Abstract
We have previously identified mgrA (rat) as a regulator of autolysis in Staphylococcus aureus. Besides its effect on autolytic activity, we recently found alterations in the expression of regulator and target virulence genes in the mgrA mutant. Northern analysis and transcription fusion assays showed that inactivation of mgrA has led to the downregulation of RNAIII of agr and hla and upregulation of sarS and spa. Although both SarA and agr are activators of α-hemolysin and a repressors of protein A synthesis, we found that the transcription of sarA was not affected in the mgrA mutant and vice versa, indicating that MgrA likely regulates hla and spa in a SarA-independent manner. Previously we have shown that SarT, a SarA homolog, is a repressor of hla and an activator of spa, presumably by activating SarS, however, analysis of the double sarT mgrA mutant for hla and spa transcription indicated that the mgrA-mediated effect is not mediated via sarT. Our results further demonstrated that the mgrA gene product regulates hla and spa expression in a dual fashion, with the first being agr dependent and the second agr independent. In the agr-independent pathway, MgrA binds directly to hla and the sarS promoter to modulate α-hemolysin and protein A expression. Thus, our studies here have defined the nature of interaction of mgrA with other regulators such as agr, sarS, and sarT and its role in regulating hla and spa transcription within the virulence regulatory network of S. aureus.
Staphylococcus aureus, a member of the family Micrococcaceae, is a gram-positive bacterium that normally colonizes the epithelial surface in 30 to 40% of humans. Despite advances in antimicrobial therapy, S. aureus remains a major cause of infections in the hospital setting. The spectrum of diseases caused by this organism is extremely wide, ranging from superficial skin infections to deep abscesses (29). Many of these infections begin locally (skin and catheters) and subsequently spread to the bloodstream, putting patients at risk of developing endocarditis and other metastatic complications (45). The capacity to cause a myriad of infections is probably attributable to the organism's capacity to colonize and survive in diverse host niches during the infection process.
The pathogenicity of S. aureus is a complex process involving the spatial-temporal production of a diverse array of virulence factors. Many cell wall components that act as adhesins (e.g., fibrinogen and fibronectin binding proteins) or contribute to the evasion of host defense (protein A) are produced primarily during the exponential phase while the production of toxins and enzymes (alpha-hemolysin) that facilitate tissue invasion occurs postexponentially (45).
Adding to this complexity in S. aureus pathogenesis is the dramatic worldwide increase in antibiotic resistance among clinical isolates. More than 90% of staphylococcal isolates are now penicillin resistant. With the introduction of methicillin in the 1960s, the percentage of methicillin-resistant S. aureus infections has gradually increased, now up to 60 to 70% in the hospital setting. In the past few years, community-acquired methicillin-resistant S. aureus infections have been reported with increased frequency (20). The increased use of vancomycin, a glycopeptide antibiotic, has led to the emergence of vancomycin-resistant strains (4, 50). This has raised the concern that resistant S. aureus infections may be difficult to treat with currently available antibiotics. Thus, there is a need to understand the pathogenetic process so that new molecular targets can be identified for the development of effective therapeutic agents.
The coordinated synthesis of cell wall proteins in the exponential phase and extracellular proteins during the postexponential phase suggests that many of these virulence determinants are governed by global regulatory elements (12). Members of these regulatory systems include the SarA protein family (8, 12) and a number of two-component regulatory systems such as AgrAC (26), SaeRS (41, 19, 51), LytRS (3), ArlRS (16), SrrAB (55), (44), YycFG (36), and VraRS (28). The sarA and agr loci comprise two critical global regulatory elements that coordinate synthesis of cell wall and extracellular virulence proteins during the exponential and postexponential phases, respectively (1, 12).
The agr locus encodes two divergent transcripts, RNAII and RNAIII, driven by two distinct promoters, P2 and P3, respectively. The P2 transcript encodes four genes, agrB, agrD, agrC, and agrA, comprising a two-component quorum-sensing system normally required for the activation of RNAII and RNAIII (38, 42). The RNAII gene product AgrD encodes a 46-residue peptide which is processed and secreted as an autoinducing peptide with the aid of the putative membrane component AgrB. This cyclic peptide carries a quorum-sensing function and, upon reaching the threshold level, binds to AgrC, the membrane sensor component of a two-component system, eventuating in the activation of the response regulator AgrA (24, 23). Presumably, phosphorylated AgrA would bind to the agr P3 promoter to promote transcription from the P3 promoter.
The P3 transcript, designated RNAIII, is the regulatory molecule of agr and acts on target genes mainly at the level of transcription and, to a lesser extent, translation (42, 22). Once RNAIII is synthesized, it somehow upregulates the transcription of exoprotein genes (e.g., hla) while downregulating genes encoding cell wall proteins (e.g., spa, encoding protein A, and fnb, encoding fibronectin binding protein) (22). However, the exact manner by which RNAIII activates target gene transcription is not clearly defined.
The agr locus is also activated by SarA, which binds to the agr P2-P3 promoter region to activate RNAII and RNAIII transcriptions (9, 13). The sarA locus is composed of three overlapping transcripts, sarA P1, P3, and P2, each encoding the 14.5-kDa SarA protein. GeneChip analysis indicated that the sarA locus affects the transcription of ≈120 genes in S. aureus (2, 15). While the sarA and agr loci are the major controlling elements for the expression of a variety of virulence proteins during the growth cycle (1, 12), a number of SarA homologs (members of the SarA protein family) have been found to participate in the sarA and agr regulatory cascade (12, 5), including SarT, a repressor of hla which is normally repressed by SarA and agr (49), SarS, an activator of protein A synthesis (10, 52), and SarU, which activates RNAIII transcription (33). Remarkably, SarT was found to activate sarS expression (48), indicating that agr may repress protein A expression by downregulating sarT and subsequently sarS.
In this study, we report the interaction of Rat/MgrA protein with agr and its effect on the sarS and hla promoters, culminating in the expression of α-hemolysin and protein A. A mutation in mgrA resulted in altered expression of RNAIII, sarS, hla, and spa expression. We also provided additional evidence that mgrA has a dual role in regulating hla and spa expression. In the agr-dependent pathway, decreased agr transcription in mgrA mutants would lead to reduced hla transcription and an increase in spa transcription. In the agr-independent pathway, direct interactions of MgrA and hla with the sarS promoter resulted in activation or repression of the respective gene. Based on these results, we propose MgrA to be an important regulator of virulence determinants in S. aureus.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Phages Φ11 and 80α were used as transducing phages for S. aureus strains. S. aureus cells were grown at 37°C with aeration in CYGP 03GL broth (39) or tryptic soy broth supplemented with antibiotics as indicated. Luria-Bertani (LB) was used for cultivating Escherichia coli. Antibiotics used for S. aureus were erythromycin, 5 μg/ml; tetracycline, 3 μg/ml; chloramphenicol, 10 μg/ml; and minocycline, 2.5 μg/ml. For E. coli, the following concentrations were used: ampicillin, 50 μg/ml; and spectinomycin, 75 μg/ml.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Comment | Reference or source |
|---|---|---|
| S. aureus | ||
| RN4220 | Mutant strain of 8325-4 that accepts foreign DNA | 39 |
| RN6390 | agr+ laboratory strain related to 8325-4, maintains hemolytic pattern when propagated on sheep erythrocytes | 39 |
| COL | Methicillin-resistant laboratory strain | 54 |
| Newman | Laboratory strain | 37 |
| FDA486 | Wild-type strain FDA486, sigB+, intact rsbU | 43 |
| RN6911 | agr mutant of RN6390 (Δagr::tetM) | 42 |
| ALC3043 | sarT mutant of RN6390 (ΔsarT::tetK) | This study |
| ALC2530 | mgrA mutant of RN6390 (ΔmgrA::ermC) | 21 |
| ALC2531 | ALC2530 complemented with the mgrA gene in single copy | 21 |
| ALC2542 | mgrA mutant of COL (ΔmgrA::ermC) | This study |
| ALC2547 | mgrA mutant of Newman (ΔmgrA::ermC) | This study |
| ALC3632 | mgrA mutant of FDA486 (ΔmgrA::ermC) | This study |
| ALC2537 | agr mgrA (deletion) double mutant of RN6390 | 21 |
| ALC3046 | sarT mgrA (deletion) double mutant of RN6390 | This study |
| ALC3188 | agr sarT (deletion) double mutant | This study |
| ALC3191 | mgrA sarT agr (deletion) triple mutant | This study |
| E. coli | ||
| XL-1 Blue | General-purpose host strain for cloning | 31 |
| InvαF′ | Host strain for the TA cloning vector (pCR2.1) | Invitrogen |
| Plasmids | ||
| pCR2.1 | E. coli PCR cloning vector | Invitrogen |
| pALC1484 | Derivative of pSK236 containing the promoterless gfpuvr gene | 25 |
| pALC1740 | pALC1484 with hla promoter fragment | 27 |
| pALC1741 | pALC1484 with spa promoter fragment | 27 |
| pALC1743 | pALC1484 with P3 agr promoter fragment | 25 |
| pALC3179 | pALC1484 with sarS promoter fragment | This study |
| pALC1594 | 235-bp hla promoter region in pCR2.1 | 13 |
| pALC2321 | sarS promoter region in pCR2.1 | 46 |
| pALC1996 | agr P2-P3 promoter region in pALC1484 | 14 |
| pRN6735 | Contains promoterless RNAIII under control of the blaZ promoter | 26 |
Genetic manipulations in E. coli and S. aureus.
For the propagation of all plasmid constructs, E. coli strain DH5α was used. Standard molecular biology and recombinant DNA techniques were followed (31). S. aureus RN4220, a restriction-deficient derivative of strain 8325-4 (39), was used as the initial recipient for the transformation of plasmid constructs by electroporation (47).
The construction of the mgrA (rat) deletion mutant in the RN6390 background to yield ALC2530 has been previously described (21). To generate mgrA deletion mutants in other genetic backgrounds, we used Φ11 and 80α phage lysates of ALC2530 to infect strains COL, Newman, FDA486, RN6911 (agr mutant), and ALC3043 (sarT mutant) (Table 1). For the construction of the sarT agr double mutant (ALC3188), a Φ11 phage lysate of the sarT mutant was used to infect the agr mutant RN6911. Similarly the triple mgrA agr sarT mutant (ALC3191) was constructed by infecting the mgrA sarT double mutant with a phage lysate of the RN6911 agr mutant.
Isolation of RNA and Northern blot hybridization.
Overnight cultures of S. aureus were diluted 1:100 in CYGP and grown to late log (optical density at 650 nm [OD650] = 1.2) phase. The cells were harvested and processed with a Trizol isolation kit (Gibco BRL, Gaithersburg, Md.) in combination with 0.1-mm sirconia-silica beads in a Biospec reciprocating shaker to yield RNA as described (7); 15 μg of each sample was electrophoresed in a 1.5% agarose-0.66 M formaldehyde gel in morpholinepropanesulfonic acid (MOPS) running buffer (20 mM MOPS, 10 mM sodium acetate, 2 mM EDTA, pH 7.0). Blotting of RNA onto Hybond N+ membranes (Amersham, Arlington Heights, Ill.) was performed with the Turboblotter alkaline transfer system (Schleicher & Schuell, Keene, N.H.). For detection of specific transcripts (RNAIII, sarA, saeRS, sarS, sarT, spa, hla, and mgrA), gel-purified DNA probes were radiolabeled with [α-32P]dCTP by the random-primed DNA labeling method (Roche Diagnostics GmbH) and hybridized under aqueous-phase conditions at 65°C (6). The blots were subsequently washed and autoradiographed as described (11).
Transcriptional fusion studies of different promoters linked to the gfpuvr reporter gene.
To confirm the effect of the mgrA mutation on promoter activities of other regulators (RNAIII and sarS) and target genes such as hla and spa, we cloned promoter fragments of these genes into the shuttle vector pALC1484 (pSK236-based plasmid containing the promoterless gfpuvr gene) upstream of the gfpuvr gene to generate transcriptional fusions. Restriction analysis and DNA sequencing confirmed the orientation and authenticity of the promoter fragments. The recombinant plasmids containing these promoters were first introduced into S. aureus strain RN4220 by electroporation (47). Plasmids purified from RN4220 transformants were then electroporated into RN6390 and its isogenic mgrA and other mutants.
After overnight culture, S. aureus strains harboring the recombinant plasmids were diluted 1:100 and grown at 37°C with shaking in tryptic soy broth with chloramphenicol (10 μg/ml). Aliquots (200 μl) were transferred hourly to microtiter plates to assay for cell density (OD650) and fluorescence for 10 h in a FL600 fluorescence spectrophotometer (BioTek Instrument, Winooski, Vt.). Promoter activities were plotted as mean fluorescence/OD650 ratio to minimize variations due to cell density, using the average values from triplicate readings.
Gel shift assays.
The purification of MgrA (Rat) protein has been described (21). To determine if the recombinant MgrA protein binds to the agr and hla promoters, a 228-bp fragment representing a region between the P2 and P3 promoters of agr (from nucleotides 1528 to 1756) (26) and a 235-bp hla promoter fragment (nucleotides 1 to 80 plus 155 bp upstream of the start codon) (13) were end-labeled with [γ-32P]ATP by using T4 polynucleotide kinase. Similarly, a 264-bp sarS promoter fragment (nucleotides 125169 to 125432) (10) obtained from plasmid pALC2321 (Table 1) was end labeled. Labeled fragments (0.1 ng) were incubated at room temperature for 20 min with various amounts of purified MgrA protein in 25 μl of binding buffer (25 mM Tris-HCl, pH 7.5, 0.1 mM EDTA, 75 mM NaCl, 1 mM dithiothreitol, and 10% glycerol) containing 0.5 μg of calf thymus DNA per ml. The reaction mixtures were analyzed in a 6.0% nondenaturing polyacrylamide gel. The band shifts were detected by exposing dried gels to X-ray film.
RESULTS
Effect of the mgrA mutation on genes involved in virulence.
In an earlier study, we have reported that MgrA (Rat) is a regulator of autolysis that shares sequence similarity with members of the SarA and MarR families (21). As SarA is part of the complex regulatory network that modulates expression of other regulators as well as target virulence genes, we wanted to determine if MgrA constitutes part of this network by examining the effect of the mgrA mutation on known regulators of virulence determinants.
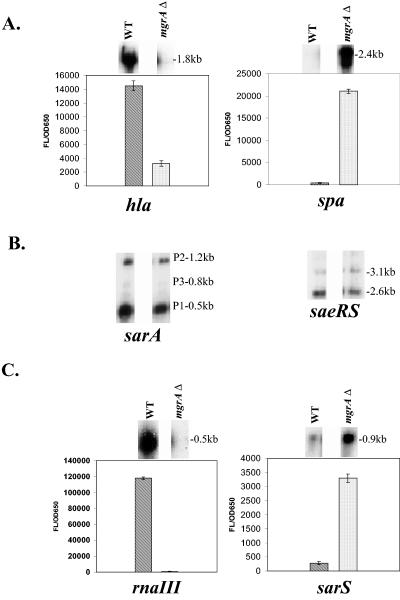
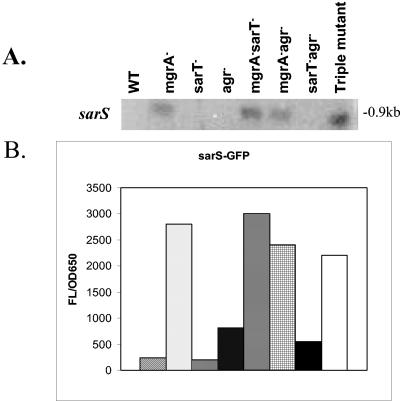
We have focused on major regulators such as agr, sae, and members of the SarA protein family (SarA, SarT, and SarS) that are involved in the control of the α-hemolysin (hla) and protein A (spa) genes, two putative virulence genes in S. aureus. Northern blot analysis revealed that the expression of hla was reduced in the mgrA mutant compared with the parent (RN6390), while the spa transcript level was increased (Fig. 1A). As sarA, sae, and agr were known to modulate hla and spa transcription, we analyzed transcript levels of these regulatory loci in the mgrA mutant of strain RN6390. Interestingly, the transcript levels of sarA and saeRS, two known global regulators of virulence, were unchanged (Fig. 1B). In contrast, there was a significant decrease in the mRNA level of RNAIII of agr in the mutant compared with the parent, while the sarS level was increased (Fig. 1C).
FIG. 1.
Effect of the mgrA mutation on the expression of genes involved in virulence. A. Northern blot analysis of hla and spa transcripts in RN6390 and isogenic mgrA mutant ALC2530. The open reading frame of each gene was used as a probe for the Northern blots. RNA was harvested from cells grown to an OD650 of ≈1.2, representing late exponential phase. The expression of GFP driven by the hla and spa promoters in RN6390 and the mgrA mutant was also measured. Promoter activation was plotted as mean fluorescence/OD650 ratio, using average values of triplicate readings at an OD650 of ≈1.4. Comparable differences were also observed at earlier points. These experiments were repeated at least three times with similar results. ▧, RN6390; ░⃞, mgrA mutant ALC2530. B. Northern blot analysis of sarA and saeRS transcripts in RN6390 and mgrA mutant ALC2530. The open reading frame of each gene was used as a probe for the Northern blots. RNA was harvested from cells grown to an OD650 of ≈1.2, representing late exponential phase. C. Northern blot analysis of agr RNAIII and sarS transcripts in RN6390 and mgrA mutant ALC2530. The probe for RNAIII was a 920-bp fragment containing the hld gene. The open reading frame of the sarS gene was used as a probe for the Northern blot. RNA was harvested from cells grown to an OD650 of ≈1.2, representing late exponential phase. The expression of GFP driven by the agr RNAIII and sarS promoters in RN6390 and the mgrA mutant was also measured. Promoter activation was plotted as mean fluorescence/OD650 ratio, using average values of triplicate readings at an OD650 of ≈1.4. Comparable differences were also observed at earlier points. These experiments were repeated at least three times with similar results. ▧, (RN6390; ░⃞, mgrA mutant ALC2530).
To confirm the effects of the mgrA mutation on RNAIII, sarS, hla, and spa transcription, we transformed strain RN6390 and its isogenic mgrA mutant with shuttle plasmids carrying the gfpuvr reporter gene driven by the RNAIII, sarS, hla, and spa promoters. Green fluorescent protein (GFP) expression levels in the mutant, expressed as fluorescence units per OD unit to minimize the effect of bacterial concentration, confirmed the Northern analysis data (Fig. 1A and 1C).
SarT is a known hla repressor that is normally repressed by agr (49). Despite the finding that hla expression was downregulated in the mgrA mutant, we were not able to detect any significant sarT transcript in the parent strain as well as in the mgrA mutant. Promoter fusion studies also verified that sarT was weakly expressed in the wild type and the mgrA mutant (data not shown). Thus, SarT either does not play a major role in the mgrA-mediated regulation of hla expression, or the low level of sarT detection precludes us from discriminating the contribution of sarT to hla regulation by mgrA (see below).
To ensure that these observations are not unique to the mgrA mutant in the RN6390 background, we also ascertained the presence of RNAIII, hla, and spa transcripts in other mgrA mutants of strains COL, Newman, and FDA486. Our results indicated that the mgrA mutants in all these genetic backgrounds displayed decreased levels of RNAIII and hla compared with the parent, while spa levels were increased (data not shown). Taken together, these results showed that the mgrA gene product modulates virulence gene expression by acting as a positive regulator for RNAIII and hla and a negative regulator of spa.
Regulation of hla expression.
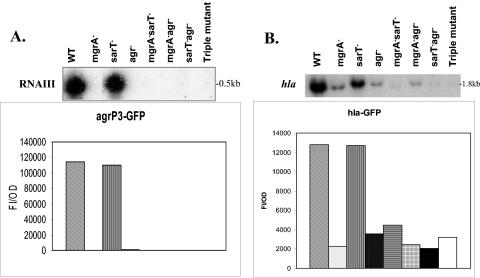
Previous studies from our laboratory have shown that SarA and RNAIII of agr could activate hla expression directly, while SarA also modulates hla transcription by repressing sarT (13, 49). Since MgrA positively affects the expression of both agr (RNAIII) and hla, we wanted to explore if this effect on hla by mgrA could be direct and/or indirect via agr or the agr sarT pathway. To dissect these possibilities, a panel of double and triple deletion mutant strains were constructed in strains RN6390 and Newman, including mgrA sarT, mgrA agr, sarT agr, and mgrA sarT agr mutants (described in Materials and Methods). All the mutants were confirmed by PCR and Southern blots. These strains were then analyzed for RNAIII and hla expression.
As expected, the mRNA levels of RNAIII were not readily detectable in the mgrA mutant and agr mutant, while RNAIII expression in the sarT mutant was similar to that in the parental strain RN6390 (Fig. 2A). Analogous to the mgrA and agr mutants, the remaining double and triple mutants did not express RNAIII well. Interestingly, the mgrA mutant, coinciding with its low agr expression, expressed hla to a level similar to that of the agr mutant. In the mgrA sarT double mutant, the hla transcript level, concordant with a low level of RNAIII, remained undetectable, indicating that activation of hla by mgrA is not mediated via sarT (Fig. 2A and 2B). In the mgrA agr double mutant, hla expression, similar to that of the single mgrA and agr mutants, was significantly lower than that of the parent (Fig. 2A and 2B). As expected, the expression of hla in the triple mgrA sarT agr mutant remained low and not readily detectable (Fig. 2A and 2B).
FIG. 2.
Transcription of agr RNAIII and hla in RN6390 and isogenic mutants. A. Northern blot analysis of agr RNAIII transcripts in RN6390 and isogenic mutants. The probe for RNAIII was a 920-bp fragment containing the hld gene. RNA was harvested from cells grown to an OD650 of ≈1.2, representing late exponential phase. The expression of GFP driven by the agr RNAIII (P3) promoter was also measured. Promoter activation was plotted as mean fluorescence/OD650 ratio, using average values of triplicate readings at an OD650 of ≈1.4. This experiment was repeated at least three times with similar results. ▧, RN6390; ░⃞, mgrA mutant ALC2530; ▥, sarT mutant ALC3043; ▩, agr mutant RN6911; ▤, sarT mgrA mutant ALC3046; , agr mgrA mutant ALC2537; ▪ agr sarT mutant ALC3188; and □ mgrA sarT agr mutant ALC3191. B. Northern blot analysis of the hla transcript in RN6390 and isogenic mutants. The probe for hla was an 800-bp fragment encompassing the open reading frame. RNA was harvested from cells grown to an OD650 of ≈1.2, representing late exponential phase. The expression of GFP driven by the hla promoter was also measured. Promoter activation was plotted as mean fluorescence/OD650 ratio, using average values of triplicate readings at an OD650 of ≈1.4. This experiment was repeated at least three times with similar results. ▧, RN6390; ░⃞, mgrA mutant ALC2530; ▥, sarT mutant ALC3043; ▩, agr mutant RN6911; ▤, sarT mgrA mutant ALC3046; , agr mgrA mutant ALC2537; ▪ agr sarT mutant ALC3188; and □ mgrA sarT agr mutant ALC3191.
GFP fusion assays also confirmed these results (Fig. 2A and 2B), leading us to hypothesize that mgrA regulates hla indirectly via agr or directly on hla or both and that there is very little cross talk, if any, between mgrA and sarT for hla expression.
Gel shift assay.
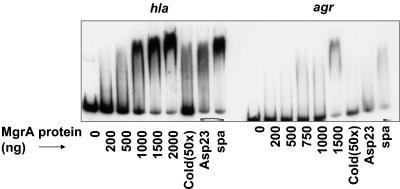
By virtue of its homology with members of the SarA and MarR protein families, we speculate that MgrA might also be a DNA-binding protein (21). Given that an mgrA mutation resulted in downregulation of RNAIII and hla, we wanted to explore if MgrA binds to these promoters to modulate gene transcription. Using purified MgrA protein and end-labeled hla promoter fragment (≈1 ng), we found that MgrA could bind to the 235-bp hla promoter fragment with 1 μg of protein (Fig. 3A), while a nearly complete shift occurred in the presence of 2.0 μg of MgrA. The protein-DNA complex could be disrupted with a 50-fold excess of unlabeled hla promoter fragment (lane 7 in Fig. 3A) but not with a 160-bp spa promoter fragment (Fig. 3A, lane 9) or a nonspecific competitor such as a 300-bp asp23 promoter fragment at comparable concentrations (Fig. 3A, lane 8).
FIG. 3.
Gel shift assays of purified MgrA with hla and agr promoter fragments. MgrA protein in increasing concentrations was incubated with end-labeled hla or agr promoter fragments. In competition assays, MgrA protein (1,500 ng) was incubated with the promoter fragment in the presence of a 50-fold excess of unlabeled specific competitor (hla or agr) (lane 7) or nonspecific competitors (≈300-bp asp23 promoter in lane 8 and ≈160-bp spa promoter fragment in lane 9).
Likewise, the mobility of the 228-bp end-labeled agr P2-P3 promoter fragment was also retarded by MgrA, with the retardation taking place at 1.5 μg of protein (Fig. 3B). As with the hla promoter fragment, unlabeled agr promoter fragment competed successfully for the binding of MgrA, whereas the spa and asp23 promoter fragments did not (Fig. 3B). Collectively, these data demonstrated that MgrA likely regulates hla and RNAIII by interacting directly with the promoters. As RNAIII of agr is known to regulate hla transcriptionally (42), these results implied that MgrA could control hla expression both directly and indirectly via agr.
Assessing hla expression by providing RNAIII in trans.
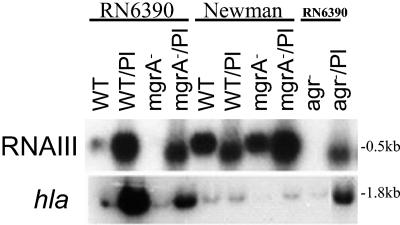
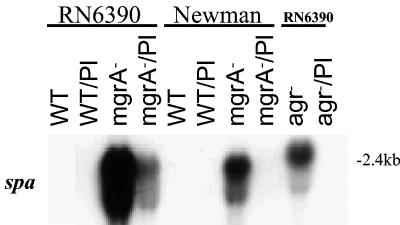
To further validate the role of agr in hla expression in mgrA mutants, we elected to complement the mgrA mutant with a plasmid carrying RNAIII (pRN6735). The cloned RNAIII fragment was under the control of a β-lactamase (blaZ) promoter and constitutively expressed in the absence of pI524, the plasmid that carried the β-lactamase repressor (26). Plasmid RN6735 was used to transform parental strains RN6390 and Newman and isogenic mgrA and agr mutants. Restriction analysis confirmed the presence of plasmid RN6735 in the resultant transformants. These transformants were then analyzed by Northern blotting to determine the levels of RNAIII and hla expression.
Our results indicated that RNAIII, as driven by the blaZ promoter, was expressed constitutively in the wild-type strain as well as in the mgrA and agr mutants (Fig. 4). Despite the presence of RNAIII in induced mgrA and parental strains, the expression of hla, while activated, was lower in the mgrA mutants than in the parents in both the RN6390 and Newman backgrounds (lanes 2 and 4 and lanes 6 and 8 in Fig. 4). Collectively, gel shift and transcriptional data with RNAIII provided in trans showed that mgrA-regulation of hla is likely under bimodal control, with both direct (hla promoter) and indirect (agr promoter) regulation.
FIG. 4.
Transcription of hla by providing RNAIII in trans. Northern blot analysis of agr RNAIII and hla transcripts in strains with and without plasmid RN6735. (Transformants are indicated as strain/Pl.) RNA was harvested from cells grown to an OD650 of ≈1.2, representing late exponential phase.
Regulation of protein A by mgrA is dependent on sarS.
As described earlier (Fig. 1A), mgrA also modulates protein A expression negatively. Previously, the expression of spa has been shown to be positively regulated by sarS (52, 10). Additional gel shift and transcriptional studies revealed that sarS is positively controlled in part by sarT and negatively controlled by agr (48, 33). As the mgrA locus positively controls agr, we wanted to ascertain if mgrA also represses sarS and spa expression via agr. To ascertain this pathway, we used the same set of strains that we used to dissect the regulation of hla.
Northern blotting data revealed that the sarS transcript level, as expected, was not readily detectable in the parent but was increased in the mgrA mutant (Fig. 5A). Interestingly, the increase in sarS promoter activity, as assessed by transcriptional fusions, was higher in the mgrA mutant than in the agr mutant, implying an additional effect on sarS by mgrA other than those of agr (Fig. 5B). As expected in the absence of sarT, there was little if any sarS transcription (48). These observations were confirmed by promoter fusion assays (Fig. 5B). Interestingly, in the double mgrA sarT mutant and triple mgrA sarT agr mutant, the sarS transcript level remained high, suggesting that the effect of mgrA on sarS is not mediated via sarT. Likewise, in the mgrA agr mutant, sarS levels continued to be elevated. In contrast, the effect of agr on sarS is sarT dependent, as exemplified by the near absence of sarS transcription in the sarT agr mutant. Collectively, these results indicate that modulation of sarS expression by mgrA is not mediated via sarT and is only partially dependent on agr.
FIG. 5.
Transcription of sarS in RN6390 and isogenic mutants. A. Northern blot analysis of the sarS transcript in RN6390 and isogenic mutants. The open reading frame of sarS was used as a probe. RNA was harvested from cells grown to an OD650 of ≈1.2, representing late exponential phase. B. The expression of GFP driven by the sarS promoter was also measured. Promoter activation was plotted as mean fluorescence/OD650 ratio, using average values of triplicate readings at an OD650 of ≈1.4. This experiment was repeated at least three times with similar results. ▧, RN6390; ░⃞, mgrA mutant ALC2530; ▥, sarT mutant ALC3043; ▩, agr mutant RN6911; ▤, sarT mgrA mutant ALC3046; , agr mgrA mutant ALC2537; ▪ agr sarT mutant ALC3188; and □ mgrA sarT agr mutant ALC3191.
To correlate sarS to spa expression, we also analyzed spa expression in the above mutants. Our results indicated that the spa mRNA level in all these strains corresponded quite nicely with the sarS levels (Fig. 6A), with upregulation in the mgrA, mgrA sarT, mgrA agr, and mgrA agr sarT mutants and downregulation or relatively unaltered levels in sarT and agr sarT mutants, using both Northern and transcriptional fusion assays (Fig. 6A and B).
FIG. 6.
Transcription of spa in RN6390 and isogenic mutants. A. Northern blot analysis of the spa transcript in RN6390 and isogenic mutants. The open reading frame of spa was used as a probe. RNA was harvested from cells grown to an OD650 of ≈1.2, representing late exponential phase. B. The expression of GFP driven by the spa promoter was also measured. Promoter activation was plotted as mean fluorescence/OD650 ratio, using average values of triplicate readings at an OD650 of ≈1.4. This experiment was repeated at least three times with similar results. ▧, RN6390; ░⃞, mgrA mutant ALC2530; ▥, sarT mutant ALC3043; ▩, agr mutant RN6911; ▤, sarT mgrA mutant ALC3046; , agr mgrA mutant ALC2537; ▪ agr sarT mutant ALC3188; and □ mgrA sarT agr mutant ALC3191.
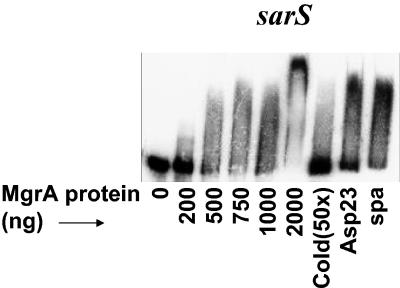
The observation that mgrA may impact on sarS independent of agr and sarT led us to speculate that a direct interaction between MgrA and the sarS promoter may occur. Gel shift assays with labeled sarS promoter (240 bp) and purified MgrA protein showed that 750 ηg of MgrA was required to retard the mobility of the sarS promoter (Fig. 7). At 2,000 ηg, there was a complete shift of the promoter DNA in the gel shift assay. The protein-DNA complex could be disrupted by 50-fold excess unlabeled promoter DNA (Fig. 7) while the nonspecific promoter DNA fragments, including the spa and asp23 promoters, did not alter the binding of MgrA to the sarS promoter (Fig. 7). Seen together, these data showed that MgrA likely acts as a negative regulator of spa by repressing sarS expression.
FIG. 7.
Gel shift assays of purified MgrA with sarS promoter fragment. MgrA protein in increasing concentrations was incubated with the end-labeled sarS promoter fragment. In competition assays, MgrA protein (1,000 ng) was incubated with end-labeled promoter in the presence of a 50-fold excess of unlabeled specific competitor (sarS) (lane 7) or the nonspecific competitor (≈300-bp asp23 promoter in lane 8 and the ≈160-bp spa promoter fragment in lane 9).
To assess if the absence of RNAIII could account for the overexpression of sarS and hence spa in the mgrA mutant, we also provided RNAIII in trans with a plasmid containing the RNAIII sequence in these strains. The strains were then evaluated for spa expression by Northern blotting. Our results indicated that overexpression of RNAIII under the β-lactamase promoter led to significant repression of spa in the mgrA mutants of strains RN6390 and Newman compared with the mgrA mutant controls (Fig. 8). These data imply that MgrA likely has a dual role in regulating spa expression, with the first pathway dependent on agr, wherein an absence of RNAIII in an mgrA mutant leads to an increase in sarS and ultimately spa expression. In the second pathway, a direct interaction between MgrA and the sarS promoter likely occurs to repress sarS transcription.
FIG. 8.
Transcription of spa by providing RNAIII in trans. Northern blot analysis of the spa transcript in strains with and without plasmid RN6735. (Transformants are indicated as strain/Pl.) The open reading frame of the spa gene was used as a probe. RNA was harvested from cells grown to an OD650 of ≈1.2, representing late exponential phase.
DISCUSSION
With the advance of genomic information coupled with transcriptional analysis, it is now recognized that a complex regulatory network exists to control growth phase-dependent expression of a number of virulence determinants (1, 15, 40). The major players within this network appear to be the two-component systems (e.g., agrAC, saeRS, arlRS, and srrAB) and members of the SarA protein family (8, 12, 16, 18, 26, 44). Based on structure and sequence alignment, the SarA protein family can be divided into three subfamilies: the single-domain proteins (e.g., SarA and SarR), the double-domain proteins (e.g., SarS and SarU), and the MarR homologs (e.g., MgrA).
In previous studies (11, 32), we have shown that members of the single-domain (SarR on sarA expression) and double-domain (SarS on spa expression) proteins participate in the modulation of virulence determinants. The first member of the third SarA subfamily, MgrA, was originally identified as an important regulator of autolytic activity in S. aureus (21, 35). We have now characterized mgrA further with regard to the expression of virulence determinants and conclude that all three SarA subfamilies likely participate in the virulence regulatory network.
We have examined in this study four prototypic S. aureus strains, including RN6390, Newman, FDA486, and COL. Inactivation of mgrA in all these strains has led to downregulation of RNAIII of agr, decreased expression of hla, and upregulation of sarS and spa (Fig. 1 and 2). Phenotypically, decreased expression of hla and upregulation of spa in an mgrA mutant is consistent with an agr mutant phenotype. However, the effect of mgrA on intermediate regulatory genes such as sarS and sarT downstream of agr as well as target genes (direct versus indirect effect) has not been previously defined. In addition, we have also found that the transcription of sarA, an important regulatory locus that partially controls agr, is not affected in the mgrA mutant, suggesting that mgrA likely regulates hla and spa in a SarA-independent manner.
Regulation of hla expression in S. aureus is not mediated by a single locus but is multifaceted. In particular, regulators such as sarA, agr, and sae have been shown to upmodulate hla expression both directly and indirectly (5, 13, 17, 22, 42, 51), while sarT downregulates it (49). Similarly, spa expression is repressed by sarA and agr and promoted by sarT and sarS ( 10, 48, 52,). Added to this list now is mgrA, which is part of the regulatory cascade that controls agr and sarS to modulate hla and spa expression.
Analysis of the double sarT mgrA mutant (ALC3046) for hla transcription indicated that the mgrA-mediated effect did not occur via sarT. Likewise, this effect is also independent of sae, as sae transcription was unaltered in the mgrA mutant (Fig. 1B). While it would seem that reduced expression of RNAIII in the mgrA mutant would account for the downmodulation in hla expression, our finding that the hla transcript level in the mgrA agr double mutant (ALC2537) was lower than in the agr mutant (Fig. 2B) would suggest a dual effect of mgrA on hla. The dual mode of regulatory control by mgrA on hla was confirmed by gel shift assays (Fig. 3) as well as transcriptional analysis of the mgrA mutant with RNAIII provided in trans (Fig. 4). Thus, the mgrA gene product acts as a positive regulator for agr to promote hla transcription as well as binds to the hla promoter to augment gene transcription.
In a previous study, Luong et al (30) also discovered mgrA in a Tn917 transposon screen, searching for mutants that had altered capsular production in cap8+ strain Becker. Contrary to our finding here, they reported upregulation of alpha toxin and protein A synthesis in an mgrA mutant of strain Becker; unfortunately, the impact of mgrA on the expression of agr RNAIII was not evaluated in detail in that study. Because of this discrepancy, we sought to analyze RNAIII and hla expression in isogenic mgrA strains in four different genetic backgrounds, including RN6390, Newman, COL, and FDA486. All of the aforementioned mgrA mutants exhibited reduced levels of agr and hla expression compared with their parental counterparts. We have also analyzed hla transcription in the mgrA mutant of Becker and found no significant increase in the transcript level compared with the parental strain; transcriptional fusion studies with the hla promoter also confirmed the Northern blot results (data not shown). In distinction to our transcriptional approach, Luong et al. studied the phenotypic effect of mgrA on alpha-toxin production by hemolytic assays on blood agar plates. Whether altered levels of V8 protease as reported in the original mgrA mutant (21) account for the increase in alpha-toxin in the Becker strain is not clear. However, in subsequent experiments in which Luong et al. transduced the mgrA mutation from strain Becker to Newman, they also found reduced levels of α-hemolysin production and elevated levels of protein A synthesis (unpublished data). Thus, differences in the genetic background may have accounted for the divergent results. Although we previously were able to complement the mgrA mutation in trans (21), we could not entirely rule out a subtle polar effect of the mutation on neighboring genes that may have altered hla translation but not transcription. Studies are now in progress to assess if the mgrA mutation may have affected neighboring genes both upstream and downstream of the mgrA mutation.
Previous studies from our laboratory have shown that SarT positively modulates sarS expression, which, in turn, upregulates spa expression (48). Although the transcript levels of sarS and spa were increased in the mgrA mutant (Fig. 5 and 6), we discovered that sarT did not play a major role in spa regulation in mgr+ strains, similar to the pattern we observed for hla regulation. However, given that the sarS transcript level in the mgrA mutant was higher than that of the agr mutant (Fig. 5B), we speculate that the effect of mgrA on spa is likely to be both agr dependent and agr independent. The dependency on agr was confirmed by the repression of spa transcription in an mgrA mutant with RNAIII provided in trans (Fig. 8). Additionally, the finding that MgrA binds directly to the sarS promoter but not the spa promoter (last lanes in Fig. 3A, 3B, and 7) is consistent with a direct effect of MgrA on sarS to downmodulate spa expression. These data thus demonstrated bimodal regulation of protein A by the mgrA gene product.
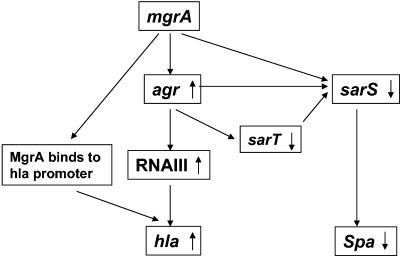
From these data, we propose that mgrA, similar to other members within the single-and double-domain SarA protein subfamilies, is an important global regulator of virulence determinants in S. aureus. We propose (Fig. 9) that the mgrA gene product likely regulates hla and spa expression bimodally, with the first mode being agr-dependent and the second mode agr independent. Interestingly, the upregulatory effect of mgrA on hla does not rely on repression of sarT, while its effect on spa is dependent on sarS and not sarT despite the known positive effect of sarT on sarS (48, 49).
FIG. 9.
Model for MgrA-mediated bimodal regulation of virulence genes in S. aureus. MgrA upregulates expression of RNAIII. RNAIII activates hla expression and downregulates sarS, leading to repression of spa. Besides the agr-dependent pathway, MgrA binds directly to the hla promoter to augment its activation. Similarly, MgrA binds to the sarS promoter to downregulate spa expression.
In our previous study, we described mgrA as a negative regulator of autolysis. Concomitantly, two other groups have described mgrA as a positive regulator of capsular polysaccharide synthesis (30) and possibly of norA, which encodes an efflux pump for mediating fluoroquinolone resistance (53). We have now added to this spectrum of effects the ability of mgrA to positively regulate agr as well as its direct effect on hla and sarS, eventually yielding what appears to be an agr-negative phenotype in an mgrA mutant. Given that mgrA is a negative regulator of autolysis and a positive regulator of agr, it seems logical that interference with MgrA may be a reasonable anti-infective strategy, since this approach would promote autolysis while minimizing the expression of agr, an important regulator of virulence determinants in S. aureus.
Acknowledgments
This work was supported by research grants AI47441 (A.L.C.) and AI54607 (C.Y.L.) from the NIH.
Editor: V. J. DiRita
REFERENCES
- 1.Arvidson, S., and K. Tegmark. 2001. Regulation of virulence determinants in Staphylococcus aureus. Int. J. Med. Microbiol. 291:159-170. [DOI] [PubMed] [Google Scholar]
- 2.Bayer, M. G., J. H. Heinrichs, and A. L. Cheung. 1996. The molecular architecture of the sar locus in Staphylococcus aureus. J. Bacteriol. 178:4563-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brunskill, E. W., and K. W. Bayles. 1996. Identification and molecular characterization of a putative regulatory locus that affects autolysis in Staphylococcus aureus. J. Bacteriol. 178:611-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang, S., D. M. Sievert, J. C. Hageman, et al. 2003. Infection with vancomycin-resistance S. aureus containing vanA resistance gene. N. Engl. J. Med. 348:1342-1347. [DOI] [PubMed] [Google Scholar]
- 5.Cheung, A. L., A. S. Bayer, G. Zhang, H. Gresham, and Y.-Q. Xiong. 2004. Regulation of virulence determinants in vitro and in vivo in Staphylococcus aureus. FEMS Microbiol. Lett. 1649:1-9. [DOI] [PubMed] [Google Scholar]
- 6.Cheung, A. L., M. G. Bayer, and J. H. Heinrichs. 1997. sar genetic determinants necessary for transcription of RNAII and RNAIII in the agr locus of Staphylococcus aureus. J. Bacteriol. 179:3963-3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheung, A. L., K. Eberhardt, and V. A. Fischetti. 1994. A method to isolate RNA from gram-positive bacteria and Mycobacteria. Anal. Biochem. 222:511-514. [DOI] [PubMed] [Google Scholar]
- 8.Cheung, A. L., J. M. Koomey, C. A. Butler, S. J. Projan, and V. A. Fischetti. 1992. Regulation of exoprotein expression in Staphylococcus aureus by a locus (sar) distinct from agr. Proc. Natl. Acad. Sci. USA 89:6462-6466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheung, A. L., and S. J. Projan. 1994. Cloning and sequencing of sarA: a gene required for the expression of agr. J. Bacteriol. 176:4168-4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheung, A. L., K. A. Schmidt, B. Bateman, and A. C. Manna. 2001. SarS, a SarA homolog repressible by agr, is an activator of protein A synthesis in S. aureus. Infect. Immun. 69:2448-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheung, A. L., and P. Ying. 1994. Regulation of α and β hemolysins by the sar locus of S. aureus. J. Bacteriol. 176:580-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheung, A. L., and G. Zhang. 2002. Regulations of virulence determinants in Staphylococcus aureus by the SarA protein family. Front. Biosci. 7:D1825-D1842. [DOI] [PubMed] [Google Scholar]
- 13.Chien, C.-T., A. C. Manna, S. J. Projan, and A. L. Cheung. 1999. SarA, a global regulator of virulence determinants in Staphylococcus aureus, binds to a conserved motif essential for sar dependent gene regulation. J. Biol. Chem. 274:37169-37176. [DOI] [PubMed] [Google Scholar]
- 14.Chien, Y., and A. L. Cheung. 1998. Molecular interactions between two global regulators, sar and agr, in Staphylococcus aureus. J. Biol. Chem. 237:2645-2652. [DOI] [PubMed] [Google Scholar]
- 15.Dunman, P. M., E. Murphy, S. Haney, D. Palacios, Tucker-Kellogg, S. Wu, E. L. Brown, R. J. Zagursky, D. Shlaes, and S. J. Projan. 2001. Transcriptional profiling based identification of S. aureus genes regualted by the agr and/or sarA loci. J. Bacteriol. 183:7341-7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fournier, B., A. Klier, and G. Rapoport,2001. The two-component system ArlS-ArlR is a regulator of virulence gene expression in Staphylococcus aureus. Mol. Microbiol. 41:247-261. [DOI] [PubMed] [Google Scholar]
- 17.Giraudo, A. T., A. L. Cheung, and R. Nagel. 1997. The sae locus of Staphylococcus aureus controls exoprotein synthesis at the transcriptional level. Arch. Microbiol. 168:53-58. [DOI] [PubMed] [Google Scholar]
- 18.Giraudo, A. T., C. Mansilla, A. Chan, C. Raspanti, and R. Nagel. 2003. Studies on the expression of regulatory locus sae in Staphylococcus aureus. Curr. Microbiol. 46:246-250. [DOI] [PubMed] [Google Scholar]
- 19.Giraudo. A. T., A. Calzolari, A. A. Cataldi, C., Bogni, and R. Nagel. 1999. The sae locus of Staphylococcus aureus encodes a two-component regulatory system. FEMS Microbiol. Lett. 177:15-22. [DOI] [PubMed] [Google Scholar]
- 20.Hiramatsu, K., H. Hanaki, T. Ino, K. Yabuta, T. Oguri, and F. C. Tenover. 1997. Methicillin resistant Staphylococcus aureus clincial strain with reduced vancomycin susceptibility. J. Antimicrob. Chemother. 40:135-136. [DOI] [PubMed] [Google Scholar]
- 21.Ingavale, S., Van W. Wamel, and A. L. Cheung. 2003. Characterization of RAT, an autolysis regulator in Staphylococcus aureus. Mol. Microbiol. 48:1451-1466. [DOI] [PubMed] [Google Scholar]
- 22.Janzon, L., and S. Arvidson. 1990. The role of the δ-hemolysin gene (hld) in the regulation of virulence genes by the accessory gene regulator (agr) in Staphylococcus aureus. EMBO.J. 9:1391-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ji, G., Beavis, R., and R. P. Novick. 1997. Bacterial interference caused by autoinducing peptide variants. Science 276:2027-2030. [DOI] [PubMed] [Google Scholar]
- 24.Ji, G., R. C. Beavis, and R. P. Novick,. 1995. Cell density control of staphylococcal virulence mediated by an octapeptide pheromone. Proc. Natl. Acad. Sci. USA 92:12055-12059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kahl, B., M. Goulian, Van W. Wamel, M. Herrmann, S. Simon, G. Kaplan, G. Peters, and A. L. Cheung. 2000. Staphylococcus aureus RN6390 replicates and induces apoptosis in a pulmonary epithelial cell line derived from a cystic fibrosis patient. Infect. Immun. 68:5385-5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kornblum, J., B. Kreiswirth, S. J. Projan, H. Ross, and R. P. Novick, 1990. Agr: a polycistronic locus regulating exoprotein synthesis in Staphylococcus aureus, p. 373-402.. In R. P. Novick (ed.), Molecular biology of the staphylococci. VCH Publishers, New York, N.Y.
- 27.Kupferwasser, L. I., M. R. Yeaman, C. C. Nast, D. Kupferwasser, Y. Q. Xiong, M. Palma, A. L. Cheung, and A. S. Bayer. 2003. Salicylic acid attenuates virulence in endovascular infections by targeting global regulatory pathways in Staphylococcus aureus. J. Clin. Investig. 112:222-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuroda, M., H. Kuroda, T. Oshima, F. Takeuchi, H. Mori, and K. Hiramatsu. 2003. Two-component system VraSR positively modulates the regulation of cell-wall biosynthesis pathway in Staphylococcus aureus. Mol. Microbiol. 49:807-821. [DOI] [PubMed] [Google Scholar]
- 29.Lowy, F. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339:520-532. [DOI] [PubMed] [Google Scholar]
- 30.Luong, T. T., S. W. Newell, and C. Y. Lee. 2003. mgr, a novel global regulator in Staphylococcus aureus. J. Bacteriology. 185:3703-3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maniatis, T., E. F. Fritsch, and J. Sambrook 1989. Molecular cloning, a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 32.Manna, A. C., and A. L. Cheung. 2001. Characterization of sarR, a modulator of sar expression in Staphylococcus aureus. Infect. Immun. 69:885-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manna, A. C., and A. L. Cheung. 2003. sarU, a sarA homolog, is repressed by SarT and regulates virulence genes in Staphylococcus aureus. Infect. Immun. 71:343-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manna, A. C., and A. L. Cheung. 2003. sarU, a sarA homolog, is repressed by SarT and regulates virulence genes in Staphylococcus aureus. Infect. Immun. 71:343-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manna, A. C., S. S. Ingavale, M. Maloney, Van W. Wamel, and A. L. Cheung. 2004. Characterization of sarV, a transcriptional regulator of the sarA family, is repressed by SarA and Rat/MgrA and regulates autolysis and virulence genes in Staphylococcus aureus. J. Bacteriol. 186:5267-5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martin, P. K., T. Li, D. sun, D. P. Biek, and M. B. Schmid. 1999. Role in cell permeability of an essential two-component system in Staphylococcus aureus. J. Bacteriol. 181:3666-3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McDevitt, D., P. Francois, P. Vaudaux, and T. J. Foster. 1994. Cloning and sequencing of the clumping factor of Staphylococcus aureus. Mol. Microbiol. 11:237-248. [DOI] [PubMed] [Google Scholar]
- 38.Novick, R. P., S. J. Projan, J. Kornblum, H. F. Ross, G. Ji, B. Kreiswirth, F. Vandenesch, and S. Moghazeh. 1995. The agr P2 operon: an autocatalytic sensory transduction system in Staphylococcus aureus. Mol. Gen. Genet. 248:446-458. [DOI] [PubMed] [Google Scholar]
- 39.Novick, R. P.1990. The staphylococcus as a molecular genetic system, p. 1-40. In R. P. Novick (ed.), Molecular biology of the staphylococci. VCH, New York, N.Y.
- 40.Novick, R. P. 2003. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 48:1429-1449. [DOI] [PubMed] [Google Scholar]
- 41.Novick, R. P., and D. Jiang. 2003. The staphylococcal saeRS system coordinates environmental signals with agr quorum sensing. Microbiology 149:2709-2717. [DOI] [PubMed] [Google Scholar]
- 42.Novick, R. P., H. F. Ross, S. J. Projan, J. Kornblum, B. Kreiswirth, and S. Moghazeh. 1993. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO.J. 12:3967-3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Palma, M., and A. L. Cheung,2001. Sigmabeta activity in Staphylococcus aureus is controlled by RsbU and an additional factor(s) during bacterial growth. Infect. Immun. 69:7858-7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pragman, A. A., J. M. Yarwood, T. J. Tripp, and P. M. Schlievert. 2004. Characterization of virulence factor regulation by SrrAB, a two-component system in Staphylococcus aureus. J. Bacteriol. 186:2430-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Projan, S. J. and R. P. Novick. 1997. The molecular basis of pathogenicity, p. 55-81. In K. B. Crossley and G. L. Archer (ed.), The staphylococci in human diseases. VCH, New York, N.Y.
- 46.Rothfork, J. M., Dessus- S. Babus, Van W. J. Wamel, A. L. Cheung, and H. D. Gresham. 2003. Fibrinogen depletion attenuates Staphyloccocus aureus infection by preventing density-dependent virulence gene upregulation. J. Immunol. 171:5389-5395. [DOI] [PubMed] [Google Scholar]
- 47.Schenk, S., and R. A. Laddaga. 1992. Improved method for electroporation of Staphylococcus aureus. FEMS Microbiol. Lett. 94:133-138. [DOI] [PubMed] [Google Scholar]
- 48.Schmidt, K. A., A. C. Manna, and A. L. Cheung. 2003. sarT influences sarS expression in Staphylococcus aureus. Infect. Immun. 71:5139-5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schmidt, K. A., A. C. Manna, S. Gill, and A. L. Cheung. 2001. SarT: a repressor of alpha-hemolysin synthesis in Staphylococcus aureus. Infect. Immun. 69:4748-4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sievert, D. M., M. L. Boulton, G. Stoltman, et al. 2002. Staphylococcus aureus resistance to vancomycin-United States 2002. Morb. Mortal. Wkly. Rep. 51:565-566. [PubMed] [Google Scholar]
- 51.Steinhuber, A., C. Goerke, M. G. Bayer, G. Doring, and C. Wolz. 2003. Molecular architecture of the regulatory locus sae of Staphylococcus aureus and its impact on expression of virulence factors. J. Bacteriol. 185:6278-6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tegmark, K., A. Karlsson, and S. Arvidson. 2000. Identification and characterization of SarH1, a new global regulator of virulence gene expression in Staphylococcus aureus. Mol. Microbiol. 37:398-409. [DOI] [PubMed] [Google Scholar]
- 53.Truong-Bolduc, Q. C., X. M. Zhang, and D. C. Hooper. 2003. Characterization of NorR protein, a multifunctional regulator of norA expression in Staphylococcus aureus. J. Bacteriol. 185:3127-3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu, S., de Lencastre, H., and A. Tomasz. 1996. Sigma-B, a putative operon encoding alternate sigma factor of Staphylococcus aureus RNA polymerase: molecular cloning and DNA sequencing. J. Bacteriol. 178:6036-6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yarwood, J. M., J. K. McCormick, and P. M. Schlievert. 2001. Identification of a novel two-component regulatory system that acts in global regulation of virulence factors of Staphylococcus aureus. J. Bacteriol. 183:1113-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]