Abstract
Nasal carriage is a major risk factor for Staphylococcus aureus infection, especially for methicillin-resistant strains (MRSA). Using a mouse model of nasal carriage, we have compared several S. aureus strains and demonstrated increased colonization levels by MRSA in cystic fibrosis transmembrane conductance regulator-deficient mice and Toll-like receptor 2 (TLR2)-deficient mice but not TLR4-deficient mice.
Methicillin-resistant Staphylococcus aureus (MRSA) has spread worldwide, primarily as a nosocomial pathogen (2). Due to multiresistance of these strains, including recent acquisition of high-level glycopeptide resistance (3), few antibiotics remain effective for MRSA infections. Recent emergence of community-acquired MRSA infections is of particular concern, with some clinical isolates harboring the Panton-Valentine leukocidin determinant responsible for lethal necrotizing pneumonia (5).
Anterior nares are the major reservoir of S. aureus: 20% of humans are persistently and asymptomatically colonized, 60% are colonized intermittently, and 20% are noncarriers (13). Subgroups, such as cystic fibrosis (CF) patients, have higher carriage rates (10). Nasal carriage is a major risk factor for staphylococcal infection (7, 25). Commensal carriage of MRSA in healthy individuals remains low (from 0.2 to 2.8%) (6) but constitutes a greater risk for subsequent infection than methicillin-susceptible S. aureus carriage (13).
Mice constitute a good animal model for S. aureus nasal colonization; most mice inoculated with 108 CFU maintain carriage for at least 20 days (12). Recently, a cotton rat model in which S. aureus persists at higher CFU for up to 6 weeks (14) demonstrated the role of S. aureus wall teichoic acid in colonization (26). A mouse model, however, permits the study of inbred and knockout strains. We have modified the previous mouse model (12) and studied various MRSA isolates and knockout mice to dissect interactions between MRSA and the nasal tissue.
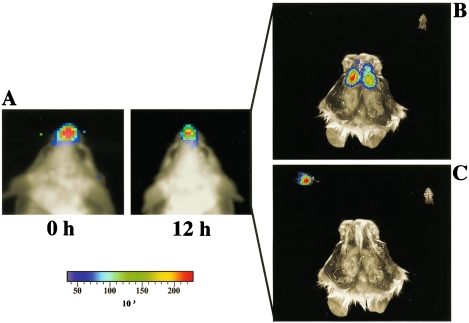
To assess anatomical location of intranasally introduced S. aureus, we used the stably bioluminescent MRSA Xen31 with a chromosomal copy of the modified Photorhabdus luminescens luxABCDE operon (Table 1). These bacteria were grown overnight in brain heart infusion medium at 37°C with shaking, washed twice, and resuspended in phosphate-buffered saline (Dulbecco) at 109 CFU/10 μl. Five- to eight-week-old female BALB/c mice (Harlan, Gannat, France) were anesthetized, and 10 μl of the bacterial suspension was introduced into both nares without touching the tip of the nose. The bioluminescent signal was strictly intranasal (Fig. 1A) and not detectable in the trachea or lungs. Bioluminescence monitored at 0, 12, 24, and 36 h after inoculation diminished during the first 24 h, disappearing by 36 h postinoculation (data not shown). Mice instilled intranasally with Xen31 were sacrificed after 12 h. The lower jaws were cut, the palate containing the nasal associated lymphoid tissue (NALT) was excised, and the nasal mucosa was exposed by cutting the head sagitally. The entire bioluminescent signal was located in the nasal mucosa, and no emission could be detected from the NALT (Fig. 1B). This is in contrast to bioluminescent group A streptococci, which were predominantly found in the NALT (16). The nasal mucosa dissected under a section microscope was exposed for bioluminetric detection; the whole bioluminescent signal was recorded from the dissected nasal tissue (Fig. 1C). Hence, this microsurgical method was used for the enumeration of CFU in the nasal cavity of mice inoculated with nonbioluminescent MRSA.
TABLE 1.
Properties of the strains
FIG. 1.
Bioluminescence from mice inoculated nasally with MRSA Xen31. (A) Bioluminescence from the entire mouse was recorded for 15 min with a backside Hamamatsu ORCA-3 camera, and the images were processed with Compix software (Compix Inc., Brandywine, Pa.). A strong bioluminescent signal was recorded from the nose of the mice upon inoculation and 12 h postinoculation. (B) Anatomical location of MRSA during colonization. Twelve hours after inoculation, mice were sacrificed and dissected as described in the text, and the palate and dissected NALT (white ovals) were exposed for bioluminetric analysis with a Xenogen 100 IVIS system. The entire bioluminescent signal was emitted from the nasal mucosa, and no signal could be detected from the NALT. (C) Upon microdissection, the whole photonic signal was recorded from the excised mucosal tissue.
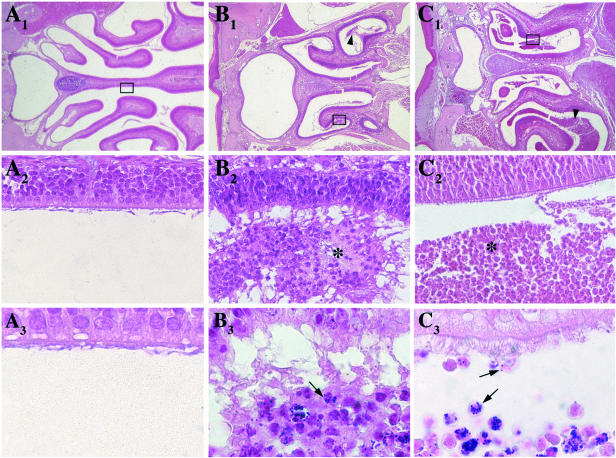
For histological study, mice inoculated intranasally with strain COL (Table 1) were sacrificed after 24 h; the heads, decalcified in 4% neutral formalin buffer with 10% trichloroacetic acid for 2 weeks, were coronally cut into five to six fragments and embedded in paraffin and 5-μm-thick sections were stained with hematoxylin-eosin or Gram stain. Cellular clumps with neutrophils and necrotic cells, undetectable in control mice (Fig. 2A), were present within the nasal lumen in COL-inoculated mice (Fig. 2B1). At higher magnification, a large number of bacteria appeared tightly attached to or inside cells (Fig. 2B3).
FIG. 2.
Histological sections of the nasal cavities of control BALB/c mice (A) and BALB/c and CFTR−/− mice 24 h after inoculation of strain COL (B and C, respectively). Hematoxylin-eosin (upper panel, 25× enlargement; middle panel, 400× enlargement of selected area) and Gram staining (lower panel, 1,000×) were performed with 5-μm-thick sections. Luminal cellular clumps spread along the nasal cornets are indicated with arrowheads (B1 and C1) and asterisks (B2 and C2). Bacteria that appeared to be located intracellularly within the cellular clumps composed of neutrophils and degenerated cells are denoted with an arrow (B3 and C3). In CFTR−/− mice, bacteria were present along the nasal epithelium (C3).
A recently described mouse model of S. aureus nasal colonization allows quantification of bacteria in the nasal mucosa of mice (12). However, this method yields highly dispersed bacterial counts ranging from 0.5 × 10 to 1 × 105 CFU/mouse at day 5 postinoculation, thus requiring many mice. In this model, the nasal tissue is excised with scissors and vortexed. Since we observed cell-associated bacteria (Fig. 2), CFU were determined on thoroughly homogenized nasal tissue obtained by careful dissection. Twenty-four BALB/c mice were inoculated with 109 CFU of strain COL and sacrificed at days 3, 7, and 14. The dissected nasal mucosa was collected in 1 ml of sterile saline and homogenized, and serial dilutions were plated on Baird-Parker agar. After overnight incubation, colonies were enumerated and replica plated on brain heart infusion agar containing 10 μl of oxacillin/ml to ensure methicillin resistance. At day 3, a mean of 4.7 × 104 CFU/mouse was obtained, with small dispersion between counts; at day 7, all mice were colonized with a mean of 1.6 × 102 CFU/mouse (Table 2 and Fig. 3). At day 14, all the mice were still colonized, but at this late time point the CFU counts were low, ranging from 1.2 × 10 to 1.8 × 10/mouse. An inoculum of 108 CFU of strain COL or of 109 CFU of strain Xen31 led to colonization of all mice at days 3 and 7 but yielded lower CFU counts than those obtained with 109 CFU (Table 2). These results indicate that, although mice are not colonized with MRSA in the long term, this model is sufficiently reproducible to study bacterial and host factors involved in MRSA nasal colonization by using small numbers of animals.
TABLE 2.
Nasal colonization by MRSA strains COL and Xen31
| Strain (inoculum) | Colonization at:
|
|||
|---|---|---|---|---|
| Day 3
|
Day 7
|
|||
| No. of carriers/total no. assayed | Mean CFU (range) | No. of carriers/total no. assayed | Mean CFU (range) | |
| COL (108) | 5/5 | 2.3 × 104 (6.3 × 103-2.5 × 104) | 5/5 | 1.5 × 102 (1 × 101-6.3 × 102) |
| COL (109) | 9/9 | 4.7 × 104 (2 × 103-3.2 × 105) | 8/8 | 1.6 × 102 (3.2 × 101-4 × 102) |
| Xen31 (109) | 5/5 | 1.3 × 103 (2.5 × 102-4 × 103) | 5/5 | 5 × 101 (2 × 101-2 × 102) |
FIG. 3.
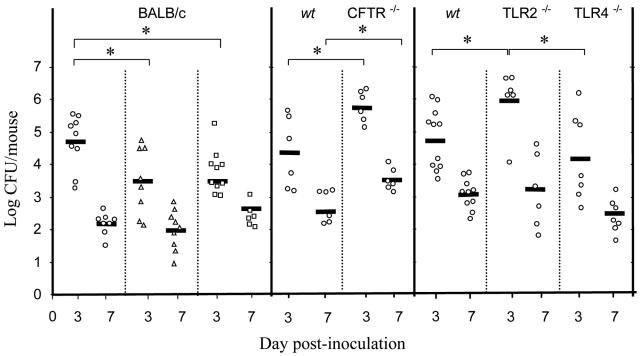
Nasal colonization of mice by MRSA. Colonization by strain COL (open circles), BM4559 (open triangles), and BM4560 (open squares) was recorded on days 3 and 7 after inoculation. Each symbol represents CFU counts in a single mouse. The mean log10 CFU per nose for each group is represented by a horizontal line. Results are from a minimum of two independent experiments. Pairwise comparison of bacterial counts from the various groups of animals was performed with the Mann-Whitney U test. A P value of <0.05 was considered significant and is denoted with an asterisk.
To study if MRSA strains differ in their colonization ability, we inoculated two epidemiologically successful clinical isolates (Table 1). Strain BM4559 is a representative of the Iberian clone, a hospital-restricted, worldwide-spread MRSA, and BM4560 is a French isolate responsible for community-acquired infections and which produces the Panton-Valentine leukocidin. On day 3, lower counts of both strains were recovered, whereas at day 7 there was no significant difference between the three MRSA strains (Fig. 3).
Methicillin resistance in S. aureus is mediated by mecA, a gene encoding additional penicillin-binding protein PBP2a with low affinity for β-lactam antibiotics. When bacteria are grown in the presence of β-lactams, the production of PBP2a is induced; PBP2a is exposed at the bacterial surface and can interact with host tissues (unpublished data). To test if PBP2a is implicated in nasal colonization, we used a mecA null COL derivative (8); colonization (mean, 4.5 × 104 CFU/mouse at day 3 and 2.3 × 102 CFU/mouse at day 7) was not significantly different from that by parental strain COL. Similarly, colonization was not significantly enhanced when strain COL was grown in the presence of oxacillin (mean, 4.9 × 104 CFU/mouse at day 3 and 8.5 × 102 CFU/mouse at day 7), indicating that PBP2a is not a colonizing factor.
S. aureus is often the first pathogen to infect CF patients, who can be colonized with the same strain for years (as reviewed in reference 18). Using the CF transmembrane conductance regulator (CFTR) knockout mouse model (21), we inoculated intranasally CFTR−/− and parental CFTR+/+ C57BL/6 mice (CNRS, Orleans, France) with MRSA COL. The CFTR−/− mice harbored increased numbers of MRSA (mean, 4.5 × 105 CFU/mouse at day 3 and 3.7 × 103 CFU/mouse at day 7) compared to the CFTR+/+ parent (mean, 2.5 × 104 CFU/mouse at day 3 [P = 0.022] and 1.9 × 102 CFU/mouse at day 7 [P = 0.01]) (Fig. 3). The difference between the two mouse strains is an underestimate, since the CFTR−/− mice are much smaller than their CFTR+/+ counterparts. Histological preparations of infected CFTR−/− mice showed bacteria entrapped in a cell-loaded intraluminal mucus mostly present within the nasal cornets (Fig. 2C). Reduced mucociliary clearance of CF epithelia (27) and reduced bactericidal activity of the airway surface fluid (20) may account for increased survival of bacteria in the mucus of these animals.
Toll-like receptors 2 and 4 (TLR2 and TLR4) mediate an inflammatory response against several gram-positive and gram-negative pathogens, respectively, and TLR2-deficient mice are highly susceptible to S. aureus sepsis (22). Although it has been shown that both receptors are expressed in the human upper airway epithelium (4), little is known about their role in the nasal mucosa. We inoculated homozygous 8- to 10-week-old TLR2−/− and TLR4−/− knockout C57BL/6 (23) and age-matched wild-type mice. At day 3, 10 times more MRSA CFU were isolated from the nasal tissue of TLR2−/− mice (mean, 9.1 × 105 CFU/mouse) than from C57BL/6 mice (mean, 6.3 × 104 CFU/mouse; P = 0.016) or from TLR4−/− mice (mean, 1.4 × 104 CFU/mouse; P = 0.032) (Fig. 3). However, at day 7, there was no significant difference between the three groups of mice. These results suggest that TLR2, but not TLR4, is involved in the early stages of the innate immune response against nasal S. aureus colonization and that other TLR/interleukin-1 family members may play a role, as demonstrated in other infection models of S. aureus (22).
Direct in vivo monitoring of biofilm formation for up to 20 days by using bioluminescent S. aureus and Pseudomonas aeruginosa in mice has been reported (11). These studies modeled foreign-body infection and used surgical subcutaneous implantation of catheters in the flanks, where bioluminescence detection is easier than through nasal bones. Under our technical conditions, bioluminescence could not be followed more than 24 h, yet strain Xen31 was still present 36 h after inoculation at a mean of 104 CFU/mouse (range, 2.4 × 103 to 1.8 × 104). Since bioluminescence production by bacteria depends on their metabolic activity (17), the constant decrease in the signal suggests that S. aureus did not multiply actively in the nasal cavity of the mice. Bioluminescence imaging under the present technical conditions will be useful to study factors involved in the early steps of colonization or to assess prevention of colonization, as shown previously in a thigh model of infection with S. aureus (9).
Humans can be colonized by a single S. aureus strain which can persist in the nasal cavity for months (10). We have demonstrated that PBP2a, responsible for methicillin resistance, does not play a role in colonization. However, only a small number of MRSA clones has been studied. Comparison of representatives of the five major pandemic clones (15) and of community-acquired MRSA (24) will allow determination of the role of other determinants encoded within or outside the staphylococcal chromosomal cassette (SCCmec) in colonization.
Acknowledgments
We thank K. Francis for the gift of strain Xen31, G. Archer for S. aureus COL and COL mecA, J. Etienne for BM4560, B. David (Hamamatsu, Massy, France) for technical support in the bioluminescence experiment, P. Ave for histological preparations, and Vaincre la Mucoviscidose for providing CFTR knockout mice.
This work was supported in part by the Programme “Microbiologie Fondamentale et Appliquée, Maladies Infectieuses, Environnement et Bioterrorisme” from the Ministère Délégué à la Recherche et aux Nouvelles Technologies.
Editor: F. C. Fang
REFERENCES
- 1.Archer, G. L., D. M. Niemeyer, J. A. Thanassi, and M. J. Pucci. 1994. Dissemination among staphylococci of DNA sequences associated with methicillin resistance. Antimicrob. Agents Chemother. 38:447-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chambers, H. F. 2001. The changing epidemiology of Staphylococcus aureus? Emerg. Infect. Dis. 7:178-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang, S., D. M. Sievert, J. C. Hageman, M. L. Boulton, F. C. Tenover, F. Pouch Downes, S. Shah, J. T. Rudrik, G. R. Pupp, W. J. Brown, D. Cardo, and S. K. Fridkin. 2003. Infection with vancomycin-resistant Staphylococcus aureus containing the vanA resistance gene. N. Engl. J. Med. 348:1342-1347. [DOI] [PubMed] [Google Scholar]
- 4.Claeys, S., T. de Belder, G. Holtappels, P. Gevaert, B. Verhasselt, P. Van Cauwenberge, and C. Bachert. 2003. Human B-defensins and toll-like receptors in the upper airway. Allergy 58:748-753. [DOI] [PubMed] [Google Scholar]
- 5.Dufour, P., Y. Gillet, M. Bes, G. Lina, F. Vandenesch, D. Floret, J. Etienne, and H. Richet. 2002. Community-acquired methicillin-resistant Staphylococcus aureus infections in France: emergence of a single clone that produces Panton-Valentine leukocidin. Clin. Infect. Dis. 35:819-824. [DOI] [PubMed] [Google Scholar]
- 6.Eady, E. A., and J. H. Cove. 2003. Staphylococcal resistance revisited: community acquired methicillin resistant Staphylococcus aureus—an emerging problem for the management of skin and soft tissue infections. Curr. Opin. Infect. Dis. 16:103-124. [DOI] [PubMed] [Google Scholar]
- 7.Fierobe, L., D. Decré, C. Muller, J. C. Lucet, J. P. Marmuse, J. Mantz, and J. M. Desmonts. 1999. Methicillin-resistant Staphylococcus aureus as a causative agent of postoperative intra-abdominal infection: relation to nasal colonization. Clin. Infect. Dis. 5:1231-1238. [DOI] [PubMed] [Google Scholar]
- 8.Finan, J. E., A. E. Rosato, T. M. Dickinson, D. Ko, and G. L. Archer. 2002. Conversion of oxacillin-resistant staphylococci from heterotypic to homotypic resistance expression. Antimicrob. Agents Chemother. 46:24-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Francis, K. P., D. Joh, C. Bellinger-Kawahara, M. J. Hawkinson, T. F. Purchio, and P. R. Contag. 2000. Monitoring bioluminescent Staphylococcus aureus infections in living mice using a novel luxABCDE construct. Infect. Immun. 68:3594-3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goerke, C., K. Kraning, M. Stern, G. Döring, K. Botzenhart, and C. Wolz. 2000. Molecular epidemiology of community-acquired Staphylococcus aureus in families with and without cystic fibrosis patients. J. Infect. Dis. 181:984-989. [DOI] [PubMed] [Google Scholar]
- 11.Kadurugamuwa, J. L., L. Sin, E. Albert, J. Yu, K. Francis, M. DeBoer, M. Rubin, C. Bellinger-Kawahara, T. R. Parr, and P. R. Contag. 2003. Direct continuous method for monitoring biofilm infection in a mouse model. Infect. Immun. 71:882-890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kiser, K. B., J. M. Cantey-Kiser, and J. C. Lee. 1999. Development and characterization of a Staphylococcus aureus nasal colonization model in mice. Infect. Immun. 67:5001-5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kluytmans, J., A. Van Belkum, and H. Verbrugh. 1997. Nasal carriage of Staphylococcus aureus: epidemiology, underlying mechanisms, and associated risks. Clin. Microbiol. Rev. 10:505-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kokai-Kun, J. F., S. M. Walsh, T. Chanturiya, and J. J. Mond. 2003. Lysostaphin cream eradicates Staphylococcus aureus nasal colonization in a cotton rat model. Antimicrob. Agents Chemother. 47:1589-1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oliveira, D. C., A. Tomasz, and H. de Lencastre. 2002. Secrets of success of a human pathogen: molecular evolution of pandemic clones of methicillin-resistant Staphylococcus aureus. Lancet Infect. Dis. 2:180-189. [DOI] [PubMed] [Google Scholar]
- 16.Park, H. S., K. P. Francis, J. Yu, and P. P. Cleary. 2003. Membranous cells in nasal-associated lymphoid tissue: a portal of entry for the respiratory mucosal pathogen group A streptococcus. J. Immunol. 171:2532-2537. [DOI] [PubMed] [Google Scholar]
- 17.Qazi, S. N. A., E. Counil, J. Morrissey, C. E. D. Rees, A. Cockayne, K. Winzer, W. C. Chan, P. Williams, and P. J. Hill. 2001. agr expression precedes escape of internalized Staphylococcus aureus from the host endosome. Infect. Immun. 69:7074-7082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saiman, L., and J. Siegel. 2004. Infection control in cystic fibrosis. Clin. Microbiol. Rev. 17:57-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Santos Sanches, I., M. Ramirez, H. Troni, M. Abecassis, M. Padua, A. Tomasz, and H. de Lencastre. 1995. Evidence for the geographic spread of a methicillin-resistant Staphylococcus aureus clone between Portugal and Spain. J. Clin. Microbiol. 33:1243-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith, J. J., S. M. Travis, E. P. Greenberg, and M. J. Welsh. 1996. Cystic fibrosis airway epithelia fail to kill bacteria because of abnormal airway surface fluid. Cell 85:229-236. [DOI] [PubMed] [Google Scholar]
- 21.Snouwaert, J. N., K. K. Brigman, A. M. Latour, N. N. Malouf, R. C. Boucher, O. Smithies, and B. H. Koller. 1992. An animal model for cystic fibrosis made by gene targeting. Science 257:1083-1088. [DOI] [PubMed] [Google Scholar]
- 22.Takeuchi, O., K. Hoshino, and S. Akira. 2000. Cutting edge: TLR2-deficient and MyD88-deficient mice are highly susceptible to Staphylococcus aureus infection. J. Immunol. 165:5392-5396. [DOI] [PubMed] [Google Scholar]
- 23.Takeuchi, O., K. Hoshino, T. Kawai, H. Sanjo, H. Takada, T. Ogawa, K. Takeda, and S. Akira. 1999. Differential roles of TLR2 and TLR4 in recognition of gram-negative and gram-positive bacterial cell wall components. Immunity 11:443-451. [DOI] [PubMed] [Google Scholar]
- 24.Vandenesch, F., T. Naimi, C. Enright, G. Lina, G. R. Nimmo, H. Heffernan, N. Liassine, M. Bes, T. Greeland, M. E. Reverdy, and J. Etienne. 2003. Community-acquired methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: worldwide emergence. Emerg. Infect. Dis. 9:978-983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Von Eiff, C., K. Becker, K. Machka, H. Stammer, and G. Peters. 2001. Nasal carriage as a source of Staphylococcus aureus bacteremia. N. Engl. J. Med. 344:11-16. [DOI] [PubMed] [Google Scholar]
- 26.Weidenmaier, C., J. F. Kokai-Kun, S. A. Kristian, T. Chanturiya, H. Kalbacher, M. Gross, G. Nicholson, B. Neumeister, J. J. Mond, and A. Peschel. 2004. Role of teichoic acids in Staphylococcus aureus nasal colonization, a major risk factor in nosocomial infections. Nat. Med. 10:243-245. [DOI] [PubMed] [Google Scholar]
- 27.Zahm, J. M., D. Gaillard, F. Dupuit, J. Hinnrasky, D. Porteous, J. R. Dorin, and E. Puchelle. 1997. Early alterations in airway mucociliary clearance and inflammation of the lamina propria in CF mice. Am. J. Physiol. 272:853-859. [DOI] [PubMed] [Google Scholar]