Abstract
A better understanding of immunity to infection is revealed from the characteristics of microbial ligands recognized by host immune responses. Murine infection with the intracellular bacterium Salmonella generates CD4+ T cells that specifically recognize Salmonella proteins expressed in bacterial surface organelles such as flagella and membrane vesicles. These natural Salmonella antigens are also ligands for Toll-like receptors (TLRs) or avidly associated with TLR ligands such as lipopolysaccharide (LPS). PhoP/PhoQ, a regulon controlling Salmonella virulence and remodeling of LPS to resist innate immunity, coordinately represses production of surface-exposed antigens recognized by CD4+ T cells and TLRs. These data suggest that genetically coordinated surface modifications may provide a growth advantage for Salmonella in host tissues by limiting both innate and adaptive immune recognition.
A successful defense against pathogenic microorganisms requires appropriate coordination between innate and adaptive immune responses. Initial recognition of microbes by dendritic cells, neutrophils, and macrophages via Toll-like receptors (TLRs) leads to recruitment and activation of B and T lymphocytes, resulting in sterilization of infected tissue and long-lasting immunological memory (50). TLRs are stimulated by conserved microbial components (35), while B- and T-cell receptors identify pathogen-specific molecules. Although distinct in temporal and functional aspects of host immunity, the connections between innate and adaptive immune responses are emerging, as recent work has identified signal transduction pathways required for both immune systems (64).
Murine infection with Salmonella enterica serovar Typhimurium causes a systemic typhoid-like disease in which bacteria replicate in the intracellular vacuoles of professional phagocytes (36, 47). Salmonellae express classical inflammatory molecules like lipopolysaccharide (LPS) and lipoproteins in their outer membranes but resist innate immune recognition by modifying the bacterial envelope through processes controlled by the PhoP/PhoQ regulatory system (18). PhoP/PhoQ is required for Salmonella virulence in infected hosts: PhoP− bacteria fail to cause disease in susceptible hosts but instead induce protective immunity (19, 21, 30, 51, 69). CD4+ T-cell responses are an essential component of immunity to salmonellae (48), and the major subunit protein of bacterial flagella, FliC, is an important antigen recognized by CD4+ T cells from both Salmonella-infected mice and humans (15, 49, 68). The innate immune response also recognizes FliC via TLR5 (29, 65).
We demonstrate that bacterial surface organelles (flagella and membrane vesicles [MVs]) stimulate TLRs and also contain the natural antigens recognized by murine CD4+ T cells responding to Salmonella infection. Thus, the host innate and adaptive immune responses preferentially target surface-exposed microbial ligands for recognition. Correspondingly, the PhoP/PhoQ virulence regulon both controls bacterial resistance to innate immunity and represses production of antigens recognized by T cells. This suggests that coordinate regulation of antigen expression and bacterial membrane modifications may contribute to Salmonella virulence and supports the notion that microbial pathogenic strategies have coevolved with the host immune system.
MATERIALS AND METHODS
Bacterial strains and antigen preparation.
Salmonella enterica serovar Typhimurium strain SL3261 (SL1344 ΔaroA) (31) was used for oral immunization of mice. Antigens were prepared from S. enterica serovar Typhimurium strains ST14028 (American Type Culture Collection [ATCC]), ST14028 pho-24 (PhoPc), ST14028 phoP102::Tn10dCam (PhoP−) (42, 51, 52), ST14028 phoP* phoQ::Tn10 (PhoP*) (14), ST14028 fliCi::Tn10 (BC118), and ST14028 ΔfliC ΔfljB (BC698). Strains were constructed by generalized transduction via P22 phage (15). Heat-killed salmonellae were prepared from bacteria grown to stationary phase in Luria broth or tryptic soy broth (BD Diagnostic Systems, Sparks, Md.) and incubated at 65°C for 1 h. Sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE)-fractionated Salmonella was prepared from ST14028 bacteria separated by SDS-16.5% PAGE (62) as previously described (15). Briefly, protein was eluted from gel sections, polyacrylamide was removed by filtration (Spin-X; Corning Inc., Corning, N.Y.), and SDS-PAGE buffer was replaced with phosphate-buffered saline by diafiltration through Microcon filtration units with a molecular weight cutoff of 10,000 (Millipore, Bedford, Mass.). Proteinase K-treated salmonellae were obtained by treating whole bacteria with 0.1 mg of proteinase K (Boehringer Mannheim Corp., Indianapolis, Ind.) per ml at 37C° for 24 h, followed by enzyme inactivation at 65°C for 15 min; addition of 0.2% SDS during proteinase K treatment solubilized bacterial membranes, allowing complete proteolysis. To purify FliC and FljB, flagella from logarithmic-phase bacteria expressing only FljB or FliC (15) were sheared off by blending (67) (Waring, East Windsor, N.J.), depolymerized at 60°C for 20 min, and passed through a Centricon filtration unit with a molecular weight cutoff of 100,000 (Millipore) to remove contaminating LPS.
Mice and immunizations.
Six- to 8-week-old female mice (C57BL/6 and C3H/HeJ; Jackson Laboratory, Bar Harbor, Maine) were used for immunization and as a source of splenocyte antigen-presenting cells (APC). Mice were immunized by oral gavage with 109 viable SL3261 bacteria (feeding needle no. 7920; Popper & Sons, Inc., New Hyde Park, N.Y.). Studies were performed in accordance with the institutional guidelines for animal use and care.
Generation of Salmonella-specific T cells.
T cells were grown in RPMI 1640 medium supplemented with l-glutamine, 50 μM β2-mercaptoethanol, and 10% fetal calf serum with penicillin, streptomycin, and gentamicin. Salmonella-immune T-cell lines were generated by stimulation of CD4+ T cells recovered from mice 90 days after immunization (SL3261) with Salmonella antigen (derived from SL1344 or ATCC 14028) presented by naive splenocyte APC as previously described (15). Alternatively, splenocytes harvested from mice infected with virulent salmonellae (containing 0.6 to 4 intracellular bacteria per splenocyte) were used directly ex vivo as APC to stimulate immune T cells. No Salmonella-specific T cells were recovered from naive mice (data not shown). Antigen-specific T-cell clones were isolated by limiting dilution, were confirmed to be CD4+ by flow-cytometric analysis, and were protein antigen specific. Major histocompatibility complex (MHC) restriction to either Ak or Ek molecules in C3H/HeJ mice was determined by (i) proliferative responses to antigen and Ak-expressing splenocyte APC from B10.4R mice (Jackson Laboratory) or (ii) inhibition of proliferative responses to antigen and APC in the presence of blocking anti-Ek antibody (clone 14-4-4S; BD Biosciences Pharmingen, San Diego, Calif.) (data not shown).
MVs and size exclusion chromatography.
ST14028 bacteria were grown in tryptic soy broth to logarithmic phase, and organisms were removed from the culture by centrifugation. Culture supernatant was filtered through 0.22-μm-pore-size filter units (Corning, Inc.) and further concentrated by diafiltration through CentriconPlus filtration units with a molecular weight cutoff of 100,000 (Millipore), yielding MV preparations. Size exclusion chromatography of MVs was performed with Sephacryl S-500 resin (Amersham Biosciences, Piscataway, N.J.).
Proliferation assays.
T-cell proliferation in response to APC plus antigen was assayed as previously described (15). Briefly, 104 T cells and 105 irradiated splenocytes plus antigen were combined in triplicate, [3H]TdR was added after 48 h, DNA was harvested after 16 h, and incorporated 3H was measured by liquid scintillation spectrophotometry.
Immunoblotting.
Salmonella antigen preparations were separated by SDS-10% PAGE, transferred to nitrocellulose, and probed with polyclonal sera specific to SecA, OmpA (kind gifts from Tina Guina, Department of Pediatrics, University of Washington), LPS (BD Diagnostic Systems), or FliC (Denka Seiken, Tokyo, Japan), followed by goat anti-rabbit immunoglobulin G conjugated to horseradish peroxidase (Santa Cruz Biotechnology, Santa Cruz, Calif.). Reactive horseradish peroxidase was detected by enhanced chemiluminescence (Amersham Biosciences).
Electron microscopy.
MVs were negatively stained with 1% phosphotungstic acid (pH 7.0) and applied directly to 0.5% Formvar-coated 300-mesh copper grids. Samples were observed with a JEM-1200EXII transmission electron microscope (JEOL). Micrographs were taken at an accelerating voltage of 80 kV.
TLR2- and TLR5-dependent NF-κB activation.
CHO K1 cells (ATCC catalog no. CRL-9618) were grown in Ham's F-12 medium supplemented with 10% fetal bovine serum, l-glutamine, penicillin, and streptomycin. CHO K1 cells were transfected by electroporation with 8 μg of either murine TLR2 or TLR5 cloned into the pEF6 V5/His TOPO expression vector (Invitrogen), 1 μg of an NF-κB-dependent firefly luciferase reporter (ELAM 1 Luc) plasmid (63A), and 0.1 μg of a control Renilla luciferase reporter (pRL-TK; Promega) plasmid. Stable cell lines were selected with 5 μg of blasticidin (Calbiochem) per ml and cloned by limiting dilution. Individual clones that demonstrated TLR2-specific recognition of synthetic bacterial lipopeptide PAM3CSK4 (Roche Biochemicals) or TLR5-specific recognition of purified bacterial flagellin (29) were chosen and used for the assays reported in this study. Clones were plated at approximately 2 × 104 cells per well in 96-well plates and, after 48 h, stimulated with bacterial products for 5 h at 37°C in 5% CO2. Firefly and Renilla luciferase activities were measured with the Dual Luciferase Assay System (Promega). Luciferase activity was calculated as a ratio of NF-κB-dependent ELAM firefly luciferase activity divided by control thymidine kinase Renilla luciferase activity (relative luciferase units). Fold induction was calculated by dividing the luciferase values for the test conditions by the relative luciferase value for the control condition.
RESULTS
Surface organelles contain natural Salmonella antigens recognized by CD4+ T cells from immune mice.
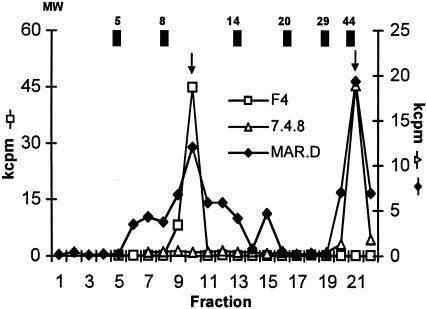
We orally infected mice (15) with S. enterica serovar Typhimurium strain SL3261 (31) and generated Salmonella-specific CD4+ T-cell lines (see Materials and Methods). SDS-PAGE-fractionated bacteria were used as a source of stimulatory antigen in proliferation assays, and the responses of the Salmonella-specific T-cell line MAR.D demonstrated recognition of proteins of various molecular weights (Fig. 1). As previously described, FliC was an important stimulatory antigen for CD4+ T cells from infected mice (15, 49) (Fig. 1, fraction 21). The second largest response was to a lower-molecular-weight fraction (15); we isolated antigen-specific CD4+ T-cell clones that only responded to SDS-PAGE fraction 10 (Fig. 1, T-cell clone F4, 1 of 32 representative clones), recognized antigen in the context of the class II MHC molecule Ek, and were no longer stimulated by Salmonella antigen treated with proteinase K in the presence of detergent (see below).
FIG. 1.
Salmonella-specific CD4+ T cells from orally immunized mice recognize multiple bacterial antigens. Salmonella-specific CD4+ T cells derived from immune mice (line MAR.D) were assayed for proliferative responses to SDS-PAGE-fractionated bacteria (see Materials and Methods). Responses of CD4+ T-cell clones 7.4.8 and F4 are also shown. Molecular weights (MW; 103) of protein standards are shown at the top; arrows indicate responses to fraction 10 and FliC (approximately 52,000). The data shown are representative of four experiments.
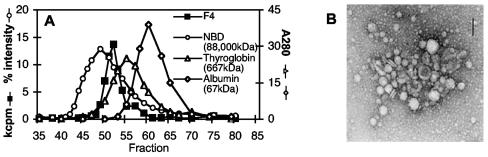
Since an important natural antigen from salmonellae, FliC, is a secreted bacterial protein, we tested bacterium-free supernatant from exponentially growing Salmonella cultures for the presence of stimulatory antigen recognized by T-cell clones like F4. Interestingly, stimulatory antigen in supernatant was completely retained by diafiltration membranes with a molecular weight cutoff of 100,000. When the supernatant was separated by size exclusion chromatography, Salmonella-specific CD4+ T-cell clone F4 recognized a single species with an apparent molecular weight between 700,000 and 88,000,000 (Fig. 2A). Electron microscopy of concentrated supernatant revealed MVs 100 to 200 nm in diameter (Fig. 2B) that were clearly distinguishable from bacteria in size (salmonellae are 0.5 to 1 by 2 μm) (56). MVs are spherical structures derived from the bacterial outer membrane and are expressed by numerous bacteria (11, 46). In addition to outer membrane proteins and LPS (11), MVs have been shown to contain numerous bacterial virulence factors (5, 20, 33, 34, 39-41, 71) and to mediate delivery of bacterial components to the cytoplasm of host cells (9, 20, 34, 38, 39, 71) by mechanisms that are independent of type I-V secretory systems (71). Because of the increasingly well-defined and specific mechanisms by which MVs contribute to bacterial pathogenesis, we propose that they be considered bacterial surface organelles, analogous to fimbriae or flagella.
FIG. 2.
CD4+ T-cell clone F4 recognizes antigen expressed in bacterial MVs. Concentrated S. enterica serovar Typhimurium culture supernatant was (i) separated by size exclusion chromatography, and fractions were used as stimulatory antigen in proliferation assays with Salmonella-specific CD4+ T-cell clone F4 (molecular weight standards: fluorescein-labeled nitrobenzoxadiazole [NBD] phosphatidylcholine vesicles [% fluorescence intensity], thyroglobulin, and albumin [A280]) (A) or (ii) examined by electron microscopy (B). Bar = 100 nm. The data shown are representative of three experiments.
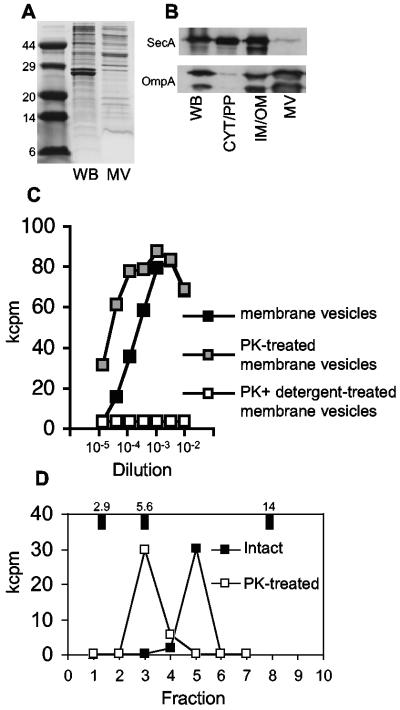
Further characterization of MVs produced by S. enterica serovar Typhimurium revealed a unique protein profile that was distinct from that of whole bacteria (Fig. 3A), lacking the cytoplasmic-inner membrane protein SecA but enriched for outer membrane proteins (Fig. 3B). The intimate association of antigen with MVs was demonstrated by destruction of stimulatory activity by proteinase K digestion only when membranes were solubilized with detergent (Fig. 3C). The association with LPS in MVs (11) was particularly avid, as demonstrated by the ability of stimulatory activity to resist extraction by phenol (6) and urea (34) (data not shown). Similar to FliC, the major subunit protein of Salmonella flagella, stimulatory antigen in MVs was surface exposed; proteinase K treatment did not destroy antigenic activity but converted it to a lower-molecular-weight species (Fig. 3D). Our data demonstrate that the adaptive immune system recognizes antigens present in bacterial MVs.
FIG. 3.
The unique composition of MVs includes surface-exposed outer membrane proteins. Concentrated MVs were compared to whole bacteria (WB) by SDS-PAGE with Coomassie staining (A) and Western analysis for the presence of cytoplasmic-inner membrane (SecA) and outer membrane (OmpA) marker proteins (isolated subcellular bacterial fractions containing cytoplasm and periplasm [CYT/PP] or inner-outer membrane [IM/OM] proteins are provided for comparison) (B) or treated with or without proteinase K (PK) in the presence or absence of detergent and used as the stimulatory antigen in F4 proliferation assays (C). (D) Surface exposure of F4 stimulatory antigen was confirmed by conversion from a 10-kDa to a 5-kDa species after proteinase K treatment of intact bacteria.
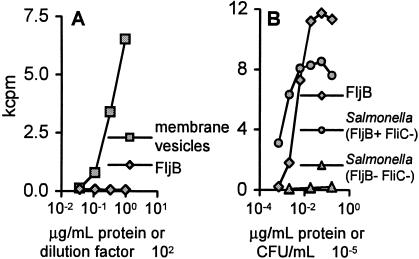
Vesicle production is triggered by in vivo growth of diverse intracellular pathogens, including S. enterica serovar Typhimurium (22) and Mycobacterium tuberculosis (8, 57, 63). Because of their association with the bacterial surface, MVs are favorably accessible to host antigen-processing machinery. We hypothesized that MVs would therefore provide a rich source of natural antigens for the adaptive immune response. To test this hypothesis, we used splenocytes from infected mice as APC directly ex vivo to generate Salmonella-specific CD4+ T-cell clones (see Materials and Methods); 73% (41 of 56) of these clones responded to vesicle proteins. As a further test, when CD4+ T-cell clones were derived from immunized mice in the absence of the dominant antigen FliC, 50% (12 of 24) proliferated in response to proteins expressed in MVs (Fig. 4A). These clones recognized vesicle antigens with apparent molecular masses of 10, 12, and 18 kDa, and individual clones only responded to single fractions from SDS-PAGE-fractionated bacteria (data not shown). Interestingly, the remaining CD4+ T-cell clones isolated in the absence of FliC (n = 12) proliferated in response to purified FljB (Fig. 4B), the alternate flagellar subunit protein that can be expressed by S. enterica serovar Typhimurium (2). Finally, 100% (14 of 14) of the clones derived from Salmonella-immune mice in the absence of both FliC and FljB recognized vesicle antigens. Thus, flagella and MVs, complex multicomponent surface organelles, contain the natural antigens most frequently recognized by CD4+ T cells isolated from protectively immunized mice.
FIG. 4.
Bacterial surface organelles contain natural antigens recognized by CD4+ T cells. CD4+ T-cell clones, derived from Salmonella-infected mice in the absence of the dominant FliC antigen, recognize either antigens in MVs (n = 12 clones) (A) or the alternate flagellin molecule FljB (n = 12 clones) (B). Proliferative responses of representative T-cell clones were measured in the presence of isolated MVs (102 dilution), purified FljB protein (micrograms per milliliter), or FljB+ FliC− or FljB− FliC− salmonellae (CFU per milliliter).
Intimate association of TLR ligands with natural Salmonella antigens.
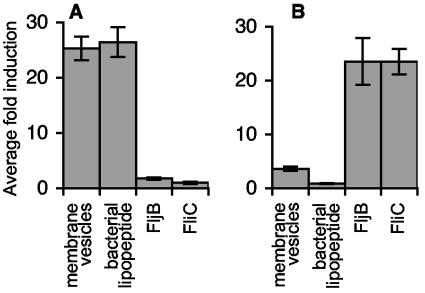
Because of the presumed interconnected functioning of the innate and adaptive immune systems and our observations that host CD4+ T-cell responses preferentially target antigens with particular attributes, we predicted that conserved surface organelles would also contain the bacterial components recognized by innate immune receptors during Salmonella infection. Consistent with this hypothesis, Salmonella vesicles activated TLR2-dependent NF-κB transcription in CHO cells at levels comparable to that of a control synthetic bacterial lipopeptide (4, 13) (Fig. 5A), indicating that MVs contain TLR2 ligands. The innate immune system also recognized FljB; compared with the known ligand FliC (29), purified FljB stimulated similar levels of TLR5-dependent NF-κB activation in CHO cells (Fig. 5B). Considering that Salmonella vesicles also contain the TLR4 ligand LPS (11, 60), these results demonstrate that natural antigens recognized by Salmonella-specific CD4+ T cells are TLR5 ligands (FliC or FljB flagellin) or are intimately associated with TLR-2 and -4 ligands (lipoprotein [Fig. 5A] and LPS [Fig. 3C], respectively) in MVs.
FIG. 5.
Bacterial surface organelles contain natural antigens recognized by TLRs. CHO cells transiently transfected with expression vectors for TLR2 (A) or TLR5 (B), together with an NF-κB luciferase reporter, were used to assess the ability of MVs and purified FljB to serve as TLR ligands. The data shown are representative of three experiments.
Genetically coordinated surface modifications reduce expression of important bacterial antigens.
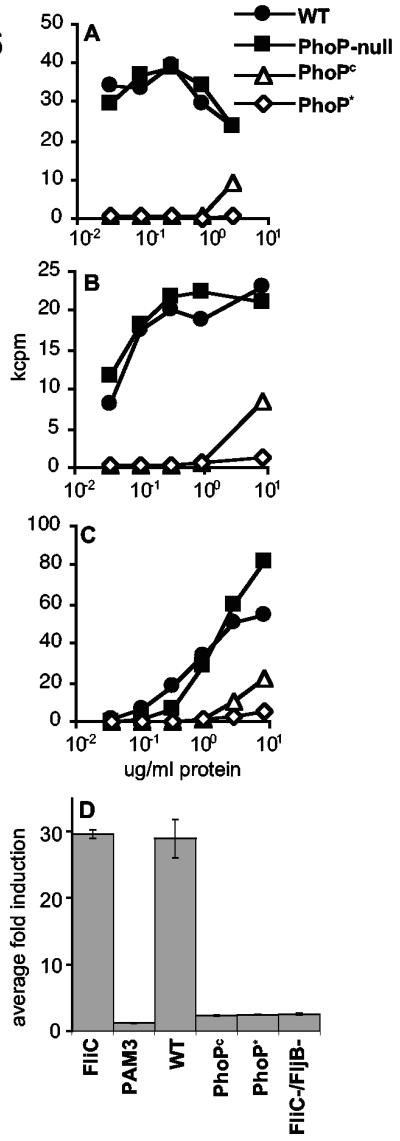
The two-component regulatory system PhoP/PhoQ, composed of the sensor kinase PhoQ and the response regulator PhoP, both activates and represses bacterial gene expression in response to environmental signals (24) such as those encountered by salmonellae during growth inside host phagocytic cells. PhoP/PhoQ activation regulates virulence and increases resistance to innate immunity (18, 24) via bacterial surface modifications that promote resistance to cationic antimicrobial peptides (19, 25, 28) and render LPS less immunostimulatory (27). Given that bacterial surface organelles contain natural antigens recognized by CD4+ T cells responding to Salmonella infection, we investigated the influence of PhoP/PhoQ on the production of those antigens. Salmonella strains with mutations that increase the active phosphorylated form of PhoP (PhoPc and PhoP*) (14, 26, 42, 52) provide a genetic means to examine antigen production by bacteria during PhoP/PhoQ activation. T-cell proliferation assays with clones specific for proteins expressed in surface organelles revealed a 100- to 10,000-fold reduction in stimulatory activity of PhoPc and PhoP* salmonellae relative to wild-type and PhoP− strains (Fig. 6A to C). Similarly, TLR5-dependent NF-κB activation by wild-type and PhoP− strains was equivalent (data not shown). However, the PhoPc and PhoP* mutations eliminated TLR5 stimulation (indistinguishable from that of a nonflagellated strain; Fig. 6D). These data were confirmed by Western analysis of the same strains for FliC production (data not shown), as well as other studies (1). This is the first demonstration of PhoP/PhoQ regulation of antigens recognized by both the innate and adaptive immune systems and suggests that salmonellae may have evolved a means to coordinately reduce production of ligands for both the innate and adaptive immune systems in order to facilitate their growth in host tissues.
FIG. 6.
PhoP/PhoQ negatively regulates the expression of natural Salmonella antigens recognized by CD4+ T cells and TLRs. Wild-type (WT) bacteria were compared with PhoP− and PhoP-activated (PhoPc, PhoP*) strains for the ability to stimulate FljB-specific clone D3D6 (A), FliC-specific clone 3A7 (B), and MV-specific T-cell clone F4 (C) or TLR5-dependent NF-κB activation in CHO cells (D). Representative data from multiple experiments are shown.
DISCUSSION
We have shown that during infection with S. enterica serovar Typhimurium, T-cell responses to natural antigens expressed in the bacterial surface organelles flagella and MVs are generated. Antigens recognized by Salmonella-specific T cells directly stimulate TLRs or are intrinsically associated with TLR ligands. In addition, salmonellae repress antigen expression via PhoP/PhoQ, the two-component regulatory system that controls resistance to innate immunity. These results suggest that genetically coordinated surface modifications enhance Salmonella growth in vivo by diminishing both innate and adaptive immune recognition.
Many bacterial pathogens express surface organelles like flagella and MVs (11, 46, 53). Recognition of such conserved surface structures is an obvious benefit to the host, but this receptor-ligand interaction may also be exploited by pathogens in various ways as a virulence strategy. For example, TLR-initiated innate immune responses could recruit cells permissive for bacterial replication (47) to the site of infection (37). Indeed, pathogens such as shigellae and mycobacteria, which infect and replicate within eukaryotic cells, actively secrete lipoproteins stimulatory for TLR2 (3, 13). Other pathogens may use TLR recognition of their surface organelles to deliver specific virulence factors without the need for dedicated, toxin-specific host receptors. For example, delivery of heat-labile enterotoxin (34) and ClyA cytotoxin (71) from Escherichia coli and packaging of virulence proteins like the hemolysins and proteases expressed by Pseudomonas aeruginosa (39, 40) rely on MVs. Finally, intoxication of host cells or inappropriate stimulation of inflammatory responses may favor bacterial colonization and replication in vivo. S. enterica serovar Typhimurium uses such a strategy by activating the caspase-1 protease in macrophages, initiating an inflammatory cell death (12) required for Salmonella infection in vivo (55). Thus, microbes appear to have evolved a subset of pathogenic mechanisms specifically to exploit host immune recognition.
As an alternative to exploitation, bacterial pathogens may also use multiple strategies to avoid innate and adaptive immune responses. For example, P. aeruginosa successfully colonizes the lungs of cystic fibrosis patients in part by growing as a biofilm, a virtual mat of microbes with increased resistance to opsonization and phagocytosis (17). LPS signaling through TLR4 has recently been shown to play an important role in the generation of both innate and adaptive immune responses to Salmonella infection (70), and persistence of Helicobacter pylori in infected hosts is thought to be facilitated by reduced signaling through TLR4 (7, 66) and TLR5 (23, 44). During malarial infection, the host humoral immune response targets malarial antigens expressed at the surface of infected red blood cells; plasmodia counter by expressing different alleles to generate antigenic variants that exhibit minimal immunological cross-reactivity (43). CD8+ T-cell responses are important for viral immunity, and herpes simplex virus, for example, has accordingly evolved mechanisms to interfere with class I MHC antigen presentation by blocking peptide translocation into the endoplasmic reticulum (72). Regulation of antigen production represents a new contribution to our understanding of the immune evasion mechanisms used by microbial pathogens. We demonstrate that activation of the PhoP/PhoQ regulon represses expression of surface antigens recognized by TLRs, as well as Salmonella-specific CD4+ T cells. Failure to repress FliC expression by PhoP− Salmonella strains could therefore contribute to the generation of protective immune responses by these strains in murine and human hosts (19, 21, 30, 51, 69). Similarly, PhoP/PhoQ-coordinated surface modifications by virulent bacteria may facilitate chronic Salmonella persistence in humans (45, 59), exemplified by Typhoid Mary (61).
Additional experimental evidence confirms the dynamic nature of the interaction of salmonellae with their hosts and supports the idea that genetically coordinated surface modifications facilitate bacterial growth in vivo by diminishing innate and adaptive immune recognition. PhoP− S. enterica serovar Typhimurium colonizes Peyer's patches after oral infection but fail to spread systemically to the liver and spleen and are attenuated for virulence (21). PhoP− strains are also unable to complete other surface modifications, such as those that reduce the immunostimulatory capacity of their LPS (27) or down-regulate expression of critical antigens recognized by T cells (as shown here). These observations underscore the importance of multiple mechanisms that contribute to the successful elimination of PhoP− salmonellae by host responses: TLR4 activation by LPS for limiting systemic Salmonella growth (58, 70), the significant proinflammatory stimulus provided by TLR5 when bacterial flagella are translocated across mucosal surfaces (73), and the recognition of flagella by the adaptive immune systems of humans (68) and mice (15, 49) orally infected with salmonellae.
In contrast to the limited dissemination of PhoP− bacteria, PhoPc S. enterica serovar Typhimurium is capable of spreading systemically in infected mice (32). Therefore the activation of PhoP/PhoQ, which directs repression and modification of surface ligands and reduces bacterial recognition by the innate and adaptive immune responses, appears to be one contributing factor that affords these mutants the ability to travel beyond the gastrointestinal mucosa after oral infection. It is also noteworthy that PhoPc mutants are attenuated and potently immunogenic and can persist in dendritic cells (32). Considering that the dendritic cell is the tour de force APC in the immunological organ system, it is no surprise that the host, with an almost infinite number of antigenic specificities among immunoglobulins and a T-cell compartment with the capacity of generating at least 1018 different receptors (16), mounts an immune response and eliminates infections caused by PhoPc S. enterica serovar Typhimurium. Further, unlike wild-type salmonellae, PhoPc bacteria fail to invade and replicate in epithelial cells (10) and macrophages (52). These host cells are less potent activators of immune responses and provide critical havens for Salmonella replication: the inability of naive susceptible mice to control infections caused by wild-type salmonellae suggests that evasion of innate and adaptive immune responses may be one reason such sites are important for the pathogenesis of virulent infections. Finally, it is clear that effective “cloaking devices” are functional and biologically relevant because wild-type salmonellae can persistently colonize the mesenteric lymph nodes of genetically resistant mice (54).
Comprehensive identification of bacterial antigens may further solidify the intriguing connection between innate and adaptive immune recognition (16). Irrespective of antigen identity, however, our results indicate that localization in surface organelles and coordinate regulation by PhoP/Q are important features of natural bacterial antigens recognized in vivo. The existence of genetically programmed responses of the pathogen to resist innate and adaptive immune recognition supports the proposal that microbial pathogenesis has been shaped by the host immune system. That is, a coevolutionary dynamic exists in which coordinated immune recognition of microbial antigens selects for pathogens capable of exploiting, modulating, or evading that recognition in order to successfully colonize and infect their hosts.
Acknowledgments
We thank Robert Alaniz for useful discussions and critical review of the manuscript.
This work was supported by Public Health Service National Research Service award T32 GM07270 from the National Institute of General Medical Sciences (M.A.B.) and by National Institutes of Health grant A147242 (B.T.C.).
Editor: V. J. DiRita
REFERENCES
- 1.Adams, P., R. Fowler, N. Kinsella, G. Howell, M. Farris, P. Coote, and C. D. O'Connor. 2001. Proteomic detection of PhoPQ- and acid-mediated repression of Salmonella motility. Proteomics 1:597-607. [DOI] [PubMed] [Google Scholar]
- 2.Aldridge, P., and K. T. Hughes. 2002. Regulation of flagellar assembly. Curr. Opin. Microbiol. 5:160-165. [DOI] [PubMed] [Google Scholar]
- 3.Aliprantis, A. O., D. S. Weiss, J. D. Radolf, and A. Zychlinsky. 2001. Release of Toll-like receptor-2-activating bacterial lipoproteins in Shigella flexneri culture supernatants. Infect. Immun. 69:6248-6255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aliprantis, A. O., R. B. Yang, M. R. Mark, S. Suggett, B. Devaux, J. D. Radolf, G. R. Klimpel, P. Godowski, and A. Zychlinsky. 1999. Cell activation and apoptosis by bacterial lipoproteins through toll-like receptor-2. Science 285:736-739. [DOI] [PubMed] [Google Scholar]
- 5.Allan, N. D., C. Kooi, P. A. Sokol, and T. J. Beveridge. 2003. Putative virulence factors are released in association with membrane vesicles from Burkholderia cepacia. Can. J. Microbiol. 49:613-624. [DOI] [PubMed] [Google Scholar]
- 6.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, and K. Struhl. 1993. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 7.Backhed, F., B. Rokbi, E. Torstensson, Y. Zhao, C. Nilsson, D. Seguin, S. Normark, A. M. Buchan, and A. Richter-Dahlfors. 2003. Gastric mucosal recognition of Helicobacter pylori is independent of Toll-like receptor 4. J. Infect. Dis. 187:829-836. [DOI] [PubMed] [Google Scholar]
- 8.Beatty, W. L., E. R. Rhoades, H. J. Ullrich, D. Chatterjee, J. E. Heuser, and D. G. Russell. 2000. Trafficking and release of mycobacterial lipids from infected macrophages. Traffic 1:235-247. [DOI] [PubMed] [Google Scholar]
- 9.Beermann, C., H. Wunderli-Allenspach, P. Groscurth, and L. Filgueira. 2000. Lipoproteins from Borrelia burgdorferi applied in liposomes and presented by dendritic cells induce CD8+ T-lymphocytes in vitro. Cell. Immunol. 201:124-131. [DOI] [PubMed] [Google Scholar]
- 10.Behlau, I., and S. I. Miller. 1993. A PhoP-repressed gene promotes Salmonella typhimurium invasion of epithelial cells. J. Bacteriol. 175:4475-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beveridge, T. J. 1999. Structures of gram-negative cell walls and their derived membrane vesicles. J. Bacteriol. 181:4725-4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brennan, M. A., and B. T. Cookson. 2000. Salmonella induces macrophage death by caspase-1-dependent necrosis. Mol. Microbiol. 38:31-40. [DOI] [PubMed] [Google Scholar]
- 13.Brightbill, H. D., D. H. Libraty, S. R. Krutzik, R. B. Yang, J. T. Belisle, J. R. Bleharski, M. Maitland, M. V. Norgard, S. E. Plevy, S. T. Smale, P. J. Brennan, B. R. Bloom, P. J. Godowski, and R. L. Modlin. 1999. Host defense mechanisms triggered by microbial lipoproteins through toll-like receptors. Science 285:732-736. [DOI] [PubMed] [Google Scholar]
- 14.Chamnongpol, S., and E. A. Groisman. 2000. Acetyl phosphate-dependent activation of a mutant PhoP response regulator that functions independently of its cognate sensor kinase. J. Mol. Biol. 300:291-305. [DOI] [PubMed] [Google Scholar]
- 15.Cookson, B. T., and M. J. Bevan. 1997. Identification of a natural T cell epitope presented by Salmonella-infected macrophages and recognized by T cells from orally immunized mice. J. Immunol. 158:4310-4319. [PubMed] [Google Scholar]
- 16.Cookson, B. T., L. A. Cummings, and S. L. Rassoulian Barrett. 2001. Bacterial antigens elicit T cell responses via adaptive and transitional immune recognition. Curr. Opin. Microbiol. 4:267-273. [DOI] [PubMed] [Google Scholar]
- 17.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 18.Ernst, R. K., T. Guina, and S. I. Miller. 2001. Salmonella typhimurium outer membrane remodeling: role in resistance to host innate immunity. Microbes Infect. 3:1327-1334. [DOI] [PubMed] [Google Scholar]
- 19.Fields, P. I., E. A. Groisman, and F. Heffron. 1989. A Salmonella locus that controls resistance to microbicidal proteins from phagocytic cells. Science 243:1059-1062. [DOI] [PubMed] [Google Scholar]
- 20.Fiocca, R., V. Necchi, P. Sommi, V. Ricci, J. Telford, T. L. Cover, and E. Solcia. 1999. Release of Helicobacter pylori vacuolating cytotoxin by both a specific secretion pathway and budding of outer membrane vesicles. Uptake of released toxin and vesicles by gastric epithelium. J. Pathol. 188:220-226. [DOI] [PubMed] [Google Scholar]
- 21.Galan, J. E., and R. Curtiss III. 1989. Virulence and vaccine potential of phoP mutants of Salmonella typhimurium. Microb. Pathog. 6:433-443. [DOI] [PubMed] [Google Scholar]
- 22.Garcia-del Portillo, F., M. A. Stein, and B. B. Finlay. 1997. Release of lipopolysaccharide from intracellular compartments containing Salmonella typhimurium to vesicles of the host epithelial cell. Infect. Immun. 65:24-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gewirtz, A. T., Y. Yu, U. S. Krishna, D. A. Israel, S. L. Lyons, and R. M. Peek, Jr. 2004. Helicobacter pylori flagellin evades toll-like receptor 5-mediated innate immunity. J. Infect. Dis. 189:1914-1920. [DOI] [PubMed] [Google Scholar]
- 24.Groisman, E. A. 2001. The pleiotropic two-component regulatory system PhoP-PhoQ. J. Bacteriol. 183:1835-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guina, T., E. C. Yi, H. Wang, M. Hackett, and S. I. Miller. 2000. A PhoP-regulated outer membrane protease of Salmonella enterica serovar Typhimurium promotes resistance to alpha-helical antimicrobial peptides. J. Bacteriol. 182:4077-4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gunn, J. S., E. L. Hohmann, and S. I. Miller. 1996. Transcriptional regulation of Salmonella virulence: a PhoQ periplasmic domain mutation results in increased net phosphotransfer to PhoP. J. Bacteriol. 178:6369-6373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo, L., K. B. Lim, J. S. Gunn, B. Bainbridge, R. P. Darveau, M. Hackett, and S. I. Miller. 1997. Regulation of lipid A modifications by Salmonella typhimurium virulence genes phoP-phoQ. Science 276:250-253. [DOI] [PubMed] [Google Scholar]
- 28.Guo, L., K. B. Lim, C. M. Poduje, M. Daniel, J. S. Gunn, M. Hackett, and S. I. Miller. 1998. Lipid A acylation and bacterial resistance against vertebrate antimicrobial peptides. Cell 95:189-198. [DOI] [PubMed] [Google Scholar]
- 29.Hayashi, F., K. D. Smith, A. Ozinsky, T. R. Hawn, E. C. Yi, D. R. Goodlett, J. K. Eng, S. Akira, D. M. Underhill, and A. Aderem. 2001. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature 410:1099-1103. [DOI] [PubMed] [Google Scholar]
- 30.Hohmann, E. L., C. A. Oletta, K. P. Killeen, and S. I. Miller. 1996. phoP/phoQ-deleted Salmonella typhi (Ty800) is a safe and immunogenic single-dose typhoid fever vaccine in volunteers. J. Infect. Dis. 173:1408-1414. [DOI] [PubMed] [Google Scholar]
- 31.Hoiseth, S. K., and B. A. Stocker. 1981. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature 291:238-239. [DOI] [PubMed] [Google Scholar]
- 32.Hopkins, S. A., F. Niedergang, I. E. Corthesy-Theulaz, and J. P. Kraehenbuhl. 2000. A recombinant Salmonella typhimurium vaccine strain is taken up and survives within murine Peyer's patch dendritic cells. Cell Microbiol. 2:59-68. [DOI] [PubMed] [Google Scholar]
- 33.Horstman, A. L., and M. J. Kuehn. 2002. Bacterial surface association of heat-labile enterotoxin through lipopolysaccharide after secretion via the general secretory pathway. J. Biol. Chem. 277:32538-32545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Horstman, A. L., and M. J. Kuehn. 2000. Enterotoxigenic Escherichia coli secretes active heat-labile enterotoxin via outer membrane vesicles. J. Biol. Chem. 275:12489-12496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Janeway, C. A., Jr., and R. Medzhitov. 2002. Innate immune recognition. Annu. Rev. Immunol. 20:197-216. [DOI] [PubMed] [Google Scholar]
- 36.Jones, B. D., and S. Falkow. 1996. Salmonellosis: host immune responses and bacterial virulence determinants. Annu. Rev. Immunol. 14:533-561. [DOI] [PubMed] [Google Scholar]
- 37.Jones, B. D., N. Ghori, and S. Falkow. 1994. Salmonella typhimurium initiates murine infection by penetrating and destroying the specialized epithelial M cells of the Peyer's patches. J. Exp. Med. 180:15-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kadurugamuwa, J. L., and T. J. Beveridge. 1998. Delivery of the non-membrane-permeative antibiotic gentamicin into mammalian cells by using Shigella flexneri membrane vesicles. Antimicrob. Agents Chemother. 42:1476-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kadurugamuwa, J. L., and T. J. Beveridge. 1997. Natural release of virulence factors in membrane vesicles by Pseudomonas aeruginosa and the effect of aminoglycoside antibiotics on their release. J. Antimicrob. Chemother. 40:615-621. [DOI] [PubMed] [Google Scholar]
- 40.Kadurugamuwa, J. L., and T. J. Beveridge. 1995. Virulence factors are released from Pseudomonas aeruginosa in association with membrane vesicles during normal growth and exposure to gentamicin: a novel mechanism of enzyme secretion. J. Bacteriol. 177:3998-4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kato, S., Y. Kowashi, and D. R. Demuth. 2002. Outer membrane-like vesicles secreted by Actinobacillus actinomycetemcomitans are enriched in leukotoxin. Microb. Pathog. 32:1-13. [DOI] [PubMed] [Google Scholar]
- 42.Kier, L. D., R. M. Weppelman, and B. N. Ames. 1979. Regulation of nonspecific acid phosphatase in Salmonella: phoN and phoP genes. J. Bacteriol. 138:155-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kyes, S., P. Horrocks, and C. Newbold. 2001. Antigenic variation at the infected red cell surface in malaria. Annu. Rev. Microbiol. 55:673-707. [DOI] [PubMed] [Google Scholar]
- 44.Lee, S. K., A. Stack, E. Katzowitsch, S. I. Aizawa, S. Suerbaum, and C. Josenhans. 2003. Helicobacter pylori flagellins have very low intrinsic activity to stimulate human gastric epithelial cells via TLR5. Microbes Infect. 5:1345-1356. [DOI] [PubMed] [Google Scholar]
- 45.Levine, M. M., R. E. Black, and C. Lanata. 1982. Precise estimation of the numbers of chronic carriers of Salmonella typhi in Santiago, Chile, an endemic area. J. Infect. Dis. 146:724-726. [DOI] [PubMed] [Google Scholar]
- 46.Li, Z., A. J. Clarke, and T. J. Beveridge. 1998. Gram-negative bacteria produce membrane vesicles which are capable of killing other bacteria. J. Bacteriol. 180:5478-5483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lowrie, D. B., V. R. Aber, and M. E. Carrol. 1979. Division and death rates of Salmonella typhimurium inside macrophages: use of penicillin as a probe. J. Gen. Microbiol. 110:409-419. [DOI] [PubMed] [Google Scholar]
- 48.Mastroeni, P., B. Villarreal-Ramos, and C. E. Hormaeche. 1993. Adoptive transfer of immunity to oral challenge with virulent salmonellae in innately susceptible BALB/c mice requires both immune serum and T cells. Infect. Immun. 61:3981-3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McSorley, S. J. , B. T. Cookson, and M. K. Jenkins. 2000. Characterization of CD4+ T cell responses during natural infection with Salmonella typhimurium. J. Immunol. 164:986-993. [DOI] [PubMed] [Google Scholar]
- 50.Medzhitov, R., and C. A. Janeway, Jr. 1998. Innate immune recognition and control of adaptive immune responses. Semin. Immunol. 10:351-353. [DOI] [PubMed] [Google Scholar]
- 51.Miller, S. I., A. M. Kukral, and J. J. Mekalanos. 1989. A two-component regulatory system (phoP phoQ) controls Salmonella typhimurium virulence. Proc. Natl. Acad. Sci. USA 86:5054-5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miller, S. I., and J. J. Mekalanos. 1990. Constitutive expression of the PhoP regulon attenuates Salmonella virulence and survival within macrophages. J. Bacteriol. 172:2485-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moens, S., and J. Vanderleyden. 1996. Functions of bacterial flagella. Crit. Rev. Microbiol. 22:67-100. [DOI] [PubMed] [Google Scholar]
- 54.Monack, D. M., D. M. Bouley, and S. Falkow. 2004. Salmonella typhimurium persists within macrophages in the mesenteric lymph nodes of chronically infected Nramp1+/+ mice and can be reactivated by IFNγ neutralization. J. Exp. Med. 199:231-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Monack, D. M., D. Hersh, N. Ghori, D. Bouley, A. Zychlinsky, and S. Falkow. 2000. Salmonella exploits caspase-1 to colonize Peyer's patches in a murine typhoid model. J. Exp. Med. 192:249-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Neidhardt, F. C., J. L. Ingraham, and M. Schaechter. 1990. Physiology of the bacterial cell: a molecular approach. Sinauer Associates, Sunderland, Mass.
- 57.Neyrolles, O., K. Gould, M. P. Gares, S. Brett, R. Janssen, P. O'Gaora, J. L. Herrmann, M. C. Prevost, E. Perret, J. E. Thole, and D. Young. 2001. Lipoprotein access to MHC class I presentation during infection of murine macrophages with live mycobacteria. J. Immunol. 166:447-457. [DOI] [PubMed] [Google Scholar]
- 58.O'Brien, A. D., D. L. Rosenstreich, I. Scher, G. H. Campbell, R. P. MacDermott, and S. B. Formal. 1980. Genetic control of susceptibility to Salmonella typhimurium in mice: role of the LPS gene. J. Immunol. 124:20-24. [PubMed] [Google Scholar]
- 59.Parry, C. M., T. T. Hien, G. Dougan, N. J. White, and J. J. Farrar. 2002. Typhoid fever. N. Engl. J. Med. 347:1770-1782. [DOI] [PubMed] [Google Scholar]
- 60.Poltorak, A., X. He, I. Smirnova, M. Y. Liu, C. V. Huffel, X. Du, D. Birdwell, E. Alejos, M. Silva, C. Galanos, M. Freudenberg, P. Ricciardi-Castagnoli, B. Layton, and B. Beutler. 1998. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science 282:2085-2088. [DOI] [PubMed] [Google Scholar]
- 61.Salyers, A. A., and D. D. Whitt. 2001. Bacterial pathogenesis: a molecular approach, second ed. ASM Press, Washington, D.C.
- 62.Schagger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 63.Schaible, U. E., K. Hagens, K. Fischer, H. L. Collins, and S. H. Kaufmann. 2000. Intersection of group I CD1 molecules and mycobacteria in different intracellular compartments of dendritic cells. J. Immunol. 164:4843-4852. [DOI] [PubMed] [Google Scholar]
- 63a.Schindler, U., and V. R. Baichwal. 1994. Three NF-κB binding sites in the human E-selectin gene required for maximal tumor necrosis factor alpha-induced expression. Mol. Cell. Biol. 14:5820-5831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schnare, M., G. M. Barton, A. C. Holt, K. Takeda, S. Akira, and R. Medzhitov. 2001. Toll-like receptors control activation of adaptive immune responses. Nat. Immunol. 2:947-950. [DOI] [PubMed] [Google Scholar]
- 65.Smith, K. D., and A. Ozinsky. 2002. Toll-like receptor-5 and the innate immune response to bacterial flagellin. Curr. Top. Microbiol. Immunol. 270:93-108. [DOI] [PubMed] [Google Scholar]
- 66.Smith, M. F., Jr., A. Mitchell, G. Li, S. Ding, A. M. Fitzmaurice, K. Ryan, S. Crowe, and J. B. Goldberg. 2003. Toll-like receptor (TLR) 2 and TLR5, but not TLR4, are required for Helicobacter pylori-induced NF-κB activation and chemokine expression by epithelial cells. J. Biol. Chem. 278:32552-32560. [DOI] [PubMed] [Google Scholar]
- 67.Stocker, B. A. D., and J. C. Campbell. 1959. The effect of non-lethal deflagellation on bacterial motility and observations on flagellar regeneration. J. Gen. Microbiol. 20:670. [DOI] [PubMed] [Google Scholar]
- 68.Sztein, M. B., S. S. Wasserman, C. O. Tacket, R. Edelman, D. Hone, A. A. Lindberg, and M. M. Levine. 1994. Cytokine production patterns and lymphoproliferative responses in volunteers orally immunized with attenuated vaccine strains of Salmonella typhi. J. Infect. Dis. 170:1508-1517. [DOI] [PubMed] [Google Scholar]
- 69.VanCott, J. L., S. N. Chatfield, M. Roberts, D. M. Hone, E. L. Hohmann, D. W. Pascual, M. Yamamoto, H. Kiyono, and J. R. McGhee. 1998. Regulation of host immune responses by modification of Salmonella virulence genes. Nat. Med. 4:1247-1252. [DOI] [PubMed] [Google Scholar]
- 70.Vazquez-Torres, A., B. A. Vallance, M. A. Bergman, B. B. Finlay, B. T. Cookson, J. Jones-Carson, and F. C. Fang. 2004. Toll-like receptor 4 dependence of innate and adaptive immunity to Salmonella: importance of the Kupffer cell network. J. Immunol. 172:6202-6208. [DOI] [PubMed] [Google Scholar]
- 71.Wai, S. N., B. Lindmark, T. Soderblom, A. Takade, M. Westermark, J. Oscarsson, J. Jass, A. Richter-Dahlfors, Y. Mizunoe, and B. E. Uhlin. 2003. Vesicle-mediated export and assembly of pore-forming oligomers of the enterobacterial ClyA cytotoxin. Cell 115:25-35. [DOI] [PubMed] [Google Scholar]
- 72.Yewdell, J. W., and A. B. Hill. 2002. Viral interference with antigen presentation. Nat. Immunol. 3:1019-1025. [DOI] [PubMed] [Google Scholar]
- 73.Zeng, H., A. Q. Carlson, Y. Guo, Y. Yu, L. S. Collier-Hyams, J. L. Madara, A. T. Gewirtz, and A. S. Neish. 2003. Flagellin is the major proinflammatory determinant of enteropathogenic Salmonella. J. Immunol. 171:3668-3674. [DOI] [PubMed] [Google Scholar]