidence of repeat variations in the three sequenced strains, it is likely that LRRs do not vary among strains, but sequencing of genes from individual strains would be needed to confirm this hypothesis. Despite the presence of an lrrG homologue in S. pyogenes, a PCR-amplified fragment was not obtained with S. pyogenes genomic DNA. A comparison of the S. pyogenes gene database entry and the lrrG primer sequences indicated that mismatches between the primers and the S. pyogenes gene were likely the cause of the failure to amplify the gene by PCR.
LrrG is a cell wall-anchored surface protein.
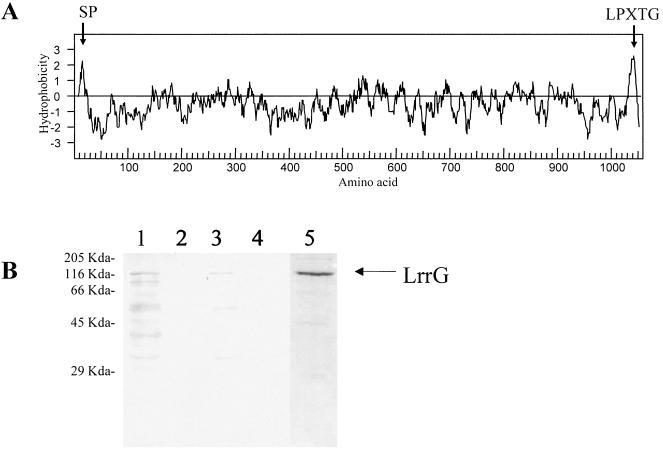
In addition to exhibiting homology to other bacterial surface proteins and characterized adhesins, the LrrG polypeptide contains motifs associated with surface proteins of other gram-positive bacteria. Analysis of the first 60 amino acids of LrrG with SignalP (see Materials and Methods) revealed a putative signal peptide with a proposed cleavage motif between residues 24 and 25 (QE-VYG) (Fig. 4A). The C terminus of LrrG also contains an LPXTG motif followed by a hydrophobic domain and a series of positively charged residues, features commonly associated with cell wall-anchored surface proteins of gram-positive bacteria (56).
FIG. 4.
Surface localization of LrrG. (A) Kyte-Doolittle plot of the deduced amino acid sequence encoded by lrrG of GBS. Hydrophobic regions are above the horizontal line; hydrophobicity is indicated on the vertical axis. Regions representing a putative secretion signal peptide (SP) and an LPXTG motif are indicated. (B) Western blot of cell protein fractions from mutanolysin-treated GBS strain 97/0099 (type III) probed with anti-LrrG antisera. Lanes: 1, cell cytosol; 2, cell membrane; 3, cell wall; 4, supernatant; 5, purified recombinant LrrG stained with Coomassie blue. Molecular mass standards are indicated on the left.
To confirm our hypothesis that LrrG is a surface antigen, Western blot analysis of proteins from different subcellular fractions of GBS strain 97/0099 was carried out with antiserum raised to recombinant LrrG purified from E. coli (Fig. 4B). A protein band with a molecular mass close to that predicted for LrrG (116 kDa) was detected in a mutanolysin-treated cell wall protein fraction, suggesting that LrrG may indeed be anchored to the cell wall. A similarly sized protein detected in the cytosolic fraction may represent uncleaved LrrG bound to the secretory apparatus prior to translocation. LrrG may also be bound to membrane vesicles present in the soluble fraction via the hydrophobic domain found near the C terminus. This feature is typical of cell wall-anchored proteins and has been proposed to anchor proteins in the cytoplasmic membrane prior to attachment to the cell wall via the sortase enzyme (52). This feature may also be an explanation for the detection in the insoluble fraction of small amounts of LrrG that are likely to comprise both cell wall and membrane components. Significantly, proteins concentrated from culture supernatants did not react with antiserum to LrrG, suggesting that LrrG is not secreted by GBS, at least during mid-exponential growth in vitro.
Immunogenicity of LrrG.
In order to investigate the immunogenicity of LrrG, the protein was expressed in E. coli, purified, and used for immunization and challenge studies with mice. The known protective antigen Rib, expressed by some but not all serotypes of GBS (80), was used for comparison and as a positive control. Gene fragments encoding the mature forms of the LrrG and Rib proteins (i.e., lacking DNA sequences encoding the signal leader peptide), were PCR amplified from GBS strain 97/0099, cloned, and expressed in E. coli BL21λ(DE3) by using His tag expression vector pET28b as described in Materials and Methods. Sequencing of the rib gene cloned from the serotype III challenge strain (97/0099) revealed that it contained only 2 (22) of up to 12 possible tandem internal repeats previously identified for the rib gene in GBS serotype III strain BM110 (84). However, immunization experiments previously carried out with mice showed that the Rib protein from strain 97/0099 conferred significant protection against a lethal challenge, thus validating the use of this Rib variant as a positive control antigen in the vaccination studies (22).
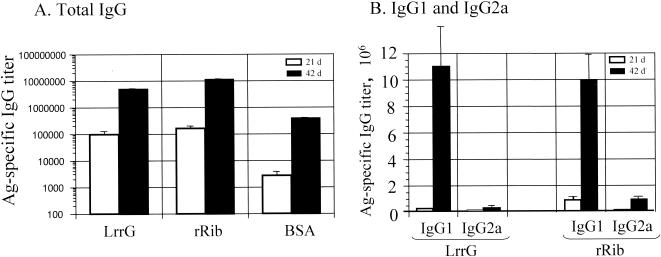
Groups of mice were immunized on days 0 and 28 with purified recombinant LrrG or recombinant Rib (rRib) or with BSA as a negative control. High levels of antigen-specific antibody responses were measured in the sera of immunized mice, and these levels were significantly increased (P < 0.01) at day 42 compared to day 21 following administration of the booster (Fig. 5A). Mice immunized with LrrG or rRib had higher levels (∼15-fold increase) of antigen-specific serum antibodies (P < 0.01) than mice vaccinated with BSA.
FIG. 5.
Serum ELISA titers of antigen (Ag)-specific total IgG (A) and antigen-specific IgG1 and IgG2a (B) antibodies following immunization with the indicated proteins. Groups of 6-week-old CBA/Ca mice were vaccinated s.c. at days 0 and 28 with 25 μg of the respective protein vaccine mixed in a 1:1 ratio with alum adjuvant. Mice were bled from the tail at days (d) 21 and 42 before bacterial challenge on day 56. Sera were analyzed by ELISAs, and the end-point titers were determined. The values represent the mean ± standard error of the mean for 12 mice.
Measurement of the antigen-specific serum IgG subclasses showed that both antigens elicited significantly higher levels (10- to 30-fold) of antigen-specific IgG1 than of IgG2a (P < 0.01) (Fig. 5B), as frequently observed by us (22) when protein antigens are administered by the parenteral route with adjuvant. The antigen-specific serum IgG1/IgG2a ratios elicited by s.c. immunization with LrrG and rRib were 30.8 and 12.9, respectively, at 42 days after the initial vaccination. The serum IgG1 antibody titers to both LrrG and rRib were significantly increased after the booster (P < 0.01), as were the IgG2a antibody titers, even though they were not of the same magnitude (P < 0.01).
Vaccination with LrrG protects mice against lethal challenge with GBS.
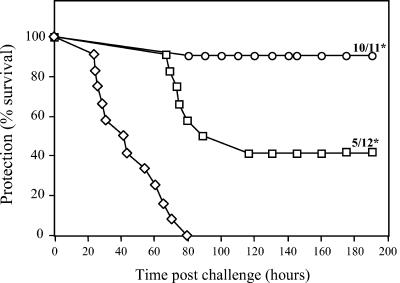
Mice were immunized and boosted with recombinant LrrG, rRib, and BSA administered in alum on days 0 and 28 and then challenged on day 49 with a lethal dose of virulent GBS. The control mice, immunized with BSA, all succumbed to disease at between 24 and 80 h after challenge. In contrast, 91 and 42% of mice immunized with LrrG and rRib, respectively, were protected from infection at 8 days after challenge, at which point the experiment was concluded (Fig. 6). Systemic GBS infection was confirmed for mice that succumbed to challenge by plating blood samples, which were all shown to be positive for GBS. The protection afforded by vaccination with LrrG and rRib was highly significant (P value for both, <0.01) compared to the results obtained for mice immunized with BSA-alum. In addition, mice immunized with LrrG were afforded significantly better protection than those immunized with rRib (P = 0.013), suggesting that LrrG may provide better efficacy as a vaccine antigen.
FIG. 6.
Vaccination with purified LrrG confers protection from challenge with GBS. Groups of 12 CBA/Ca mice aged between 6 and 7 weeks were immunized s.c. with 25 μg of protein antigen in a 1:1 mixture with alum adjuvant (final volume, 200 μl). Mice were immunized on day 0 and boosted on day 28. Mice were challenged intraperitoneally 21 days later with a lethal dose of GBS (3.5 × 106 CFU). Deaths were recorded hourly, and the percent survival was determined for up to 7 days following GBS challenge. The final ratios (number of surviving mice/number of mice challenged) are indicated for groups immunized with rRib and recombinant LrrG. Statistically significant differences (P < 0.01) in survival between experimental groups and the control group (BSA-alum-vaccinated mice) are indicated by asterisks and were calculated by using the Mann-Whitney U test. Symbols: ◊, BSA-alum; □, rRib-alum; ○, LrrG-alum.
Adherence of LrrG to epithelial cells.
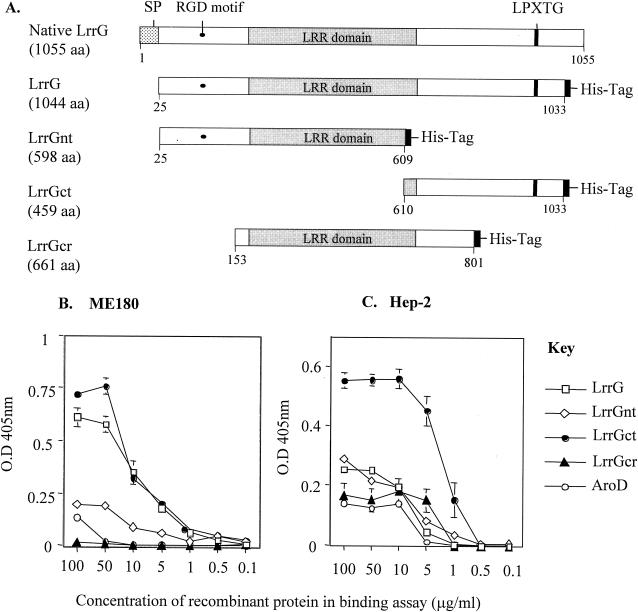
GBS colonize the genital tract of about 15 to 40% of pregnant women and increase the risk that infants born to these women will become colonized with GBS and subsequently develop severe early-onset disease (EOD) (75). It is postulated that EOD infection from GBS results from the aspiration of infected amniotic fluid, followed by adhesion and colonization of the lung airways of the newborn, leading in some cases to invasive disease (66, 67). The specific molecular interactions involved in the colonization of epithelial cells by GBS are unknown, although binding studies with protease-treated and untreated GBS suggest that surface proteins are indeed involved (81). Because LrrG was identified as a surface protein and contained LRR motifs commonly associated with protein-protein interactions, the binding of LrrG to human lung epithelial cell line HEp-2 and cervical epithelial cell line ME180 was investigated. Recombinant LrrG and truncated versions of the protein (i.e., LrrGct, LrrGnt, and LrrGcr) were expressed in and purified from E. coli and tested for their abilities to adhere to confluent monolayers of HEp-2 and ME180 cells as described in Materials and Methods. The efficacy of adherence of LrrG and its defined domains was determined by detecting bound LrrG proteins with antibodies specific for the His tag present on all of the purified recombinant proteins. As a control, recombinant His-tagged AroD protein from GBS was also purified and compared to the LrrG proteins in these binding assays. The AroD enzyme has been well characterized for several gram-negative bacteria and is known to be an intracellular enzyme that catalyzes the conversion of 3-dehydroquinate to 3-dehydroshikimate in the shikimate pathway; it is therefore not expected to bind to a specific receptor on eukaryotic cells (24).
All recombinant proteins tested adhered in a dose-dependent and saturable fashion to glutaraldehyde-fixed HEp-2 and ME180 cells (Fig. 7B and C); similar results were also found in preliminary experiments with unfixed cells (data not shown). When tested over a range of concentrations, the mature form of the whole protein (i.e., LrrG) and the C-terminal region (i.e., LrrGct) bound ME180 cells in substantially larger amounts than the other truncated LrrG derivatives (LrrGnt and LrrGcr) or the AroD control protein. Surprisingly, only LrrGct and not LrrG bound to HEp-2 cells in significantly larger amounts than the AroD control protein. Indeed, all purified LrrG derivatives other than LrrGct displayed binding curves similar to those of the AroD control protein when incubated with HEp-2 cells. Taken together, these results suggest that while the C-terminal region of LrrG is capable of binding to both cell lines, the mature full-length LrrG protein appears to preferentially bind to cervical epithelial cell line ME180 only.
FIG. 7.
LrrG binding to epithelial cells. LrrG and truncated versions of the antigen (A) were purified by affinity chromatography and incubated in a dose-dependent manner with ME180 cells (B) and HEp-2 cells (C) for 2 h at 37°C. Bound recombinant proteins was detected with antibody specific to the His tag present on each of the recombinant proteins followed by alkaline phosphatase-coupled anti-mouse IgG. The inclusion of the AroD protein served as a negative control and to record nonspecific protein-host cell interactions. Binding experiments were carried out in triplicate, and data represent the mean ± standard error of the mean. Numerals below the maps indicate the number of amino acid (aa) residues in each region. SP, signal peptide; O.D 405nm, optical density at 405 nm.
DISCUSSION
Although prenatal screening for GBS and the administration of intrapartum antibiotics to “at-risk” individuals have greatly reduced the incidence of invasive disease, GBS remain a leading cause of pregnancy-related morbidity in the United States. Approaches to the development of an efficient vaccine against GBS have focused on the use of CPSs or surface proteins. Recent studies suggested that the evolution of serotype distributions in GBS populations is likely to have a major impact on the efficacy of serotype-specific conjugate vaccines and that a vaccine targeting up to seven serotypes will be needed to provide effective coverage in both the United States and other parts of the world (39, 72). Similar issues concern the formulation of vaccines based on GBS surface proteins, such as Rib, the C protein alpha subunit, Fbs, and R28, which also show serotype- or strain-specific patterns of expression (2, 16, 27, 80). This situation has made it of interest to characterize other surface proteins of GBS as potential cross-protective targets.
In the course of carrying out a genetic screen for secreted GBS proteins based on the use of the Staphylococcus aureus nuclease (nuc) as a reporter (22), we identified the region of a GBS gene encoding a signal peptide with features found in other bacterial signal peptides and a motif recognized by signal peptidases (51, 57). Full-length sequencing of the predicted gene, designated lrrG, was done for strain 97/0099 (serotype III) and revealed that LrrG possesses features in common with numerous other cell wall-anchored proteins in gram-positive bacteria. LrrG possesses a putative signal peptide, an LPXTG motif near the C terminus, a hydrophobic domain, and positively charged residues at the C terminus, suggesting that LrrG is an extracellular protein processed by a sortase and covalently attached to the cell wall peptidoglycan (56, 59). Further evidence that LrrG is anchored in the cell wall of GBS was obtained by cellular fractionation and immunoblotting experiments with LrrG antiserum; these experiments showed that LrrG was not detected in the supernatants of exponentially growing cultures but could be released from the cell wall of osmotically stabilized cells by treatment with mutanolysin.
The predicted coding sequence of lrrG encodes 15 LRR sequences (each 22 amino acids long) in the N-terminal half of the protein that match the consensus sequence for other bacterial LRR proteins (35). LRRs are found in at least six families of eukaryotic proteins previously characterized on the basis of variations in the consensus motif and the repeat length (29). LRR-containing proteins found in eukaryotes have been shown to be involved in diverse functions involving protein-protein interactions, e.g., signal transduction, enzyme inhibition, cell adhesion, and cellular trafficking (10, 29, 48). In prokaryotes, LRR proteins are exclusively exported virulence factors of pathogenic bacteria (29), the most well characterized being internalins InlA and InlB of Listeria monocytogenes (9, 53). InlB has been shown to stimulate phosphatidylinositol 3-kinase, membrane ruffling, and the entry of Listeria into host epithelium (9, 77). InlA, another LRR protein of L. monocytogenes, is involved in adhesion to epithelial cells and in mediating uptake via the human E-cadherin surface receptor (42, 53). The LRR consensus sequence for LrrG places the protein in a new LRR family of bacterial LRR proteins, designated TpLRR (30), based on the LRR surface antigen of T. pallidum, which possesses the consensus sequence XLXXIXXLPXXLXXIXXAFXX. Other members of the TpLRR family include BspA, a surface protein from B. forsythus (76), PcpA from S. pneumoniae (69), and an uncharacterized hypothetical surface protein of group A S. pyogenes (accession no. AAK33772) to which LrrG has the highest overall similarity (81%) and identity (71%) over the entire coding sequence (Fig. 1).
Since this work commenced, the complete genome sequences of two S. agalactiae strains have been published: S. agalactiae 2603V/R (serotype III) (19) and NEM316 (serotype V) (82). Compared to that in strain 97/0099, the genetic organization of the lrrG locus was the same in strains 2603V/R and NEM316. Moreover, the translated products of their corresponding lrrG genes (accession no. AAM99327 and CAD46100, respectively) shared 100% identity with LrrG in strain 97/0099, indicating that this antigen is likely to be well conserved. This notion was further confirmed by Southern blot analysis, which detected a single lrrG DNA fragment varying in size from 6.1 to 9.1 kb in genomic digests from all strains representing each of the nine serotypes of GBS. In addition, an lrrG gene fragment of ∼3 kb was successfully amplified by PCR from all but one of the strains tested. These data would indicate that the fragments of different lengths detected by Southern blotting were due to restriction fragment length polymorphisms occurring outside the lrrG coding sequence. Therefore, in contrast to the Rib protein and the C protein alpha subunit of GBS, which were not present in all of the strains tested and which demonstrated strain-dependent variations in the numbers of repeat motifs at the gene level, lrrG was well conserved in all of the strains and serotypes of GBS tested.
While GBS have been shown to adhere efficiently and invade a variety of tissue-specific epithelial and endothelial cell lines, including those of placental and respiratory origins, at present little is known about the molecular basis of these host-microbe interactions. To demonstrate the potential role of LrrG in GBS colonization, adherence assays were carried out with recombinant LrrG (mature form) and its truncated variants and with the ME180 (cervical) and HEp-2 (lung) epithelial cell lines. Purified LrrG and LrrGct bound ME180 epithelial cells in substantially larger amounts than the other truncated LrrG proteins or control protein AroD. Surprisingly, however, only LrrGct and not the mature form of LrrG adhered to Hep-2 epithelial cells in amounts larger than those observed with the other truncated LrrG proteins or control protein AroD. Because these data suggest that the mature form of LrrG preferentially binds to a host receptor found on the cervical cell line but not on the lung cell line, the binding of LrrGct to both cell types was unexpected. One possible explanation for this finding could be that the expression of the LrrGct polypeptide allows protein interactions with host cells that are not possible in the context of the full-length folded LrrG protein, at least when expressed in and purified from E. coli. While these data indicate a role for LrrG in adhesion to host cells, further research is needed to confirm the specificity of LrrG binding to different receptors on host epithelium and to define the molecular basis of these interactions. A structural feature that could be involved in binding to host cells is an integrin-binding RGD motif (14) identified in the amino-terminal region of LrrG (amino acids 143 to 145). However, the low level of binding demonstrated by LrrGnt suggests that this motif is probably not involved in cell adhesion. It may also be significant that this RGD motif is not conserved in the close homologue found in S. pyogenes, where an asparagine residue replaces the aspartic acid residue (RGD→RGN), further endorsing the possibility that this motif is not functionally important in LrrG.
Immune defense against GBS, like that against other gram-positive bacteria, depends on clearance of the bacteria by phagocytic leukocytes, a mechanism consistently demonstrated to be greatly enhanced by opsonic antibodies to surface antigens or CPSs (7, 79). Given that LrrG was identified as a conserved surface protein, we investigated the vaccine potential of this surface antigen. s.c. immunization of groups of mice with LrrG and alum as an adjuvant elicited high levels of total IgG antibodies (end-point titer, ∼5,000,000). Similarly, immunization with rRib, a previously described immunogen, elicited comparable levels of antigen-specific IgG. In addition, mice immunized with LrrG were significantly protected against challenge with virulent GBS (91% survival) (P < 0.01), whereas control mice immunized with BSA all succumbed to disease within 24 to 80 h. A significant level of protection was also obtained following immunization with rRib (42% survival) (P < 0.01), although the level of protection was significantly lower than that obtained with LrrG. It should be noted that higher levels of protection have been achieved against challenge with GBS serotype III strain BM110 by use of native Rib protein isolated from this bacterium (80). As those experiments were also carried out with a different strain of mice (C3H/HeN), the results described here cannot be compared directly with those results. That LrrG is a good vaccine immunogen candidate was also confirmed in further challenge experiments with larger groups of mice (66% protection in a group of 15 mice) (data not shown).
The development of an LrrG subunit vaccine against GBS might be a good alternative approach to the use of CPSs as vaccines. Bacterial CPSs are T-cell-independent antigens, and experiments carried out with both animal and human subjects have shown that immune responses to purified bacterial CPSs fail to induce immunological memory (7). Moreover, other potential drawbacks of CPS vaccines include geographical variations in serotype prevalence and the possibility that these vaccines will ultimately select for increased prevalence of serotypes not included in the vaccines. For S. pneumoniae, vaccination with conjugate vaccines has already been shown to cause an increase in the prevalence of serotypes not present in the vaccines (58). A highly conserved protein antigen, such as LrrG, might also be used as a carrier protein for a restricted number of CPSs in order to provide greater coverage of the vaccine against clinically less important serotypes. This approach has been explored experimentally with the C protein alpha subunit and C5a peptidase of GBS conjugated to type III CPS; both of these conjugates induced protective immune responses in murine infection models (12, 21). A type III GBS CPS-tetanus toxoid conjugate induced predominantly the IgG2a subclass when administered to healthy nonpregnant women (32). Furthermore, this conjugate promoted opsonophaocytosis and killing of GBS and, after maternal immunization, protected neonatal mice against lethal challenge with type III GBS.
In conclusion, we have shown that LrrG, a novel LRR surface protein, is a highly promising candidate antigen for use in a GBS vaccine. Immunization of mice with purified LrrG and alum confers significant protection in a mouse model of invasive disease, and the level of protection was higher than that obtained with another promising but serotype-restricted vaccine antigen, Rib. The lrrG gene was shown to be well conserved among all nine clinically important serotypes, indicating its potential to afford broad cross-protection. Although the exact role of the LRR region in LrrG remains unclear, preliminary studies suggest that LrrG may be an important virulence factor that plays a role in the adhesion of GBS to epithelial cells.
Acknowledgments
We gratefully acknowledge financial support from Provalis Ltd. for the research carried out at both the University of Cambridge and the Institute of Food Research.
We thank Gunnar Lindahl (Department of Medical Microbiology and Immunology, Göteborg University, Göteborg, Sweden), Laurence Paoletti (Channing Laboratory, Department of Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, Mass.), Androulla Efstratiou (Respiratory and Systemic Infection Laboratory, Central Public Health Laboratory, London, United Kingdom), and Jitka Motlova (National Reference Laboratory for Streptococci and Enterococci, Prague, Czech Republic) for kindly providing us with strains and clinical isolates.
Editor: J. N. Weiser
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Areschoug, T., M. Stalhammar-Carlemalm, C. Larsson, and G. Lindahl. 1999. Group B streptococcal surface proteins as targets for protective antibodies: identification of two novel proteins in strains of serotype V. Infect. Immun. 67:6350-6357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2a.Badcock, D. 2002. Ph.D. thesis. University of Cambridge, Cambridge, United Kingdom.
- 3.Baker, C. J., and M. S. Edwards. 2003. Group B streptococcal conjugate vaccines. Arch. Dis. Child. 88:375-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baker, C. J., M. S. Edwards, B. J. Webb, and D. L. Kasper. 1982. Antibody-independent classical pathway-mediated opsonophagocytosis of type Ia, group B streptococcus. J. Clin. Investig. 69:394-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baker, C. J., M. A. Rench, M. Fernandez, L. C. Paoletti, D. L. Kasper, and M. S. Edwards. 2003. Safety and immunogenicity of a bivalent group B streptococcal conjugate vaccine for serotypes II and III. J. Infect. Dis. 188:66-73. [DOI] [PubMed] [Google Scholar]
- 6.Baker, C. J., M. A. Rench, and P. McInnes. 2003. Immunization of pregnant women with group B streptococcal type III capsular polysaccharide-tetanus toxoid conjugate vaccine. Vaccine 21:3468-3472. [DOI] [PubMed] [Google Scholar]
- 7.Baker, C. J., B. J. Webb, D. L. Kasper, and M. S. Edwards. 1986. The role of complement and antibody in opsonophagocytosis of type II group B streptococci. J. Infect. Dis. 154:47-54. [DOI] [PubMed] [Google Scholar]
- 8.Berner, R. 2002. Group B streptococci during pregnancy and infancy. Curr. Opin. Infect. Dis. 15:307-313. [DOI] [PubMed] [Google Scholar]
- 9.Braun, L., F. Nato, B. Payrastre, J. C. Mazie, and P. Cossart. 1999. The 213-amino-acid leucine-rich repeat region of the Listeria monocytogenes InlB protein is sufficient for entry into mammalian cells, stimulation of PI 3-kinase and membrane ruffling. Mol. Microbiol. 34:10-23. [DOI] [PubMed] [Google Scholar]
- 10.Buchanan, S. G., and N. J. Gay. 1996. Structural and functional diversity in the leucine-rich repeat family of proteins. Prog. Biophys. Mol. Biol. 65:1-44. [DOI] [PubMed] [Google Scholar]
- 11.Centers for Disease Control and Prevention. 2000. Early-onset group B streptococcal disease, United States, 1998-1999. Morb. Mortal. Wkly. Rep. 49:793-796. [Google Scholar]
- 12.Cheng, Q., S. Debol, H. Lam, R. Eby, L. Edwards, Y. Matsuka, S. B. Olmsted, and P. P. Cleary. 2002. Immunization with C5a peptidase or peptidase-type III polysaccharide conjugate vaccines enhances clearance of group B streptococci from lungs of infected mice. Infect. Immun. 70:6409-6415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davies, H. D., S. Raj, C. Adair, J. Robinson, and A. McGeer. 2001. Population-based active surveillance for neonatal group B streptococcal infections in Alberta, Canada: implications for vaccine formulation. Pediatr. Infect. Dis. J. 20:879-884. [DOI] [PubMed] [Google Scholar]
- 14.D'Souza, S. E., M. H. Ginsberg, and E. F. Plow. 1991. Arginyl-glycyl-aspartic acid (RGD): a cell adhesion motif. Trends Biochem. Sci. 16:246-250. [DOI] [PubMed] [Google Scholar]
- 15.Farley, M. M. 2001. Group B streptococcal disease in nonpregnant adults. Clin. Infect. Dis. 33:556-561. [DOI] [PubMed] [Google Scholar]
- 16.Ferrieri, P., and A. E. Flores. 1997. Surface protein expression in group B streptococcal invasive isolates. Adv. Exp. Med. Biol. 418:635-637. [DOI] [PubMed] [Google Scholar]
- 17.Finlay, B. B., and S. Falkow. 1997. Common themes in microbial pathogenicity revisited. Microbiol. Mol. Biol. Rev. 61:136-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flores, A. E., and P. Ferrieri. 1989. Molecular species of R-protein antigens produced by clinical isolates of group B streptococci. J. Clin. Microbiol. 27:1050-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glaser, P., C. Rusniok, C. Buchrieser, F. Chevalier, L. Frangeul, T. Msadek, M. Zouine, E. Couve, L. Lalioui, C. Poyart, P. Trieu-Cuot, and F. Kunst. 2002. Genome sequence of Streptococcus agalactiae, a pathogen causing invasive neonatal disease. Mol. Microbiol. 45:1499-1513. [DOI] [PubMed] [Google Scholar]
- 20.Gravekamp, C., D. S. Horensky, J. L. Michel, and L. C. Madoff. 1996. Variation in repeat number within the alpha C protein of group B streptococci alters antigenicity and protective epitopes. Infect. Immun. 64:3576-3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gravekamp, C., D. L. Kasper, L. C. Paoletti, and L. C. Madoff. 1999. Alpha C protein as a carrier for type III capsular polysaccharide and as a protective protein in group B streptococcal vaccines. Infect. Immun. 67:2491-2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanniffy, S. B., R. Seepersaud, P. J. W. Mayne, P. Sizer, R. W. F. LePage, and J. M. Wells. Unpublished data.
- 23.Harrison, L. H., J. A. Elliott, D. M. Dwyer, J. P. Libonati, P. Ferrieri, L. Billmann, A. Schuchat, et al. 1998. Serotype distribution of invasive group B streptococcal isolates in Maryland: implications for vaccine formulation. J. Infect. Dis. 177:998-1002. [DOI] [PubMed] [Google Scholar]
- 24.Haslam, E. 1993. Shikimic acid: metabolism and metabolites. John Wiley & Sons, Inc., New York, N.Y.
- 25.Isaacs, D., J. A. Royle, et al. 1999. Intrapartum antibiotics and early onset neonatal sepsis caused by group B streptococcus and by other organisms in Australia. Pediatr. Infect. Dis. J. 18:524-528. [DOI] [PubMed] [Google Scholar]
- 26.Jelinkova, J., and J. Motlova. 1985. Worldwide distribution of two new serotypes of group B streptococci: type IV and provisional type V. J. Clin. Microbiol. 21:361-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson, D. R., and P. Ferrieri. 1984. Group B streptococcal Ibc protein antigen: distribution of two determinants in wild-type strains of common serotypes. J. Clin. Microbiol. 19:506-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kajava, A. V. 2001. Review: proteins with repeated sequence-structural prediction and modeling. J. Struct. Biol. 134:132-144. [DOI] [PubMed] [Google Scholar]
- 29.Kajava, A. V. 1998. Structural diversity of leucine-rich repeat proteins. J. Mol. Biol. 277:519-527. [DOI] [PubMed] [Google Scholar]
- 30.Kajava, A. V., and B. Kobe. 2002. Assessment of the ability to model proteins with leucine-rich repeats in light of the latest structural information. Protein Sci. 11:1082-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kasper, D. L., C. J. Baker, B. Galdes, E. Katzenellenbogen, and H. J. Jennings. 1983. Immunochemical analysis and immunogenicity of the type II group B streptococcal capsular polysaccharide. J. Clin. Investig. 72:260-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kasper, D. L., L. C. Paoletti, M. R. Wessels, H. K. Guttormsen, V. J. Carey, H. J. Jennings, and C. J. Baker. 1996. Immune response to type III group B streptococcal polysaccharide-tetanus toxoid conjugate vaccine. J. Clin. Investig. 98:2308-2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kilstrup, M., and K. N. Kristiansen. 2000. Rapid genome walking: a simplified oligo-cassette mediated polymerase chain reaction using a single genome-specific primer. Nucleic Acids Res. 28:E55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kling, D. E., C. Gravekamp, L. C. Madoff, and J. L. Michel. 1997. Characterization of two distinct opsonic and protective epitopes within the alpha C protein of the group B streptococcus. Infect. Immun. 65:1462-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kobe, B., and A. V. Kajava. 2001. The leucine-rich repeat as a protein recognition motif. Curr. Opin. Struct. Biol. 11:725-732. [DOI] [PubMed] [Google Scholar]
- 36.Kogan, G., J. R. Brisson, D. L. Kasper, C. von Hunolstein, G. Orefici, and H. J. Jennings. 1995. Structural elucidation of the novel type VII group B streptococcus capsular polysaccharide by high resolution NMR spectroscopy. Carbohydr. Res. 277:1-9. [DOI] [PubMed] [Google Scholar]
- 37.Kogan, G., D. Uhrin, J. R. Brisson, L. C. Paoletti, A. E. Blodgett, D. L. Kasper, and H. J. Jennings. 1996. Structural and immunochemical characterization of the type VIII group B streptococcus capsular polysaccharide. J. Biol. Chem. 271:8786-8790. [DOI] [PubMed] [Google Scholar]
- 38.Lachenauer, C. S., R. Creti, J. L. Michel, and L. C. Madoff. 2000. Mosaicism in the alpha-like protein genes of group B streptococci. Proc. Natl. Acad. Sci. USA 97:9630-9635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lachenauer, C. S., D. L. Kasper, J. Shimada, Y. Ichiman, H. Ohtsuka, M. Kaku, L. C. Paoletti, P. Ferrieri, and L. C. Madoff. 1999. Serotypes VI and VIII predominate among group B streptococci isolated from pregnant Japanese women. J. Infect. Dis. 179:1030-1033. [DOI] [PubMed] [Google Scholar]
- 40.Lancefield, R. C. 1938. Two serological types of group B hemolytic streptococci with related, but not identical, type-specific substances. J. Exp. Med. 67:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Larsson, C., M. Stalhammar-Carlemalm, and G. Lindahl. 1996. Experimental vaccination against group B streptococcus, an encapsulated bacterium, with highly purified preparations of cell surface proteins Rib and alpha. Infect. Immun. 64:3518-3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lecuit, M., H. Ohayon, L. Braun, J. Mengaud, and P. Cossart. 1997. Internalin of Listeria monocytogenes with an intact leucine-rich repeat region is sufficient to promote internalization. Infect. Immun. 65:5309-5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin, F. Y., J. D. Clemens, P. H. Azimi, J. A. Regan, L. E. Weisman, J. B. Philips III, G. G. Rhoads, P. Clark, R. A. Brenner, and P. Ferrieri. 1998. Capsular polysaccharide types of group B streptococcal isolates from neonates with early-onset systemic infection. J. Infect. Dis. 177:790-792. [DOI] [PubMed] [Google Scholar]
- 44.Liu, G. Y., and V. Nizet. 2004. Extracellular virulence factors of group B streptococci. Front. Biosci. 9:1794-1802. [DOI] [PubMed] [Google Scholar]
- 45.Madoff, L. C., S. Hori, J. L. Michel, C. J. Baker, and D. L. Kasper. 1991. Phenotypic diversity in the alpha C protein of group B streptococci. Infect. Immun. 59:2638-2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Madoff, L. C., J. L. Michel, E. W. Gong, D. E. Kling, and D. L. Kasper. 1996. Group B streptococci escape host immunity by deletion of tandem repeat elements of the alpha C protein. Proc. Natl. Acad. Sci. USA 93:4131-4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marai, W. 2001. Lower genital tract infections among pregnant women: a review. East Afr. Med. J. 78:581-585. [PubMed] [Google Scholar]
- 48.Marcotte, E. M., M. Pellegrini, T. O. Yeates, and D. Eisenberg. 1999. A census of protein repeats. J. Mol. Biol. 293:151-160. [DOI] [PubMed] [Google Scholar]
- 49.Marino, M., L. Braun, P. Cossart, and P. Ghosh. 2000. A framework for interpreting the leucine-rich repeats of the Listeria internalins. Proc. Natl. Acad. Sci. USA 97:8784-8788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martin, D., S. Rioux, E. Gagnon, M. Boyer, J. Hamel, N. Charland, and B. R. Brodeur. 2002. Protection from group B streptococcal infection in neonatal mice by maternal immunization with recombinant Sip protein. Infect. Immun. 70:4897-4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martoglio, B., and B. Dobberstein. 1998. Signal sequences: more than just greasy peptides. Trends Cell Biol. 8:410-415. [DOI] [PubMed] [Google Scholar]
- 52.Mazmanian, S. K., H. Ton-That, and O. Schneewind. 2001. Sortase-catalysed anchoring of surface proteins to the cell wall of Staphylococcus aureus. Mol. Microbiol. 40:1049-1057. [DOI] [PubMed] [Google Scholar]
- 53.Mengaud, J., H. Ohayon, P. Gounon, R. M. Mege, and P. Cossart. 1996. E-cadherin is the receptor for internalin, a surface protein required for entry of L. monocytogenes into epithelial cells. Cell 84:923-932. [DOI] [PubMed] [Google Scholar]
- 54.Michel, J. L., L. C. Madoff, K. Olson, D. E. Kling, D. L. Kasper, and F. M. Ausubel. 1992. Large, identical, tandem repeating units in the C protein alpha antigen gene, bca, of group B streptococci. Proc. Natl. Acad. Sci. USA 89:10060-10064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Musser, J. M., S. J. Mattingly, R. Quentin, A. Goudeau, and R. K. Selander. 1989. Identification of a high-virulence clone of type III Streptococcus agalactiae (group B streptococcus) causing invasive neonatal disease. Proc. Natl. Acad. Sci. USA 86:4731-4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Navarre, W. W., and O. Schneewind. 1999. Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol. Mol. Biol. Rev. 63:174-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. A neural network method for identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Int. J. Neural Syst. 8:581-599. [DOI] [PubMed] [Google Scholar]
- 58.Obaro, S. K., R. A. Adegbola, W. A. Banya, and B. M. Greenwood. 1996. Carriage of pneumococci after pneumococcal vaccination. Lancet 348:271-272. [DOI] [PubMed] [Google Scholar]
- 59.Pallen, M. J., A. C. Lam, M. Antonio, and K. Dunbar. 2001. An embarrassment of sortases—a richness of substrates? Trends Microbiol. 9:97-102. [DOI] [PubMed] [Google Scholar]
- 60.Paoletti, L. C. 2001. Potency of clinical group B streptococcal conjugate vaccines. Vaccine 19:2118-2126. [DOI] [PubMed] [Google Scholar]
- 61.Paoletti, L. C., and D. L. Kasper. 2002. Conjugate vaccines against group B streptococcus types IV and VII. J. Infect. Dis. 186:123-126. [DOI] [PubMed] [Google Scholar]
- 62.Paoletti, L. C., R. A. Ross, and K. D. Johnson. 1996. Cell growth rate regulates expression of group B streptococcus type III capsular polysaccharide. Infect. Immun. 64:1220-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Poquet, I., S. D. Ehrlich, and A. Gruss. 1998. An export-specific reporter designed for gram-positive bacteria: application to Lactococcus lactis. J. Bacteriol. 180:1904-1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Regan, J. A., M. A. Klebanoff, R. P. Nugent, D. A. Eschenbach, W. C. Blackwelder, Y. Lou, R. S. Gibbs, P. J. Rettig, D. H. Martin, R. Edelman, et al. 1996. Colonization with group B streptococci in pregnancy and adverse outcome. Am. J. Obstet. Gynecol. 174:1354-1360. [DOI] [PubMed] [Google Scholar]
- 65.Robinson, K., L. M. Chamberlain, K. M. Schofield, J. M. Wells, and R. W. Le Page. 1997. Oral vaccination of mice against tetanus with recombinant Lactococcus lactis. Nat. Biotechnol. 15:653-657. [DOI] [PubMed] [Google Scholar]
- 66.Rubens, C. E., H. V. Raff, J. C. Jackson, E. Y. Chi, J. T. Bielitzki, and S. L. Hillier. 1991. Pathophysiology and histopathology of group B streptococcal sepsis in Macaca nemestrina primates induced after intraamniotic inoculation: evidence for bacterial cellular invasion. J. Infect. Dis. 164:320-330. [DOI] [PubMed] [Google Scholar]
- 67.Rubens, C. E., S. Smith, M. Hulse, E. Y. Chi, and G. van Belle. 1992. Respiratory epithelial cell invasion by group B streptococci. Infect. Immun. 60:5157-5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed., p. 1659-1711. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 69.Sanchez-Beato, A. R., R. Lopez, and J. L. Garcia. 1998. Molecular characterization of PcpA: a novel choline-binding protein of Streptococcus pneumoniae. FEMS Microbiol. Lett. 164:207-214. [DOI] [PubMed] [Google Scholar]
- 70.Schrag, S., R. Gorwitz, K. Fultz-Butts, and A. Schuchat. 2002. Prevention of perinatal group B streptococcal disease. Revised guidelines from CDC. Morb. Mortal. Wkly. Rep. Recomm. Rep. 51:1-22. [PubMed] [Google Scholar]
- 71.Schrag, S. J., S. Zywicki, M. M. Farley, A. L. Reingold, L. H. Harrison, L. B. Lefkowitz, J. L. Hadler, R. Danila, P. R. Cieslak, and A. Schuchat. 2000. Group B streptococcal disease in the era of intrapartum antibiotic prophylaxis. N. Engl. J. Med. 342:15-20. [DOI] [PubMed] [Google Scholar]
- 72.Schuchat, A. 1998. Epidemiology of group B streptococcal disease in the United States: shifting paradigms. Clin. Microbiol. Rev. 11:497-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schuchat, A. 2001. Group B streptococcal disease: from trials and tribulations to triumph and trepidation. Clin. Infect. Dis. 33:751-756. [DOI] [PubMed] [Google Scholar]
- 74.Schuchat, A., and J. D. Wenger. 1994. Epidemiology of group B streptococcal disease. Risk factors, prevention strategies, and vaccine development. Epidemiol. Rev. 16:374-402. [DOI] [PubMed] [Google Scholar]
- 75.Schuchat, A., S. S. Zywicki, M. J. Dinsmoor, B. Mercer, J. Romaguera, M. J. O'Sullivan, D. Patel, M. T. Peters, B. Stoll, and O. S. Levine. 2000. Risk factors and opportunities for prevention of early-onset neonatal sepsis: a multicenter case-control study. Pediatrics 105:21-26. [DOI] [PubMed] [Google Scholar]
- 76.Sharma, A., H. T. Sojar, I. Glurich, K. Honma, H. K. Kuramitsu, and R. J. Genco. 1998. Cloning, expression, and sequencing of a cell surface antigen containing a leucine-rich repeat motif from Bacteroides forsythus ATCC 43037. Infect. Immun. 66:5703-5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shen, Y., M. Naujokas, M. Park, and K. Ireton. 2000. InIB-dependent internalization of Listeria is mediated by the Met receptor tyrosine kinase. Cell 103:501-510. [DOI] [PubMed] [Google Scholar]
- 78.Shevchenko, D. V., D. R. Akins, E. Robinson, M. Li, T. G. Popova, D. L. Cox, and J. D. Radolf. 1997. Molecular characterization and cellular localization of TpLRR, a processed leucine-rich repeat protein of Treponema pallidum, the syphilis spirochete. J. Bacteriol. 179:3188-3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shyur, S. D., H. V. Raff, J. F. Bohnsack, D. K. Kelsey, and H. R. Hill. 1992. Comparison of the opsonic and complement triggering activity of human monoclonal IgG1 and IgM antibody against group B streptococci. J. Immunol. 148:1879-1884. [PubMed] [Google Scholar]
- 80.Stalhammar-Carlemalm, M., L. Stenberg, and G. Lindahl. 1993. Protein rib: a novel group B streptococcal cell surface protein that confers protective immunity and is expressed by most strains causing invasive infections. J. Exp. Med. 177:1593-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tamura, G. S., J. M. Kuypers, S. Smith, H. Raff, and C. E. Rubens. 1994. Adherence of group B streptococci to cultured epithelial cells: roles of environmental factors and bacterial surface components. Infect. Immun. 62:2450-2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tettelin, H., V. Masignani, M. J. Cieslewicz, J. A. Eisen, S. Peterson, M. R. Wessels, I. T. Paulsen, K. E. Nelson, I. Margarit, T. D. Read, L. C. Madoff, A. M. Wolf, M. J. Beanan, L. M. Brinkac, S. C. Daugherty, R. T. DeBoy, A. S. Durkin, J. F. Kolonay, R. Madupu, M. R. Lewis, D. Radune, N. B. Fedorova, D. Scanlan, H. Khouri, S. Mulligan, H. A. Carty, R. T. Cline, S. E. Van Aken, J. Gill, M. Scarselli, M. Mora, E. T. Iacobini, C. Brettoni, G. Galli, M. Mariani, F. Vegni, D. Maione, D. Rinaudo, R. Rappuoli, J. L. Telford, D. L. Kasper, G. Grandi, and C. M. Fraser. 2002. Complete genome sequence and comparative genomic analysis of an emerging human pathogen, serotype V Streptococcus agalactiae. Proc. Natl. Acad. Sci. USA 99:12391-12396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wastfelt, M., M. Stalhammar-Carlemalm, A. M. Delisse, T. Cabezon, and G. Lindahl. 1996. Identification of a family of streptococcal surface proteins with extremely repetitive structure. J. Biol. Chem. 271:18892-18897. [DOI] [PubMed] [Google Scholar]
- 85.Wells, J. M., P. W. Wilson, and R. W. Le Page. 1993. Improved cloning vectors and transformation procedure for Lactococcus lactis. J. Appl. Bacteriol. 74:629-636. [DOI] [PubMed] [Google Scholar]
- 86.Wessels, M. R., J. L. DiFabio, V. J. Benedi, D. L. Kasper, F. Michon, J. R. Brisson, J. Jelinkova, and H. J. Jennings. 1991. Structural determination and immunochemical characterization of the type V group B streptococcus capsular polysaccharide. J. Biol. Chem. 266:6714-6719. [PubMed] [Google Scholar]