Abstract
Helicobacter pylori causes disease in the human stomach and in mouse and gerbil stomach models. Previous results have shown that motility is critical for H. pylori to colonize mice, gerbils, and other animal models. The role of chemotaxis, however, in colonization and disease is less well understood. Two genes in the H. pylori chemotaxis pathway, cheY and tlpB, which encode the chemotaxis response regulator and a methyl-accepting chemoreceptor, respectively, were disrupted. The cheY mutation was complemented with a wild-type copy of cheY inserted into the chromosomal rdxA gene. The cheY mutant lost chemotaxis but retained motility, while all other strains were motile and chemotactic in vitro. These strains were inoculated into gerbils either alone or in combination with the wild-type strain, and colonization and inflammation were assessed. While the cheY mutant completely failed to colonize gerbil stomachs, the tlpB mutant colonized at levels similar to those of the wild type. With the tlpB mutant, there was a substantial decrease in inflammation in the gerbil stomach compared to that with the wild type. Furthermore, there were differences in the numbers of each immune cell in the tlpB-mutant-infected stomach: the ratio of lymphocytes to neutrophils was about 8 to 1 in the wild type but only about 1 to 1 in the mutant. These results suggest that the TlpB chemoreceptor plays an important role in the inflammatory response while the CheY chemotaxis regulator plays a critical role in initial colonization. Chemotaxis mutants may provide new insights into the steps involved in H. pylori pathogenesis.
Helicobacter pylori infects gastric tissue in humans and can lead to peptic ulcers, gastric adenocarcinoma, and mucosa-associated lymphoid tumors (25). Many people worldwide are infected, but only a subset develops disease. H. pylori faces formidable challenges upon entering the stomach. The organism must overcome innate defenses including gastric acid, gastric epithelial cell exfoliation, and peristalsis. To overcome these obstacles, H. pylori responds rapidly to chemical signals provided by the host. Some of these poorly defined signals are proposed to direct the organism to swim to the mucous layer and gastric epithelial cell surface and then promote bacterial attachment; failure to carry out these pivotal steps may result in clearance of the organism from the stomach. In support of this model, motility and flagella are essential for H. pylori infection of piglet, mouse, and gerbil stomachs (11, 13, 23, 29).
Chemotaxis, the ability of microorganisms to move in response to chemical cues, is widespread within the prokaryotic world, but its role in mammalian infection is poorly understood (19, 30). Motility and chemotaxis both promote initial H. pylori colonization, but nonchemotactic mutants are able to infect, albeit with various degrees of attenuation (3, 11, 15, 29, 39). The study of chemotaxis in vivo, therefore, may allow us to better dissect the steps of pathogenesis, because nonchemotactic mutants may be deficient in a specific subset of pathogenesis mechanisms.
Chemotaxis has been extensively studied in Escherichia coli (4, 6). In this microbe, chemoreceptors (also known as methyl-accepting chemotaxis proteins) sense environmental cues such as amino acids and transmit the ligand-binding information to a signal transduction cascade that affects the direction of flagellar rotation. The core signal transduction proteins consist of the receptor-kinase coupling and adapter protein (CheW), the kinase (CheA), and the CheY response regulator, which interacts with the flagellar motor in its phosphorylated state. When an attractant ligand binds to the chemoreceptor, the kinase activity of CheA is squelched and unphosphorylated CheY predominates. In this form, CheY fails to interact with the flagellar motor, the flagella rotate counterclockwise, and the bacteria swim straight. When no ligand is bound to the chemoreceptor, the CheA kinase is active and CheY is phosphorylated (CheY-P) and interacts with the flagellar motor. In the presence of CheY-P, the flagellar motor rotates clockwise and the bacteria randomly reorient in space by using a behavior called tumbling.
H. pylori chemotaxis appears to be comprised of some similar components, as well as unique features, compared with E. coli (2, 28, 40). H. pylori has four chemoreceptors: TlpA, TlpB, TlpC, and HlyB. TlpA appears to sense arginine and bicarbonate (7), but the ligands of the other three chemoreceptors are unknown. H. pylori has CheW, CheA, and CheY homologues, and all of these have the predicted chemotaxis phenotypes based on the E. coli model (5, 15, 31). H. pylori additionally has three proteins that are hybrids of CheW and CheY, called CheVs. While Bacillus subtilis CheV is redundant with CheW (32), none of the H. pylori CheVs are redundant with its CheW (31), suggesting that H. pylori may have a number of unique features in its chemotaxis pathway.
Analysis of nonchemotactic mutants has shown that chemotaxis is important for H. pylori animal infection, although the degree that these mutants are impaired varies between different reports. One group reported that mutants lacking cheA or cheY were totally unable to infect mice and piglets (15), while some of us (39) have observed that mutants lacking cheA, cheY, or cheW infect mice but not as well as the wild type. To attempt to resolve these differences, a third model was sought to examine how H. pylori mutants defective in different parts of the chemotaxis pathway would behave. Gerbils have become a popular model for H. pylori pathogenesis studies because infection in these animals mimics the human-like spectrum of symptoms, including severe gastritis, ulcers, and cancer (17, 42). We show here that nonchemotactic cheY mutants are unable to infect gerbils, while cheY mutants complemented with a wild-type copy of cheY regain infection ability. In addition, a tlpB chemoreceptor mutant infects gerbils to levels that are indistinguishable from the wild type but causes significantly less gastritis.
MATERIALS AND METHODS
H. pylori strains, growth conditions, and general molecular biology.
For these studies, we used H. pylori strain SS1 for genetics, chemotaxis, and animal studies (21) or E. coli DH10B for plasmid cloning and propagation. For solid-medium culture during mutant construction, H. pylori was grown on Columbia blood agar (Difco) plates with 5% defibrinated horse blood (Hemostat Labs, Davis, Calif.), 5 μg of trimethoprim/ml, 8 μg of amphotericin B/ml, 10 μg of vancomycin/ml, 50 μg of cycloheximide/ml, 5 μg of cefsulodin/ml, 2.5 μg of polymyxin B/ml, and 0.2% (wt/vol) β-cyclodextrin (Sigma) (CHBA) at 37°C under 7 to 10% O2, 10% CO2, and 80 to 83% N2. All antibiotics were from Sigma or ISC Bioexpress. Where appropriate, chloramphenicol was used at 5 to 10 μg/ml (H. pylori) or 20 μg/ml (E. coli) and metronidazole was used at 9 μg/ml.
Plasmids were prepared using QIAGEN kits. For preparation of genomic DNA, DNeasy kits (QIAGEN) or Wizard Genomic kits (Promega) were used. All restriction and DNA modification enzymes were from New England Biolabs or Invitrogen. Amplification of DNA was carried out using Pfu-Turbo polymerase (Stratagene) or Taq polymerase (a generous gift of D. Kellogg). All DNA sequencing was performed at the University of California at Berkeley sequencing facility.
For gerbil inoculation, wild-type SS1, the isogenic tlpB and cheY mutants, and the cheY/cheY+-complemented strain were passaged an equal number of times from the frozen stock onto Campylobacter agar (Becton Dickinson, Sparks, Md.) containing 10% defibrinated sheep blood (Quad Five, Ryegate, Mont.). Strains were then grown overnight in a T25 cm2 tissue culture flask in a 5% CO2 atmosphere in humidified air without aeration in 5 ml of Ham's F-12 medium (Invitrogen) containing 2% heat-inactivated fetal bovine serum (FBS) plus vancomycin (10 μg/ml) and flucytosine (5 μg/ml). Strains were then diluted 1:40 in 40 ml of F-12 medium plus serum (2%), flucytosine (5 μg/ml), bacitracin (30 μg/ml), amphotericin B (5 μg/ml), vancomycin, and trimethoprim (10 μg/ml) in a T75 cm2 tissue culture flask and allowed to grow an additional 16 to 18 h. All strains grew equally well under these conditions and displayed motility. Bacteria were harvested by centrifugation, washed with 1× phosphate-buffered saline (PBS; Invitrogen), resuspended in 1 ml of PBS, and used for the inoculation of animals.
Assessment of motility and chemotaxis.
Motility was determined by phase-microscopic inspection of cultures (Olympus IMT-2). Motility was further evaluated using soft-agar plates composed of brucella broth, 5% FBS, 0.35% agar, and H. pylori-selective antibiotics. A small portion of the strain to be tested was stabbed about three-fourths of the way into the thickness of the agar, and the diameter of the bacterial halo was measured each day for 7 days. Formation of the halo in this assay depends on both motility and chemotaxis, although the chemotactic component of this medium is unknown.
Mutagenesis of cheY
The cheY gene (hp1067) was mutagenized using inverse PCR (iPCR) to create an in-frame deletion of the majority of cheY on pKO126 by using primers 126del1 and 126del2 (3). All plasmids are listed in Table 1, and primers are listed in Table 2. Because there are several genes downstream of cheY, we used a cat gene lacking transcriptional terminators to minimize polar effects. The terminatorless cat gene (cat-m) was digested from pCat-mut (39) with SmaI and HincII and ligated with the iPCR cheY product to create pKO127, in which the cat gene is oriented in the same direction as cheY. Plasmid pKO127 was verified by restriction enzyme analysis and DNA sequencing.
TABLE 1.
Plasmids used in this studya
| Plasmid | Characteristics | Reference or source |
|---|---|---|
| pBS | Cloning plasmid; Ap | Stratagene |
| pKO126 | pBS::cheYSS1; Ap | 3 |
| pBS-cat | pBS::cat; Ap Cm | 36 |
| pCat-Mut | pBS-cat-m; can separate cat from its transcriptional terminators; Ap Cm | 39 |
| pKO127 | pKO126 ΔcheY::cat-m; Ap Cm | 39 |
| pRdxA | pBC-SK with 5′ and 3′ regions of H. pylori rdxA locus flanking a polylinker; Cm | 38; J. Kusters |
| pLC292 | Ap-resistant pRdxA; Ap | 39 |
| pKO140 | pLC292::cheYSS1 | 39 |
| pTC-B101 | pBS::tlpBSS1 | This study |
| pTC-B112 | pTC-B101 ΔtlpB::cat | This study |
pBS, pBluescript KS.
TABLE 2.
Oligonucleotide primers used in this study
| Primer | Sequence (all 5′-3′) |
|---|---|
| 126del1 | CCAGTAGTTTCAAAGTGCTTC |
| 126del2 | CCAACGATTGAGTGTTAAAGCC |
| cheY1 | GAAGGGATCCTTACAAATAAGAACGCTC |
| LCcheY2 | GCTCTAGATCAATCGTTTGTCCCTAAAACAACC |
| rdxAstart | CGCCATTCTTGCAAGATGTTTG |
| rdxAend | CTCGCTTCTGCCACCCTCTT |
| cheY3 | GGAAGCTGCAGGTTTCTTTATCGTCAAACGC |
| cheY4 | GCTCATTGAACGCTCCATTTAGC |
| tlpB-10 | CGTGGGGTGCGCTGGCACTAC |
| tlpB-20 | CCGCCGGACAAAGGATTATC |
| tlpB-30 | CCCCCCTAAACCTAAAAGAGC |
| tlpB-40 | CATCGCTACGCATGTGAGTGG |
| HP102-1 | CGGGGCTAGCACGGATAGC |
| HP102-2 | CGCTCGCGCTAACCCCACC |
Plasmid pKO127 was used to transform H. pylori SS1 by natural transformation as described previously (10, 36). PCR was used to verify the chromosomal ΔcheY::cat-m architecture of chloramphenicol-resistant transformants (data not shown). In addition, reverse transcription-PCR showed that a gene downstream of cheY and transcribed from the same promoter, copA (5), was still transcribed at wild-type levels in the ΔcheY::cat-m mutants, confirming that this mutation is nonpolar (data not shown).
Complementation of cheY.
The cheY coding region and its promoter were amplified from pKO126 with primers cheY1 and LCcheY2. This PCR fragment was digested with BamHI and XbaI and cloned into BamHI- and XbaI-digested pLC292, an ampicillin-resistant version of pRdxA (38), to create pKO140. Plasmid pKO140 was verified by restriction enzyme analysis. This plasmid directs the integration of cheY to the rdxA locus (hp0954), where disruption is selectable by metronidazole resistance. Plasmid pKO140 treated with cell-free H. pylori SS1 wild-type extract (to methylate the DNA) (10) was used to transform H. pylori wild-type SS1 to metronidazole resistance. Genomic DNA from SS1 rdxA::cheY was used to transform SS1 ΔcheY::cat-m to metronidazole resistance as described above to create strain SS1 cheY/cheY+. Verification of the rdxA::cheY insertion and retention of the cheY::cat-m was done by PCR using primers that flank the insertion sites (rdxAstart with rdxAend and cheY3 with cheY4).
Cloning and mutagenesis of tlpB.
The tlpB gene (hp0103) with flanking sequences was amplified from H. pylori SS1 genomic DNA with primers tlpB-10 and tlpB-20 and with Pfu-Turbo polymerase. The 2.8-kb PCR product was ligated with EcoRV-cut pBluescript KS to create pTC-B101. Restriction enzyme analysis, DNA sequencing, and database comparison were used to confirm the construction of pTC-B101.
To generate the deletion of tlpB, we used primers tlpB-30 and tlpB-40 to perform iPCR, yielding a 4.1-kb product with a coding sequence for only 27 amino-terminal amino acids and 20 carboxy-terminal amino acids remaining. This product was ligated to cat from pBS-cat created by digestion with HincII to create pTC-B112 (cat has the same orientation as tlpB).
Plasmid pTC-B112 was transformed into H. pylori wild-type SS1, and chloramphenicol-resistant transformants containing ΔtlpB::cat were verified by PCR analysis (data not shown). Western blotting with an antichemoreceptor antibody demonstrated that SS1 tlpB mutants had lost an immunoreactive band (data not shown). This antibody was generated to the cytoplasmic domain of H. pylori TlpA and recognizes all H. pylori chemoreceptors (including TlpB) in the wild type (T. M. Andermann and K. M. Ottemann, unpublished data). Additionally, we verified that the tlpB mutation is not polar by determining that the downstream gene, hp0102, is expressed using reverse transcription-PCR analysis with primers HP102-1 and HP102-2 (data not shown).
Inoculation of gerbils, tissue processing, and recovery of H. pylori.
Animal experiments were carried out at the University of South Alabama with the approval of the Institutional Animal Care and Use Committee. A total of 42 female 3-month-old Mongolian gerbils (Meriones unguiculatus; Charles River Laboratories) were used in this study. Six animals per group were employed. Animals were inoculated with SS1 wild type, the tlpB mutant, the cheY mutant, the cheY-complemented strain (cheY/cheY+), 1:1 mixtures of wild type with the tlpB mutant, or 1:1 mixtures of wild type and the cheY mutant (coinfection experiments). Each animal was orally inoculated with ∼5 × 108 viable CFU/ml in a volume of 50 μl (exact doses inoculated into gerbils are shown in figure legends). Six animals received 50 μl of PBS to serve as uninfected controls. At 4 weeks postinfection, animals were euthanized by cervical dislocation. Stomachs were removed and dissected longitudinally along the greater curvature, and the chyme was removed. A small section of the antrum was removed for histology. The rest of the stomach was excised into multiple slices, weighed, and then homogenized (10 to 15 s at a setting of 3; Ultra-Turrax T25, IKA Works, Inc.) in 1.5 ml of sterile PBS. Stomach homogenates and dilutions thereof in PBS were plated for viable counts in duplicate on Campylobacter blood agar plates containing flucytosine (5 μg/ml), vancomycin (10 μg/ml), amphotericin B (5 μg/ml), bacitracin (30 μg/ml), polymyxin B (10 U/ml), and trimethoprim (10 μg/ml) to suppress normal flora. Plates were incubated for 5 days in an anaerobic jar containing CampyPak Plus (Becton Dickinson). Data are presented as CFU/g of stomach.
Histology.
A small portion of the distal end of the antrum was excised and fixed in 10% formalin. The tissue was embedded in paraffin, sectioned (5 μm thick; three consecutive sections), stained with hematoxylin and eosin stain, and evaluated in a blind fashion by a pathologist (J.E.C.). Each slide was read twice (independent sections) to ensure reproducibility, and identical grades were obtained in all cases. Grading of the antrum was conducted by two established methods. First, the method of Rugge et al. was used to assess the degree of gastric atrophy (34). Scores are as follows: 0, absent; 1, indefinite; 2, present. Second, the method of Eaton et al. was used to assess the degree of lymphocyte inflammation (12). Scores are as follows: 0, no infiltrates; 1, mild, multifocal infiltration; 2, mild, widespread infiltration; 3, mild, widespread, and moderate, multifocal infiltration; 4, moderate, widespread infiltration; 5, moderate, widespread, and severe, multifocal infiltration. Neutrophil infiltration was scored as present or absent. Pictures were taken using an Olympus C-5050 digital camera attached to an Olympus IMT-2 inverted phase-contrast microscope. Pictures were oriented apical side up, and all pictures were identically adjusted with brightness and contrast settings in Adobe Photoshop.
Stomach sections (from the above processing) from animals infected with the wild type or the tlpB mutant were analyzed further by counting 100 consecutive inflammatory cells in three fields per sample (magnification, ×400) and scoring each cell as a neutrophil, lymphocyte, or eosinophil. Fields containing inflammation were selected at random, and the histologist was blind to the genotype of the infecting strain. The microscope used was an Olympus BX40. Other inflammatory cell types were never or very rarely observed.
Statistical analysis of data.
For animals infected with the wild type or the tlpB mutant, the mean inflammatory scores were assessed only from animals that had been colonized with H. pylori to prevent undue skewing of the data. Statistical analysis of histology grading by atrophy and by inflammation was calculated by the Mann-Whitney U test using Instat 3.03 software (GraphPad Software, San Diego, Calif.). P < 0.05 was considered statistically significant.
The alternative Welch's two-tailed t test was used to determine whether a statistically significant difference occurred in the ratios of neutrophils to lymphocytes in wild-type versus tlpB mutant-infected gerbil stomach sections.
Colonization levels were analyzed statistically using the two-tailed t test (unpaired) at http://www.graphpad.com/quickcalcs/ttest1.cfm.
RESULTS
The SS1 cheY mutant is nonchemotactic, while the tlpB mutant and cheY/cheY+-complemented strain have normal chemotactic abilities
To analyze the role(s) of chemotaxis in a gerbil model of infection, we created mutants lacking either the cheY-encoded response regulator or one of the four chemoreceptors, TlpB. We chose to analyze strains lacking cheY because this mutant has previously been shown to be nonchemotactic (5) and thus would address the role of chemotaxis in gerbil infection. cheY mutants are nonchemotactic because they have uncoupled the chemoreceptors from the flagellar motor; the flagella still rotate and the bacteria still swim, but the microbes cannot change direction. We chose to analyze a mutant lacking tlpB because we were unable to find a phenotype associated with loss of this chemoreceptor in a murine model of infection (Y.-T. Chen and K. M. Ottemann, unpublished data) and thus wanted to see whether it might play a role in infection in another animal model. Both mutants have the majority of their corresponding open reading frames deleted and replaced by cat either with (tlpB) or without (cheY) the weak cat transcriptional terminator. These strains are called SS1 tlpB and SS1 cheY. In addition, we complemented the cheY mutant by placing a wild-type copy of cheY, with its own promoter and Shine-Dalgarno sequence, into the rdxA locus, as accomplished with other genes (38). This strain is called SS1 cheY/cheY+. We verified that both the tlpB and cheY mutations did not affect expression of the downstream gene, hp0102 or copA, respectively, by using reverse transcription-PCR (data not shown). All mutants retained wild-type motility as assessed by light and phase-contrast microscopy.
Using brucella broth plus FBS soft agar, we confirmed that mutants lacking cheY are nonchemotactic whereas SS1 cheY/cheY+ and tlpB retain wild-type chemotactic abilities (Fig. 1). Because all these mutants retain motility, the defect in colonial expansion is consistent with the cheY mutant having a defect in chemotaxis.
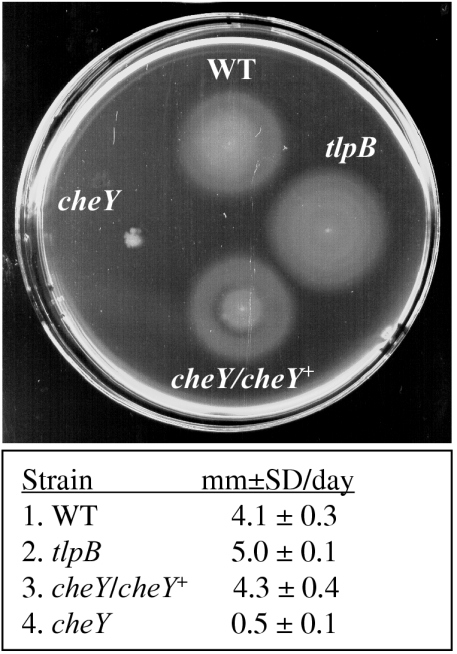
FIG. 1.
Chemotactic abilities of SS1, SS1 tlpB, SS1 cheY/cheY+, and SS1 cheY. Scanned image of a 5-day-old soft-agar plate. The colonial diameters (in millimeters [mm]) are as follows: for SS1, 21.0 ± 1.2; for SS1 tlpB, 25.75 ± 1; for SS1 cheY/cheY+, 21.25 ± 1.7; for SS1 cheY, 3.25 ± 0.5. The table below the image shows the colonial expansion rate over 5 days; data are the averages (in millimeters) ± standard deviations (SD) of three separate assays. WT, wild type.
The tlpB mutant retains the ability to infect at wild-type levels, while loss of cheY prevents infection.
We next examined the role of chemotaxis genes in the gerbil model of infection. Three-month-old female gerbils were infected with approximately 5 × 108 CFU of single strains (SS1 wild type or its isogenic cheY, tlpB, or cheY/cheY+ strains) or with coinfections of wild type plus cheY or wild type plus tlpB. After 4 weeks, the animals were euthanized, and the number of H. pylori in each stomach was enumerated. The cheY mutant was unable to infect any gerbils, consistent with chemotaxis being critical in this infection model (Fig. 2). Complementation of cheY restored infection to levels similar to those for the wild type, confirming that mutation of cheY is responsible for the infection defect. The mutant lacking tlpB infected gerbils to levels that were similar to those of wild type, suggesting that this receptor is not needed under these conditions.
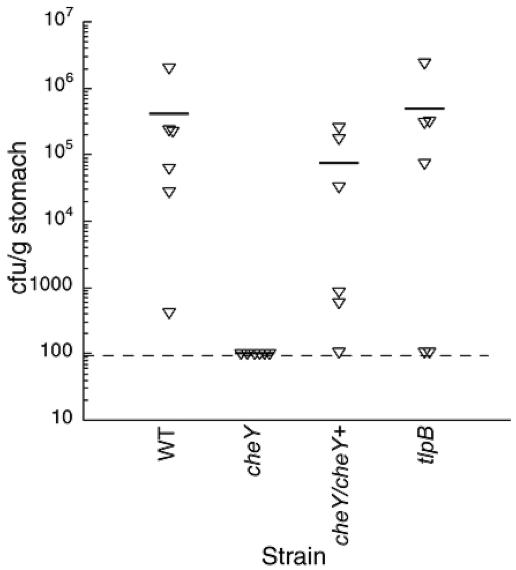
FIG. 2.
Colonization levels of nonchemotactic mutants after 4-week infection of gerbils. Gerbils (n = six per group, one experimental replicate) were infected with wild-type SS1 (WT) (3.2 × 108 CFU), SS1 cheY (1.6 × 108 CFU), SS1 tlpB (1.3 × 108 CFU), or SS1 cheY/cheY+ (3.3 × 108 CFU). Each animal is represented by an inverted triangle. The average, which includes the uninfected animals, is shown as a horizontal bar, and the limit of detection (∼100 CFU/g) is shown as a broken horizontal line. The number infected in each group was as follows: of WT, 6; of cheY, 0; of cheY/cheY+, 5; of tlpB, 4. There was no statistical difference between the wild type and tlpB or the wild type and cheY/cheY+.
We also examined the phenotypes of these mutants when coinfected with the wild type, because this form of infection has been shown to amplify subtle phenotypes (3, 26, 36). The tlpB mutant was recovered as often as the wild-type strain, suggesting that this chemoreceptor has no significant role in allowing H. pylori to achieve full colonization levels (Fig. 3). In contrast, we detected only the wild-type strain, never the cheY mutant, when these two strains were coinfected, suggesting that the wild-type strain does not alter the infection abilities of the mutant and the mutant does not affect infection by the wild type.
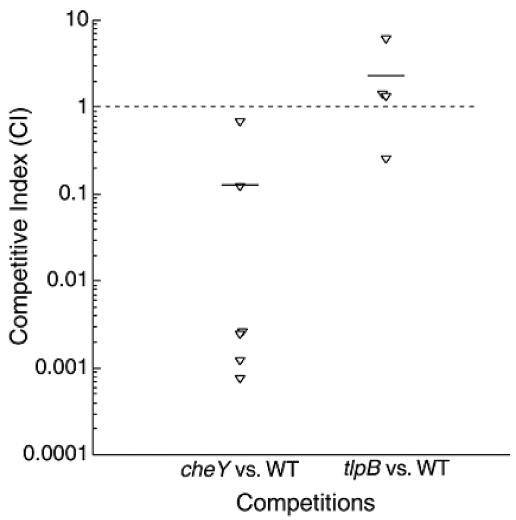
FIG. 3.
Competition results from gerbils coinfected with the wild type (WT) plus cheY or tlpB. Gerbils (n = six per group, one experimental replicate) were infected with mixtures of cheY plus the wild type (mutant/WT ratio = [8.0 × 107]/[1.6 × 108] = 0.5) or tlpB plus wild type (mutant/WT ratio = [6.5 × 107]/[1.6 × 108] = 0.4). Average competitive index (horizontal bars) is the output ratio (mutant/wt) divided by the input ratio (mutant/wt). Each animal is represented by an inverted triangle. The broken horizontal line represents equally competitive strains. In the cheY coinfection group, none of the animals were infected with cheY but all were infected with the wild type. In the tlpB coinfection group, four of six animals were infected, and all of these had both tlpB and the wild type. We carried out statistical analyses by comparing each mutant's competitive index to 1, the number expected for a strain equal to the wild type, and found that the cheY mutant is significantly different from 1 (P < 0.01) and the tlpB mutant is not significantly different from 1 (P = 0.4).
The tlpB mutant induces less inflammation in gerbils, despite colonizing at similar levels to wild-type H. pylori.
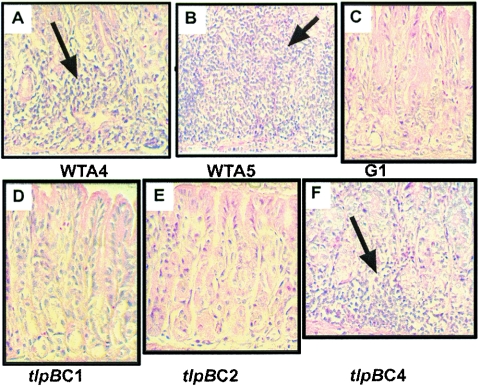
To assess the degree of gastric atrophy and inflammation in gerbils infected with H. pylori chemotaxis mutants, sections of the antrum were processed as described in Materials and Methods and graded in a blind fashion by a pathologist. There were significantly greater inflammatory scores in gerbils infected with wild-type H. pylori than those infected with either the cheY or tlpB mutant (Table 3). In all animals there was very little evidence of gastric atrophy, and there were no significant atrophy differences between the wild type, tlpB mutant, and uninfected controls. In all animals, there was no ulceration, metaplasia, or hyperplasia observed in any sample, as seen by others during short-term infections (1). Histology of representative animals is shown in Fig. 4. We were particularly intrigued by the significant reduction in inflammation in gastric sections from tlpB mutant-infected gerbils, given that this strain infected gerbils to wild-type levels. We found that the majority of gerbils infected with this mutant, animals C1, C2, C3, C5, and C6, displayed very little or no inflammation (animals C1 and C2 [Fig. 4D and E, respectively]). One tlpB-infected animal, C4, showed inflammation levels similar to one of the wild-type infected animals (A4) but lower than another wild-type-infected animal (A5) (compare Fig. 4A, B, and F). In wild-type or tlpB mutant-colonized animals, there was no significant correlation between the amount of bacteria recovered from the stomach and degree of inflammation. Additionally, animals coinfected with the wild type and the tlpB mutant displayed wild-type inflammation, suggesting that the tlpB mutant does not dominantly suppress the immune response (Table 3). Animals that were inoculated with the cheY mutant, that were not colonized with H. pylori (e.g., C2 and C6), or that had served as uninfected controls (G1 to G6), however, exhibited no inflammation (Table 3 and Fig. 4C and E).
TABLE 3.
Histology of antrums from uninfected gerbils and gerbils inoculated with wild-type SS1, the cheY mutant, or the tlpB mutant or with both the wild type and the tlpB mutant
| Animal sample | Log CFU/g in stomach | Atrophy grade | Inflammation grade (with neutrophils, as indicated) | Other notable observations (plasias, ulcers, etc.) |
|---|---|---|---|---|
| Wild-type SS1a | ||||
| A4 | 2.6 | 0 | 4 (+ neutrophils) | |
| A5 | 4.4 | 1 | 5 (+ neutrophils) | |
| A6 | 5.3 | 0 | 3 (+ neutrophils) | One focus of moderate inflammation with the remainder being slight to mild |
| Avg (colonized animals only)b | 4.0 ± 1.0 | |||
| cheY mutant: B1, B2, B3, B4, B5, B6 | 0.0 | 0 | 1 (+ neutrophils) | |
| Avg (all) | 0 ± 0 | 0 | ||
| tlpB mutant | ||||
| C1 | 5.5 | 0 | 1 (+ neutrophils) | |
| C2, C6 | 0.0 | 0 | 0 | |
| C3 | 4.9 | 0 | 0 | Scattered lymphocytes, but within normal range |
| C4 | 6.4 | 1 | 4 (+ neutrophils) | |
| C5 | 5.5 | 0 | 2 (+ neutrophils) | |
| Avg (colonized animals only)a | 1.75 ± 1.7c | |||
| WT/tlpB mutant coinfected | ||||
| F1, F4 | d | 0 | 3 (+ neutrophils) | |
| F2 | d | 1 | 4 (+ neutrophils) | |
| F3 | d | 1 | 5 (+ neutrophils) | |
| F5 | d | 0 | 2 (+ neutrophils) | |
| F6 | d | 0 | 1 (+ neutrophils) | |
| Avg | 3.0 ± 1.4 | |||
| Uninfected | ||||
| G1, G2, G3, G4, G6 | 0 | 0 | 0 | |
| G5 | 0 | 0 | 0 | Chronic inflammation in mesenteric tissue |
| Avg | 0 ± 0 |
Antrums from only three of the six gerbils inoculated with wild-type SS1 were available for tissue sectioning, staining, and histology.
The average histology scores were determined only from animals in which H. pylori was recovered (i.e., colonized animals). Means ± standard deviations are given.
P < 0.05 by Mann-Whitney U test compared to that for animals infected with the wild type.
The colonization data for coinfected animals are shown in Fig. 3.
FIG. 4.
Histology of antrum from uninfected gerbils or from gerbils inoculated with the wild type or the tlpB mutant of H. pylori. All sections were stained with hematoxylin and eosin. Magnification, ×200. Arrows point to areas of lymphocytic inflammation. (A) Antrum from gerbil A4 inoculated with wild-type H. pylori strain SS1. (B) Antrum from gerbil A5 inoculated with wild-type H. pylori strain SS1. (C) Antrum from gerbil G1 inoculated with sterile buffer (uninfected control). (D) Antrum from gerbil C1 inoculated with the tlpB mutant. (E) Antrum from gerbil C2 inoculated with the tlpB mutant. (F) Antrum from gerbil C4 inoculated with the tlpB mutant.
Gastric sections from gerbils infected with the tlpB mutant exhibit an altered neutrophil-to-lymphocyte ratio compared to that for the wild type.
Stomach sections from three gerbils infected with wild-type H. pylori and three infected with the tlpB mutant were analyzed for the presence of specific inflammatory cells by counting 100 consecutive inflammatory cells in three fields per sample. In the wild-type-infected stomach sections, 82 to 90% of the total inflammatory cells were lymphocytes, 10 to 17% were neutrophils, and the rest were eosinophils (Table 4). Strikingly, the tlpB mutant-infected stomach sections showed a dramatic shift in the neutrophil-to-lymphocyte ratio, with 47 to 67% lymphocytes and 33 to 53% neutrophils (Table 4). There was no significant difference in eosinophils in the wild-type versus tlpB mutant stomach sections (P = 0.87).
TABLE 4.
Mice infected with the tlpB mutant show altered stomach neutrophil-to-lymphocyte ratios
| Animal no. | Infected with: | Cell counta
|
||
|---|---|---|---|---|
| Neutrophils | Lymphocytes | Eosinophils | ||
| A4 | Wild type | 17.3 ± 4.2 | 81.7 ± 3.5 | 1.0 ± 1.0 |
| A5 | Wild type | 10.3 ± 4.7 | 89.3 ± 5.1 | 0.3 ± 0.6 |
| A6 | Wild type | 9.7 ± 8.7 | 90.3 ± 8.7 | 0.0 ± 0.0 |
| Avgb | Wild type | 12.4 ± 6.5 | 87.1 ± 6.8 | 0.4 ± 0.7 |
| C1 | tlpB | 52.7 ± 9.3 | 47.3 ± 9.3 | 0 ± 0 |
| C4 | tlpB | 33.3 ± 12.9 | 63.3 ± 18.6 | 0 ± 0 |
| C5 | tlpB | 43.0 ± 11.5 | 57.0 ± 11.5 | 0 ± 0 |
| Avgb | tlpB | 43.0 ± 12.9c | 55.9 ± 13.8c | 0.0 ± 0.0d |
Three fields per sample were chosen at random, and 100 consecutive inflammatory cells were counted per field. Magnification used for counting was ×400. Values are given as means ± standard deviations.
Average of neutrophils plus lymphocytes plus eosinophils does not total exactly 100 due to rounding.
P < 0.0001 compared to that for the wild type.
P = 0.87 compared to that for the wild type.
DISCUSSION
In this report, we demonstrate that gerbil stomach colonization by H. pylori requires chemotaxis mediated by the CheY response regulator but not chemotactic response by the TlpB chemoreceptor. We furthermore show that loss of chemotaxis causes the gerbil infection defect by complementing the cheY mutation with a copy of cheY at the rdxA chromosomal locus, the first genetic complementation carried out in gerbils. The finding that cheY mutants fail to colonize even in the presence of the wild-type strain suggests that the wild type cannot supply in trans a factor to complement the mutation, similar to findings with cheA mutants in gerbils during a signature-tagged mutagenesis screen (20).
Loss of the TlpB chemoreceptor does not affect the ability of H. pylori to colonize gerbils, a phenotype that is similar to that observed with tlpB mutants in mice (Chen and Ottemann, unpublished data). H. pylori is predicted to have four chemoreceptors that govern its chemotactic response. In contrast to TlpB, loss of either the TlpA or TlpC chemoreceptors causes a mouse colonization defect when the tlpA or tlpC mutants are coinfected with the wild type (3). The ability of the tlpB mutant to colonize gerbils even in the presence of the wild type strongly suggests that TlpB's function is not required for H. pylori growth or maintenance in vivo. Presumably, TlpB directs H. pylori's response to some environmental cues, but these cues are not needed for colonization in the gerbil. The ligand-binding portion of TlpB shares similarity only with uncharacterized proteins, and thus nothing is known about the types of signals sensed by this or related proteins. Furthermore, it is unknown whether TlpB-related proteins play roles in other bacterial-host interactions.
Consistent with their colonization abilities in gerbils, tlpB mutants display no chemotaxis defect in vitro. Loss of tlpB has no effect on the ability of H. pylori to chemotax in rich media in laboratory analyses, presumably because the bacterium has three other chemoreceptors that carry out redundant functions in this medium. Loss of any one of the other H. pylori chemoreceptors similarly has no effect in this assay (3; S. M. Williams and K. M. Ottemann, unpublished data).
Even though tlpB mutants colonize gerbil stomachs well, they do not produce the same type and level of inflammation. H. pylori induces inflammation in the gastric mucosa by causing the production of proinflammatory cytokines (14, 24, 37, 41). Several cells produce the cytokines, including gastric epithelial cells, Th1 lymphocytes, and other immune cells. Some Helicobacter spp. molecules that stimulate cytokine manufacture are known from studies of H. pylori or the related Helicobacter felis. For example, proteins encoded by the cag pathogenicity island (cag PAI) trigger epithelial cells to secrete the proinflammatory cytokines interleukin-8 (IL-8), IL-6, and tumor necrosis factor alpha (41). The same cytokines are released by macrophages confronted with H. pylori urease, while a different protein, OipA, has been shown to cause cultured gastric epithelial cells to produce IL-8 (43). Neutrophil-activating protein activates neutrophils such that they produce more reactive oxygen species and thus enhance inflammation. Lastly, lipopolysaccharide can stimulate Toll-like receptor 4 molecules to cause severe atrophic gastritis (35). Helicobacter proteins that directly stimulate Th1 cells are not yet known, although it is well known that these cells are critical for the development of gastritis (33). The strain used here, SS1, induces a strong inflammatory response in both gerbils and mice (9, 23), but is proposed to have an incomplete cag PAI (8). Whether the inflammation observed in gerbils in this study is cag dependent warrants further investigation.
While the possibility exists that the tlpB mutation could render polar effects on downstream genes possibly involved in inflammation, we found in vitro that expression of the downstream gene, encoding a predicted glycosyltransferase (hp0102), is similar in both the wild type and the tlpB mutant. Therefore, the polarity possibility is unlikely.
Other mutants are known to colonize gerbils well but lead to lessened inflammation, similar to the tlpB mutant reported here. In H. pylori strain TN2, mutants lacking the cag PAI lead to decreased inflammation while colonizing at wild-type levels (1, 18, 27). The cag PAI is known to trigger the secretion of proinflammatory cytokines from gastric epithelial cells, so it is easy to imagine why this mutant causes less inflammation (25). It is not known, however, why loss of a chemoreceptor leads to decreased inflammation. One hypothesis is that H. pylori cells missing TlpB fail to reach the proper stomach location to initiate inflammation, either by failure to deliver proteins to host cells or by failure to receive signals that induce the expression of bacterial genes involved in inducing inflammation. It is known that direct contact with the gastric epithelium enhances the severity of inflammation (16), and thus, one possibility is that TlpB guides H. pylori to contact epithelial cells. Another possibility is that tlpB mutants do not populate the antrum efficiently, and thus, there are fewer bacteria in that region to trigger inflammation. Such an idea comes from other studies showing that completely nonchemotactic mutants (lacking cheW) have a severe defect in antrum colonization and a mild defect in overall colonization of the mouse stomach (39). Unlike these fully nonchemotactic mutants, tlpB mutants infect the stomach to normal levels, and thus, the tlpB mutant defect in inflammation is probably not due to decreased numbers of H. pylori cells in the antrum.
In bacteria such as Vibrio cholerae, virulence gene regulation is controlled by the chemotaxis system (22). In this microbe, mutants in several chemotaxis genes, including cheA, cheY, cheZ, and one chemoreceptor (VC2161), do not induce expression of key virulence genes at the proper time. VC2161 has a similar architecture as TlpB (two predicted transmembrane domains) but is not homologous in the ligand-binding region and its ligand is unknown.
In H. pylori-infected stomachs, polymorphonuclear leukocytes such as neutrophils are the first inflammatory cells at the site, followed later by mononuclear cells such as T lymphocytes (37). The finding that stomach sections from the tlpB mutant-infected gerbils exhibit an altered ratio of neutrophils to lymphocytes, with higher neutrophils than the wild type, suggests that the tlpB mutants are still in the early neutrophil-heavy inflammatory gastritis phase, while wild-type-infected gerbils have already progressed to the latter lymphocyte-dominant phase. These observations suggest there is a significant delay in the inflammatory response in tlpB mutant-infected animals, and this delay may coincide with the reduced overall inflammation observed in these gerbils. Future studies will examine the inflammatory response at later infection times to see whether the tlpB mutant-derived inflammation remains defective or progresses to the more advanced inflammation observed in gastric tissue from wild-type-infected gerbils.
This study underscores the critical role H. pylori chemotaxis plays in animal infection and expands the possible roles for chemotaxis to include the induction of inflammation. Thus, the study of genes from the chemotaxis pathway may lead us to uncover different aspects of H. pylori pathogenesis. Deciphering the host signal transduction mechanisms behind the attenuated inflammation observed with the tlpB mutant will no doubt give us a better understanding of the complex and dynamic interactions of H. pylori with the host.
Acknowledgments
We especially thank Sangeetha Bathala, Traci L. Testerman, and Ryan J. Viator for assistance with animal handling, tissue processing, and consultations; Judy King for assistance with the preparation of tissue specimens for histology; Andy Woodruff for constructing pCat-mut; Lynn Connolly for constructing pLC292; and Johannes Kusters for providing pRdxA.
This work was supported by Public Health Service grants R01 CA101931 (to D.J.M.) and RO1 AI050000 (to K.M.O.) from the National Institutes of Health.
Editor: D. L. Burns
REFERENCES
- 1.Akanuma, M., S. Maeda, K. Ogura, Y. Mitsuno, Y. Hirata, T. Ikenoue, M. Otsuka, T. Watanabe, Y. Yamaji, H. Yoshida, T. Kawabe, Y. Shiratori, and M. Omata. 2002. The evaluation of putative virulence factors of Helicobacter pylori for gastroduodenal disease by use of a short-term Mongolian gerbil infection model. J. Infect. Dis. 185:341-347. [DOI] [PubMed] [Google Scholar]
- 2.Alm, R. A., L.-S. L. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. Dejonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. [DOI] [PubMed] [Google Scholar]
- 3.Andermann, T. M., Y.-T. Chen, and K. M. Ottemann. 2002. Two predicted chemoreceptors of Helicobacter pylori promote stomach infection. Infect. Immun. 70:5877-5881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Armitage, J. P. 1999. Bacterial tactic responses. Adv. Microb. Physiol. 41:229-289. [DOI] [PubMed] [Google Scholar]
- 5.Beier, D., G. Spohn, R. Rappuoli, and V. Scarlato. 1997. Identification and characterization of an operon of Helicobacter pylori that is involved in motility and stress adaptation. J. Bacteriol. 179:4676-4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blair, D. F. 1995. How bacteria sense and swim. Annu. Rev. Microbiol. 49:489-522. [DOI] [PubMed] [Google Scholar]
- 7.Cerda, O., A. Rivas, and H. Toledo. 2003. Helicobacter pylori strain ATCC700392 encodes a methyl-accepting chemotaxis receptor protein (MCP) for arginine and sodium bicarbonate. FEMS Microbiol. Lett. 224:175-181. [DOI] [PubMed] [Google Scholar]
- 8.Crabtree, J., R. Ferrero, and J. Kusters. 2002. The mouse colonizing Helicobacter pylori strain SS1 may lack a functional cag pathogenicity island. Helicobacter 7:139-140. [DOI] [PubMed] [Google Scholar]
- 9.Crabtree, J. E., M. Court, M. A. Aboshkiwa, A. H. T. Jeremy, M. F. Dixon, and P. A. Robinson. 2004. Gastric mucosal cytokine and epithelial cell responses to Helicobacter pylori infection in Mongolian gerbils. J. Pathol. 202:197-207. [DOI] [PubMed] [Google Scholar]
- 10.Donahue, J. P., D. A. Israel, R. M. J. Peek, M. J. Blaser, and G. G. Miller. 2000. Overcoming the restriction barrier to plasmid transformation of Helicobacter pylori. Mol. Microbiol. 37:1066-1074. [DOI] [PubMed] [Google Scholar]
- 11.Eaton, K. A., D. R. Morgan, and S. Krakowka. 1992. Motility as a factor in the colonisation of gnotobiotic piglets by Helicobacter pylori. J. Med. Microbiol. 37:123-127. [DOI] [PubMed] [Google Scholar]
- 12.Eaton, K. A., M. J. Radin, and S. Krakowka. 1995. An animal model of gastric ulcer due to bacterial gastritis in mice. Vet. Pathol. 32:489-497. [DOI] [PubMed] [Google Scholar]
- 13.Eaton, K. A., S. Suerbaum, C. Josenhans, and S. Krakowka. 1996. Colonization of gnotobiotic piglets by Helicobacter pylori deficient in two flagellin genes. Infect. Immun. 64:2445-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferrero, R. L., and J. G. Fox. 2001. In vivo modeling of Helicobacter-associated gastrointestinal diseases, p. 565-582. In H. L. T. Mobley, G. L. Mendz, and S. L. Hazell (ed.), Helicobacter pylori: physiology and genetics. ASM Press, Washington, D.C. [PubMed]
- 15.Foynes, S., N. Dorrell, S. J. Ward, R. A. Stabler, A. A. McColm, A. N. Rycroft, and B. W. Wren. 2000. Helicobacter pylori possesses two CheY response regulators and a histidine kinase sensor, CheA, which are essential for chemotaxis and colonization of the gastric mucosa. Infect. Immun. 68:2016-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guruge, J. L., P. G. Falk, R. G. Lorenz, M. Dans, H.-P. Wirth, M. J. Blaser, D. E. Berg, and J. I. Gordon. 1998. Epithelial attachment alters the outcome of Helicobacter pylori infection. Proc. Natl. Acad. Sci. USA 95:3925-3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Honda, S., T. Fujioka, M. Tokieda, R. Satoh, A. Nishizono, and M. Nasu. 1998. Development of Helicobacter pylori-induced gastric carcinoma in Mongolian gerbils. Cancer Res. 58:4255-4259. [PubMed] [Google Scholar]
- 18.Israel, D. A., N. Salama, C. N. Arnold, S. F. Moss, T. Ando, H.-P. Wirth, K. T. Tham, M. Camorlinga, M. J. Blaser, S. Falkow, and R. M. Peek, Jr. 2001. Helicobacter pylori strain-specific differences in genetic content, identified by microarray, influence host inflammatory responses. J. Clin. Investig. 107:611-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Josenhans, C., and S. Suerbaum. 2002. The role of motility as a virulence factor in bacteria. Int. J. Med. Microbiol. 291:605-614. [DOI] [PubMed] [Google Scholar]
- 20.Kavermann, H., B. P. Burns, K. Angermüller, S. Odenbreit, W. Fischer, K. Melchers, and R. Haas. 2003. Identification and characterization of Helicobacter pylori genes essential for gastric colonization. J. Exp. Med. 197:813-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee, A., J. O'Rourke, M. C. De Ungria, B. Robertson, G. Daskalopoulos, and M. F. Dixon. 1997. A standardized mouse model of Helicobacter pylori infection: introducing the Sydney strain. Gastroenterology 112:1386-1397. [DOI] [PubMed] [Google Scholar]
- 22.Lee, S. H., S. M. Butler, and A. Camilli. 2001. Selection for in vivo regulators of bacterial virulence. Proc. Natl. Acad. Sci. USA 98:6889-6894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McGee, D. J., C. Coker, T. L. Testerman, J. M. Harro, S. V. Gibson, and H. L. Mobley. 2002. The Helicobacter pylori flbA flagellar biosynthesis and regulatory gene is required for motility and virulence and modulates urease of H. pylori and Proteus mirabilis. J. Med. Microbiol. 51:958-970. [DOI] [PubMed] [Google Scholar]
- 24.Michetti, P., and A. M. Svennerholm. 2003. Helicobacter pylori—inflammation, immunity and vaccines. Helicobacter 8(Suppl. 1):31-35. [DOI] [PubMed] [Google Scholar]
- 25.Montecucco, C., and R. Rappuoli. 2001. Living dangerously: how Helicobacter pylori survives in the human stomach. Nat. Rev. Mol. Cell Biol. 2:457-466. [DOI] [PubMed] [Google Scholar]
- 26.Nolan, K. J., D. J. McGee, H. M. Mitchell, T. Kolesnikow, J. M. Harro, J. O'Rourke, J. E. Wilson, S. J. Danon, N. D. Moss, H. L. T. Mobley, and A. Lee. 2002. In vivo behavior of a Helicobacter pylori SS1 nixA mutant with reduced urease activity. Infect. Immun. 70:685-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ogura, K., S. Maeda, M. Nakao, T. Watanabe, M. Tada, T. Kyutoku, H. Yoshida, Y. Shiratori, and M. Omata. 2000. Virulence factors of Helicobacter pylori responsible for gastric diseases in Mongolian gerbil. J. Exp. Med. 192:1601-1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Toole, P. W., M. C. Lane, and S. Porwollik. 2000. Helicobacter pylori motility. Microbes Infect. 2:1207-1214. [DOI] [PubMed] [Google Scholar]
- 29.Ottemann, K. M., and A. C. Lowenthal. 2002. Helicobacter pylori uses motility for initial colonization and to attain robust infection. Infect. Immun. 70:1984-1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ottemann, K. M., and J. F. Miller. 1997. Roles for motility in bacterial-host interactions. Mol. Microbiol. 24:1109-1117. [DOI] [PubMed] [Google Scholar]
- 31.Pittman, M. S., M. Goodwin, and D. J. Kelly. 2001. Chemotaxis in the human gastric pathogen Helicobacter pylori: different roles for CheW and the three CheV paralogues, and evidence for CheV2 phosphorylation. Microbiology 147:2493-2504. [DOI] [PubMed] [Google Scholar]
- 32.Rosario, M. M., K. L. Fredrick, G. W. Ordal, and J. D. Helmann. 1994. Chemotaxis in Bacillus subtilis requires either of two functionally redundant CheW homologs. J. Bacteriol. 176:2736-2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roth, K. A., S. B. Kapadia, S. M. Martin, and R. G. Lorenz. 1999. Cellular immune responses are essential for the development of Helicobacter felis-associated gastric pathology. J. Immunol. 163:1490-1497. [PubMed] [Google Scholar]
- 34.Rugge, M., P. Correa, M. F. Dixon, R. Fiocca, T. Hattori, J. Lechago, G. Leandro, A. B. Price, P. Sipponen, E. Solcia, H. Watanabe, and R. M. Genta. 2002. Gastric mucosal atrophy: interobserver consistency using new criteria for classification and grading. Aliment. Pharmacol. Ther. 16:1249-1259. [DOI] [PubMed] [Google Scholar]
- 35.Sakagami, T., J. Vella, M. F. Dixon, J. O'Rourke, F. Radcliff, P. Sutton, T. Shimoyama, K. Beagley, and A. Lee. 1997. The endotoxin of Helicobacter pylori is a modulator of host-dependent gastritis. Infect. Immun. 65:3310-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salama, N. R., G. Otto, L. Tompkins, and S. Falkow. 2001. Vacuolating cytotoxin of Helicobacter pylori plays a role during colonization in a mouse model of infection. Infect. Immun. 69:730-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shimoyama, T., and J. E. Crabtree. 1998. Bacterial factors and immune pathogenesis in Helicobacter pylori infection. Gut 43(Suppl. 1):S2-S5. [PMC free article] [PubMed] [Google Scholar]
- 38.Smeets, L. C., J. J. E. Bijlsma, S. Y. Boomkens, C. M. J. E. Vandenbroucke-Grauls, and J. G. Kusters. 2000. comH, a novel gene essential for natural transformation of Helicobacter pylori. J. Bacteriol. 182:3948-3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Terry, K., S. M. Williams, L. Connolly, and K. M. Ottemann. 2005. Chemotaxis plays multiple roles in Helicobacter pylori mouse infection. Infect. Immun. 73:803-811. [DOI] [PMC free article] [PubMed]
- 40.Tomb, J.-F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzgerald, N. Lee, M. D. Adams, E. K. Kichey, D. E. Berg, J. D. Gocayne, T. R. Utterback, J. D. Person, J. Kelley, M. D. Cotton, J. M. Weidman, C. Fujii, C. Bowman, L. Wathey, E. Wallin, W. S. Hayes, M. Borodovsky, P. D. Karp, H. O. Smith, C. M. Fraser, and J. C. Venter. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 338:539-547. [DOI] [PubMed] [Google Scholar]
- 41.Wang, J., T. G. Blanchard, and P. B. Ernst. 2001. Host inflammatory response to infection, p. 471-480. In H. L. T. Mobley, G. L. Mendz, and S. L. Hazell (ed.), Helicobacter pylori: physiology and genetics. ASM Press, Washington, D.C.
- 42.Wirth, H.-P., M. H. Beins, M. Yang, K. T. Tham, and M. J. Blaser. 1998. Experimental infection of Mongolian gerbils with wild-type and mutant Helicobacter pylori strains. Infect. Immun. 66:4856-4866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamaoka, Y., D. H. Kwon, and D. Y. Graham. 2000. A Mr 34,000 proinflammatory outer membrane protein (oipA) of Helicobacter pylori. Proc. Natl. Acad. Sci. USA 97:7533-7538. [DOI] [PMC free article] [PubMed] [Google Scholar]