Abstract
An important step in Salmonella enterica serovar Typhimurium virulence is the ability to invade the intestinal epithelium. The invasion process requires a large number of genes encoded on Salmonella pathogenicity island 1 (SPI-1) at centisome 63 as well as genes located in other positions throughout the chromosome. Expression of the invasive phenotype is tightly regulated by environmental cues that are processed by a complex regulatory scheme. A central player in the invasion regulatory pathway is the HilA protein, which is transcriptional activator belonging to the OmpR/ToxR family. A number of positive regulators (hilC, hilD, fis, sirA/barA, csrAB, phoBR, fadD, envZ/ompR, and fliZ) and negative regulators (hha, hilE, lon, ams, phoPc and pag) have been identified that are able to alter expression of hilA transcription. Recent work has found that hilA transcription requires the HilD protein for activation. Other work has emphasized the importance of HilE as a negative regulator of hilA. Overexpression of hilE superrepresses hilA transcription, as well as the invasive phenotype. Two-hybrid experiments suggest that HilE exerts its regulatory influence on hilA through protein-protein interactions with HilD as the protein does not bind to the hilA promoter nor does it affect hilD transcription. As it seems likely that hilE plays an important role in translating environmental signals into invasion gene regulation, we have attempted to identify how the hilE gene itself is regulated. Our results indicate that the fimYZ genes, response regulatory proteins involved in type 1 fimbrial gene expression and recently implicated in motility gene regulation, are important activators of hilE expression. These findings indicate that invasion gene expression is coregulated with motility and adherence and provide experimental evidence that the expression of these virulence phenotypes is a subset of the overall regulation of bacterial physiology.
Pathogenic Salmonella species are an important cause of disease throughout the world and cause infections ranging from self-limiting gastroenteritis to life threatening typhoid fever. Efforts to understand the mechanisms by which Salmonella spp. establish infection in a human host have identified several interactions that include adherence, invasion, and the ability of the bacteria to grow within host cells. Collectively, the various virulence mechanisms of the bacteria determine the ability to interact with and colonize the host in the process known as infectious disease.
Adherence to tissue culture cells is mediated in Salmonella spp. by mannose-sensitive type 1 fimbriae (12, 13). Type 1 fimbrial structures are assembled by transporting the structural proteins through the bacterial membrane in the chaperone-usher pathway, followed by assembly into the rigid appendages that facilitate adherence to host cells (25, 49). The structural components of Salmonella type 1 fimbriae are homologous to type 1 fimbriae of Escherichia coli, but the regulatory mechanisms governing expression of Salmonella type 1 fimbriae appear to be unique. The Salmonella fim gene cluster contains three genes, fimW, fimY, and fimZ, that affect expression of the structural subunit gene fimA. Genetic experiments have demonstrated that both FimY and FimZ are necessary for expression of the fim gene cluster, but DNA binding experiments have demonstrated that FimZ is able to bind to the fimA promoter in the absence of the FimY protein (47, 55). Interestingly, fimZ belongs to the response regulator family of proteins with highest homology to bvgA of Bordetella pertussis, but no sensor kinase has been identified as its partner (55).
Salmonella invasion into host cells occurs through a process by which effector proteins are transferred into cells via the action of a type III secretion system (15, 16). The expression of the invasive phenotype is tightly regulated by a variety of positive and negative regulatory genes, in response to environmental conditions. The hilA gene is a member of the OmpR/ToxR family of transcriptional activators that play a critical role in the transcription of genes encoding components of the type III secretion system as well as the invF gene, which specifically activates genes encoding effector proteins involved in the invasion process (4, 29). In addition to the hilA-dependent pathway of invasion gene activation that many laboratories have described and characterized, other groups have reported hilA-independent invasion gene activation pathways (1, 20, 42). That work has demonstrated that AraC-like proteins (RtsA/B, HilC, and HilD) can activate a subset of invasion genes in the absence of the hilA gene. However, the hilD and hilC genes (19, 42, 43), which encode AraC-like transcriptional activators, are also important positive regulatory elements of hilA. Mutation of hilD leads to a significant decrease in hilA expression (14-fold) and a >50-fold decrease in invasiveness. Recent work has demonstrated that both HilD and HilC bind to sequences upstream of the hilA promoter (39, 44) and that the presence of HilD is required for hilA activation even in the absence of several negative regulatory elements (9). Several other genes have been identified as being important in activation of hilA expression, including csrAB, sirA/barA, phoBR, fadD, envZ, fliZ, and fis (2, 3, 26, 32, 53). The mechanism of action of these genes in invasion regulation is under investigation.
Several elements that negatively affect hilA expression have also been identified. The phoP(Con) mutant carries a point mutation in the phoQ gene that results in constitutive phosphorylation of phoP. This event has a dominant negative effect on hilA expression and results in a noninvasive phenotype (5, 7). The hha gene has been identified as a negative modulator of hilA expression as hha mutations increase hilA expression and overexpression of hha significantly decreases hilA expression and the invasiveness of Salmonella spp. (23). A mutation in the Lon protease has also recently been shown to decrease both hilA expression and Salmonella invasiveness (48).
A search for repressors of hilA identified ams, pag, and hilE as negative regulatory elements of hilA (22). Mutation of each of these genes results in overexpression of hilA. Recently, more extensive characterization of the hilE gene has been performed (6). That work demonstrated that overexpression of hilE completely represses hilA and results in a noninvasive phenotype. Efforts to identify the mechanism of regulation failed to show that the HilE protein binds to the hilA promoter or that hilE regulates hilD transcription. However, bacterial two-hybrid experiments demonstrated that HilD and HilE bind to one another, suggesting that a possible mechanism of action for HilE is to disrupt HilD activation of hilA by disrupting its function. Finally, the hilE gene was found to reside in a region of the Salmonella chromosome that resembles a pathogenicity island and is apparently unique to Salmonella strains.
Due to the evidence indicating that hilE plays a role in controlling hilA expression, we have performed experiments aimed at understanding how HilE is involved in the regulatory cascades that upregulate or downregulate expression of the SPI-1 Salmonella invasion genes. Those experiments have revealed that the fimYZ genes regulate hilE expression. These findings indicate that fimYZ are important components of a global regulatory network, as work by others has demonstrated that fimZ (and fimY) also regulates both motility and adherence (14).
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are shown in Table 1. Bacteria were routinely grown in Lennox broth (Gibco-BRL) containing the appropriate antibiotics added at the following concentrations: ampicillin, 100 μg/ml; kanamycin, 25 μg/ml; streptomycin, 100 μg/ml; chloramphenicol, 10 μg/ml; and tetracycline, 25 μg/ml. For the β-galactosidase assays, bacterial cultures were shaken at 250 rpm at 37°C to an optical density at 600 nm of ≈0.4 to 0.5, which corresponds to ≈4 × 108 to 5 × 108 CFU/ml. For the invasion assays, low-oxygen conditions were created by inoculating 5 ml of Lennox broth with 10 μl of a stationary-phase culture and incubating statically overnight at 37°C until an optical density at 600 nm of 0.4 to 0.5 was reached (27, 41). Plasmid purifications were performed with kits manufactured by Qiagen Inc., and molecular manipulations were performed with standard protocols (34).
TABLE 1.
Bacterial strains and plasmids used in the study
| Strain or plasmid | Descriptiona | Reference(s) or source |
|---|---|---|
| Escherichia coli DH12S | mcrA Δ(mrr-hsdRMS-mcrBC) F′ lacIq ΔM15 | Invitrogen |
| Salmonella enterica serovar Typhimurium | ||
| BJ2710 | SL1344 derivative containing LT2 fimH gene | This work |
| BJ2710 fimZ-kan | fimZ::kan in BJ2710, Kanr | This work |
| BJ2462 | hilE::cam in SL1344, Camr | 6 |
| LB5010 | Strain LT2 containing a complete fim gene cluster | 56 |
| LBZ100 | LB5010 fimZ::kan, Kanr | 56 |
| SL1344 | Wild-type virulent strain | 54 |
| SL1344 fimZ-kan | fimZ::kan in SL1344 transduced from LBZ100, Kanr | This work |
| SL1344H3 | SL1344 strain containing a fimH::kan insertion, Kanr | 8 |
| Plasmids | ||
| pACYC184 | Camr Tetr | 10 |
| pISF182 | fimYZ cloned into pACYC184, Camr | 55 |
| pLS31 | pRW50 vector encoding −497 to +420 of hilA fused to lacZY, Tetr | 43 |
| pKD3 | pANTSγ vector containing the chloramphenicol template gene cloned from pSC140, Ampr | 17 |
| pKD46 | Temperature-sensitive red helper plasmid expressing araC-ParaB and γβexo from λ phage, Ampr | 17 |
| pMAB54 | pGEM-T vector encoding hilE promoter from −886 to +121, Ampr | This work |
| pMAB55 | pGEM-T vector encoding hilE promoter from −886 to +121 and promoterless lacZY genes downstream of the promoter, Ampr | This work |
| pMAB56 | pZC320 vector encoding hilE promoter from −886 to +121, Ampr | This work |
| pMAB69 | hilE promoter −886 to +121 cloned in pRW50, Tetr | This work |
| pMAB95 | pGEM-T encoding the hilE promoter fragment −191 to +121, Ampr | This work |
| pMAB97 | pGEM-T vector encoding the hilE promoter fragment −886 to −271, Ampr | This work |
| pMAB98 | pGEM-T vector encoding the hilE promoter from −886 to +121 minus the −271 to −191 region, Ampr | This work |
| pMAB99 | pMAB98 ligated into pRW50, Ampr Tetr | This work |
| pMAB102 | pRW50 lacZY-expressing vector with the hilE promoter missing −271 to −191 relative to the putative translation start site inserted upstream to lacZY, Tetr | This work |
| pMRP9-1 | GFP-expressing plasmid, Camr | E. P. Greenberg |
| pRTP::Tn5 | Tn5 transposon inserted onto plasmid pRTP1 which carries the rpsL gene (Strs) Ampr Kanr | 27, 46 |
| pRW50 | lacZ reporter vector, Tetr | 31 |
| pZC320 | Mini-F, Ampr | 45 |
Tetr, tetracycline resistant; Ampr, ampicillin resistant; Kanr, kanamycin resistant; Camr, chloramphenicol resistant.
Plasmid and strain construction.
A hilE reporter plasmid, pMAB56, was created by amplifying the region from −886 to +121 in relation to the putative hilE translation start site from the chromosome with the primers hilE5′ (5′-GGATCCTTTGCGGATTACTGCCGTT-3′) and hilE3B (5′-AAGCTTCTTCAATACCGTCCAGTT-3′) and ligating the DNA fragment upstream of the lacZY genes to form plasmid pMAB55. The hilE-lacZY fragment was then removed from pMAB55 with NsiI and SphI and cloned into pZC320 (45) to form the single-copy reporter pMAB56. Sequencing of the insert confirmed the presence and sequence of the hilE promoter. The hilE reporter pMAB69 was created by amplifying the hilE promoter (−886 to +121) from the chromosome with the primers hilE5′/Eco (5′-GAATTCTTTGCGGATTACTGCCGTT-3′) and hilE3B/Bam (5′-GGATCCCTTCAATACCGTCCAGTT-3′) and cloning the fragment upstream of the lacZY genes in the vector pRW50 (31).
The hilE reporter pMAB102, with the putative fimZ binding sites from −271 to −191 deleted relative to the translation start site, was constructed by amplifying the hilE promoter fragment from −191 to +121 with the primers hilEdel695 (5′-GAATTCGATATTTCTTTTTTGATATGGTTC-3′) and hilE3B/Bam. The resulting fragment was ligated into pGEM-T to create pMAB95. The hilE promoter fragment from −886 to −271 was amplified with the primers hilE5′/Eco and hilE15 (5′-AGATCTCCTTTTCACATCAATGGGTTTT-3′), and the resulting fragment was ligated into pGEM-T to form pMAB97.
By overlapping PCR, the fragments from pMAB95 and pMAB97 were used to create the deleted hilE promoter fragment (30). Briefly, pMAB95 was amplified with the primers hilE5′Eco and hilE21 (5′-TAAGAACCATATCAAAAAAGATGGGTTTTTTAGACTTTTG-3′), while in a separate reaction pMAB97 was amplified with the primers hilE20 (5′-CAAAAGTCTAAAAAACCCATCTTTTTTGATATGGTTC-3′) and hilE3B/Bam. Primers hilE20 and hilE21 contain a tail that is homologous to the other PCR fragments on either side of the −271 to −191 deleted region. After amplification, the two PCR products were mixed and amplified with the primers hilE5′Eco and hilE3B/Bam. This PCR product was then ligated into pGEM-T, creating pMAB98. The reporter vectors pRW50 and pMAB98 were then digested with BamHI followed by ligation to create pMAB99. Finally, the pGEM-T portion of pMAB99 was removed by digesting the plasmid with EcoRI, followed by religation of the ends to create pMAB102, which contains the hilE promoter missing the −271 to −191 region relative to the putative translation start site, fused to a lacZY reporter.
Plasmid pISF187 is a derivative of plasmid pISF182 in which a universal translational terminator was introduced into a unique EcoRV site to inactivate the fimY gene (55). Subsequent work revealed that the plasmid carries a functional fimZ gene, as it could complement a fimZ mutant. Plasmid pISF189 is a derivative of plasmid pISF182 in which a universal translational terminator was introduced into a unique PvuI site to inactivate the fimZ gene (55). Subsequent work revealed that the plasmid carries a functional fimY gene, as it could complement a fimY mutant.
Tissue culture conditions and cell invasion assays.
HEp-2 tissue culture cells (38) were maintained in RPMI 1640 medium containing 10% fetal bovine serum. The cells were passaged every 2 to 4 days as required. Invasion assays were conducted with the bacteria being grown under inducing conditions with previously described protocols (27, 41).
β-Galactosidase assays.
β-Galactosidase assays were conducted on bacterial cultures with the standard method described by Miller (36).
P22-mediated transductions.
Phage P22 HT int was used to move antibiotic resistance gene insertions between strains as described previously (18). Transductants were selected on Lennox agar containing the necessary antibiotic and 10 mM EGTA to prevent P22 reinfection. Transductants were purified twice on Lennox EGTA agar prior to use of the colonies.
Tn5 transposon mutagenesis.
In order to screen for mutations that increased hilE expression, a previously described protocol was used (22). Briefly, EE251 cells were transformed with pRTP1::Tn5. This plasmid contains a Tn5 transposon (kanamycin resistance), the wild-type rpsL gene, which confers streptomycin sensitivity to the bacteria, and an ampicillin resistance marker. The bacteria were plated onto Lennox agar with ampicillin and kanamycin at a concentration of 500 CFU per plate. Colonies were replica plated onto Lennox streptomycin kanamycin agar and grown overnight at 37°C. Growth on Lennox agar with streptomycin and kanamycin indicated that Tn5 had moved randomly from pRTP1 into the S. enterica serovar Typhimurium chromosome and that the delivery plasmid, pRTP1, had been lost. Next, pMAB69, a hilE-lacZY reporter plasmid, was transformed into the pools of Tn5 mutants followed by plating on MacConkey plates with tetracycline and kanamycin.
Colonies that exhibited an increased red colony phenotype compared to wild-type Salmonella colonies were isolated. To identify the site of the Tn5 insertion, chromosomal DNA was isolated from colonies with increased hilE expression. Chromosomal DNA was digested with BamHI to completion, and fragments 2 to 5 kb in size were gel purified. The vector pACYC184 (10) was digested with BamHI, and the isolated chromosomal fragments were ligated into the vector. The ligated DNA was transformed into DH12S and plated on Lennox-kanamycin agar to identify a clone carrying the kanamycin resistance gene from Tn5 and flanking chromosomal DNA. To identify the junction between the Tn5 sequence and the chromosome, the oxregA primer (5′-TTCAGGACGCTACTTGTG-3′) was used to sequence the cloned DNA. The sequence obtained was compared to the annotated S. enterica serovar Typhimurium LT2 genomic sequence (35) to identify the Tn5 insertion site.
RNA isolation and primer extension.
To determine where hilE transcription begins, we first isolated RNA by incubating DH12S/pMAB69/pISF182 and DH12S/pMAB102/pISF182 by growing the two E. coli strains overnight in 125 ml of Lennox broth. The bacteria were pelleted, and the total RNA was isolated (11). For primer extension analysis, the primer hilE23 (5′-GAACGTTCCATTTTCCAGCCA-3′) was end labeled with [γ-32P]ATP, and the primer extension reaction was performed with the Primer Extension System avian myeloblastosis virus reverse transcriptase kit (Promega).
To determine the exact nucleotide start point for hilE transcription, the previously labeled primer hilE23 was used in sequencing reactions performed with the Fmol DNA cycle sequencing system (Promega). The products for the primer extension reactions and the products from the sequencing reactions were electrophoresed on a 6% acrylamide-7 M urea sequencing gel. The sequencing gel was dried onto filter paper and visualized after exposure to film.
RESULTS
HilE-lacZY expression in S. enterica serovar Typhimurium and in Escherichia coli.
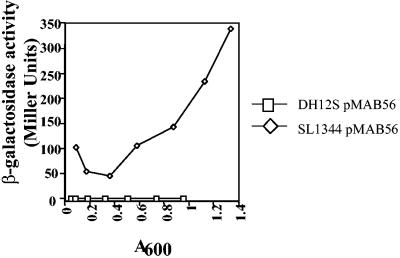
The hilE-lacZY reporter plasmid pMAB56 was created to measure hilE expression throughout the growth cycle in Salmonella enteritidis serovar Typhimurium and additionally whether hilE could be expressed within Escherichia coli. Previous searches have shown that hilE is a Salmonella-specific gene (6). When the reporter carried by S. enterica serovar Typhimurium strain SL1344 was assayed for activity, it was clear that hilE expression was regulated, since expression varied significantly over the course of the growth curve. At inoculation, β-galactosidase levels were relatively high and then decreased as the culture entered the early stages of logarithmic growth (Fig. 1). It is likely that the levels of β-galactosidase at early time points are residual from the stationary phase of the starting culture and that the subsequent decrease in activity is a result of cellular division. The hilE-lacZY levels were the lowest (45.5 Miller units) when the culture density was between optical densities at 600 nm of 0.17 and 0.36. Measurements of β-galactosidase levels throughout the rest of the growth cycle showed a continual increase in hilE-lacZY expression into the stationary phase.
FIG. 1.
Expression profile of a hilE-lacZY reporter throughout the growth curves of E. coli and S. enterica serovar Typhimurium. Strains were grown with shaking overnight in Lennox broth and then reinoculated into 20 ml of Lennox broth. Samples were taken every 40 min after the cultures began actively growing. Levels of β-galactosidase activity were determined for each sample taken. Plasmid pMAB56 is a single-copy mini-F plasmid that carries 1,000 bp of the putative hilE promoter fused upstream of a promoterless lacZY gene. Experiments were performed at least three times.
Analysis of the expression of hilE-lacZY from plasmid pMAB56 indicated that the hilE gene is virtually not expressed in E. coli throughout the growth curve. This result suggests that E. coli lacks regulatory factors necessary for hilE expression. Unpublished work from our laboratory is consistent with this idea since BLAST searches of the genome databases have consistently failed to identify any gene with significant similarity to hilE. Current information indicates that hilE is a Salmonella-specific gene, and genes that control its expression may also have regulatory properties unique to Salmonella spp.
Identification of the response regulators fimYZ as activators of the hilE repressor gene.
As our results suggest that hilE is controlled by a Salmonella-specific factor, we began a screen for S. enterica serovar Typhimurium genes that affect hilE-lacZY expression by performing Tn5 mutagenesis (22). We identified several mutants that displayed increased hilE-lacZY expression. Two isolates that overexpressed hilE-lacZY from the plasmid reporter sevenfold more than the S. enterica serovar Typhimurium parent strain were selected for further characterization. Identification of the insertion sites of each of the Tn5 insertions, by cloning and sequencing, revealed that each was located in a different position upstream of the fimY and fimZ genes, which are involved in type 1 fimbrial gene regulation (47, 50, 56) and regulation of Salmonella motility (14).
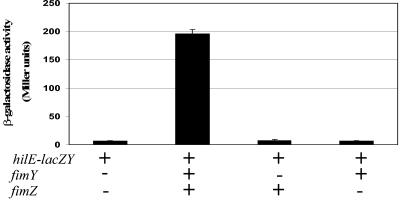
To test the possibility that the Tn5 chromosomal insertions were increasing fimY and fimZ expression, we introduced a medium-copy plasmid (p15A replicon) that expresses fimYZ into E. coli and assayed what effect increasing the expression of fimYZ would have on hilE-lacZY expression. As shown in Fig. 2, the presence of the fimYZ-overexpressing plasmid, pISF182, in DH12S/pMAB69 increased expression of the hilE-lacZY reporter 31-fold. Next, we examined whether fimY only, fimZ only, or both fimY and fimZ were required for hilE activation. It has been shown previously that FimZ can bind to the promoter of the fimA gene cluster but that FimY is also required to activate gene expression of fimA (47, 55). Accordingly, we assayed hilE-lacZY expression in E. coli with the fimY fimZ+ plasmid pISF187 and the fimY+ fimZ plasmid pISF189. Both the E. coli strain DH12S/pMAB69/pISF187 (fimY fimZ+) and the E. coli strain DH12S/pMAB69/pISF189 (fimY+ fimZ) expressed hilE-lacZY at levels comparable to that of the E. coli strain containing the hilE-lacZY reporter alone (Fig. 2). These results indicate that both fimY and fimZ are required to induce hilE-lacZY expression in E. coli.
FIG. 2.
Overexpression of fimYZ activates hilE-lacZY expression in E. coli. Strains were grown with shaking in Lennox broth to the late stationary phase. Each strain carried plasmid pMAB69, which is a low-copy-number hilE reporter carried on the pRW50 vector. Plasmid pISF182 has the fimYZ genes cloned into pACYC184 and expressed from the tetracycline promoter. Plasmids pISF187 (fimZ+ fimY) and pISF189 (fimZ fimY+) are identical to pISF182 except for the insertion of a transcriptional terminator (see Materials and Methods). E. coli carrying only pISF187 or only pISF189 was not induced for hilE-lacZY expression, indicating that both fimY and fimZ are necessary for the expression of hilE. Expression levels were determined by measuring β-galactosidase activity. Experiments were performed at least three times.
We next determined if overexpression of fimYZ increases hilE-lacZY expression in S. enterica serovar Typhimurium. Analysis of S. enterica serovar Typhimurium strain SL1344 containing the hilE reporter pMAB69 showed that hilE-lacZY expression increased 5.1-fold (55.3 ± 2.5 units versus 280.5 ± 6.5 units) by introduction of the fimYZ-expressing plasmid pISF182 (Fig. 3). Additionally, deletion of the fimZ gene affected hilE-lacZY expression. A strain lacking a functional fimZ gene, BJ2710 fimZ-kan, had 2.7-fold-reduced expression of β-galactosidase from pMAB69 compared to the parent strain (Fig. 3). These results indicate that the fimYZ genes function to activate expression of the Salmonella invasion gene repressor hilE.
FIG. 3.
Overexpression of fimYZ in S. enterica serovar Typhimurium leads to activation of a hilE-lacZY reporter. SL1344 (wild type), SL1344/pISF182 (fimYZ overexpressing), and SL1344 fimZ-kan, each containing the hilE-lacZY reporter pMAB69, were grown with shaking in Lennox broth to the late stationary phase. Levels of expression were measured by β-galactosidase activity. Each assay was performed at least three times.
Overexpression or deletion of fimYZ affects the expression of the invasion regulator hilA in S. enterica serovar Typhimurium.
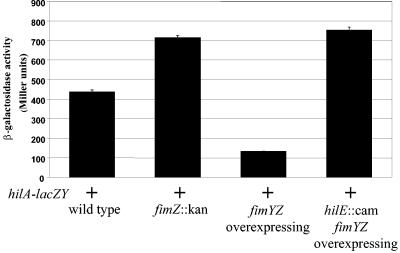
Previously, our research group has shown that variations in hilE expression have a direct effect on the level of hilA expression in S. enterica serovar Typhimurium (6). To extend those observations, we wanted to determine if deletion or overexpression of the fimYZ genes would have corresponding effects on hilA expression. We conducted a β-galactosidase assay with the hilA-lacZY reporter pLS31. When wild-type S. enterica serovar Typhimurium SL1344 was assayed, it expressed 437.4 ± 13.5 Miller units from the hilA-lacZY reporter (Fig. 4). Chromosomal deletion of fimZ in the SL1344 strain increased hilA::lacZY expression 1.6-fold (715 ± 11.5 Miller units). Overexpression of the fimYZ genes from plasmid pISF182 decreased hilA-lacZY expression 3.3-fold (131.8 ± 3.2 Miller units) compared to the wild-type SL1344/pLS31 (437.4 ± 13.5 Miller units), which is consistent with effects observed on the hilE-lacZY reporter.
FIG. 4.
Regulatory influence of fimYZ on hilA-lacZY expression. Deletion of fimZ increased hilA-lacZY expression, while overexpression of fimYZ within S. enterica serovar Typhimurium repressed an hilA-lacZY reporter. A deletion of the hilE gene eliminated the effect of fimYZ overexpression on hilA expression. Cultures were grown to the late stationary phase in Lennox broth. Levels of lacZ activity were measured by the β-galactosidase assay. Each assay was performed in triplicate and repeated three times.
To demonstrate in a more direct manner that the hilE gene is mediating the effects of fimZ on hilA, an hilE mutant Salmonella strain was constructed by the method developed by Datsenko and Wanner (17). The expression of the hilA-lacZY reporter was then measured in the hilE mutant strain when fimYZ was expressed from pISF182. Repression by fimYZ was lost in this strain, as β-galactosidase activity from hilA-lacZY was 1.7-fold higher than from the wild-type strain with hilA-lacZY, which was nearly identical to expression levels when a fimZ mutation was present. These results provide additional evidence that the fimYZ genes are acting as negative regulatory elements on hilA transcription via the activity of the HilE invasion gene repressor.
Overexpression or deletion of fimYZ affects the ability of S. enterica serovar Typhimurium to invade tissue culture cells in vitro.
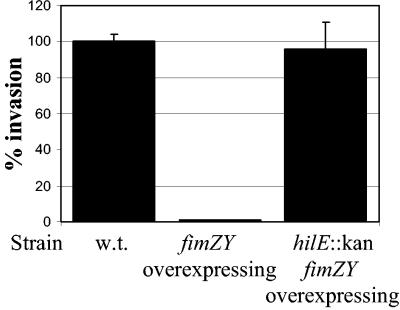
The experiments described above have demonstrated that increasing expression (by an overexpressing plasmid) or reducing expression (by deletion) of fimYZ has effects on both hilE and hilA expression levels. We next examined whether alterations in regulatory gene expression correlated with changes in the ability of S. enterica serovar Typhimurium to invade HEp-2 tissue culture cells. The S. enterica serovar Typhimurium strain BJ2710 was used as the invasive parental control, and invasiveness was arbitrarily set at 100% ± 4.2% when the bacteria were grown in invasion-inducing conditions (Fig. 5). Strain BJ2710/pISF182, which overexpresses fimYZ, had dramatically reduced levels of invasiveness (≈100-fold). This reduction in cellular entry indicates that the activities of fimYZ are required for determining whether S. enterica serovar Typhimurium will express the invasion machinery. Finally, the requirement for hilE in the repressing activity of fimYZ on S. enterica serovar Typhimurium invasion was assessed. The invasiveness of strain SL1344 hilE/pISF182 was measured and found to restore cellular entry to 96% ± 14.6%. Therefore, the repression of invasion observed in the S. enterica serovar Typhimurium strain carrying the fimYZ plasmid pISF182 requires a functional hilE gene.
FIG. 5.
Overexpression of fimYZ leads to repression of invasion of HEp-2 cells. Strains were grown under low-oxygen and high-osmolarity conditions. The deletion of hilE restored S. enterica serovar Typhimurium invasiveness in HEp-2 cells even when fimYZ were overexpressed. Values are presented as a percentage of invasion by the wild-type strain, with the invasiveness of that strain being set to 100%.
Deletion of sequences in the hilE promoter eliminates the ability of fimYZ to induce hilE expression in Escherichia coli.
As a result of our findings, we began an analysis of the hilE promoter in an effort to determine how the fimYZ genes exert their influence on the transcription of hilE. Published work has shown that the FimZ protein binds to the fimA promoter at a DNA region with a tandem repeat of the motif AATAAGA separated by 20 bp (56). Analysis of the hilE promoter identified the same AATAAGA sequence as that observed in the fimA promoter, although the repeats were separated by 47 bp. This motif was found to be 202 bp upstream of the putative translational start site of the hilE protein. Experiments were initiated to determine whether deletion of the putative FimZ binding site would eliminate the ability of fimYZ gene products to induce hilE expression.
We first deleted the putative FimYZ binding sequences in the hilE promoter with an overlapping PCR protocol described previously (30). The hilE-lacZY reporter plasmid pMAB102 is identical to the wild-type hilE-lacZY reporter plasmid pMAB69 except for the deletion of 80 bp encompassing the putative FimYZ binding site. Following construction of pMAB102, we performed experiments to measure the importance of the putative FimYZ binding sites in activation of hilE transcription.
E. coli carrying the hilE-lacZY reporter plasmid pMAB69 had typically low expression of the fusion reporter, which was induced 26.4-fold by the presence of the fimYZ plasmid pISF182 (Fig. 6A). E. coli carrying the hilE-lacZY fusion plasmid with the promoter deletion, pMAB102, expressed 135 ± 5.9 Miller units of β-galactosidase from hilE. Introduction of pISF182 (fimYZ) did not increase expression from the hilE promoter but decreased it slightly to 116.9 ± 5.4 Miller unit of β-galactosidase. Apparently, the deletion of the putative FimYZ binding sites had the unanticipated consequence of increasing the basal level expression of the hilE promoter in E. coli. Since basal expression of the hilE promoter, carrying the deletion of the putative FimYZ binding sites, increased in E. coli, we examined the behavior of the hilE-lacZY reporter plasmids in S. enterica serovar Typhimurium. As shown in Fig. 6B, the fimYZ-overexpressing plasmid pISF182 increased hilE-lacZY expression greater than fivefold in S. enterica serovar Typhimurium. However, the hilE promoter deletion construct was unresponsive to overexpression of fimYZ, and the levels of expression of hilE in the pMAB102 deletion plasmid remained low when present in S. enterica serovar Typhimurium. We therefore conclude that the AATAAGA tandem repeat in the hilE promoter is required for fimYZ induction of hilE transcription in S. enterica serovar Typhimurium.
FIG. 6.
FimYZ activation of the hilE promoter requires the presence of sequences in the hilE promoter in E. coli and S. enterica serovar Typhimurium. Expression values for the hilE-lacZY reporter plasmids in E. coli are shown in panel A. The E. coli strains DH12S/pMAB69, DH12S/pMAB69/pISF182, DH12S/pMAB102, and DH12S/pMAB102/pISF182 were grown with shaking overnight in Lennox broth. Expression values for hilE-lacZY reporter plasmids in S. enterica serovar Typhimurium are shown in panel B. The S. enterica serovar Typhimurium strains SL1344 ΔhilE/pMAB69, SL1344 ΔhilE/pMAB69/pISF182, SL1344 ΔhilE/pMAB102, and SL1344 ΔhilE/pMAB102/pISF182 were grown with shaking overnight in Lennox broth. Plasmid pMAB69 encodes the hilE-lacZY reporter. Plasmd pMAB102 encodes the hilE-lacZY reporter with putative FimYZ binding sites deleted from the promoter. Plasmid pISF182 encodes functional fimY and fimZ genes. β-Galactosidase assays were performed at least three times.
Mapping of the hilE transcriptional start site.
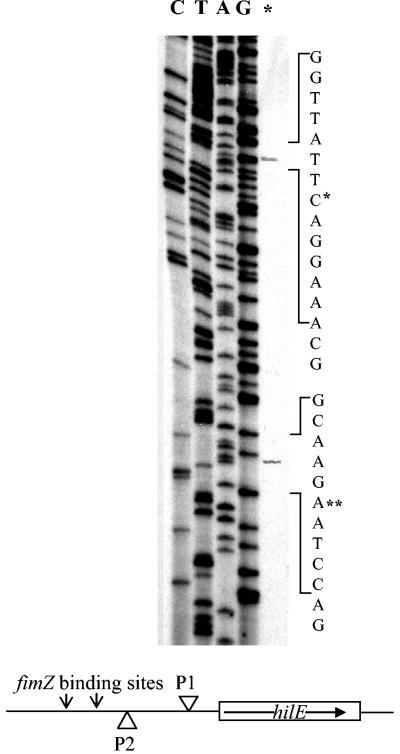
With the above information, we wanted to confirm that the hilE transcriptional start site was in close proximity to the putative FimYZ binding sites. With a primer extension assay, we mapped the transcriptional start site of the hilE mRNA. As shown in Fig. 7, the hilE transcript initiates at two independent sites, designated P1 and P2. The first transcript, designated P1, initiates at an adenosine which is 55 bp upstream of the putative ATG start codon. The P1 transcript starts 147 bp downstream of the putative FimYZ binding site motif. With the FimYZ binding site being quite far upstream of the P1 transcript, it seemed unlikely that the binding of FimYZ would play a significant role in activating transcription of the P1 hilE mRNA transcript. While speculative, it is possible that the increase in hilE expression seen in the pMAB102 hilE promoter deletion plasmid may be due to the increased availability of this site for hilE mRNA initiation.
FIG. 7.
Mapping the hilE transcriptional start site in relation to the putative translation start site for hilE and the putative FimYZ binding site. The primer extension products from pMAB69 and pMAB102 were primed with hilE23 and run alongside dideoxy sequencing reactions (lanes marked CTAG). The 5′ end of the hilE mRNA appears to initiate at two positions. The first mRNA initiates at an adenosine 55 bp upstream of the putative ATG start codon. The second transcription start site initiates at an adenosine 148 bp upstream of the putative ATG start codon. This second transcript starts 54 bp downstream of the putative FimYZ binding sites.
The second mRNA transcript, designated P2, initiates at an adenosine that is 148 bp upstream of the putative ATG start codon. The P2 mRNA transcript starts 54 bp downstream of the putative fimZ binding site motif. The positioning of the putative FimYZ binding site in relation to the P2 transcriptional mRNA start site would seem to make it more likely to be the FimYZ binding site that is involved in activating hilE transcription.
To test this hypothesis, we performed primer extension analysis on mRNA isolated from an E. coli strain carrying pMAB102, the hilE promoter deletion plasmid. The results of this assay seem to confirm our hypothesis. The primer extension product from the P1 hilE mRNA template indicated that mRNA was transcribed at a low level from this promoter, but no primer extension product was visible from the P2 promoter template on the sequencing gel (data not shown). With the results from the β-galactosidase assay and from mapping the hilE transcription start sites, our data indicate that FimYZ do bind to the hilE promoter and act to initiate the transcription of the hilE mRNA, leading to increased hilE expression within the bacterial cell.
DISCUSSION
The ability of Salmonella enterica serovar Typhimurium to invade occurs through the combined action of the secreted effector proteins being transported through a functional type III secretion system (15, 16). Since the process of invasion requires the expression of many different proteins, the bacteria require that the expression of these proteins be tightly regulated. The invasive phenotype is regulated by a variety of environmental conditions. A significant amount of work has been devoted to studying the genes that control the regulation of SPI-1. Characterization of hilA and invF has shown that the products of these genes are critical to the regulation of invasion as they are the transcriptional activators of the type III secretion system and secreted effector proteins (4, 29). Other genes within SPI-1, hilD and hilC, have also been identified as important regulators of hilA (19, 42, 43). Research has continued to identify other genes that play roles in the ability to activate or repress the transcription of hilA.
We have identified the hilA repressor, hilE, as being important in the control of hilA expression (6, 22). In this study, we performed experiments in an effort to identify genes that play a role in the activation of hilE expression and the subsequent repression of hilA expression and Salmonella invasiveness. This search has identified the fimbrial activator genes fimYZ as important transcriptional activators of hilE (55).
In this paper, we have found that the hilE gene is not expressed in E. coli but is significantly induced in S. enterica serovar Typhimurium, indicating that a specific factor is required for its activation. We observed that expression of hilE-lacZY is lowest as the bacterial culture begins logarithmic growth. The early logarithmic stage of growth has been shown by several groups, including our own, to be the point at which invasion gene expression is maximally induced by the hilA transcriptional activator (4, 5, 29, 41). Subsequently, hilE expression is induced as the bacteria approach the stationary phase of growth. While it is unclear why S. enterica serovar Typhimurium represses invasion gene expression at later stages of the growth cycle, this observation is also consistent with published work. Combined, these observations suggest that hilE-repressing activity is likely to be controlled by signals generated during the late mid-log and stationary phases of growth.
Subsequent work revealed that the fimYZ genes are factors that regulate hilE transcription. Overexpression of fimYZ induced hilE transcription 31-fold in E. coli and 5.1-fold in S. enterica serovar Typhimurium. Both fimY and fimZ are required for hilE activation. Assays were performed in which only fimY or fimZ was overexpressed, and in neither instance did hilE-lacZY levels increase. These findings are consistent with other research results in which the expression of the type 1 fimbrial gene operon did not occur without both fimY and fimZ being present (50, 56).
Work on the fimY gene has found that a fimY S. enterica serovar Typhimurium mutant is nonfimbriate (50). Furthermore, transcriptional studies have revealed that fimA, fimY, and fimZ all require functional fimY and fimZ genes for transcription, indicating that both are positive regulators (50, 56). Interestingly, fimZ has also been implicated in regulating bacterial motility. In those experiments, overexpression of fimZ alone significantly decreased S. enterica serovar Typhimurium motility (14). To ensure that fimYZ-mediated effects on hilE were transmitted through the invasion regulatory cascade, we examined what effects the overexpression or deletion of fimYZ would have on hilA expression. β-Galactosidase assays showed that increased activation of fimYZ led to a 3.3-fold decrease in hilA reporter expression. The deletion of fimZ from S. enterica serovar Typhimurium increases hilA reporter expression by 1.6-fold. Deletion of hilE alleviated the effects of fimYZ overexpression on the hilA reporter. Furthermore, when fimYZ was overexpressed, Salmonella invasiveness was reduced 100-fold, which was reversed by deletion of hilE.
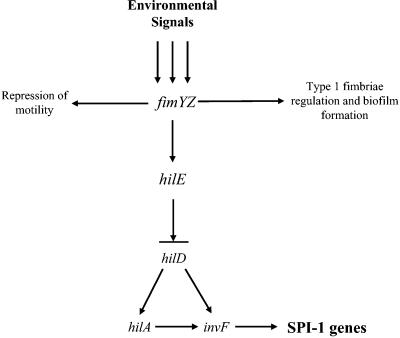
Our results indicate that fimYZ play an active role in the control of S. enterica serovar Typhimurium pathogenesis, and we propose a working model for Salmonella invasion gene regulation that incorporates these newly described functions of fimYZ (Fig. 8). The fimYZ genes were originally identified as regulators that are required for expression of type 1 fimbriae (47, 55). More recently, it has been shown that the level of fimZ activation is inversely related to the motility of S. enterica serovar Typhimurium (14). Our work now shows that the fimYZ genes also control the expression of hilE. Induction of hilE transcription, via fimYZ, leads to repression of hilA expression with corresponding repression of downstream invasion genes. These findings highlight that fimYZ are components of a global regulon in S. enterica serovar Typhimurium that potentially controls the expression of a wide range of phenotypes. Future efforts in the laboratory will be directed at characterizing the role of these regulators in pathogenic S. enterica serovar Typhimurium.
FIG. 8.
Model for processing environmental signals that affect SPI-1 invasion gene expression. An environmental signal activates (or deactivates) FimYZ, which then activate (or deactivate) hilE transcription. Activation of fimYZ also leads to expression of type 1 fimbriae and an adhesive phenotype. The activation of hilE transcription produces HilE protein, which then leads to repression of hilA expression and subsequent expression of the other SPI-1 genes through its interaction with the HilD regulatory protein.
Analysis of the FimZ amino acid sequence reveals some features about the protein. First, FimZ is homologous to known response regulators. However, unlike most other two-component regulator systems, a sensor kinase protein partner has not been identified for FimZ (56). FimZ has sequences that resemble phosphorylation sites described for other response regulators, although the protein has not been proven to be phosphorylated. Past studies on S. enterica serovar Typhimurium pathogenesis have identified other two-component regulatory systems that have the ability to regulate hilA expression. A phoQ mutation (pho-24) that causes the phoPQ two-component system to be hyperactivated has been shown to superrepress hilA expression and invasion (5, 37, 40). The phoPQ two-component system responds to magnesium and calcium concentrations within the environment. When magnesium levels are at micromolar levels, PhoQ becomes activated via phosphorylation and subsequently transfers phosphate to PhoP in a phosphorelay reaction to activate the response regulator (24, 28, 51). The mechanism by which the pho-24 mutation exerts its effect on hilA transcription is currently unknown.
The phoBR system has also been identified as a two-component regulatory system that alters hilA expression levels (32). This system responds to extracellular phosphate levels. In low-phosphate conditions, the pho regulon becomes upregulated, leading to increased expression of many different gene operons (52). It has been speculated that phosphate levels within the small intestine are elevated which leads to repression of the phoBR system and the upregulation of hilA (32). Analysis of the gene regulated by the phoBR two-component system has shown that PhoB binds to a site known as the pho box and that, in many cases, binding increases when the PhoB protein is phosphorylated (21, 33). A screen of the fimZ promoter has found, upstream of the fimZ transcriptional start site, a sequence that is 78% identical to the pho box (unpublished data). This suggests that phoBR may be involved in regulating fimZ. Activation of fimZ by phoBR could lead to increased hilE expression and subsequent repression of hilA. Future research will be directed at determining what effects, if any, the phoBR and phoPQ two-component systems have on fimZ and hilE expression.
Acknowledgments
We thank Jennifer Boddicker and Nate Ledeboer for careful review of the manuscript.
Editor: J. B. Bliska
REFERENCES
- 1.Akbar, S., L. M. Schechter, C. P. Lostroh, and C. A. Lee. 2003. AraC/XylS family members, HilD and HilC, directly activate virulence gene expression independently of HilA in Salmonella typhimurium. Mol. Microbiol. 47:715-728. [DOI] [PubMed] [Google Scholar]
- 2.Altier, C., M. Suyemoto, and S. D. Lawhon. 2000. Regulation of Salmonella enterica serovar Typhimurium invasion genes by csrA. Infect. Immun. 68:6790-6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altier, C., M. Suyemoto, A. I. Ruiz, K. D. Burnham, and R. Maurer. 2000. Characterization of two novel regulatory genes affecting Salmonella invasion gene expression. Mol. Microbiol. 35:1872-1882. [DOI] [PubMed] [Google Scholar]
- 4.Bajaj, V., C. Hwang, and C. A. Lee. 1995. hilA is a novel ompR/toxR family member that activates the expression of Salmonella typhimurium invasion genes. Mol. Microbiol. 18:715-727. [DOI] [PubMed] [Google Scholar]
- 5.Bajaj, V., R. L. Lucas, C. Hwang, and C. A. Lee. 1996. Co-ordinate regulation of Salmonella typhimurium invasion genes by environmental and regulatory factors is mediated by control of hilA expression. Mol. Microbiol. 22:703-714. [DOI] [PubMed] [Google Scholar]
- 6.Baxter, M. A., T. F. Fahlen, R. L. Wilson, and B. D. Jones. 2003. HilE interacts with HilD and negatively regulates hilA transcription and expression of the Salmonella enterica serovar Typhimurium invasive phenotype. Infect. Immun. 71:1295-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Behlau, I., and S. I. Miller. 1993. A PhoP-repressed gene promotes Salmonella typhimurium invasion of epithelial cells. J. Bacteriol. 175:4475-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boddicker, J. D., N. A. Ledeboer, J. Jagnow, B. D. Jones, and S. Clegg. 2002. Differential binding to and biofilm formation on, HEp-2 cells by Salmonella enterica serovar Typhimurium is dependent upon allelic variation in the fimH gene of the fim gene cluster. Mol. Microbiol. 45:1255-1265. [DOI] [PubMed] [Google Scholar]
- 9.Boddicker, J. D., B. M. Knosp, and B. D. Jones. 2003. Transcription of the Salmonella invasion gene activator, hilA, requires HilD activation in the absence of negative regulators. J. Bacteriol. 185:525-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang, A. C., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chuang, S. E., D. L. Daniels, and F. R. Blattner. 1993. Global regulation of gene expression in Escherichia coli. J. Bacteriol. 175:2026-2036. [DOI] [PMC free article] [PubMed]
- 12.Clegg, S., and G. F. Gerlach. 1987. Enterobacterial fimbriae. J. Bacteriol. 169:934-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clegg, S. and D. L. Swenson. 1994. Fimbriae, p. 105-114. In P. Klemm (ed.), Adhesion, genetics, biogenesis, and vaccines. CRC Press, Boca Raton, Fla.
- 14.Clegg, S., and K. T. Hughes. 2002. FimZ is a molecular link between sticking and swimming in Salmonella enterica serovar Typhimurium. J. Bacteriol. 184:1209-1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Collazo, C. M., and J. E. Galán. 1997. The invasion-associated type III system of Salmonella typhimurium directs the translocation of Sip proteins into the host cell. Mol. Microbiol. 24:747-756. [DOI] [PubMed] [Google Scholar]
- 16.Darwin, K. H., and V. L. Miller. 1999. Molecular basis of the interaction of Salmonella with the intestinal mucosa. Clin. Microbiol. Rev. 12:405-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davis, R. W., D. Botstein, and J. R. Roth. 1980. Advanced Bacterial Genetics: A manual for genetic engineering. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 19.Eichelberg, K., W. D. Hardt, and J. E. Galán. 1999. Characterization of SprA, an AraC-like transcriptional regulator encoded within the Salmonella typhimurium pathogenicity island 1. Mol. Microbiol. 33:139-152. [DOI] [PubMed] [Google Scholar]
- 20.Ellermeier, C. D., and J. M. Slauch. 2003. RtsA and RtsB coordinately regulate expression of the invasion and flagellar genes in Salmonella enterica serovar Typhimurium. J. Bacteriol. 185:5096-5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ellison, D. W., and W. R. McCleary. 2000. The unphosphorylated receiver domain of PhoB silences the activity of its output domain. J. Bacteriol. 182:6592-6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fahlen, T. F., N. Mathur, and B. D. Jones. 2000. Identification and characterization of mutants with increased expression of hilA, the invasion gene transcriptional activator of Salmonella typhimurium. FEMS Immunol. Med. Microbiol. 28:25-35. [DOI] [PubMed] [Google Scholar]
- 23.Fahlen, T. F., R. W. Wilson, J. D. Boddicker, and B. D. Jones. 2001. Hha is a negative modulator of hilA transcription, the Salmonella typhimurium invasion gene transcriptional activator. J. Bacteriol. 183:6620-6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Groisman, E. A. 2001. The pleiotropic two-component regulatory system PhoP-PhoQ. J. Bacteriol. 183:1835-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hultgren, S. J., S. Normark, and S. N. Abraham. 1991. Chaperone-assisted assembly and molecular architecture of adhesive pili. Annu. Rev. Microbiol. 45:383-415. [DOI] [PubMed] [Google Scholar]
- 26.Johnston, C., D. A. Pegues, C. J. Hueck, A. Lee, and S. I. Miller. 1996. Transcriptional activation of Salmonella typhimurium invasion genes by a member of the phosphorylated response-regulator superfamily. Mol. Microbiol. 22:715-727. [DOI] [PubMed] [Google Scholar]
- 27.Jones, B. D., and S. Falkow. 1994. Identification and characterization of a Salmonella typhimurium oxygen-regulated gene required for bacterial internalization. Infect. Immun. 62:3745-3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kato, A., H. Tanabe, and R. Utsumi. 1999. Molecular characterization of the PhoP-PhoQ two-component system in Escherichia coli K-12: identification of extracellular Mg2+-responsive promoters. J. Bacteriol. 181:5516-5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee, C. A., B. D. Jones, and S. Falkow. 1992. Identification of a Salmonella typhimurium invasion locus by selection for hyperinvasive mutants. Proc. Natl. Acad. Sci. USA 89:1847-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lefebvre, B., P. Formstecher, and P. Lefebvre. 1995. Improvement of the gene splicing overlap (SOE) method. BioTechniques 19:186-188. [PubMed] [Google Scholar]
- 31.Lodge, J., J. Fear, S. Busby, P. Gunasekaran, and N. R. Kamini. 1992. Broad host range plasmids carrying the Escherichia coli lactose and galactose operons. FEMS Microbiol. Lett. 74:271-276. [DOI] [PubMed] [Google Scholar]
- 32.Lucas, R. L., C. P. Lostroh, C. C. DiRusso, M. P. Spector, B. L. Wanner, and C. A. Lee. 2000. Multiple factors independently regulate hilA and invasion gene expression in Salmonella enterica serovar typhimurium. J. Bacteriol. 182:1872-1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Makino, S., C. Sasakawa, K. Kamata, T. Kurata, and M. Yoshikawa. 1986. A genetic determinant required for continuous reinfection of adjacent cells on large plasmid in S. flexneri 2a. Cell 46:551-555. [DOI] [PubMed] [Google Scholar]
- 34.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 35.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 36.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 37.Miller, S. I., A. M. Kurkal, and J. J. Mekalanos. 1989. A two-component regulatory system (phoP phoQ) controls Salmonella typhimurium virulence. Proc. Natl. Acad. Sci. USA 86:5054-5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moore, A. E., L. Sabachewsky, and H. W. Toolan. 1955. Culture characteristics of four permanent lines of human cancer cells. Cancer Res. 15:598. [PubMed] [Google Scholar]
- 39.Olekhnovich, I. N., and R. J. Kadner. 2002. DNA-binding activities of the HilC and HilD virulence regulatory proteins of Salmonella enterica serovar Typhimurium. J. Bacteriol. 184:4148-4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pegues, D. A., M. J. Hantman, I. Behlau, and S. I. Miller. 1995. PhoP/PhoQ transcriptional repression of Salmonella typhimurium invasion genes: evidence for a role in protein secretion. Mol. Microbiol. 17:169-181. [DOI] [PubMed] [Google Scholar]
- 41.Penheiter, K. L., N. Mathur, D. Giles, T. Fahlen, and B. D. Jones. 1997. Non-invasive Salmonella typhimurium mutants are avirulent because of an inability to enter and destroy M cells of ileal Peyer's patches. Mol. Microbiol. 24:697-709. [DOI] [PubMed] [Google Scholar]
- 42.Rakeman, J. L., H. R. Bonifield, and S. I. Miller. 1999. A HilA-independent pathway to Salmonella typhimurium invasion gene transcription. J. Bacteriol. 181:3096-3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schechter, L. M., S. M. Damrauer, and C. A. Lee. 1999. Two AraC/XylS family members can independently counteract the effect of repressing sequences upstream of the hilA promoter. Mol. Microbiol. 32:629-642. [DOI] [PubMed] [Google Scholar]
- 44.Schechter, L. M., and C. A. Lee. 2001. AraC/XylS family members, HilC and HilD, directly bind and derepress the Salmonella typhimurium hilA promoter. Mol. Microbiol. 40:1289-1299. [DOI] [PubMed] [Google Scholar]
- 45.Shi, J., and D. P. Biek. 1995. A versatile low-copy-number cloning vector derived from plasmid F. Gene 164:55-58. [DOI] [PubMed] [Google Scholar]
- 46.Stock, J. B., A. J. Ninfa, and A. M. Stock. 1989. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol. Rev. 53:450-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Swenson, D. L., and S. Clegg. 1992. Identification of ancillary fim genes affecting fimA expression in Salmonella typhimurium. J. Bacteriol. 174:7697-7704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takaya, A., T. Tomoyasu, A. Tokumitsu, M. Morioka, and T. Yamamoto. 2002. The ATP-dependent lon protease of Salmonella enterica serovar Typhimurium regulates invasion and expression of genes carried on Salmonella pathogenicity island 1. J. Bacteriol. 184:224-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thanassi, D. G., and S. J. Hultgren. 2000. Assembly of complex organelles: pilus biogenesis in gram-negative bacteria as a model system. Methods 20:111-126. [DOI] [PubMed] [Google Scholar]
- 50.Tinker, J. K., and S. Clegg. 2000. Characterization of FimY as a coactivator of type 1 fimbrial expression in Salmonella enterica serovar Typhimurium. Infect. Immun. 68:3305-3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vescovi, E. G., Y. M. Ayala, C. E. Di, and E. A. Groisman. 1997. Characterization of the bacterial sensor protein PhoQ. Evidence for distinct binding sites for Mg2+ and Ca2+. J. Biol. Chem. 272:1440-1443. [DOI] [PubMed] [Google Scholar]
- 52.Wanner, B. L. 1996. Phosphorus assimilation and control of the phosphate regulon, p. 1357-1381. In. F. C. Neidhardt, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology. American Society for Microbiology, Washington, D.C.
- 53.Wilson, R. L., S. J. Libby, A. M. Freet, J. D. Boddicker, T. F. Fahlen, and B. D. Jones. 2001. Fis, a DNA nucleoid-associated protein, is involved in Salmonella typhimurium SPI-1 invasion gene expression. Mol. Microbiol. 39:79-88. [DOI] [PubMed] [Google Scholar]
- 54.Wray, C., and W. J. Sojka. 1978. Experimental Salmonella typhimurium in calves. Res. Vet. Sci. 25:139-143. [PubMed] [Google Scholar]
- 55.Yeh, K. S., L. S. Hancox, and S. Clegg. 1995. Construction and characterization of a fimZ mutant of Salmonella typhimurium. J. Bacteriol. 177:6861-6865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yeh, K. S., J. K. Tinker, and S. Clegg. 2002. FimZ binds the Salmonella typhimurium fimA promoter region and may regulate its own expression with FimY. Microbiol. Immunol. 46:1-10. [DOI] [PubMed] [Google Scholar]