Abstract
The autotrophic ammonia-oxidizing bacteria in a eutrophic freshwater lake were studied over a 12-month period. Numbers of ammonia oxidisers in the lakewater were small throughout the year, and tangential-flow concentration was required to obtain meaningful estimates of most probable numbers. Sediments from littoral and profundal sites supported comparatively large populations of these bacteria, and the nitrification potential was high, particularly in summer samples from the littoral sediment surface. In enrichment cultures, lakewater samples nitrified at low (0.67 mM) ammonium concentrations only whereas sediment samples exhibited nitrification at high (12.5 mM) ammonium concentrations also. Enrichments at low ammonium concentration did not nitrify when inoculated into high-ammonium medium, but the converse was not true. This suggests that the water column contains a population of ammonia oxidizers that is sensitive to high ammonium concentrations. The observation of nitrification at high ammonium concentration by isolates from some winter lakewater samples, identified as nitrosospiras by 16S rRNA probing, is consistent with the hypothesis that sediment ammonia oxidizers enter the water column at overturn. With only one exception, nested PCR amplification enabled the detection of Nitrosospira 16S rDNA in all samples, but Nitrosomonas (N. europaea-eutropha lineage) 16S rDNA was never obtained. However, the latter were part of the sediment and water column communities, because their 16S rRNA could be detected by specific oligonucleotide probing of enrichment cultures. Furthermore, a specific PCR amplification regime for the Nitrosomonas europaea ammonia monooxygenase gene (amoA) yielded positive results when applied directly to sediment and lakewater samples. Patterns of Nitrosospira and Nitrosomonas detection by 16S rRNA oligonucleotide probing of sediment enrichment cultures were complex, but lakewater enrichments at low ammonium concentration were positive for nitrosomonads and not nitrosospiras. Analysis of enrichment cultures has therefore provided evidence for the existence of subpopulations within the lake ammonia-oxidizing community distinguishable on the basis of ammonium tolerance and possibly showing a seasonal distribution between the sediment and water column.
Nitrification is a key microbiological process of the nitrogen cycle in freshwater lakes. The dynamic interaction of microbiological reactions within the nitrogen cycle renders the analysis of different processes very difficult. Since nitrite does not generally accumulate to high concentrations, ammonia oxidation is considered to be the rate-limiting process of nitrification.
Lakes in temperate regions undergo seasonal stratification, which results in the compartmentalization of microbial communities. In eutrophic lakes, the effects of temperature, oxygen, and chemical gradients interact to concentrate the ammonia-oxidizing bacteria at the oxycline. Upon overturn, the nitrifying populations are dispersed through the water column and nitrification recedes. Ammonia oxidizers cannot be directly enumerated on agar plates, and information on their occurrence and distribution in freshwater lakes is derived from most-probable-number (MPN) determinations based on measurements of nitrite formation in vitro (19). This method can also be applied to sediments, where ammonia oxidizer populations are larger and less affected by seasonal variation (4). MPN determinations can be augmented by specific nitrification rate measurements, but the range of methods available and the requirement for laboratory incubation makes these data difficult to interpret in population ecology terms.
The autotrophic ammonia-oxidizing species that mediate the process can be detected and identified by enrichment culture. To this can be added the development of fluorescent-antibody techniques (26, 29, 32, 33, 36) and in situ rRNA hybridization (15, 31), which enable the direct observation of species. The latter has been developed from 16S rDNA sequence information on ammonia-oxidizing bacteria (8), which has also provided PCR primer and oligonucleotide probe sequences for recovery and identification of ammonia oxidizer DNA from freshwater-lake samples and/or enrichment cultures (10). These molecular biological techniques have also been applied to the study of ammonia oxidizers in other environments, including soils (24), estuarine and marine habitats (12, 13, 23), and sewage treatment plants (31). This continues to be underpinned by 16S rDNA amplification and cloning experiments that further elucidate the phylogeny of ammonia-oxidizing bacteria and expand the sequence database for the design of new primers and probes (13, 24, 27, 28). An additional molecular target for these organisms can now be added in the form of the ammonia monooxygenase gene (14), which has been amplified directly from environmental samples (7, 18, 21) and can be used to support the 16S rDNA data (18).
To date, molecular biological techniques have been applied primarily to demonstrate the considerable genetic diversity of ammonia oxidizer communities in different environments (12, 13, 24) and to determine the relationship between these communities and those generated by enrichment cultures. However, there are comparatively few studies in which molecular biological techniques have been applied to determine the effects of environmental parameters on community structure (7, 12, 34). Ward et al. (34) recovered ammonia oxidizer 16S rDNA along the depth profile of a freshwater lake and were able to comment on the relative distribution of specific nitrosospiras and nitrosomonads. In this paper, we describe the results of a seasonal study in which the occurrence of nitrosomonads and nitrosospiras was monitored with time in water column and sediment samples. This was achieved by the application of 16S rDNA primers and probes to both environmental samples and enrichment cultures at different ammonium concentrations. These data are related to the nitrification process by measurements of nitrification potential and MPN determinations. Nitrosomonad DNA has previously been detected in soil and seawater samples by amplification of the amoA gene (7, 21), and this is also attempted here with samples of lake water and sediment.
MATERIALS AND METHODS
Bacterial strains.
Pure cultures of ammonia-oxidizing bacteria were maintained in the medium described by Watson and Mandel (35). The strains of ammonia-oxidizing bacteria used in this study were obtained from the University of Liverpool Culture Collection and were Nitrosomonas europaea C-31 and Nitrosospira multiformis C-71. Nucleic acids were extracted from these cultures as described previously (8) and used as controls for the specificity of 16S rDNA- and amoA-specific PCR primers and oligonucleotide probes throughout.
Environmental sampling.
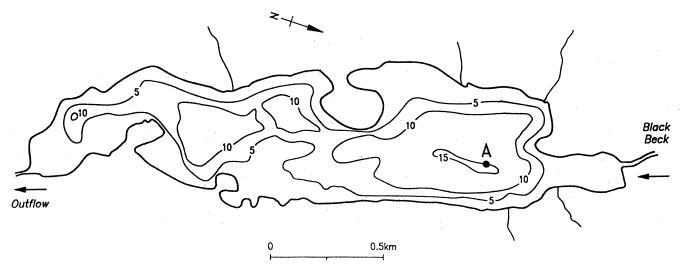
The study site was Esthwaite Water, a productive lake in the English Lake District whose basic limnology has been described by Heaney et al. (9). The lakewater sampling site was at the deepest part of the lake (Fig. 1). Temperature and oxygen concentration profiles were measured on each occasion with a combination oxygen meter and thermistor (model 57; Yellow Springs Instruments, Yellow Springs, Ohio). Water samples were obtained from the oxycline or from a depth of 9 m when the lake was fully mixed. Untreated water samples were collected in a 1-liter Friedinger bottle, and other samples were concentrated from 80 liters by tangential-flow filtration (pore diameter, 0.22 μm) to a volume of ca. 1 liter (3). Sediment core samples were obtained from depths of 15 m (profundal site) and 5 m (littoral site) with a Jenkins surface sediment sampler. The overlying water was eliminated and the surface 0.5 cm of sediment was removed by using the apparatus described by Ohnstad and Jones (16). Lakewater and sediment samples were taken at approximately monthly intervals throughout 1993.
FIG. 1.
Depth contour map of Esthwaite Water (meters) and position of sampling point (A).
Determination of nitrification potentials and ammonia oxidizer MPN values.
The microtechnique of Rowe et al. (19) was used for MPN determinations. Serial dilutions of lakewater and sediment samples were prepared as eightfold replicates in 96-well microtiter plates in duplicate with the medium of Watson and Mandel (35) containing either 0.67 mM (low) or 12.5 mM (high) (NH4)2SO4. Cultures were incubated in the dark at 30°C for 8 weeks before nitrite and nitrate determination by the method of Bendschneider and Robinson (1). Data were evaluated by using statistical tables (6). Sediment samples were also subjected to analysis for nitrification potential by incubating a 10-g sample overnight in 90 ml of 10 mM phosphate buffer (pH 7.0) supplemented to 10 mg of NH4-N per liter and determining the amounts of nitrite and nitrate produced by ammonia oxidation (5).
Extraction and amplification of DNA from environmental samples.
The method of Bruce et al. (2) was used for the extraction of DNA from sediments, and the method of Schmidt et al. (20) was used for the extraction of DNA from lakewater samples. Substances inhibitory to Taq polymerase were removed from DNA preparations with Centricon C-100 spin columns (Amicon) as specified by the manufacturer. All amplification reactions were performed in 0.2-ml thin-walled microtubes (Sarstedt), with the reaction mixture covered by a layer of sterile paraffin oil, in a model 480 thermal cycler (Perkin-Elmer Cetus). The reaction mixtures (100 μl) contained 1× PCR buffer (10× buffer is 100 mM Tris-HCl [pH 8.8], 15 mM MgCl2, 500 mM KCl, 1% Triton X-100, 0.1% [vol/vol] gelatin), 200 μM each deoxynucleoside triphosphate, 20 pM each primer, 50 ng of template DNA, and 1 U of Taq polymerase (HT Biotechnologies, Cambridge, United Kingdom). Amplification primers specific for eubacteria, nitrosospiras, and nitrosomonads were those described previously (10). Initially, some DNA samples were subjected to PCR amplification by direct application of Nitrosomonas and Nitrosospira primers, but all the samples were processed by a nested-amplification method summarized as follows. The reaction thermal profile for amplification with eubacterial 16S rDNA-directed primers was 95°C for 7 min and then 80°C for the addition of Taq polymerase, followed by 25 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 2 min. A 15-min extension at 72°C was performed after the final cycle. Nested primer amplification was performed on products of eubacteria-specific PCR with primers Nm75f and Nm1007r, specific for Nitrosomonas spp. (N. europaea-eutropha lineage) (17) and primers Ns85f and Ns1009r, specific for Nitrosospira spp. Annealing temperatures of 63 and 62°C were used for these genus-specific primers, respectively, with denaturation and extension temperatures as described above.
In addition to 16S rDNA-directed amplification, DNA preparations were subjected to an amoA-directed, nested PCR involving primers AMOF1 and AMOR2 and then AMOF2 and AMOR2R (7), using annealing temperatures of 57 and 55°C respectively, with other thermal parameters as described above. The template for nested PCR was obtained by diluting first-round amplification products appropriately with HiPerSolv water (BDH, Poole, United Kingdom). All amplification products were resolved by electrophoresis of 10-μl aliquots of the reaction mixtures on a 0.8% (wt/vol) horizontal agarose gel run in 1× TAE buffer (40 mM Tris-acetate, 1 mM EDTA). PCR product immobilization on Hybond-N+ nylon membrane (Amersham) from agarose was performed by Southern blotting.
16S rDNA oligonucleotide probing.
Oligonucleotides were end labelled with [32P]ATP-γS (ICN Pharmaceuticals, Irvine, Calif.), using T4 polynucleotide kinase (Boehringer) in 10-μl volumes with the following components: 1 μl of 10× kinase buffer (0.5 M Tris-HCl [pH 7.6], 0.1 M MgCl2, 50 mM dithiothreitol, 1 mM spermidine, 1 mM EDTA), 10 pmol of oligonucleotide, 370 kBq of [γ-32P]dATP, 6 μl of HiPerSolv water, and 5 U of T4 polynucleotide kinase. Labelling mixes were incubated at 37°C for 1 h, and then unincorporated nucleotide was removed with Sephadex G-50 NICK columns (Pharmacia) and the incorporation percentage was determined. The oligonucleotide probe sequence for beta-subgroup ammonia oxidizer 16S rDNA (AAO258r) has been described previously (10); this sequence is widespread among beta-subdivision ammonia oxidizers (24), and although it occurs in a number of other bacteria, it is a useful internal confirmatory probe for 16S rDNA that has been amplified with ammonia oxidizer-specific primers. Membranes were prehybridized at 45°C for 1 h in a solution containing 2% (wt/vol) blocking reagent solution (Boehringer), 5× SSPE (20× SSPE is 3.6 M NaCl, 0.2 M NaH2PO4 [pH 7.7], and 20 mM EDTA), 20% (vol/vol) deionized formamide, 0.02% (wt/vol) sodium dodecyl sulfate (SDS), and 0.1% (wt/vol) N-lauroylsarcosine prepared in distilled water. Hybridization was performed at the appropriate temperature in 20 ml of fresh solution (blocking reagent omitted) containing 10 pmol of the labelled oligonucleotide. After overnight incubation, the membranes were washed at hybridization temperature in fresh solution for 5 min, the washing step was repeated as necessary, and the membranes were exposed to X-ray film for autoradiographic development.
amoA gene probe labeling and hybridization.
Approximately 50 ng of genomic DNA from a pure culture of N. europaea ATCC 25978 was amplified under the stringent conditions detailed above, with primers AMOF2/R2R in a 50-μl reaction mixture containing 2.5 μl of [α-32P]CTP (ICN Pharmaceuticals) (370 kBq.μl−1). To determine whether there had been adequate incorporation of radiolabel into synthesized DNA fragments, 1 μl of PCR product was electrophoresed in 0.8% (wt/vol) agarose, transferred to a nylon membrane by Southern blotting, and exposed to X-ray film for 6 h at −80°C. Unincorporated [α-32P]CTP was removed from the remaining reaction mix with Sephadex G-50 NICK columns (Pharmacia Biotech), and radiolabelled amplification products were heat denatured for use as amoA gene probes. Membranes for gene probing were prehybridized in 25 ml of prehybridization solution (6× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate; pH 7.0], 5× Denhardt’s solution, and 0.5% SDS made up to 25 ml with distilled water containing 0.5 ml of a 1-mg · ml−1 solution of calf thymus DNA, denatured by heating to 100°C for 5 min, and chilled on ice) for at least 1 h at 65°C with gentle shaking. Probe incubation was performed overnight in 100 ml of hybridization buffer (6× SSC, 5× Denhardt’s solution, 0.5% SDS) with shaking at 65°C. After hybridization, the membranes were washed in 50 ml of 2× SSC at 65°C for 15 min. A second wash was performed in 50 ml of 2× SSC–0.1% SDS at 65°C for 30 min, and a final wash was performed in 50 ml of 0.1× SSC at 65°C for 10 min. The membranes were air dried and exposed to X-ray film for autoradiography.
Enrichment of ammonia-oxidizing bacteria.
Environmental samples were inoculated in duplicate (1%, wt/vol [sediment] and 1%, vol/vol [lake water]) in 200 ml of Watson and Mandel medium (35) containing 0.67 or 12.5 mM (NH4)2SO4 (pH 8.0), incubated at 30°C with shaking (150 rpm) in the dark, and monitored at 5-day intervals for NH4-N and nitrite by Quantofix Test sticks (Camlab, Cambridge, United Kingdom). The pH of the enrichment cultures was monitored and adjusted to pH 8.0 as necessary by the addition of sterile 5% (wt/vol) sodium carbonate. Enrichments were subcultured (1%, vol/vol) into the appropriate medium when nitrification was indicated by a decrease in pH and the NH3-N concentration combined with a concomitant increase in the nitrite concentration. This was repeated three times, and 30 μl of the third subculture was transferred to 3 ml of fresh medium in a sterile 3.5-in. glass tube, mixed, and serially diluted to a factor of 10−7. The dilutions were sealed with Parafilm to prevent evaporation and incubated with monitoring of nitrification as above. The highest dilution to show nitrification over a period of 12 weeks was itself serially diluted as described above. This was repeated four times, and 1 ml of culture from the highest dilution showing nitrification in the fourth dilution series was finally inoculated into 200 ml of fresh medium. This final culture was used for the extraction of nucleic acid and determination of the presence of nitrosospiras and/or nitrosomonads as described below. Such an exhaustive procedure is normally applied to the isolation of autotrophic ammonia-oxidizing bacteria (22).
After three enrichment subcultures as described above, those in which there was evidence of ammonia oxidation were inoculated (1%, vol/vol) into both high- and low-ammonium sulfate media, incubated, and monitored at 5-day intervals up to 60 days for nitrification.
Cell blots.
All nitrifying enrichment cultures, including those cross-inoculated into media containing higher or lower substrate concentrations, were processed. Cells from 100-ml aliquots of enrichment media were applied to nylon membranes under vacuum by using a Minifold-II manifold (Schleicher & Schuell). Cell lysis was performed in the assembled manifold by enzymatic attack at room temperature for 30 min in lysozyme solution (100 mM Tris-HCl [pH 7.6], 150 mM NaCl, 5 mM MgCl2, 1.5% [wt/vol] bovine serum albumin, 40 mg of lysozyme [Sigma] per ml). Lysis was continued by incubation at room temperature for 15 min in detergent solution (2% [wt/vol] SDS, 1 mM EDTA [pH 8.0]). The manifold slots were washed with two 0.5-ml volumes of 10× SSPE. Membrane-bound nucleic acids were denatured by laying the membrane on 3MM Whatman paper soaked in denaturing solution for 7 min and neutralized by transferring the membrane for 1 min to Whatman paper soaked in neutralization solution. Finally, nucleic acids were cross-linked to the membrane by exposure to UV light for 45 s. All the reagents and apparatus were pretreated with diethylpyrocarbonate. Membranes were probed with the Nitrosospira-specific (NS85r) and Nitrosomonas-specific (Nm75r) 16S rRNA oligonucleotides defined by Hiorns et al. (10).
RESULTS
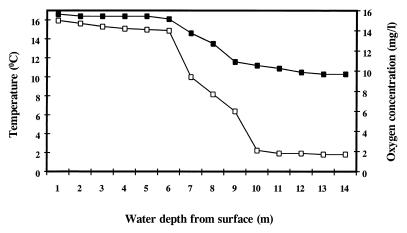
Esthwaite Water is generally regarded to be the most productive lake in the English Lake District. This work was undertaken in 1993, and data on the physicochemical characterization of the lake over the 12-month period demonstrated that the lake had become stratified during the period from April to October (Fig. 2). The maximum concentration of chlorophyll a in the surface water was 74 mg m−3. The water temperature varied between 3 and 16°C over the year; the pH of the lake water at a depth of 5 m was fairly constant at ca. pH 7, with isolated peaks of pH 8 to 9 in summer, when biological activity was high.
FIG. 2.
Oxygen (□) and temperature (■) profiles of the water column of Esthwaite Water recorded at stratification (July) during 1993.
Nitrification in Esthwaite Water.
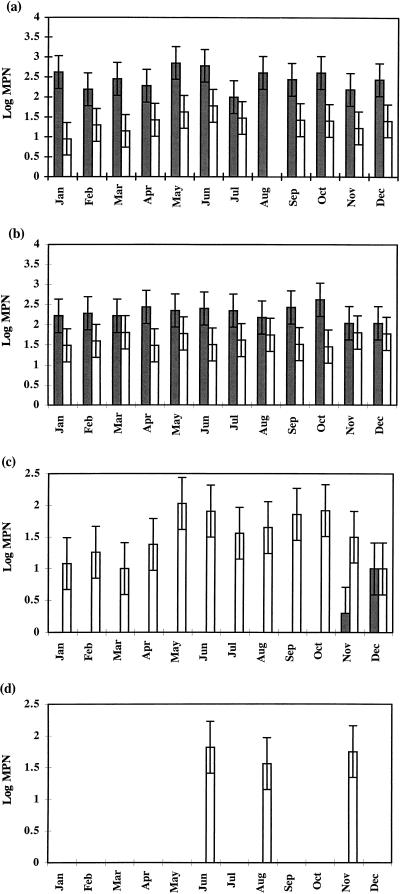
At monthly intervals, samples of lake water and littoral and profundal sediment were inoculated into two media, differing in ammonium sulfate concentration, for MPN determinations of ammonia-oxidizing bacteria. The data are presented in Fig. 3 and demonstrate that ammonia oxidation in sediments differs fundamentally from that in the water column. First, MPNs were an order of magnitude greater in sediment samples than in lake water, and this is further emphasized by the requirement for tangential-flow concentration of water samples before their inoculation into the MPN media. When unconcentrated lake water samples were used as the inoculum, ammonia oxidation was undetectable for 9 of the 12 months sampled, and only <70 cells ml of inoculum−1 were detected for the remaining 3 months. Profundal and littoral sediment samples yielded MPN counts in the range of 100 to 700 cells g (dry weight) of inoculum−1, with profundal sediment showing less variation throughout the year than littoral sediment, for which there was a trend toward increased activity during the summer months (Fig. 3). The most interesting observation was that ammonia oxidation by lake water could be demonstrated only in MPN tubes containing a low (0.67 mM) ammonium sulfate concentration, whereas for sediment samples an increased concentration (12.5 mM) strongly stimulated ammonia oxidation. The November and December MPN determinations for concentrated lakewater samples are exceptional in that they were the only samples for which ammonia oxidation (<10 cells ml−1) was detected at the high ammonium concentration, and this is supported by data from enrichment cultures (see below).
FIG. 3.
MPN values for ammonia-oxidizing bacteria cultured in 12.5 mM ( ) and 0.67 mM (□) NH4(SO4)2 medium from littoral sediment (a), profundal sediment (b), a concentrated lakewater sample (c) and an untreated lakewater sample (d). Thin bars indicate 95% confidence limits.
) and 0.67 mM (□) NH4(SO4)2 medium from littoral sediment (a), profundal sediment (b), a concentrated lakewater sample (c) and an untreated lakewater sample (d). Thin bars indicate 95% confidence limits.
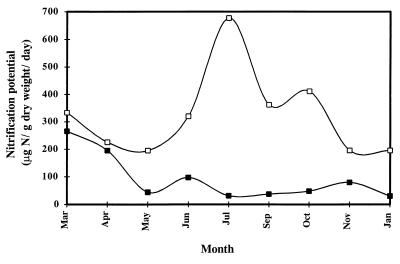
Unlike direct measurements of different oxidation states of nitrogen in sediment and water samples, determination of nitrification potential can provide a specific assessment of the size and viability of the nitrifying community. This was applied here to sediment samples to complement the data on MPN determinations of ammonia-oxidizing bacteria. Both nitrite and nitrate concentrations were determined in the nitrification potential incubations, and although there was considerable temporal variation (Fig. 4), this could not be simply correlated with environmental and biological factors. However, within both littoral and profundal sediments, the nitrification potential was always significantly greater at the sediment surface and was dominated by nitrate as the end product (data not shown). Furthermore, nitrification potentials in profundal sediment samples were always lower than those in littoral sediment samples, especially when the lake was stratified in the summer months (Fig. 4).
FIG. 4.
Nitrification potentials of littoral (□) and profundal (■) sediment of Esthwaite Water throughout 1993.
Analysis of ammonia oxidizer DNA recovered directly from lake water and sediments.
Application of the eubacterial 16S rDNA primers pAf and pHr generated 1.5-kb DNA fragments from all the lakewater and sediment samples examined. In a few cases, amplification products were not clearly visible on agarose gels, probably due to the persistence of inhibitory humic substances despite the purification procedures applied. PCR for Nitrosomonas and Nitrosospira 16S rDNA with primer pairs Nm75f/1007r and Ns85f/1009r, respectively, failed to yield products from any of the environmental DNA samples. These primer pairs were designed and applied previously (10) based on interrogation of a limited number of full 16S rRNA gene alignments (8), but subsequent examination of the current database has revealed that they have limitations. The Nitrosospira primers retain their broad utility for amplification of members of this group, whereas the Nitrosomonas primers must now be regarded as targeting only representatives of the N. europaea-eutropha lineage (17). The potential amplification of 16S rDNA from bacteria other than ammonia oxidizers is obviated by our routine application of the internal probe AAO258 (10) to verify the fidelity of all the amplification products obtained. The widespread occurrence of this sequence among beta-subdivision ammonia oxidizer 16S rRNA genes has been confirmed (24), and although the sequence is present in a range of other bacteria, its application as a confirmatory probe for amplification products generated by primers that are specific is valid.
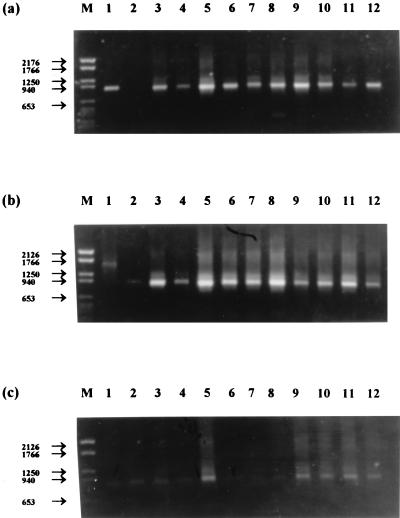
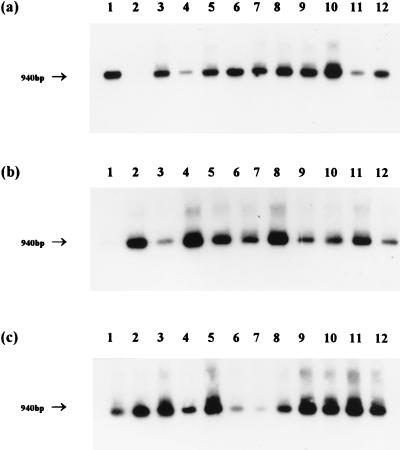
When these primers were applied to the pAf/pHr amplification products in a nested reaction, ethidium bromide-stained bands of DNA of the expected size were usually obtained with Nitrosospira primers but were never obtained with the Nitrosomonas primers (Fig. 5). The specificity of the amplification products was confirmed by hybridization to the internal oligonucleotide probe AAO258, and this also revealed the presence of Nitrosospira 16S rDNA in samples that did not produce visible bands on agarose gels (Fig. 6). The littoral sediment sampled in February was the only sample of the 36 tested in which Nitrosospira 16S rDNA was detected, but we do not believe that this is significant. However, the universal absence of Nitrosomonas 16S rDNA amplification products was confirmed by an inability to detect any hybridization signals with this probe. It is difficult to interpret PCR data quantitatively, particularly when repeated nested amplifications are applied. However, the comparative intensity of ethidium bromide-stained bands of amplified Nitrosospira 16S rDNA from sediment and lakewater samples (Fig. 5) supports the clear quantitative distinction between MPN determinations of ammonia oxidizers in sediments and lake water (Fig. 3).
FIG. 5.
Detection of Nitrosospira 16S rDNA in littoral sediment (a), profundal sediment (b), and lakewater samples (c) by PCR followed by ethidium bromide-stained agarose gel electrophoresis. Lanes: M, DNA molecular size markers (base pairs); 1 to 12, January to December samples, respectively.
FIG. 6.
Hybridization pattern after probing of Nitrosospira 16S rDNA PCR products amplified from littoral sediment (a), profundal sediment (b), and lakewater samples (c) with oligonucleotide AAO258. Lanes: 1 to 12, January to December samples, respectively. The approximate size of fragments is indicated.
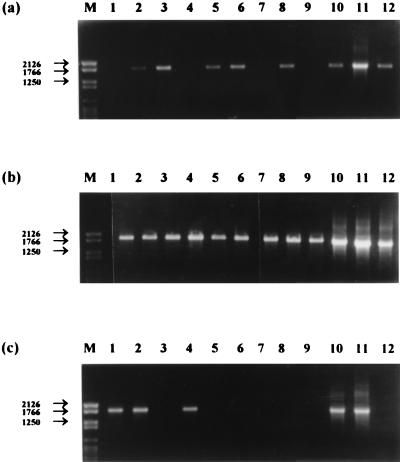
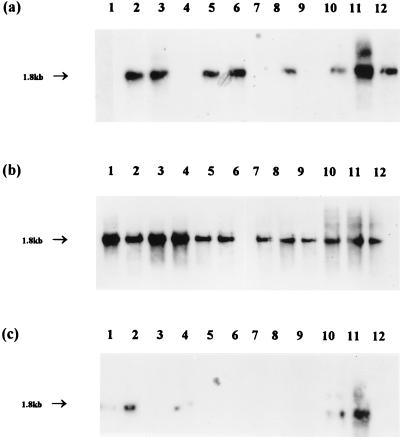
Application of PCR amplification primers specific for the amoA gene of N. europaea (AMOF1 and AMOR2) to DNA extracted from sediment and lakewater samples failed to yield visible bands on gels. The reaction mixtures were diluted 10-fold and reamplified with the internal primer pair AMOF2 and AMOR2R; this resulted in amplification products of the expected size (1.8 kb) from a number of samples (Fig. 7). All the gels were Southern blotted and hybridized with the labelled amoA gene as the probe. This confirmed the identity of the amplified products in all cases (Fig. 8), and no hybridization signals were obtained for lanes which did not contain visible DNA bands. The distribution pattern of positive results for this gene target could not readily be correlated with spatial and temporal parameters. A surprisingly large number of positive amplifications were obtained with lakewater samples in view of the comparative data on MPN determinations for sediment and water samples (Fig. 3) and the failure of the Nitrosomonas 16S rDNA primers to yield amplification products. Similarly, profundal sediment samples compared favorably with littoral samples across the year (Fig. 7 and 8), but ammonia oxidizer populations were generally higher in the latter (Fig. 3). However, these data appear to demonstrate the presence of N. europaea and/or closely related organisms in the ammonia-oxidizing communities of both the water column and sediments, which the 16S rDNA primers failed to achieve.
FIG. 7.
Detection of N. europaea amoA DNA in littoral sediment (a), profundal sediment (b), and lakewater samples (c) by PCR followed by ethidium bromide-stained agarose gel electrophoresis. Lanes: M, DNA molecular size markers (base pairs); 1 to 12, January to December samples, respectively.
FIG. 8.
Hybridization pattern after probing of N. europaea amoA DNA PCR products amplified from littoral sediment (a), profundal sediment (b), and lakewater samples (c) with the amoA gene probe. Lanes: 1 to 12, January to December samples, respectively. The approximate size of fragments is indicated.
Analysis of enrichment cultures.
Monthly samples (April to December) of lake water, filtered lake water, littoral sediment, and profundal sediment were inoculated into 0.67 mM (low) and 12.5 mM (high) ammonium sulfate medium, and the mixtures were incubated at 30°C. These enrichment cultures were monitored for nitrifying activity by measuring nitrite, nitrate, and pH, and they were appropriately subcultured three times. Where nitrification had occurred, cells were harvested from the final enrichments and nucleic acid extracts were hybridized to genus-specific 16S rRNA oligonucleotide probes for Nitrosospira and Nitrosomonas. Untreated lake water failed to provide nitrifying enrichment cultures in the high-ammonium medium but yielded positive enrichments in the low-ammonium medium for samples obtained between July and December. In all cases, hybridization signals were obtained with the Nitrosomonas probe but not the Nitrosospira probe. When lakewater filtered by tangential flow was used as the inoculum, almost identical results were obtained for the low-ammonium enrichments. In addition, positive enrichments were recorded in high-ammonium medium for the November and December samples, both of which gave hybridization signals for Nitrosospira but not Nitrosomonas probes. These data are summarized in Table 1.
TABLE 1.
Genus-specific oligonucleotide probe hybridization results for ammonia-oxidizing bacteria enriched from littoral sediment, profundal sediment, concentrated lake water, and untreated lake water by using 12.5 and 0.67 mM NH4(SO4)2-containing medium
| Type of sample and time of collection | Hybridization signal from:
|
|
|---|---|---|
| 12.5 mM NH4+ medium | 0.67 mM NH4+ medium | |
| Littoral sediment | ||
| April | Ns+, Nm++ | Ns++ |
| May | Ns+, Nm+++ | − |
| June | Ns+++, Nm+ | Ns+ |
| July | Ns++, Nm+++ | − |
| August | − | − |
| September | Ns+, Nm++ | Ns+, Nm+ |
| October | Nm+ | Ns+ |
| November | Ns+, Nm++ | Ns+, Nm+ |
| December | ND | Nm+ |
| Profundal sediment | ||
| April | Nm+++ | Ns++ |
| May | Ns+, Nm+++ | − |
| June | − | − |
| July | Ns+, Nm++ | Ns+ |
| August | Ns++ | Nm++ |
| September | Nm+ | Ns++ |
| October | Nm+ | Nm+ |
| November | Ns+, Nm++ | Nm+ |
| December | Nm+ | Nm+ |
| Concentrated lake water | ||
| April | − | − |
| May | − | − |
| June | − | − |
| July | − | Nm+ |
| August | − | Nm+ |
| September | − | Nm+ |
| October | − | Nm+ |
| November | Ns+++ | − |
| December | Ns++ | Nm+ |
| Untreated lake water | ||
| April | − | − |
| May | − | − |
| June | − | − |
| July | − | Nm+ |
| August | − | Nm+ |
| September | − | Nm+ |
| October | − | Nm+ |
| November | − | Nm+ |
| December | − | Nm+ |
+, weak; ++, moderate; +++, strong; −, no growth; ND not determined; Ns, Nitrosospira; Nm, Nitrosomonas.
The pattern of positive enrichments and genus-specific hybridizations for sediment samples was more complex than that described above for the water column (Table 1). Enrichments of both profundal and littoral sediment samples were, in contrast to those of lake water, generally successful in high-ammonium medium and often comprised populations of both nitrosospiras and nitrosomonads as revealed by oligonucleotide probing. The strengths of the hybridization signals could be compared to give an indication of the predominance of one genus over another in individual enrichments, and these results varied (Table 1). There were also examples of high-ammonium sediment enrichments in which only one of these genera could be detected by probe hybridization. Enrichments from sediment samples inoculated into low-ammonium medium gave slightly fewer positive results than did those inoculated into high-ammonium medium, but unlike the data for lakewater enrichments, where only nitrosomonads were detected, members of both genera were detected either alone or in combination (Table 1).
This difference between high- and low-ammonium enrichment cultures was further examined for any evidence that Esthwaite Water contained subpopulations of ammonia oxidizers defined by substrate affinity. This was done by subculturing July (summer) and November (winter) high-ammonium enrichments into low-ammonium medium and vice versa. The results are presented in Table 2. None of the enrichment cultures that exhibited nitrification in low-ammonium medium continued to nitrify when subcultured into the high-ammonium medium. Furthermore, none of these enrichments were reactivated by subsequent inoculation from the high-ammonium back into the low-ammonium medium. In contrast, apart from the high-ammonium enrichment culture for the July profundal sediment sample, high-ammonium enrichment cultures subcultured into low-ammonium medium continued to nitrify. Of the five high-ammonium enrichment cultures used as inocula, four contained both nitrosomonads and nitrosospiras on the basis of genus-specific oligonucleotide hybridization to extracted nucleic acid. This mixed population appeared to be maintained in one subculture, whereas only the nitrosomonad component could be detected in the other two low-ammonium enrichment subcultures. The November lakewater enrichment culture in high-ammonium medium, which was unusual in showing nitrification at this ammonium concentration, appeared to contain only nitrosospiras, and these continued to nitrify when subcultured into low-ammonium medium.
TABLE 2.
Data from cross-inoculation of ammonia oxidizers enriched in either high- or low-ammonium medium into the other medium
| Time of collection and type of sample | Low-NH4+ medium to high-NH4+ medium
|
High-NH4+ medium to low-NH4+ medium
|
||||
|---|---|---|---|---|---|---|
| Nitrificationa | Hybridization signalb in:
|
Nitrification | Hybridization signal in:
|
|||
| Original | Subculture | Original | Subculture | |||
| July | ||||||
| Lake water | − | Nm+ | − | ND | ND | ND |
| Littoral sediment | − | ND | ND | + | Ns++ | Ns+ |
| − | Nm+++ | Nm+ | ||||
| Profundal sediment | − | Ns+ | − | − | Ns+ | ND |
| Nm++ | ||||||
| November | ||||||
| Lake water | − | Nm+ | − | + | Ns+++ | Ns+ |
| Littoral sediment | − | Ns+ | − | + | Ns+ | Nm++ |
| Nm+ | Nm++ | |||||
| Profundal sediment | − | Nm+ | − | + | Ns+ | Nm+ |
| Nm++ | ||||||
Nitrification is indicated to have ceased (−) or continued (+) after inoculation into the contrasting medium.
Organisms continuing to nitrify in culture were identified by genus-specific oligonucleotide probing. Hybridization signal: +, weak; ++, moderate; +++, strong; Ns, Nitrosospira; Nm, Nitrosomonas.
DISCUSSION
Esthwaite Water demonstrated typical seasonal stratification in 1993, with compartmentalization of biological activity in relation to temperature and oxygen gradients. It therefore presented a useful system in which to investigate nitrification as influenced by changes in the water column structure and its interaction with sediment. Monitoring populations of ammonia-oxidizing bacteria by MPN determinations was straightforward and demonstrated the presence of significantly larger numbers of these organisms in sediments where ammonia generation is known to be higher than in the water column (4). Ammonia oxidation is not usually associated with anaerobic environments, and consequently the activity of ammonia oxidizers in lake sediments will be stimulated by oxygenation. This accords with our data, which revealed larger numbers of ammonia oxidizers and higher nitrification potentials in littoral than in profundal sediment sites (Fig. 3). The susceptibility of littoral sediment to variation, which is probably temperature induced, is illustrated by the fluctuation in ammonia oxidizer MPNs compared with the more uniform data for profundal sediment over the 12-month period (Fig. 3a and b). Populations of ammonia oxidizers in the water column were low, almost certainly due to low concentrations of nutrients including ammonium, and meaningful MPN determinations became possible only by applying tangential-flow filtration as a pretreatment to concentrate the cells (Fig. 3c and d).
That sediment and water column populations have become adapted to prevailing concentrations of ammonia is indicated by the influence of ammonium concentration on MPN determinations. Thus, water column samples would nitrify only when inoculated into low-ammonium medium, in contrast to sediment samples, which exhibited the expected stimulation of ammonia oxidation by increasing their ammonium concentration (Fig. 3). This hypothesis that distinct populations have evolved in relation to ammonium concentration derives support from the observation that some winter lakewater samples inoculated into high-ammonium medium nitrified, possibly indicating the introduction of sediment ammonia oxidizers into the water column at overturn. Enrichment of different populations of ammonia-oxidizing bacteria by manipulating the substrate ammonium concentration has been reported previously (25), but only for sewage treatment systems. The data on enrichment culture of Esthwaite Water ammonia oxidizers in relation to ammonium concentration (Table 1) are also supportive. Sediment samples yielded positive enrichments at both ammonium concentrations, whereas lakewater samples proved positive for nitrification only at the low ammonium concentration. Again, November and December lakewater enrichments were the exception, as described for the MPN determinations above, showing nitrification at the higher ammonium concentration preferred by sediment samples. The lakewater population of ammonia-oxidizing bacteria is probably dominated by species that are sensitive to inhibition at high ammonium concentrations, as evidenced by the consistent failure of low-ammonium enrichments to act as a source of viable inoculum for subculture to high-ammonium medium (Table 2). These enrichment cultures contained sufficiently large populations of ammonia-oxidizing bacteria for direct detection by 16S rRNA oligonucleotide probing without the PCR amplification applied to environmental samples. Probes for nitrosospiras and nitrosomonads (N. europaea-eutropha lineage) were applied to determine whether the distribution of these groups was correlated with the ammonium concentration. It has been previously suggested that nitrosospiras are ubiquitous whereas nitrosomonads occur only in enrichment cultures or in environmental situations where nutrient concentrations are high (7, 10, 15, 24). In this study of Esthwaite Water, sediment sample enrichments gave positive hybridizations to either or both without any discernible pattern. Unexpectedly, however, all of the lakewater enrichments at low ammonium concentration comprised only nitrosomonads, with the exception again of the winter enrichments at high ammonium concentration, which appeared to comprise only nitrosospiras (Table 1). This population of ammonium-sensitive nitrosomonads is an excellent target for 16S rDNA and amoA sequence analysis to ascertain its genotypic composition and hence its origin.
Direct PCR amplification of environmental DNA revealed the distribution and relative abundance of autotrophic ammonia oxidizers throughout Esthwaite Water. Despite their crucial role in the nitrogen cycle, populations of autotrophic ammonia-oxidizing bacteria are low in the environment, and this is reflected in the cell densities that can be obtained in laboratory culture (<105 ml−1). Thus, the low MPN values recorded here are in agreement with our inability to amplify either 16S rDNA or amoA DNA in a single-stage reaction, necessitating the application of a nested PCR approach throughout. The requirement for this nested amplification strategy has been previously reported for different primer pairs specific for beta-subdivision ammonia oxidizers applied to a variety of environments (7, 10, 30, 34). However, there are reports of other beta-subdivision ammonia oxidizer 16S rDNA primers yielding amplification products from environmental samples after a single round of PCR amplification (12, 24). While multistage PCR would in theory be more likely to yield products from minority members of communities, we still failed to amplify Nitrosomonas 16S rDNA, although these organisms were detected in enrichment cultures and by amoA-directed amplification. It is important to stress that the 16S rDNA primer pairs used here for Nitrosomonas will amplify only the closely related species N. europaea and N. eutropha, but, in contrast to some reports (11, 17), these organisms do not appear to have a restricted environmental distribution; they are simply outnumbered by nitrosospiras. It is difficult to comment on the specificity of the amoA primers because so few sequences have been determined, but we do know that the primers used in the present study under the conditions described above do not amplify the amoA gene of cultured Nitrosospira spp. or the pmmo (particulate methane monooxygenase) gene of methanotrophs (7). Indeed, the failure of these amoA primers to amplify DNA from pure cultures of the closely related species N. eutropha (7) supports the suggestion that N. europaea has been specifically detected by this approach. amoA sequence diversity has recently been analyzed (18), but the regions sequenced do not encompass our primer sites. However, Rotthauwe et al. (18) did amplify amoA DNA directly from freshwater lake samples and identified it as originating from members of the Nitrosospira lineage. This approach supported a range of previous studies in which it was suggested that Nitrosospira spp. are more common in terrestrial and freshwater environments than are Nitrosomonas spp. (7, 10, 12, 24). Here, we have again shown, in a systematic study of a single freshwater lake system, the reproducible and consistent detection of Nitrosospira 16S rDNA in lakewater and sediment samples. It has been argued that amoA as a functional gene target is advantageous over the 16S rDNA approach to studying ammonia oxidizer community structure (18) because the latter approach has reduced specificity and coamplification of 16S rDNA sequences from nonnitrifying bacteria occurs (24). In this study, we have used an internal oligonucleotide probe to verify that the 16S rDNA amplified from environmental samples does indeed contain only nitrifier DNA.
Direct amplification of ammonia oxidizer 16S rDNA from lakewater and sediment samples throughout a 12-month period is a useful method of detecting the presence of these organisms. This is certainly true for nitrosospiras, whose presence in lake water in particular was revealed even when MPN determinations of the ammonia-oxidizing community as a whole were very low (<10 cells ml−1 [Fig. 3c]). Although these data are certainly not quantitative, it is clear that amplified bands of Nitrosospira 16S rDNA were more intensely stained in sediment samples than in concentrated-lakewater samples (Fig. 5), as would be suggested by comparison of the ammonia oxidizer MPN values for lake water versus profundal and littoral sediments (Fig. 3). The same comparison does not apply to amplified N. europaea amoA DNA, which was at least as readily amplified from lakewater samples as from sediment samples (Fig. 7). This is particularly impressive in view of the failure of the N. europaea 16S rDNA primers to yield amplification products from any samples. The explanation may be technical; the first round of 16S rDNA amplification uses universal primers, and the Nitrosomonas template must compete with other 16S rDNA genes, thus limiting the enrichment effect, in comparison with amoA, which was subjected to two specific and noncompetitive rounds of amplification. Alternatively, the 16S rDNA primers used here for nitrosomonads are specific to the N. europaea-eutropha lineage and the sequences do not occur in a number of other Nitrosomonas spp. (17). If these and closely related species have strongly homologous amoA gene sequences, the data could be indicating their presence in Esthwaite Water. Unfortunately, Rotthauwe et al. (18) did not include nitrosomonads other than N. europaea and N. eutropha in their comparative partial sequencing of amoA genes, so this explanation cannot be verified. Furthermore, the amoA primers used by us failed to amplify amoA from N. eutropha, a species that is very closely related to N. europaea on the basis of 16S rRNA sequences (8).
This study emphasises the value of using independent gene targets wherever possible to support (7) or, as shown here, to produce a more accurate description of community structure. In particular it will be possible to describe sediment and water column genotypes by sequence analysis of amplified DNA and elucidate their relationship to one another and the influence of spatial and temporal parameters. The foundations laid in this study will allow us to evaluate nutritionally diverse lake systems with a view to identifying both common aspects and specific features of freshwater ammonia oxidizer ecology.
ACKNOWLEDGMENT
This work was supported by the Natural Environment Research Council of the United Kingdom.
REFERENCES
- 1.Bendschneider K, Robinson R J. A new spectrophotometric determination of nitrite in seawater. J Mar Res. 1952;2:87–96. [Google Scholar]
- 2.Bruce K D, Hiorns W D, Hobman J L, Osborn A M, Strike P, Ritchie D A. Amplifications of DNA from native populations of soil bacteria by using the polymerase chain reaction. Appl Environ Microbiol. 1992;58:3413–3416. doi: 10.1128/aem.58.10.3413-3416.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Giovannoni S J, DeLong E F, Schmidt T M, Pace N R. Tangetial-flow filtration and preliminary phylogenetic analysis of marine picoplankton. Appl Environ Microbiol. 1990;56:2572–2575. doi: 10.1128/aem.56.8.2572-2575.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hall G H. Nitrification in lakes. In: Prosser J I, editor. Nitrification. Oxford, England: IRL Press; 1986. p. 99. [Google Scholar]
- 5.Hall G H, Jeffries C. The contribution of nitrification in the water column and profundal sediments to the total oxygen deficit of the hypolimnion of a mesotrophic lake (Grasmere, English Lake District) Microb Ecol. 1984;10:37–46. doi: 10.1007/BF02011593. [DOI] [PubMed] [Google Scholar]
- 6.Harris R F, Sommers L E. Plate-dilution frequency technique for assay of microbial ecology. Appl Microbiol. 1968;16:330–334. doi: 10.1128/am.16.2.330-334.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hastings R C, Ceccherini M T, Nerino M, Saunders J R, Bazzicalupo M, McCarthy A J. Direct molecular biological analysis of ammonia oxidising bacteria populations in cultivated soil plots treated with swine manure. FEMS Microb Ecol. 1997;23:45–54. [Google Scholar]
- 8.Head I M, Hiorns W D, Embley T M, McCarthy A J, Saunders J R. The phylogeny of autotrophic ammonia-oxidizing bacteria as determined by analysis of 16S ribosomal RNA gene sequences. J Gen Bacteriol. 1993;139:1147–1153. doi: 10.1099/00221287-139-6-1147. [DOI] [PubMed] [Google Scholar]
- 9.Heaney S I, Smyly W J P, Talling J F. Interactions of physical, chemical and biological processes in depth and time within as productive English lake during summer stratification. Int Rev Gesamten Hydrobiol. 1986;17:441–494. [Google Scholar]
- 10.Hiorns W D, Hastings R C, Head I M, McCarthy A J, Saunders J R, Pickup R W, Hall G H. Amplification of 16S ribosomal RNA genes of the autotrophic ammonia-oxidizing bacteria demonstrates the ubiquity of nitrosospiras in the environment. Microbiology. 1995;141:2793–2800. doi: 10.1099/13500872-141-11-2793. [DOI] [PubMed] [Google Scholar]
- 11.Koops H-P, Bottcher B, Moller U C, Pommerening-Roser A, Stehr G. Classification of eight new species of ammonia-oxidizing bacteria: Nitrosomonas communis sp. nov., Nitrosomonas ureae sp. nov., Nitrosomonas aestuarii sp. nov., Nitrosomonas marina sp. nov., Nitrosomonas nitrosa sp. nov., Nitrosomonas eutropha sp. nov., Nitrosomonas oligotropha sp. nov., and Nitrosomonas halophila sp. nov. J Gen Bacteriol. 1991;137:1689–1699. [Google Scholar]
- 12.Kowalchuk G A, Stephen J R, DeBoer W, Prosser J I, Embley T M, Woldendorp J W. Analysis of ammonia-oxidizing bacteria of the β subdivision of the class Proteobacteria in coastal sand dunes by denaturing gradient gel electrophoresis and sequencing of PCR-amplified 16S ribosomal DNA fragments. Appl Environ Microbiol. 1997;63:1489–1497. doi: 10.1128/aem.63.4.1489-1497.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCaig A E, Embley T M, Prosser J I. Molecular analysis of enrichment cultures of marine ammonia-oxidizers. FEMS Microbiol Lett. 1994;120:363–368. doi: 10.1111/j.1574-6968.1994.tb07059.x. [DOI] [PubMed] [Google Scholar]
- 14.McTavish H, Fuchs J A, Hooper A B. Sequence of the gene coding for ammonia monooxygenase in Nitrosomonas europaea. J Bacteriol. 1993;175:2436–2444. doi: 10.1128/jb.175.8.2436-2444.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mobarry B K, Wagner M, Urbain V, Rittman B E, Stahl D A. Phylogenetic probes for analyzing abundance and spatial organization of nitrifying bacteria. Appl Environ Microbiol. 1996;62:2156–2162. doi: 10.1128/aem.62.6.2156-2162.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ohnstad F R, Jones J G. The Jenkins surface mud sampler. User manual. Occasional publication 15. Cumbria, United Kingdom: Freshwater Biological Association, Windermere Laboratories; 1982. [Google Scholar]
- 17.Pommerening-Roser A, Rath G, Koops H-P. Phylogenetic diversity within the genus Nitrosomonas. Syst Appl Microbiol. 1996;19:344–351. [Google Scholar]
- 18.Rotthauwe J-H, Witzel K-P, Liesack W. The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl Environ Microbiol. 1997;63:4704–4712. doi: 10.1128/aem.63.12.4704-4712.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rowe R, Todd R, Waide J. Microtechnique for most-probable number analysis. Appl Environ Microbiol. 1977;33:675–680. doi: 10.1128/aem.33.3.675-680.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmidt T M, DeLong E F, Pace N R. Analysis of a marine picoplankton community by 16S rRNA gene cloning and sequencing. J Bacteriol. 1991;173:4371–4378. doi: 10.1128/jb.173.14.4371-4378.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sinigalliano C D, Kuhn D N, Jones R D. Amplification of the amoA gene from diverse species of ammonia-oxidizing bacteria and from an indigenous bacterial population from seawater. Appl Environ Microbiol. 1995;61:2702–2706. doi: 10.1128/aem.61.7.2702-2706.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soriano S, Walker N. Isolation of ammonia-oxidizing autotrophic bacteria. J Appl Bacteriol. 1968;31:493–497. doi: 10.1111/j.1365-2672.1968.tb00397.x. [DOI] [PubMed] [Google Scholar]
- 23.Stehr G, Bottcher B, Dittberner P, Rath G, Koops H-P. The ammonia-oxidizing nitrifying populations of the River Elbe estury. FEMS Microbiol Ecol. 1995;17:177–186. [Google Scholar]
- 24.Stephen J R, McCaig A E, Smith Z, Prosser J I, Embley T M. Molecular diversity of soil and marine 16S rRNA gene sequences related to β-subgroup ammonia-oxidizing bacteria. Appl Environ Microbiol. 1996;62:4147–4154. doi: 10.1128/aem.62.11.4147-4154.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suwa Y, Imamura Y, Suzuki T, Tashiro T, Urushigawa Y. Ammonia-oxidizing bacteria with different sensitivities to (NH4)2SO4 in activated sludges. Water Res. 1994;28:1523–1532. [Google Scholar]
- 26.Szwerinski H, Gaiser S, Bardtke D. Immunofluorescence for the quantitative determination of nitrifying bacteria: interference of the test in biofilm reactors. Appl Environ Microbiol. 1985;21:125–128. [Google Scholar]
- 27.Teske A, Regan J M, Toze S, Rittman B E, Stahl D A. Evolutionary relationships among ammonia and nitrite-oxidizing bacteria. J Bacteriol. 1994;176:6623–6630. doi: 10.1128/jb.176.21.6623-6630.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Utaker J B, Bakken L, Jiang Q Q, Nes I F. Phylogenetic analysis of seven new isolates of ammonia-oxidizing bacteria based on 16S rRNA gene sequences. Syst Appl Microbiol. 1995;18:549–559. [Google Scholar]
- 29.Volsch A, Nader W F, Geiss H K, Nebe G, Birr C. Detection and analysis of two serotypes of ammonia-oxidizing bacteria in sewage plants by flow cytometry. Appl Environ Microbiol. 1990;56:2430–2435. doi: 10.1128/aem.56.8.2430-2435.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Voytek M A, Ward B B. Detection of ammonia-oxidizing bacteria of the beta-subclass of the class Proteobacteria in aquatic samples with the PCR. Appl Environ Microbiol. 1995;61:1444–1450. doi: 10.1128/aem.61.4.1444-1450.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wagner M, Rath G, Amann R, Koops H-P, Schleifer K-H. In situ identification of ammonia-oxidizing bacteria. Syst Appl Microbiol. 1995;18:251–264. [Google Scholar]
- 32.Ward B B. Oceanic distribution of ammonium-oxidizing bacteria determined by immunofluorescence assay. J Mar Res. 1982;40:1155–1172. [Google Scholar]
- 33.Ward B B, Carlucci A F. Marine ammonia- and nitrite-oxidizing bacteria: serological diversity determined by immunofluorescence in culture and in the environment. Appl Environ Microbiol. 1985;50:194–201. doi: 10.1128/aem.50.2.194-201.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ward B B, Voytek M A, Witzel K-P. Phylogenetic diversity of natural populations of ammonia-oxidizers investigated by specific PCR amplification. Microb Ecol. 1997;33:87–96. doi: 10.1007/s002489900011. [DOI] [PubMed] [Google Scholar]
- 35.Watson S W, Mandel M. Comparisons of the morphology and deoxyribonucleic acid components of 27 strains of nitrifying bacteria. J Bacteriol. 1971;107:563–569. doi: 10.1128/jb.107.2.563-569.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zaccone R, Caruso G, Azzaro M. Detection of Nitrosococcus oceanus in a Mediterranean lagoon by immunofluorescence. J Appl Bacteriol. 1996;80:611–616. doi: 10.1111/j.1365-2672.1996.tb03265.x. [DOI] [PubMed] [Google Scholar]